Abstract
The molecular mechanism that leads to pregnancy-induced hypertension (PIH), a pregnancy-specific syndrome, remains poorly understood. It has been suggested that microRNAs (miRNAs) may be potentially useful biomarkers for severe preeclampsia (PE), which is an important condition associated with PIH. The aim of the present study was to identify miR-204 by verifying differentially expressed serum miRNAs in patients with PIH during pregnancy compared with normal controls. Subsequently, the effects of miR-204 on proliferation and apoptosis of human choriocarcinoma (JAR) cells in hypoxic microenvironment were investigated. Previous studies indicated a number of miRNA candidates and the present study validated the expression of eight miRNAs in serum samples using reverse transcription-quantitative polymerase chain reaction (RT-qPCR). A higher expression of miR-204 was identified in patients with PIH. To assess the impact of miR-204 inhibition on hypoxic JAR cells function in vitro, cell proliferation was detected using a Cell Counting Kit-8 assay. The rate of apoptosis and cell cycle progression was then examined by flow cytometry. RT-qPCR confirmed that serum miR-204-5p is more highly expressed in patients with PIH. Further statistical analysis indicated that the survival ratio of JAR cells in hypoxic microenvironments was increased in the miR-204-5p inhibitor group. However, the miR-204-5p inhibitor protected hypoxic JAR cells from apoptosis. The analysis of cell-cycle status demonstrated that the percentage of cells in the G2/G1 phase was larger compared with the control group. The results of the present study suggest that low levels of miR-204-5p may increase cell proliferation and reduce cell apoptosis with cell cycle changes in vitro. Therefore, serum miR-204-5p may be used as a notable biomarker for the diagnosis, prevention and treatment of PIH.
Keywords: pregnancy-induced hypertension, preeclampsia, miR-204-5p, cell proliferation, cell apoptosis
Introduction
Pregnancy-induced hypertension (PIH) is a syndrome that occurs during pregnancy. PIH is the primary cause of maternal, neonatal and fetal mortality and morbidity (1). PIH is defined by a diastolic blood pressure (DBP) >90 mmHg and systolic blood pressure (SBP) >140 mmHg, and causes complications in 6–10% of pregnancies. Therefore, PIH is classified as either mild (SBP, 140–149 mmHg; and DBP, 90–99 mmHg), moderate (SBP, 150–159 mmHg; and DBP, 100–109 mmHg) or severe (SBP, ≥160mmHg; and DBP, ≥110 mmHg) (2,3). According to the Canadian Hypertension Society (4), an important condition associated with PIH is gestational hypertension and preeclampsia (PE), a common health problem with adverse effects for fetus and mother (5). It has been indicated that patients with PIH may be at a higher long-term risk of other medical conditions in later life, including diabetes mellitus, hypertension, kidney disease and cardiovascular disease (6). Indeed, a positive association between PIH and hypertensive diseases was observed in a study of 3,593 women with PE during their first singleton pregnancy (7).
It has been suggested that PIH is impacted by endothelial cell injury occurring during maternal and blood vessel formation disorder in the placenta, which may result from different cell factors generated in placental hypoxia (8). However, the pathogenic molecular mechanism of PIH is not yet fully understood and therefore there is difficulty in monitoring disease progress. PIH is a genetic disease that develops through a multi-step process (9,10). Significant progress in the development of novel therapeutic agents may improve the understanding of the molecular basis of gene-disease interactions. Although a number of challenges remain regarding the prediction, prevention and management of PIH, previous studies have suggested that serum-based microRNAs (miRNAs) may be potential biomarkers or effective therapy for early detection, diagnosis and follow-up of severe PE (11,12). This may provide novel suggestions for the management of PIH.
The discovery of miRNAs was a landmark milestone in molecular biology and pharmacology. miRNAs are a series of small (18–24 nucleotides) endogenous noncoding single-stranded RNAs, which can post-transcriptionally regulate target mRNAs by a non-perfect pairing of 6–8 nucleotides (13). Target mRNAs are subsequently degraded by the formation of RNA-induced silencing complex, which suggests that miRNAs may control a range of different biological functions including cellular differentiation, proliferation and apoptosis (14). A higher expression of miRNA has been identified in the placenta of patients with PIH, suggesting that they may have a function in the placenta of patients with severe PE. These miRNAs include miR-92b, miR-342-3p, miR-197, miR-25, miR-296-5p, miR-26a, miR-202, miR-198, miR-95 and miR-204-5p (15–17), and alterations in miRNA expression may serve a critical role in disease progression due to the negative transcriptional or post-transcriptional regulation of important target genes (18). The present study aimed to examine whether the aforementioned miRNAs that underwent significant alterations in expression during the course of PIH are regulated by miRNAs.
In the present study, differentially expressed miRNAs were investigated in the serum of patients with PIH using reverse transcription-quantitative polymerase chain reaction (RT-qPCR). The results indicate that miR-204-5p may affect cell proliferation, cell apoptosis and cell cycle progression in the hypoxic model in vitro. The results of the present study may be therefore be the basis for further studies to examine the pathophysiological mechanism and identify PIH biomarkers in order to improve the diagnosis, prevention and treatment of PIH.
Materials and methods
Patient characteristics, clinical features and serum harvest
For the identification of PIH-specific serum miRNAs, a total of 40 patients were selected: 20 healthy female volunteers as controls and 20 female patients with PIH, were recruited from the Third Affiliated Hospital, Sun Yat-sen University (Guangzhou, China; Table I) between December 2014 and March 2015, having given informed consent to be included in the present study. Differentially expressed miRNAs were directly validated using RT-qPCR according to previous research (19). As presented in Table I, the exclusion criteria for both groups included: Patients with kidney disease or essential hypertension, a history of alcohol or drug abuse, and illegal drug addiction within the 6 months prior to signing the informed consent. Furthermore, the PIH patients were all pathologically diagnosed by doctors and all blood samples were collected prior to any surgery, chemotherapy and/or radiation treatment.
Table I.
Characteristics of study subjects in the control and PIH groups.
| Onset | |||||
|---|---|---|---|---|---|
| Group | Age, years | Height, cm | Weight, kg | GA, weeks | BP, mmHg |
| Control | 25.84±3.56 (range, 23–34) | 161.54±13.28 | 67.35±7.68 | 35.32±2.79 | 140.23±18.32 |
| PIH | 27.23±6.42 (range, 21–34) | 165.32±17.45 | 67.29±6.83 | 34.92±1.98 | 200.35±12.91 |
Data are presented as mean ± standard deviation. PIH, pregnancy-induced hypertension; GA, gestational age; BP, blood pressure.
From each patient, 5 ml venous blood was collected on first admission to the hospital. Blood was drawn into a sterile tube without anticoagulant to harvest cell-free serum. The tube was left in a standing position for 20 min prior to centrifugation at 20°C and 1,500 × g for 10 min. The supernatant serum was quickly removed by pipette and stored immediately at −80°C until analysis. The present study was approved by the Ethics Committee from the Third Affiliated Hospital, Sun Yat-sen University and a signed informed consent form was obtained from each participant prior to the study.
Cell culture and treatment
The human choriocarcinoma (JAR) cell line was obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). JAR cells were cultured for 24 h in growth media containing high glucose-Dulbecco's modified Eagle's medium and supplemented with 10% fetal bovine serum (Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) and 1% penicillin/streptomycin (Mediatech, Inc., Manassas, VA, USA) in a humidified atmosphere of 5% CO2 and a temperature of 37°C.
Following dilution into single cell suspensions and seeding into 96-well plates (1×104 cells/well), a JAR cell hypoxic model was induced using an AnaeroPack® system (Mitsubushi Gas Chemical America, Inc., New York, NY, USA) for 48 and 72 h, respectively, prior to harvesting for total RNA isolation (20).
Total RNA isolation and reverse transcription
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) was used to prepare total RNA and subsequently, 75% ethanol replaced isopropanol for RNA precipitation, according to the manufacturer's protocol. RNA quality was determined using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Inc., Wilmington, DE, USA). A total of 1 µg RNA was reverse-transcribed into cDNA using a DBI Bestar® qPCR RT kit (DBI Bioscience, Ludwigshafen, Germany) according to the manufacturer's protocol.
Quantitative polymerase chain reaction (qPCR) of mature miRNAs
RT-qPCR was performed using a 7500 Fast Real-Time PCR System Light Cycler (Applied Biosystems; Thermo Fisher Scientific, Inc.). The 20 µl PCR reaction included 1 µl reverse transcription product (1:5), 0.5 µl sense primer, 0.5 µl universal reverse primer and 10 µl DBI-2043 Bestar® Real time PCR Master Mix (DBI Bioscience). The reactions were incubated at 94°C for 2 min in a 96-well optical plate, followed by 40 cycles of 94°C for 20 sec, 8°C for 20 sec and 72°C for 20 sec. All reactions were completed in triplicate and primer sequences are listed in Table II. mRNAs were quantified using the 2−ΔΔCq formula (21).
Table II.
Sequences of primers used to validate selected miRNAs.
| Gene | Sequence (5′-3′) |
|---|---|
| miR-197-3p | F: ACACTCCAGCTGGGTTCACCACCTTCT |
| miR-26a-5p | F: ACACTCCAGCTGGGTTCAAGTAATCCA |
| miR198 | F: ACACTCCAGCTGGGGGTCCAGAGG |
| miR-204-5p | F: ACACTCCAGCTGGGTTCCCTTTGTCA |
| miR-92 | F: TATTGCACTCGTCCCGGCCTCC |
| miR-95-5p | F: ACACTCCAGCTGGGTCAATAAATGTC |
| miR-202-3p | F: ACACTCCAGCTGGGAGAGGTATAGG |
| miR-296-5p | F: ACACTCCAGCTGGGAGGGCCCCCC |
| For all | R: TGGTGTCGTGGAGTCG |
| U6 | F: CTCGCTTCGGCAGCACA |
| R: AACGCTTCACGAATTTGCGT |
miR, microRNA. F, forward; R, reverse.
Cell proliferation detection
miR-204-5p inhibitor (Shanghai GenePharma, Ltd., Shanghai, China) was transfected into the hypoxic JAR cells using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) prior to incubation in DMEM without FBS at 37°C for 48 h. JAR cells were then transfected with 100 nM miR-204-5p inhibitor and subjected to the hypoxia precondition for 48, 72 and 96 h respectively. Then, 100 µl Cell Counting Kit-8 solution (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was added to each well and incubation was completed for 1 h at 37°C. Absorbance was measured at 450 nm using a microplate reader.
Apoptosis assay
Following transfection of the miRNA inhibitor in the hypoxic environment for 48 h, quantification of the apoptotic cells was completed using the Annexin V-FITC/PI apoptosis detection kit (Merck Millipore, Darmstadt, Germany). JAR cells were collected by trypsin digestion method (22), washed with phosphate buffered saline (PBS) and re-suspended in 200 µl binding buffer containing 5 µl Annexin V (10 µg/ml) in DMEM with FBS at 37°C for 10 min in the dark. The cells then underwent incubation with 10 µl PI (20 µg/ml) for 15 min and samples were analyzed using an EPICS® XL™ flow cytometer (Beckman Coulter, Inc., Brea, CA, USA). Data acquisition and analysis were performed using CellQuest™ software version 5.1 (BD Biosciences, Franklin Lakes, NJ, USA) (23,24).
Cell cycle assay
Following transfection of the miRNA inhibitor into the hypoxic environment and incubation at 37°C for 48 h, JAR cells were collected by trypsin digestion method and washed with PBS prior to re-suspension in 250 µl DMEM. Cold (4°C) dehydrated ethanol (99%) was added to this buffer and incubated overnight at 4°C. Following treatment, cells were collected and incubated with 200 µl PI (20 µg/ml) using a cell cycle assay kit (Vazyme Biotech, Co., Ltd., Nanjing, China) at 37°C for 15 min. Samples were immediately analyzed using flow cytometry (EPICS® XL™; Beckman Coulter, Inc.). Data acquisition and analysis were performed using CellQuest software version 5.1 (BD Biosciences) (23,24).
Data analysis
For RT-qPCR data analysis, the relative quantification method was used to determine the changes in the expression of the target miRNAs. U6 RNA was used to normalize the expression and change in amplification. The fold change in expression was calculated for each sample using 2−ΔΔCq, where ΔΔCq=(Cq target gene-CqU6) PIH-(Cq target gene-CqU6) control (25). A value of 2−ΔΔCq >1.5 or <0.67 was considered to represent differentially expressed miRNA. The Welch t-test was used to assess the differential expression of miRNA measured by RT-qPCR.
For other data analysis, Statistical analysis was performed using SPSS, version 17.0 (SPSS, Inc., Chicago, IL, USA). One-way analysis of variance was used to compare log10-transformed relative quantities of target miRNAs between all groups. Bartlett's test was used to assess the differences in variance between genes. P<0.05 was considered to represent a statistically significant difference for all experiments.
Results
Validation of eight serum miRNAs in patients with PIH
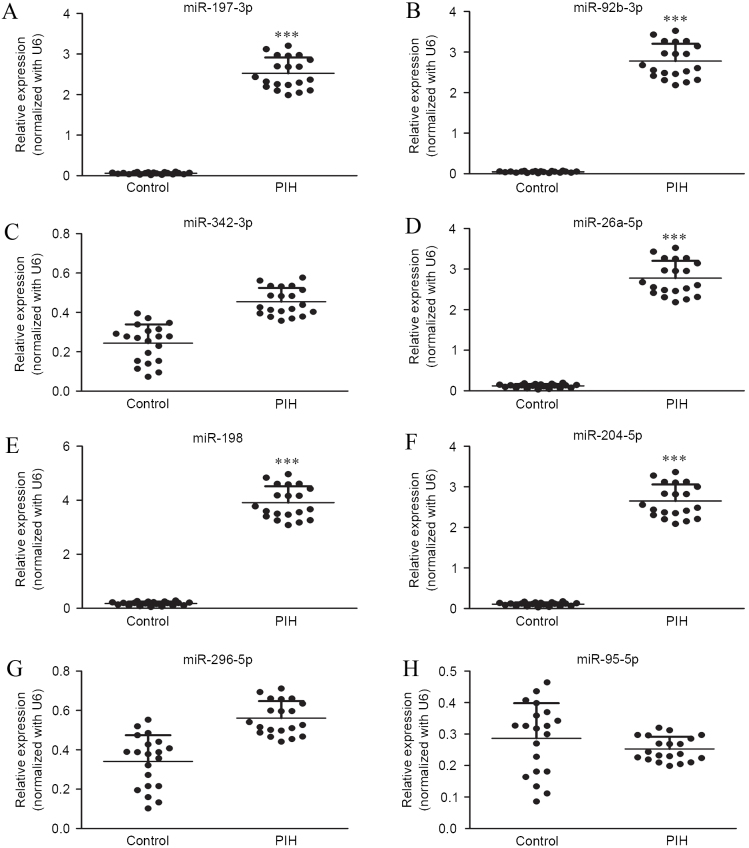
The expression profile of eight serum microRNAs were selected, based on a previous study, (under review, data not shown), for further examination in 18 PIH patients that represented a significant 2-fold change using RT-qPCR. Expression levels of miR-197-3p, miR-92b-3p, miR-342-3p, miR-26a-5p, miR-198, miR-204-5p, miR-296-5p and miR-95-5p were measured in the serum samples of 12 patients with PIH and 6 normal controls (Fig. 1). The results showed significantly elevated expression of miR-197-3p, miR-92b-3p, miR-26a-5p, miR-198 and miR-204-5p in patients with PIH compared with controls (P<0.001; Fig. 1A, B and D-F). However, levels of miR-342-3p, miR-296-5p and miR-95-5p not differ significantly between PIH patients and controls (Fig. 1C and G-H).
Figure 1.
Expression of miRNAs in serum samples of healthy controls and patients with PIH, detected using reverse transcription-quantitative polymerase chain reaction. Expression of (A) miR-197-3p, (B) miR-92b-3p, (C) miR-342-3p, (D) miR-26a-5p, (E) miR-198, (F) miR-204-5p, (G) miR-296-5p and (H) miR-95-5p in serum samples of healthy normal controls and patients with PIH. Data are presented as the mean ± standard deviation. All experiments were repeated independently three times. ***P<0.001 vs. control group. miR, microRNA; PIH, pregnancy-induced hypertension.
Expression of miRNAs in the hypoxic environment in vitro
The expression of miR-197-3p, miR-92b-3p, miR-26a-5p, miR-198 and miR-204-5p were selected to be assessed in vitro, on the basis of the results of the aforementioned clinical sample examination. Only miR-204-5p expression increased significantly in a time-dependent manner in hypoxic JAR cells (0, 48 and 72 h; Fig. 2).
Figure 2.
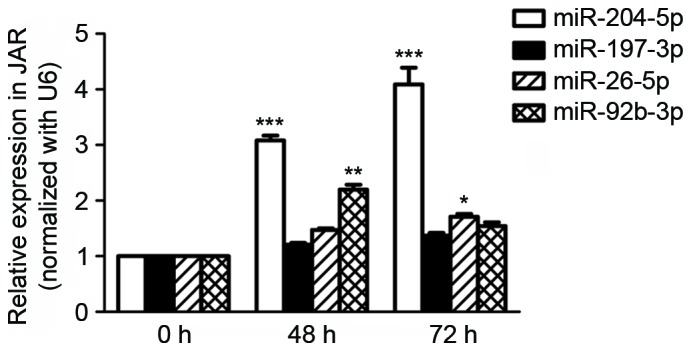
Expression of miR-197-3p, miR-92b-3p, miR-26a-5p, miR-198 and miR-204-5p in human choriocarcinoma JAR cells following hypoxia pre-treatment for 0, 48 and 72 h respectively, detected using reverse transcription-quantitative polymerase chain reaction. Data are presented as the mean ± standard deviation. All experiments were repeated independently three times. *P<0.05; **P<0.01; ***P<0.001 vs. 0 h. miR, microRNA.
Effect of miR-204-5p inhibitor on cell proliferation
The effect of miR-204-5p inhibitor on JAR cell proliferation was subsequently assessed. In hypoxia pre-treatment JAR cells, cellular proliferation was enhanced significantly in a time-independent manner in the miR-204-5p inhibitor group, as indicated by the number of cells detected at each time point (P<0.001; Fig. 3).
Figure 3.
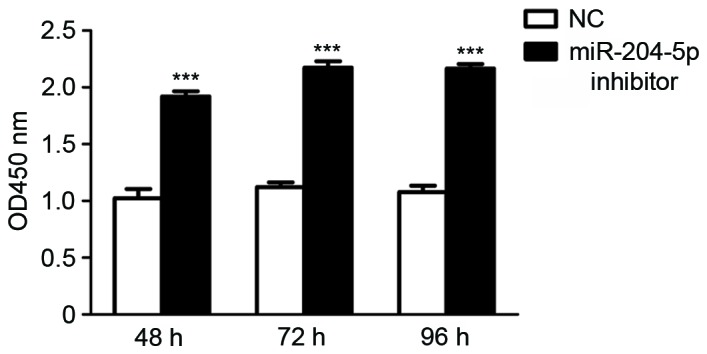
miR-204-5p inhibitor protects growth of JAR cells in the hypoxic environment in vitro. Cell viability was measured using a Cell Counting Kit-8 assay. Cells were counted at 48, 72 and 96 h. Data are presented as the mean ± standard deviation. All experiments were repeated independently three times. ***P<0.001 vs. NC group. NC, normal control group, OD, optical density; miR, microRNA.
Effects of miR-204-5p inhibitor on cell apoptosis and cell cycle progression
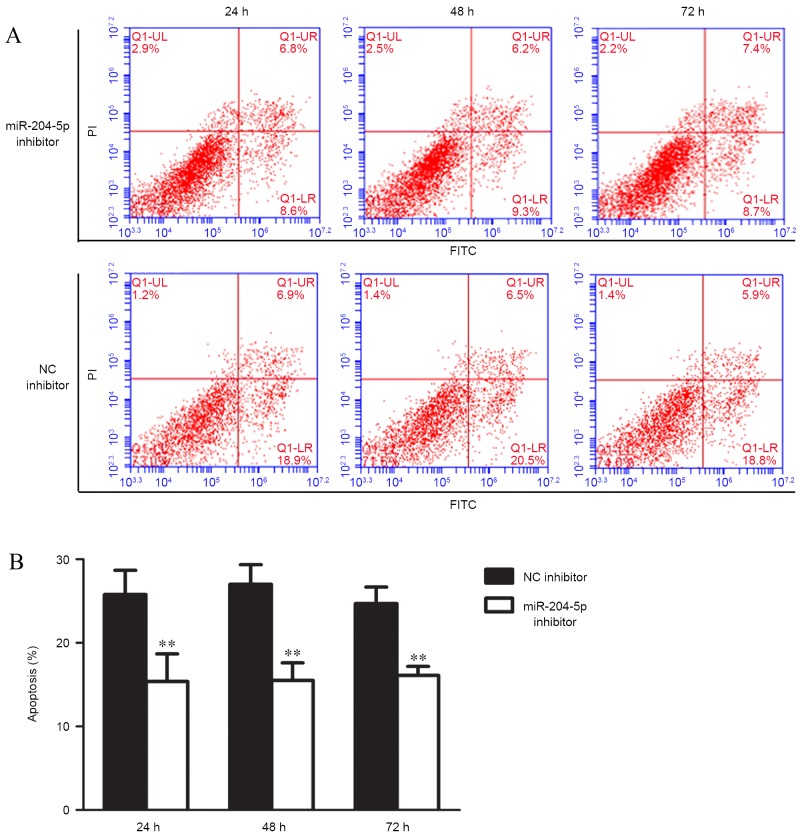
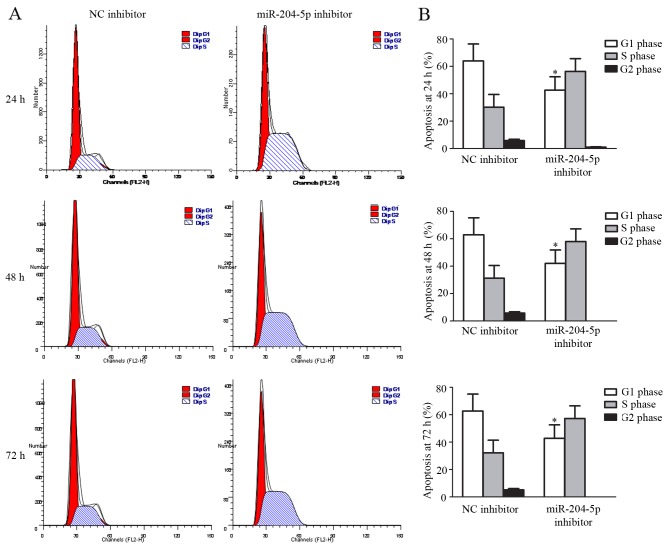
Apoptosis and cell cycle distribution were analyzed using flow cytometry following the transfection of miR-204-5p inhibitor and hypoxia pre-treatment for 48 h. Compared with the control group, JAR cell exposure to miR-204-5p inhibitor exhibited typical protection from apoptotic morphology with nuclear fragmentation, cell shrinkage and cellular rupture into debris. Apoptosis occurred at a significantly higher rate in cells treated with the negative control compared with the group treated with miR-204-5p inhibitor (P<0.01; Fig. 4). Assessment of the cell cycle indicated that the ratio of cells in the G2/G1 phase increased, although this increase was not significant. However, G1 cell cycle arrest was significantly reduced following transfection with the miR-204-5p inhibitor (P<0.05; Fig. 5).
Figure 4.
Induction of JAR apoptosis. (A) Cell apoptosis following hypoxia treatment for 24, 48 and 72 h. (B) Percentage of JAR cells undergoing apoptosis at different time points during hypoxia. Data are presented as the mean ± standard deviation. All experiments were repeated independently three times. **P<0.01 vs. NC inhibitor group. NC, normal control; PI, propidium iodide; FITC, fluorescein isothiocyanate; miR, microRNA.
Figure 5.
Induction of JAR cell cycle change. (A) Cell cycle distribution following hypoxia treatment for 24, 48 and 72 h. (B) Percentage of JAR cells in cycle in S, G1 and G2 phases following 48 h of hypoxia. Data are presented as the mean ± standard deviation. All experiments were repeated independently three times. *P<0.05 vs. G1 phase arrest in NC group at the same time point. NC, normal control; miR, microRNA.
Discussion
To the best of our knowledge, the molecular mechanism during PIH pathogenesis, which is responsible for the occurrence and progression of the syndrome, remains unknown. miRNAs may target a large number of genes and participate in numerous cellular events that may be important in maintaining homoeostasis in a number of organs, including the placenta (26). In the present study, due to the developing understanding of miRNA expression in PIH, the expression of eight miRNAs was identified in the serum samples of healthy controls and patients with PIH using RT-qPCR. The results of the present study identified that five of the eight miRNAs (miR-197-3p, miR-92b-3p, miR-26a-5p, miR-198 and miR-204-5p) are more highly expressed in patients with PIH (Fig. 1). This indicates that a number of miRNAs may impact the course of PIH. Further detection of these miRNAs using a hypoxia pre-treatment cell model demonstrated that only miR-204-5p expression increased in a time-dependent manner in hypoxic JAR cells (Fig. 2), which suggests that miR-204-5p reduced JAR cell proliferation as a suppressor in the hypoxic environment. Effects of the miR-204-5p inhibitor on cell function were also examined, the results of which suggested that the miR-204-5p inhibitor may enhance the cellular population of JAR cells in the hypoxic environment (Fig. 3) and protect JAR cells from apoptosis (Fig. 4) by reducing G1 cell cycle arrest (Fig. 5). The data collected in the present study indicates that miR-204-5p may be an important therapeutic target for the improved prediction, prevention and treatment of PIH.
The present study produced evidence that expression of miR-204-5p, which acts as a potent cell survival suppressor in vitro, is somatically increased in patients with PIH. This is consistent with the results from a previous study, which indicated that miR-204 is also upregulated in the serum of endometrial carcinoma patients (27). Other previous studies have demonstrated that miR-204-5p expression is decreased in several types of solid tumors (28–32). It has been previously determined, using a human miRNA microarray, that miR-204-5p is markedly downregulated in endometrioid adenocarcinoma tissues (32), inhibiting tumor growth in renal clear cell carcinoma (33,34), and that it suppresses invasion I endometrial cancer (35), gastric cancer (36), and head and neck tumor (37). This implies that a change of miR-204-5p may be common in tumorigenesis and reinforces the complexity of miR-204 regulation. There have been limited studies focusing on the association between PIH and miR-204-5p, as a potential marker of this syndrome. The results of the present study support the possibility that miR-204-5p may be a potential biomarker for good prognosis of PIH. Thus, therapeutic approaches targeting elevated levels of miR-204-5p should be investigated as a novel approach to improve clinical outcomes for patients with PIH.
miRNA typically targets the 3′ untranslated region of genes and leads to a significant reduction of full-length proteins (38). It has previously been suggested that the ratio of fms-like tyrosine kinase-1 (sFlt-1) and placental growth factor (PIGF) may be an additional diagnostic or predictive tool for PE (39,40). A meta-analysis showed increased concentrations of placental sFlt1 and decreased concentrations of PIGF in pregnancies in which the mother developed PE (41). Furthermore, the level of vascular endothelial growth factor was lower in women who developed PE, compared with those that did not (42). One previous study produced a series of gene profiles in human peripheral blood mononuclear cells and investigated their association with PE (43). However, the molecular mechanism of miR-204-5p of targeting these important genes during PIH or PE requires further study.
In conclusion, the present study demonstrated that the expression of miR-204-5p is significantly upregulated in the clinical serum samples of patients with PIH. Suppression of miR-204-5p by its specific inhibitor induces JAR cell growth, decreases cell apoptosis and changes the cell cycle in vitro. Therefore, miR-204-5p may be considered as a potential unfavorable progression indicator for patients with PIH and may serve as a therapeutic target in the future.
Acknowledgements
The present study was supported by funds from the Social development project of science and Technology Department of Guangdong (no. 2011B031800).
References
- 1.Li S, Xiong X, Harville E, Zhang T, Sun D, Fernandez C, Krousel-Wood M, Chen W, Whelton PK. Childhood risk factors and pregnancy-induced hypertension: The bogalusa heart study. Am J Hypertens. 2016;29:1206–1211. doi: 10.1093/ajh/hpw057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kintiraki E, Papakatsika S, Kotronis G, Goulis DG, Kotsis V. Pregnancy-induced hypertension. Hormones (Athens) 2015;14:211–223. doi: 10.14310/horm.2002.1582. [DOI] [PubMed] [Google Scholar]
- 3.Magee LA, Pels A, Helewa M, Rey E, von Dadelszen P. Canadian Hypertensive Disorders of Pregnancy Working Group: Diagnosis, evaluation, and management of the hypertensive disorders of pregnancy: Executive summary. J Obstet Gynaecol Can. 2014;36:416–441. doi: 10.1016/S1701-2163(15)30588-0. [DOI] [PubMed] [Google Scholar]
- 4.Caetano M, Ornstein MP, von Dadelszen P, Hannah ME, Logan AG, Gruslin A, Willan A, Magee LA. A survey of Canadian practitioners regarding diagnosis and evaluation of the hypertensive disorders of pregnancy. Hypertens Pregnancy. 2004;23:197–209. doi: 10.1081/PRG-120028295. [DOI] [PubMed] [Google Scholar]
- 5.Magee LA, Helewa M, Moutquin JM, von Dadelszen P. Hypertension Guideline Committee; Strategic Training Initiative in Research in the Reproductive Health Sciences (STIRRHS) Scholars: Diagnosis, evaluation, and management of the hypertensive disorders of pregnancy. J Obstet Gynaecol Can. 2008;30:S1–S48. doi: 10.1016/S1701-2163(16)32776-1. (Suppl 3) [DOI] [PubMed] [Google Scholar]
- 6.Tranquilli AL. Hypertension during pregnancy is associated with increased risk of later cardiovascular disease, kidney disease and diabetes. Evid Based Nurs. 2014;17:36–37. doi: 10.1136/eb-2013-101322. [DOI] [PubMed] [Google Scholar]
- 7.Kitson R, Williams V, Howell C. Caesarean section in a parturient with type III spinal muscular atrophy and pre-eclampsia. Anaesthesia. 2004;59:94–95. doi: 10.1111/j.1365-2044.2004.03593.x. [DOI] [PubMed] [Google Scholar]
- 8.Heimrath J, Czekanski A, Krawczenko A, Dus D. The role of endothelium in the pathogenesis of pregnancy-induced hypertension. Postepy Hig Med Dosw (Online) 2007;61:48–57. [PubMed] [Google Scholar]
- 9.Padmanabhan S, Caulfield M, Dominiczak AF. Genetic and molecular aspects of hypertension. Circ Res. 2015;116:937–959. doi: 10.1161/CIRCRESAHA.116.303647. [DOI] [PubMed] [Google Scholar]
- 10.Chen Z, Xu F, Wei Y, Liu F, Qi H. Angiotensin converting enzyme insertion/deletion polymorphism and risk of pregnancy hypertensive disorders: A meta-analysis. J Renin Angiotensin Aldosterone Syst. 2012;13:184–195. doi: 10.1177/1470320311427755. [DOI] [PubMed] [Google Scholar]
- 11.Lycoudi A, Mavreli D, Mavrou A, Papantoniou N, Kolialexi A. miRNAs in pregnancy-related complications. Expert Rev Mol Diagn. 2015;15:999–1010. doi: 10.1586/14737159.2015.1053468. [DOI] [PubMed] [Google Scholar]
- 12.Ura B, Feriotto G, Monasta L, Bilel S, Zweyer M, Celeghini C. Potential role of circulating microRNAs as early markers of preeclampsia. Taiwan J Obstet Gynecol. 2014;53:232–234. doi: 10.1016/j.tjog.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 13.Zou Q, Li J, Hong Q, Lin Z, Wu Y, Shi H, Ju Y. Prediction of MicroRNA-disease associations based on social network analysis methods. Biomed Res Int. 2015;2015:810514. doi: 10.1155/2015/810514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Wynsberghe PM, Chan SP, Slack FJ, Pasquinelli AE. Analysis of microRNA expression and function. Methods Cell Biol. 2011;106:219–252. doi: 10.1016/B978-0-12-544172-8.00008-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi SY, Yun J, Lee OJ, Han HS, Yeo MK, Lee MA, Suh KS. MicroRNA expression profiles in placenta with severe preeclampsia using a PNA-based microarray. Placenta. 2013;34:799–804. doi: 10.1016/j.placenta.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 16.Wu L, Zhou H, Lin H, Qi J, Zhu C, Gao Z, Wang H. Circulating microRNAs are elevated in plasma from severe preeclamptic pregnancies. Reproduction. 2012;143:389–397. doi: 10.1530/REP-11-0304. [DOI] [PubMed] [Google Scholar]
- 17.Yu Y, Wang L, Liu T, Guan H. MicroRNA-204 suppresses trophoblast-like cell invasion by targeting matrix metalloproteinase-9. Biochem Biophys Res Commun. 2015;463:285–291. doi: 10.1016/j.bbrc.2015.05.052. [DOI] [PubMed] [Google Scholar]
- 18.Lauschke VM, Mkrtchian S, Ingelman-Sundberg M. The role of microRNAs in liver injury at the crossroad between hepatic cell death and regeneration. Biochem Biophys Res Commun. 2016 Oct 24; doi: 10.1016/j.bbrc.2016.10.084. (Epub ahead of print). doi: 10.1016/j.bbrc.2016.10.084. [DOI] [PubMed] [Google Scholar]
- 19.Abe E, Matsubara K, Ochi H, Ito M, Oka K, Kameda K. Elevated levels of adhesion molecules derived from leukocytes and endothelial cells in patients with pregnancy-induced hypertension. Hypertens Pregnancy. 2003;22:31–43. doi: 10.1081/PRG-120016793. [DOI] [PubMed] [Google Scholar]
- 20.Kamiya T, Kwon AH, Kanemaki T, Matsui Y, Uetsuji S, Okumura T, Kamiyama Y. A simplified model of hypoxic injury in primary cultured rat hepatocytes. In Vitro Cell Dev Biol Anim. 1998;34:131–137. doi: 10.1007/s11626-998-0095-9. [DOI] [PubMed] [Google Scholar]
- 21.Cikos S, Bukovská A, Koppel J. Relative quantification of mRNA: Comparison of methods currently used for real-time PCR data analysis. BMC Mol Biol. 2007;8:113. doi: 10.1186/1471-2199-8-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kobata A, Takeuchi M. Structure, pathology and function of the N-linked sugar chains of human chorionic gonadotropin. Biochim Biophys Acta. 1999;1455:315–326. doi: 10.1016/S0925-4439(99)00060-5. [DOI] [PubMed] [Google Scholar]
- 23.Abu-Absi NR, Zamamiri A, Kacmar J, Balogh SJ, Srienc F. Automated flow cytometry for acquisition of time-dependent population data. Cytometry A. 2003;51:87–96. doi: 10.1002/cyto.a.10016. [DOI] [PubMed] [Google Scholar]
- 24.Holmes KL, Otten G, Yokoyama WM. Flow cytometry analysis using the Becton Dickinson FACS Calibur. Curr Protoc Immunol Chapter. 2002;5 doi: 10.1002/0471142735.im0504s49. Unit 5.4. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Mouillet JF, Mishima T, Paffaro AM, Parks TW, Ziegler JA, Chu T, Sadovsky Y. The expression and post-transcriptional regulation of FSTL1 transcripts in placental trophoblasts. Placenta. 2015;36:1231–1238. doi: 10.1016/j.placenta.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jia W, Wu Y, Zhang Q, Gao G, Zhang C, Xiang Y. Identification of four serum microRNAs from a genome-wide serum microRNA expression profile as potential non-invasive biomarkers for endometrioid endometrial cancer. Oncol Lett. 2013;6:261–267. doi: 10.3892/ol.2013.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tarasov VA, Matishov DG, Shin EF, Boiko NV, Timoshkina NN, Makhotkin MA, Lomonosov AM, Kirpii AA, Kit OI, Maksimov AY. Coordinated aberranit expression of miRNAs in colon cancer. Genetika. 2014;50:1232–1244. (In Russian) [PubMed] [Google Scholar]
- 29.Liu L, Wang J, Li X, Ma J, Shi C, Zhu H, Xi Q, Zhang J, Zhao X, Gu M. MiR-204-5p suppresses cell proliferation by inhibiting IGFBP5 in papillary thyroid carcinoma. Biochem Biophys Res Commun. 2015;457:621–626. doi: 10.1016/j.bbrc.2015.01.037. [DOI] [PubMed] [Google Scholar]
- 30.Zhang B, Yin Y, Hu Y, Zhang J, Bian Z, Song M, Hua D, Huang Z. MicroRNA-204-5p inhibits gastric cancer cell proliferation by downregulating USP47 and RAB22A. Med Oncol. 2015;32:331. doi: 10.1007/s12032-014-0331-y. [DOI] [PubMed] [Google Scholar]
- 31.Sümbül AT, Göğebakan B, Ergün S, Yengil E, Batmacı CY, Tonyalı Ö, Yaldız M. miR-204-5p expression in colorectal cancer: An autophagy-associated gene. Tumour Biol. 2014;35:12713–12719. doi: 10.1007/s13277-014-2596-3. [DOI] [PubMed] [Google Scholar]
- 32.Bao W, Wang HH, Tian FJ, He XY, Qiu MT, Wang JY, Zhang HJ, Wang LH, Wan XP. A TrkB-STAT3-miR-204-5p regulatory circuitry controls proliferation and invasion of endometrial carcinoma cells. Mol Cancer. 2013;12:155. doi: 10.1186/1476-4598-12-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mikhaylova O, Stratton Y, Hall D, Kellner E, Ehmer B, Drew AF, Gallo CA, Plas DR, Biesiada J, Meller J, Czyzyk-Krzeska MF. VHL-regulated MiR-204 suppresses tumor growth through inhibition of LC3B-mediated autophagy in renal clear cell carcinoma. Cancer Cell. 2012;21:532–546. doi: 10.1016/j.ccr.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Atala A. Re: VHL-Regulated miR-204 suppresses tumor growth through inhibition of LC3B-mediated autophagy in renal clear cell carcinoma. J Urol. 2012;188:2434. doi: 10.1016/j.juro.2012.08.067. [DOI] [PubMed] [Google Scholar]
- 35.Chung TK, Lau TS, Cheung TH, Yim SF, Lo KW, Siu NS, Chan LK, Yu MY, Kwong J, Doran G, et al. Dysregulation of microRNA-204 mediates migration and invasion of endometrial cancer by regulating FOXC1. Int J Cancer. 2012;130:1036–1045. doi: 10.1002/ijc.26060. [DOI] [PubMed] [Google Scholar]
- 36.Qiu YH, Wei YP, Shen NJ, Wang ZC, Kan T, Yu WL, Yi B, Zhang YJ. miR-204 inhibits epithelial to mesenchymal transition by targeting slug in intrahepatic cholangiocarcinoma cells. Cell Physiol Biochem. 2013;32:1331–1341. doi: 10.1159/000354531. [DOI] [PubMed] [Google Scholar]
- 37.Lee Y, Yang X, Huang Y, Fan H, Zhang Q, Wu Y, Li J, Hasina R, Cheng C, Lingen MW, et al. Network modeling identifies molecular functions targeted by miR-204 to suppress head and neck tumor metastasis. PLoS Comput Biol. 2010;6:e1000730. doi: 10.1371/journal.pcbi.1000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pule GD, Mowla S, Novitzky N, Wonkam A. Hydroxyurea down-regulates BCL11A, KLF-1 and MYB through miRNA-mediated actions to induce γ-globin expression: Implications for new therapeutic approaches of sickle cell disease. Clin Transl Med. 2016;5:15. doi: 10.1186/s40169-016-0092-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nevo O, Lee DK, Caniggia I. Attenuation of VEGFR-2 expression by sFlt-1 and low oxygen in human placenta. PLoS One. 2013;8:e81176. doi: 10.1371/journal.pone.0081176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kleinrouweler CE, Wiegerinck MM, Ris-Stalpers C, Bossuyt PM, van der Post JA, von Dadelszen P, Mol BW, Pajkrt E. EBM CONNECT Collaboration: Accuracy of circulating placental growth factor, vascular endothelial growth factor, soluble fms-like tyrosine kinase 1 and soluble endoglin in the prediction of pre-eclampsia: A systematic review and meta-analysis. BJOG. 2012;119:778–787. doi: 10.1111/j.1471-0528.2012.03311.x. [DOI] [PubMed] [Google Scholar]
- 41.Liu Y, Zhao Y, Yu A, Zhao B, Gao Y, Niu H. Diagnostic accuracy of the soluble Fms-like tyrosine kinase-1/placental growth factor ratio for preeclampsia: A meta-analysis based on 20 studies. Arch Gynecol Obstet. 2015;292:507–518. doi: 10.1007/s00404-015-3671-8. [DOI] [PubMed] [Google Scholar]
- 42.Dubova EA, Pavlov KA, Lyapin VM, Shchyogolev AI, Sukhikh GT. Vascular endothelial growth factor and its receptors in the placental villi of pregnant patients with pre-eclampsia. Bull Exp Biol Med. 2013;154:792–795. doi: 10.1007/s10517-013-2058-8. [DOI] [PubMed] [Google Scholar]
- 43.Martinez-Fierro ML, Garza-Veloz I, Carrillo-Sanchez K, Martinez-Gaytan V, Cortes-Flores R, Ochoa-Torres MA, Guerrero GG, Rodriguez-Sanchez IP, Cancela-Murrieta CO, Zamudio-Osuna M, et al. Expression levels of seven candidate genes in human peripheral blood mononuclear cells and their association with preeclampsia. Hypertens Pregnancy. 2014;33:191–203. doi: 10.3109/10641955.2013.853777. [DOI] [PMC free article] [PubMed] [Google Scholar]