Abstract
Traditional treatments for spinal tuberculosis (TB) involve chemotherapy and surgery. In the present study, it has been identified that chemotherapy lasting <6 months [ultra-short course chemotherapy (UCCT)], rather than the 6–18 months of the traditional regimen, is effective in sustaining TB clearance following complete surgical debridement. This current study aims to compare the changes in peripheral blood gene expression prior to and following UCCT, subsequent to complete debridement of spinal TB lesions. The study includes 5 patients without TB and 27 patients with spinal, divided into three groups: Group 1 (untreated group, n=8); group 2 (UCCT treatment group, n=9); and group 3 (UCCT treatment 1 year follow-up group, n=10). Gene changes were detected using DNA microarray analysis, confirmed through reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and the results were examined using the DAVID Knowledgebase to identify the pathways and functions of differentially expressed genes. TB lesions were active in group 1, while groups 2 and 3 showed no signs of active TB, as indicated by clinical manifestations and imaging. Comparison of the transcription profiles of the control and study groups showed that treatment of spinal TB resulted in upregulation of genes that are associated with immune response pathways; RT-qPCR produced similar findings. In conclusion, these results indicate that UCCT is an effective treatment against TB following complete surgical debridement. Furthermore, DNA microarray analysis proved a useful tool to evaluate the effects of spinal TB treatment on the expression of genes associated with immune response pathways.
Keywords: spinal tuberculosis, surgical debridement, ultra short-course chemotherapy, DNA microarray, DAVID pathway analysis
Introduction
Tuberculosis (TB) is a lethal disease caused by Mycobacterium tuberculosis that is commonly diagnosed worldwide. In 2012 there were 8.6 million new TB cases and 1.3 million mortalities resulting from TB (1). TB primarily infects the lung; however, 1% of all TB cases involve the spinal column (2). Traditional treatments for spinal TB involve chemotherapy and surgery. The chemotherapy regimen involves 2–4 drugs, such as isoniazid and rifampicin, with the duration of usage ranging between 6 and 18 months (3). This long duration of chemotherapy can have adverse effects on patients and may lower patient compliance. Ultra-short course chemotherapy (UCCT), chemotherapy lasting <6 months, can be used as a treatment in patient's with spinal TB to counter these issues. In the present study, the blood transcript signatures of patients with spinal TB prior to and following treatment with UCCT, subsequent to complete debridement of TB lesions, were identified. The chemotherapy included streptomycin (S), isoniazid (H), rifampicin (R) and pyrazinamide (Z). Results of previous studies have demonstrated that UCCT with a mean duration of 5.5 months is effective in treating patients with spinal TB (4–6).
Although surgical treatment and UCCT improve spinal TB, the pathways involved in clearing spinal TB are uncharacterized. Therefore, the present study utilized DNA microarray analysis to measure the gene expression profiles from the whole blood of patients with spinal TB prior to and following treatment. Spinal TB varies in the severity of infection and prognosis (7). DNA microarray analysis can identify the pathways associated with spinal TB infection and provide insight into the effectiveness of the UCCT regimen used.
Materials and methods
Patient selection
A total of 38 patients with TB that underwent complete debridement of lesions and UCCT were recruited from the Department of Spinal Surgery at the General Hospital of Ningxia Medical University (Yinchuan, China) between October 2010 and June 2011. Peripheral blood samples were collected from these patients at various treatment periods. Out of the recruited 38 patients, only 27 patients had verified spinal TB by RNA quality testing, this consisted of 13 males and 14 females, ranging between 17–72 years old (average 38.8±15.6 years). Tuberculous lesions were found in the lumbar region (n=11), thoracic region (n=8), thoracolumbar region (n=5), lumbosacral segment region (n=2), and thoracic and lumbar damage of the region (n=1). In all cases, complete debridement of the lesions was performed, followed by interbody graft fusion and internal fixation between the focal vertebras (4,5,7). Complete debridement, based on traditional debridement, involves tissues including the edge of lesions, the multi-cavity and bony bridge being cleaned (4,5).
In the control group of 7 patients without TB, 5 peripheral blood samples were confirmed to not have spinal TB by RNA quality testing. Table I provides the study group descriptions. The Ethics Commission (General Hospital of Ningxia Medical University, Yinchuan, China) approved all the clinical cases and all patients involved in the study provided informed consent.
Table I.
Study group descriptions.
| Group | Group description | Patient condition | Number of individuals | Age range | Gender | ESR (mm/h) | CRP (mg/l) |
|---|---|---|---|---|---|---|---|
| Control | No TB infection | No TB detected | 5 | 35–50 | 3F, 2M | 2–10 | 0.23–1.23 |
| Study group 1 | TB infection, no medical treatment | Active TB | 8 | 21–72 | 5F, 3M | 1–79 | 0.49–74.49 |
| Study group 2 | Complete debridement and UCCT | Inactive TB | 9 | 20–52 | 6F, 3M | 3–15 | 0.58–8.58 |
| Study group 3 | 1 year follow-up (post-treatment) | Inactive TB | 10 | 17–58 | 5F, 5M | 2–10 | 0.17–5.17 |
TB, tuberculosis; UCCT, ultra-short chemotherapy; ESR, erythrocyte sedimentation rate; CRP, C-reactive protein; F, female; M, male.
The inclusion criteria were as follows: Patients who had recovered from spinal TB with complete removal of spinal lesions and UCCT treatment. Patients with other infections, tumors, trauma, metabolic disease and/or immunological deficiency were excluded.
Chemotherapy regimen
Following complete debridement of spinal TB lesions, all patients received 2SHRZ/2-4HR (Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) for 4–6 months (mean, 5.5 months). Based on the different therapeutic periods, the peripheral blood samples of the 27 patients in the study group were divided into three groups (Table I). Group 1 consisted of 8 untreated, TB-infected individuals that received no treatment prior to hospitalization, with clinical data indicating active spinal TB. Group 2 consisted of 9 TB-infected individuals that had thorough clearance of focal lesions and UCCT for 4–6 months (mean, 5.5 months). Their clinical data indicated lesion healing and fusion of the interbody graft. Group 3, the 1 year follow-up group, consisted of 10 individuals that had not been taking medications for 5–6 months following initial UCCT. The clinical results of groups 2 and 3 showed a similar recovery of spinal function and interbody healing. Additionally, a control group consisting of 5 uninfected individuals was used for this study.
In the control group, there were 5 cases of carpal tunnel syndrome (2 males and 3 females, aged between 35 and 50 years old, mean age 40.6±7.4 years old). All individuals in the control group were free of TB and other related diseases, with normal erythrocyte sedimentation rates, and negative C-reactive protein and TB skin tests.
Imaging analysis
The study group of 27 patients with TB that underwent complete debridement of lesions and UCCT were subsequently scanned by computed topography (CT) and magnetic resonance imaging (MRI; both, General Electric Company, Fairfield, CT, USA).
Blood sample collection and preparation
Fasting peripheral blood was collected from patients with spinal TB and anti-coagulated with EDTA. Whole blood (250 µl) was collected and placed into a 1.5 ml centrifuge tube that contained TRI pure RNA preserving liquid (RNA long preservation solution; RP2101; Beijing Biotech, Inc., Beijing, China). Following mixing, the samples were stored at −80°C.
Extraction of total RNA from peripheral blood and quality testing
Total RNA was extracted from peripheral blood with an RNA extraction kit (RP4002; Beijing Biotech, Inc.) and purified using the RNAClean kit (ORP1801; Beijing Biotech, Inc.). Quantitative detection of RNA was performed with a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Inc., Wilmington, DE, USA). Denaturing agarose gel electrophoresis (1.3 g agarose gel/100 ml 1X TAE; 120 V; 25 min) was employed to determine the integrity and quality of the RNA.
Preparation of labeled genes
Total RNA (100 ng) was reverse transcribed into double-stranded DNA using the RevertAid First strand cDNA Synthesis kit (K1622; Thermo Fisher Scientific, Inc., Waltham, MA, USA), followed by in vitro transcription to prepare biotin-conjugated complementary RNA (cRNA) using the cRNA Amplification and Biotin Labeling kit (C5201s; ORANGENE, China). Next, the purified cRNA underwent fragmentation in a reaction system (total, 25 µl) containing: 5 µl cRNA (1,000 ng), 2.5 µl klenow buffer (5X), 1 µl klenow enzyme, 2.5 µl DNTP (2.5 mM), 14 µl ddH2O. Reaction conditions were as follows: 37°C for 1.5 h and 70°C for 10 min using DNA Polymerase I Large Klenow Fragment (M0210L; New England Biolabs Inc., Ipswich, UK).
Hybridization, washing and scanning of the microarrays
The molecular hybridization instrument used in the present study was a Affymetrix Genchip Scan System, which included a GeneChip Scanner 3000, GeneChip Hybridization Oven 645 and Fluidics Station 450 (Affymetrix, Inc., Santa Clara, CA, USA) along with a GeneChip Human Genome U133 Plus 2.0 Array (Affymetrix, Inc.). Briefly, the fragmented cRNA was used to prepare a hybridization solution, which was added to the microarray, followed by overnight hybridization at 45°C. The microarray was then placed in a fluid workstation for washing and staining by GeneChip Hybridization, Wash, and Stain kit (900720; Affymetrix, Inc.). Scanning and imaging of the microarrays following hybridization was performed using the Affymetrix GeneChip System 3000Dx (version 2; Affymetrix, Inc.).
Reverse transcription-quantitative PCR (RT-qPCR)
Representative genes with high expression levels were selected from the differentially expressed genes identified. Following the design and synthesis of primers (Table II), total RNA was extracted as described. Total RNA (100 ng) was reverse transcribed into double-stranded DNA using the RevertAid First strand cDNA Synthesis kit (K1622; Thermo Fisher Scientific, Inc.). qPCR was performed to amplify these target genes using Fast SYBR-Green Master Mix (4385610; Thermo Fisher Scientific, Inc.) by ABI 7900HT system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The reaction system (total, 20 µl) contained: 1 µl total RNA (200 ng), 0.5 µl sense primer, 0.5 µl reverse primer, 8 µl ddH2O and 10 µl SYBR Real-time PCR Mix. Thermal cycling conditions were as follows: One cycle of 94°C for 3 min, followed by 35 cycles of 94°C for 1 min, 56°C for 40 sec and 72°C for 40 sec; and final extension at 72°C for 5 min. Following 1.5% agarose gel electrophoresis, the PCR products were compared with their anticipated consequences. GAPDH was used as the internal control (Table II). Expression levels were analyzed using the 2-ΔΔCq method (8).
Table II.
Quantitiative polymerase chain reaction primer sequences and size of products (bp).
| Gene | Primer (5′-3′) | Product (bp) |
|---|---|---|
| TMX4 | F: AAGTCATCCCAGCCCTCTGAATCT | 104 |
| R: AGTGAAGAGTGTGGAGAGCCTGAA | ||
| FGL2 | F: AATGTCAAGGCAGGCAGATCACTT | 171 |
| R: CCTCTCAGGTTCAAGCGATTCTCC | ||
| GNLY | F: CACCTTGTCCTGTGGAAGAAGCA | 109 |
| R: GGAGACTGGAGAGTGGATTCTGGA | ||
| PF4V1 | F: GAGATGCTGTTCTTGGCGTTGCT | 189 |
| R: CGTGGCTATGAGTTGGGCAGTG | ||
| USP33 | F: GTTCAAGCGATTCTCCTGCCTCA | 188 |
| R: TCACACCTGTAATCCCAGCACTTT | ||
| GAPDH | F: CAGCAAGAGCACAAGAGGAAGAG | 109 |
| R: GGTCTACATGGCAACTGTGAGGAG | ||
| GBP1 | F: AGAAGAAGAAGTGAAGGCGGGAAT | 183 |
| R: AGTCTGGTCTGTCTGGAGAATTGC | ||
| LY96 | F: GCCGAGGATCTGATGACGATTACT | 190 |
| R: GGTGTAGGATGACAAACTCCAAGC | ||
| GAPDH | F: CCAGCAAGAGCACAAGAGGAAGAG | 109 |
| R: GGTCTACATGGCAACTGTGAGGAG |
TMX; thioredoxin related transmembrane protein 4; FGL2, fibrinogen like 2; GNLY, granulysin; PF4V1, platelet factor 4 variant 1; USP33, ubiquitin specific peptidase 33; GBP1, guanylate binding protein 1; LY96, lymphocyte antigen 96; bp, base pairs; F, forward; R, reverse.
Data analysis
The microarray data was obtained from both groups significance analysis of microarrays was performed using Affymetrix Expression Console Software 1.3 (Affymetrix, Inc.) and Microsoft Office Excel 2013 (Microsoft Corporation, Redmond, WA, USA), followed by analysis with the robust multi-array average method (9). Relative expression between genes was calculated, where a difference of ≤0.5 or ≥2 fold (P<0.05) and a false discovery rate of 0.05 was used to determine differentially expressed genes (10). To evaluate pathways associated with the microarray data, gene cluster analysis was performed using DAVID Bioinformatics Resources (11). Briefly, gene cluster identification numbers were uploaded onto DAVID and the KEGG Pathway (12) was selected to identify the pathways associated with the microarray data.
Results
Spinal CT and MRI
CT and MRI scans were taken prior to and following surgical debridement in patients with spinal TB, as shown in Fig. 1. For example, the example CT scan of a patient with spinal TB displayed a lost bone at lumbar vertebrae 1–2 (L1-2; Fig. 1A). MRI confirmed this loss of bone (Fig. 1B and C). The diseased area of the spine was removed through complete debridement and an interbody graft fusion was made at L1-2. CT and MRI scans 1 year following surgical debridement and UCCT confirmed interbody graft fusion and lesion healing (Fig. 1D and E).
Figure 1.
Spinal CT and MRI films prior to and following complete surgical debridement internal fixation of focal vertebrae and UCCT of L1-2. (A) Vertebral bone destruction and inter vertebral space disappearance of L1-2 revealed by CT exam prior to surgery. (B) T1-weighted MRI images. (C) T2-weighted MRI images. Severe destruction of L1-2 revealed by MRI exam prior to surgery. (D) Interbody graft fusion of L1-2 revealed by CT 1 year following surgery. (E) T1-weighted MRI images. (F) T2-weighted MRI images. Healing of the lesion of L1-2 revealed by MRI exam 1 year following surgery. CT, computerized topography; MRI, magnetic resonance imaging; UCCT, ultra-short course chemotherapy; L1-2, lumbar vertebrae 1–2.
Differentially expressed genes between the study and control groups
Following analysis for the microarray data, from the 54,675 genes evaluated, 2,971 statistically differentially expressed genes (fold-change ≤0.5 or ≥2; P<0.05) were found between the study groups and the control (Fig. 2). This suggests that over the course of spinal TB treatment, there was an upregulation of genes.
Figure 2.

Results of cluster analysis between the study groups (Groups 1, 2 and 3) and healthy controls. The color key shows the gene expression fold change in the study groups compared with healthy controls, respectively: −2 represents a decrease in expression; 0 represents equivalent expression levels; and +2 represents an increase in expression.
Whole blood transcriptional profiling of patients with active spinal TB
Patients with active TB were compared with healthy controls. A total of 125 differential genes were identified by gene cluster analysis (Fig. 3). Of these genes, 79 were upregulated and 46 were downregulated when comparing the control group with group 1 (untreated, TB-infected). Using the DAVID knowledgebase, analysis of the microarray data for pathways identified 2 pathways that were significantly different between patients with active TB and healthy controls: The antigen processing and presentation (APP) pathway and a small cell lung cancer related pathway. Furthermore, it showed that genes that were upregulated in the controls were found to be downregulated or unexpressed in group 1 (Fig. 3).
Figure 3.
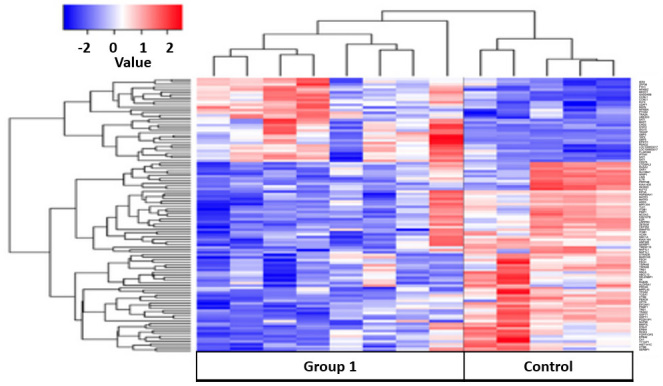
Results of cluster analysis between healthy control and untreated tuberculosis patients (Group 1). The color key shows the gene expression fold change in the healthy control group compared with Group 1, respectively: −2 represents a decrease in expression; 0 represents equivalent expression levels; and +2 represents an increase in expression.
Whole blood transcriptional profiling of patients following treatment
Spinal TB patients were treated with UCCT following complete debridement. Gene expression profiles were measured from these patients and compared with healthy controls. Cluster analysis identified 1,550 differential genes (Fig. 4). Of these genes, 66 genes were upregulated, while 1,484 genes were downregulated when comparing the control group with group 2 (treated, TB-infected). Analysis of the microarray data for pathways involved, using the DAVID knowledgebase, identified 35 pathways that differed between group 2 and the control group (Table III). These included immunoregulatory and inflammatory pathways, such as chemokine signaling pathways, B and T cell receptor pathways, and leukocyte transendothelial migration. The majority of genes that were downregulated in the healthy controls were upregulated in group 2 (Fig. 4). Additionally, changes in genes that were upregulated in the control groups were downregulated in group 2 (Fig. 4).
Figure. 4.

Results of cluster analysis between healthy control and treated tuberculosis patients (Group 2). The color key shows the gene expression fold change in the healthy control group compared with Group 2, respectively: −2 represents a decrease in expression; 0 represents equivalent expression levels; and +2 represents an increase in expression.
Table III.
Pathway analysis between the control group vs. group 2.
| Pathway | Count | P-value | Q-value |
|---|---|---|---|
| Natural killer cell mediated cytotoxicity | 31 | 8.40E-08 | 1.40E-05 |
| Spliceosome | 28 | 1.10E-06 | 6.30E-05 |
| B cell receptor signaling | 27 | 1.90E-04 | 3.90E-03 |
| Chemokine signaling | 27 | 3.00E-03 | 2.90E-02 |
| Endocytosis | 23 | 3.20E-02 | 2.10E-01 |
| Antigen processing and presentation | 22 | 1.10E-06 | 8.80E-05 |
| T cell receptor signaling | 21 | 2.50E-04 | 4.50E-03 |
| Lysosome | 20 | 1.80E-03 | 2.20E-02 |
| Toll-like receptor signaling | 19 | 8.00E-04 | 1.20E-02 |
| Leukocyte transendothelial migration | 16 | 4.40E-02 | 2.50E-01 |
| Cell adhesion molecules | 16 | 9.60E-02 | 3.80E-01 |
| RIG-I-like receptor signaling | 11 | 5.10E-02 | 2.70E-01 |
| NOD-like receptor signaling | 10 | 5.30E-02 | 2.80E-01 |
RIG, retinoic acid-inducible gene; NOD, nucleotide-binding oligomerization domain.
Whole blood gene transcriptional profiling of patients with spinal TB 1 year post-treatment
Patients with spinal TB were treated with UCCT following complete debridement. Peripheral blood samples were collected and gene expression profiles of these patients were compared with healthy controls. Cluster analysis identified 400 differentially expressed genes (Fig. 5). Of these genes, 65 were upregulated and 335 were downregulated when comparing the control group with group 3 (1 year follow-up). Bioinformatical pathway analysis, using the DAVID knowledgebase, identified 9 pathways that differed between the control group and the majority of group 3 (Table IV). A number of notable pathways that differed were as follows: Natural killer cell mediated cytotoxicity signaling, p53 signaling, T cell receptor signaling and apoptosis. Additionally, genes that were downregulated in the control group were upregulated in group 3 (Fig. 5).
Figure 5.
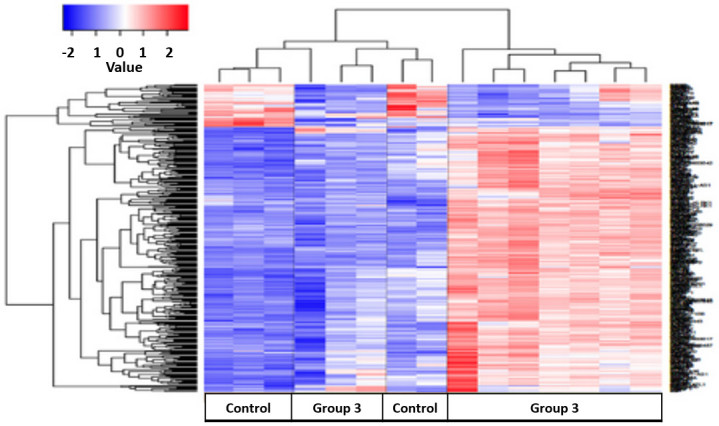
Results of cluster analysis between healthy control and 1-year follow-up, treated tuberculosis patients (Group 3). The color key shows the gene expression fold change in the healthy control group compared with Group 3, respectively: −2 represents a decrease in expression; 0 represents equivalent expression levels; and +2 represents an increase in expression.
Table IV.
Pathway analysis between the control group vs. group 3.
| Pathway | Count | P-value | Q-value |
|---|---|---|---|
| Ribosome | 10 | 5.60E-05 | 5.20E-03 |
| Natural killer cell mediated cytotoxicity | 9 | 5.20E-03 | 2.20E-01 |
| Spliceosome | 7 | 4.20E-02 | 5.50E-01 |
| p53 signaling pathway | 6 | 1.20E-02 | 3.10E-01 |
| Apoptosis | 6 | 3.10E-02 | 5.20E-01 |
| T cell receptor signaling | 6 | 6.70E-02 | 5.60E-01 |
| Graft-vs.-host disease | 4 | 4.40E-02 | 5.00E-01 |
| Type I diabetes mellitus | 4 | 5.20E-02 | 5.10E-01 |
| GPI-anchor biosynthesis | 3 | 9.00E-02 | 6.30E-01 |
GPI, glycosylphosphatidylinositol.
Comparison of whole blood gene transcriptional profiles of untreated vs. treated spinal TB patients. Gene expression profiles were compared between untreated (group 1) and treated (group 2) spinal TB patients
A total of 1,989 differentially expressed genes were identified (Fig. 6). Of these genes, 24 genes were upregulated and 1,965 genes were downregulated when comparing group 1 with group 2. Pathway analysis using the DAVID knowledgebase identified 41 pathways that differed between these two groups (Table V). The pathways identified were associated with immunoregulation, such as toll-like receptor signaling and FcγR receptor-mediated phagocytosis, and inflammation, such as chemokine and p53 signaling. A variety of cancer-associated pathways were activated, such as cell cycle signaling and pathways associated with non-small cell lung cancer, prostate cancer, colorectal cancer and glioma. In addition, ribosomal, proteasomal and apoptotic pathways were activated, which are associated with TB infection. The majority of group 2 displayed an upregulation of genes that were downregulated in group 1 (Fig. 6). There were a number of exceptions; one untreated patient with TB displayed an excessive upregulation of these genes, while two treated patients with TB did not show an increase in the upregulation of these genes.
Figure 6.

Results of cluster analysis between untreated TB patients (Group 1) and treated TB patients (Group 2). The color key shows the gene expression fold change in Group 1 compared with Group 2, respectively: −2 represents a decrease in expression; 0 represents equivalent expression levels; and +2 represents an increase in expression. TB, tuberculosis.
Table V.
Pathway analysis between group 1 vs. group 2.
| Pathway | Count | P-value | Q-value |
|---|---|---|---|
| Spliceosome | 33 | 3.40E-07 | 2.90E-05 |
| Regulation of actin cytoskeleton | 33 | 1.20E-02 | 1.00E-01 |
| Ribosome | 31 | 3.10E-10 | 5.40E-08 |
| Endocytosis | 28 | 2.40E-02 | 1.60E-01 |
| Chemokine signaling | 27 | 4.70E-02 | 2.40E-01 |
| Natural killer cell mediated cytotoxicity | 27 | 5.00E-04 | 2.10E-02 |
| Leukocyte transendothelial migration | 24 | 1.10E-03 | 3.10E-02 |
| T cell receptor signaling | 20 | 9.10E-03 | 1.10E-01 |
| Antigen processing and presentation | 17 | 6.70E-03 | 1.00E-01 |
| Toll-like receptor signaling | 16 | 7.20E-02 | 3.00E-01 |
| NOD-like receptor signaling | 12 | 4.00E-02 | 2.20E-02 |
| Graft-vs.-host disease | 9 | 3.50E-02 | 2.10E-02 |
NOD, nucleotide-binding oligomerization domain.
Comparison of whole blood gene transcriptional profiles of untreated vs. 1 year post-treatment patients with spinal TB
Gene expression profiles were compared between group 1 (untreated) and group 3 (1 year post-treatment) patients with spinal TB. A total of 824 differentially expressed genes were identified (Fig. 7). Of these genes, 46 were upregulated and 778 were downregulated when comparing group 1 with group 3. Pathway analysis using the DAVID knowledgebase identified 5 pathways that differed between these two groups (Table VI). These pathways were associated with the ribosome, spliceosome, APP, retinoic acid-inducible gene (RIG)-I-like receptor signaling and RNA degradation. The majority of group 3 displayed an upregulation of genes that were downregulated in group 1 (Fig. 7).
Figure 7.
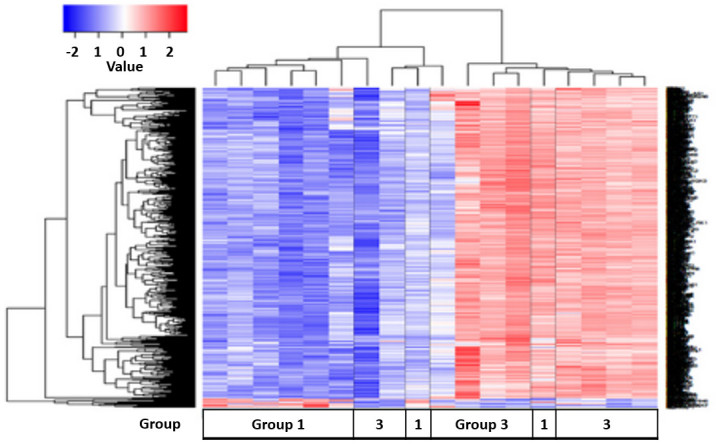
Results of cluster analysis between untreated TB patients (Group 1) and 1-year followed-up, treated TB patients (Group 3). The color key shows the gene expression fold change in Group 1 compared with Group 3, respectively: −2 represents a decrease in expression; 0 represents equivalent expression levels; and +2 represents an increase in expression. TB, tuberculosis.
Table VI.
Pathway analysis between group 1 vs. group 3.
| Pathway | Count | P-value | Q-value |
|---|---|---|---|
| Ribosome | 42 | 2.70E-35 | 3.40E-33 |
| Spliceosome | 16 | 1.40E-04 | 9.10E-03 |
| Antigen processing and presentation | 8 | 4.90E-02 | 8.80E-01 |
| RIG-I-like receptor signaling | 7 | 6.50E-02 | 8.90E-01 |
| RNA degradation | 6 | 7.80E-02 | 8.80E-01 |
RIG, retinoic acid-inducible gene.
Comparison of whole blood gene transcriptional profiles of treated vs. 1 year post-treatment patients with spinal TB
The gene expression profiles of group 2 (treated) and group 3 (1 year post-treatment) patients with spinal TB were compared. A total of 27 differentially expressed genes were upregulated when comparing group 2 with group 3 (Fig. 8). Pathway analysis using the DAVID knowledgebase identified 1 pathway that differed between these two groups; the RIG-I-like receptor signaling pathway (P=2.80×102; Q=5.50×102). The majority of group 3 displayed a downregulation of the genes that were upregulated in group 2 (Fig. 8).
Figure 8.
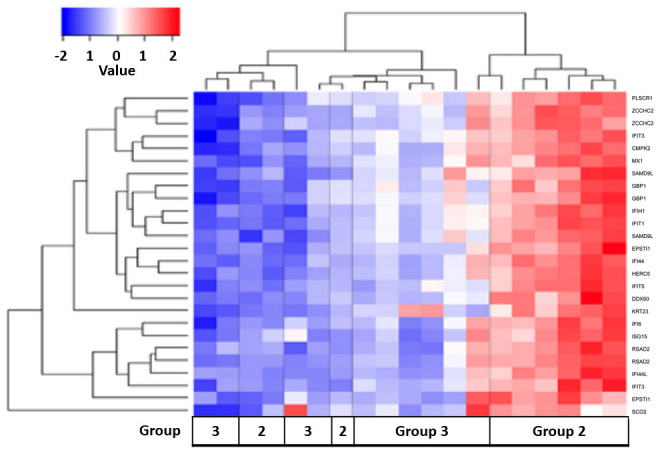
Results of cluster analysis between treated tuberculosis patients (Group 2) and 1-year followed-up, treated TB patients (Group 3). The color key shows the gene expression fold change in Group 2 compared with Group 3, respectively: −2 represents a decrease in expression; 0 represents equivalent expression levels; and +2 represents an increase in expression.
RT-PCR validation
RT-PCR validations were run on several genes. From the total microarray data, the upregulated genes TMX4 (thioredoxin related transmembrane protein 4), FGL2 (fibrinogen like 2) and GNLY (granulysin), and the downregulated genes PF4V1 (platelet factor 4 variant 1) and USP33 (ubiquitin specific peptidase 33), were selected for RT-PCR validation with GAPDH as the internal control (Table II). In a separate analysis, GBP1 (guanylate binding protein 1) and LY96 (lymphocyte antigen 96) expression levels, with GAPDH as an internal reference, were validated with RT-PCR (Table II). The findings from this RT-PCR were similar to those observed in the microarray assays.
Discussion
The Affymetrix GeneChip has ~50,000 probes for examining gene expression. Considering the differences between individuals, peripheral blood samples were collected from patients at various treatment periods. The consistency of the study and clinical results demonstrated the reliability of this detection method. Previous studies reported that UCCT lasting <6 months has proved effective in the initial treatment of pulmonary TB (10–13). In addition, complete debridement and UCCT for spinal TB have proven to be clinically effective (4–6). In the present study, it is demonstrated that using DNA microarray technology to determine whole blood transcriptional profiles at various stages of spinal TB treatment is a promising adjunctive therapeutic tool. Transcriptional profiling can be used to measure the changes in gene expression prior to and following treatment, so it can be used to indirectly evaluate the therapeutic efficacy of treatment and accurately determine the type and severity of infection (10,14–17).
DNA microarray technology and gene cluster analysis using the DAVID knowledgebase were used to detect the gene expression profiles from the peripheral blood of patients with spinal TB at the three different stages: Prior to spinal TB treatment (group 1), following the completion of UCCT treatment (group 2) and 1 year post-UCCT treatment (group 3). These profiles were then compared with those of healthy controls. As shown in the heat map of all the groups, a transition from the downregulation of certain genes to upregulation of certain genes was identified over the course of spinal TB treatment. It was observed that following the removal of TB lesions with surgical debridement and subsequent UCCT, the expression of certain genes increased in the peripheral blood of the treated groups (group 2 and group 3). This heat maps provide an overview of the changes in the gene expression from the microarray data. The 2,971-transcriptional profile can be used to observe whether the treatment has an effect on spinal TB.
Furthermore, analysis of the groups in comparison to one another provided an overview of the effects of spinal TB treatment on changes in gene expression. For example, although few changes were identified between the control group and group≈1, a separate analysis identified 125 differentially expressed genes between the two groups. Profiling expression of these 125 genes may prove useful in separating TB-infected individuals from uninfected individuals. In addition, transcriptional profiling provided an indication of the infection phases of TB. Bioinformatical pathway analysis using the DAVID knowledgebase between TB infected patients and healthy controls identified 2 pathways that are associated with TB infection: Small cell lung cancer associated signaling and the APP pathway. These pathway changes are consistent with TB infection, since TB primarily infects the lung. The changes observed in small cell lung cancer-associated signaling indicated the infectious phase of TB spread from the lungs to the spine. Observed activity in the APP pathway indicated that TB infection was triggering an immune response. One explanation for the downregulation of genes that were upregulated in the control group, is that TB infection is downregulating the expression of genes that would interfere with its spread. Further studies are needed to explore the genes that are downregulated following TB infection.
In patients with spinal TB who underwent UCCT treatment, 1,550 differentially expressed genes were identified from cluster analysis. It was observed that genes that were downregulated in the control group were upregulated in UCCT-treated patients. This transcriptional profile was found to be associated with several immune response-related pathways including the following: Autophagy, inflammation, apoptosis and protein degradation. This shows that the clearance of TB and UCCT treatment serve a primary role in inducing an immune response. This 1,550-gene transcriptional profile reflects the changes in gene expression between individuals exposed to UCCT compared with healthy controls. The activation of apoptosis and protein degradation pathways are indicative of the toxic side effects of chemotherapy. However, activation of B and T cell receptors, APP and natural killer cell-mediated cytotoxicity pathways were additionally noted. This suggests that treatment with UCCT and complete debridement supports the clearance of spinal TB.
In patients who had completed treatment (1 year follow-up), there was a change in the expression of 400 genes and a reduced number of altered cell death pathways compared to healthy controls. A continued upregulation in these genes in comparison to the control group was observed. The upregulation of these genes is not attributable to UCCT, as treatment was 1 year ago. Pathway analysis identified that, compared with the control group, a lesser number of immune response, inflammation and apoptosis associated pathways were changed. These changes demonstrate that patients are recovering both from TB and the chemotherapy treatment as spinal tuberculosis itself is an inflammatory reaction; as the inflammatory response after one year follow-up indicators are decreased, the treatment is effective.
The present study identified transcriptional profiles that changed over the course of spinal TB treatment. In addition to evaluating the profile changes, the transcriptional profiles of untreated and treated spinal TB groups were compared, along with comparing the 1 year follow-up group with the treated TB group. The transcriptional profiles of 1,989 genes were found to differ between the untreated and treated spinal TB patients. Pathway analysis identified 41 pathways that differed between the two groups. These pathways were associated with cancer, inflammation, the immune response and apoptosis. The heat map confirmed that the treated patient displayed an upregulation of genes associated with those pathways. Interestingly, when comparing the untreated and the 1-year post-treatment of patients with spinal TB, the number of differentially expressed genes reduced to 824, which were associated with protein degradation and the immune system. Although the genes associated with these pathways are upregulated in group 3, the decreased number of upregulated genes compared with the untreated group showed that the individuals were recovering.
The difference in the transcriptional profiles between the treated spinal TB group and the 1 year post-treatment group was examined to evaluate the changes in gene expression following UCCT and complete debridement. There was no increase in the number of differentially expressed gene. Instead, it was observed that there were only 27 differentially expressed genes between group 2 and group 3. These genes are associated with the RIG-1 like receptor pathway. These receptors recognize viruses, which trigger an innate immune response. The reduced number of differentially expressed genes showed that patient's gene expression levels are returning to normal 1 year post-treatment. The heat maps showed that the differentially expressed genes were downregulated when comparing group 3 with group 2. In addition, the heat maps confirmed that following therapy, a number of genes are downregulated to similar levels of those in the control group.
The present study shows that the gene expression in the peripheral blood of spinal TB patients changes over the course of treatment. The gene expression levels measured are consistent with the clinical results in the current study, indicating that complete surgical debridement and 5–6 months of UCCT have therapeutic efficacy on spinal TB. The complete debridement of TB lesions removes the barriers that prevent the medication from targeting the infected area (9). Analysis of the upregulated differentially expressed genes found that they are involved in immune response pathways. These changes at the different stages of treatment indicate the clearance of TB with UCCT and complete debridement. The removal of TB from the lesion relieved the host immune system and promoted expression of regulatory factors that are involved in clearing the bacteria (18).
In conclusion, to the best of our knowledge, the present study has demonstrated for the first time that DNA microarray technology can detect and monitor the changes in gene expression in the peripheral blood cells of patients with spinal TB at different therapeutic stages. This is shown through the consistency of the study results, clinical results and the differentially expressed genes associated with immune responses between groups. Evaluating the therapeutic efficacy of spinal TB treatments could prove an important application of DNA microarray technology. In addition, the microarray data allowed the use of heat maps to monitor changes during TB infection and treatment with UCCT. These heat maps can be used to determine how effective TB treatments are. Additionally, the present study provides an important insight into novel genes involved in the immune response against TB. The next step is to screen for UCCT-specific gene targets, in order to elucidate genes involved in the chemotherapeutic response.
Acknowledgements
The present study was funded by the National Natural Science Foundation of China (grant no. 81260282) and the Ningxia Natural Science Foundation (grant nos. NZ11195 and NZ11275).
References
- 1.Onozaki I. National surveys of the prevalence of tuberculosis disease-overview, progress and lessons learnt. Kekkaku. 2013;88:777–783. (In Japanese) [Google Scholar]
- 2.Rasouli MR, Mirkoohi M, Vaccaro AR, Yarandi KK, Rahimi-Movaghar V. Spinal tuberculosis: Diagnosis and management. Asian Spine J. 2012;6:294–308. doi: 10.4184/asj.2012.6.4.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karaeminogullari O, Aydinli U, Ozerdemoglu R, Ozturk C. Tuberculosis of the lumbar spine: Outcomes after combined treatment of two-drug therapy and surgery. Orthopedics. 2007;30:55–59. doi: 10.3928/01477447-20070101-15. [DOI] [PubMed] [Google Scholar]
- 4.Wang Z, Shi J, Geng G, Qiu H. Ultra-short-course chemotherapy for spinal tuberculosis: Five years of observation. Eur Spine J. 2013;22:274–281. doi: 10.1007/s00586-012-2536-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin W, Wang Z. Clinical evaluation of the stability of single-segment short pedicle screw fixation for the reconstruction of lumbar and sacral tuberculosis lesions. Arch Orthop Trauma Surg. 2012;132:1429–1435. doi: 10.1007/s00402-012-1575-7. [DOI] [PubMed] [Google Scholar]
- 6.Wang Z, Ge Z, Jin W, Qiao Y, Ding H, Zhao H, Lin Z, Chen J, Yang W. Treatment of spinal tuberculosis with ultrashort-course chemotherapy in conjunction with partial excision of pathologic vertebrae. Spine J. 2007;7:671–681. doi: 10.1016/j.spinee.2006.07.016. [DOI] [PubMed] [Google Scholar]
- 7.Wang Z. Complete removal of the lesions of spinal tuberculosis and its related issues opinion. Chinese Journal of Spine and Spinal Cord. 2008;18:568–570. [Google Scholar]
- 8.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 9.Ge Z, Wang Z, Wei M. Measurement of the concentration of three antituberculosis drugs in the focus of spinal tuberculosis. Eur Spine J. 2008;17:1482–1487. doi: 10.1007/s00586-008-0778-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. doi: 10.1186/gb-2003-4-9-r60. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y, Wang Y, Teng Z, Zhang X, Ding M, Zhang Z, Chen J, Xu Y. DNA microarray analysis in screening features of genes involved in spinal cord Injury. Med Sci Monit. 2016;22:1571–1581. doi: 10.12659/MSM.895889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Young DB, Perkins MD, Duncan K, Barry CE., III Confronting the scientific obstacles to global control of tuberculosis. J Clin Invest. 2008;118:1255–1265. doi: 10.1172/JCI34614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mistry R, Cliff JM, Clayton CL, Beyers N, Mohamed YS, Wilson PA, Dockrell HM, Wallace DM, van Helden PD, Duncan K, Lukey PT. Gene-expression patterns in whole blood identify subjects at risk for recurrent tuberculosis. J Infect Dis. 2007;195:357–365. doi: 10.1086/510397. [DOI] [PubMed] [Google Scholar]
- 15.Jacobsen M, Repsilber D, Gutschmidt A, Neher A, Feldmann K, Mollenkopf HJ, Ziegler A, Kaufmann SH. Candidate biomarkers for discrimination between infection and disease caused by Mycobacterium tuberculosis. J Mol Med (Berl) 2007;85:613–621. doi: 10.1007/s00109-007-0157-6. [DOI] [PubMed] [Google Scholar]
- 16.Ockenhouse CF, Bernstein WB, Wang Z, Vahey MT. Functional genomic relationships in HIV-1 disease revealed by gene-expression profiling of primary human peripheral blood mononuclear cells. J Infect Dis. 2005;191:2064–2074. doi: 10.1086/430321. [DOI] [PubMed] [Google Scholar]
- 17.Bleharski JR, Li H, Meinken C, Graeber TG, Ochoa MT, Yamamura M, Burdick A, Sarno EN, Wagner M, Röllinghoff M, et al. Use of genetic profiling in leprosy to discriminate clinical forms of the disease. Science. 2003;301:1527–1530. doi: 10.1126/science.1087785. [DOI] [PubMed] [Google Scholar]
- 18.Cooper AM. Cell-mediated immune responses in tuberculosis. Annu Rev Immunol. 2009;27:393–422. doi: 10.1146/annurev.immunol.021908.132703. [DOI] [PMC free article] [PubMed] [Google Scholar]



