Abstract
Context:
Turner syndrome (TS) is due to a complete or partial loss of an X chromosome in female patients and is not currently part of newborn screening (NBS). Diagnosis is often delayed, resulting in missed crucial diagnostic and therapeutic opportunities.
Objectives:
This study sought to determine if whole-exome sequencing (WES) as part of a potential NBS program could be used to diagnose TS.
Design, Setting, Patients:
Karyotype, chromosomal microarray, and WES were performed on blood samples from women with TS (n = 27) enrolled in the Personalized Genomic Research study at the National Institutes of Health. Female control subjects (n = 37) and male subjects (n = 27) also underwent WES. Copy number variation was evaluated using EXCAVATOR2 and B allele frequency was calculated from informative single nucleotide polymorphisms. Simulated WES data were generated for detection of low-level mosaicism and complex structural chromosome abnormalities.
Results:
We detected monosomy for chromosome X in all 27 TS samples, including 1 mosaic for 45,X/46,XX and another with previously unreported material on chromosome Y. Sensitivity and specificity were both 100% for the diagnosis of TS with no false-positive or false-negative results. Using simulated WES data, we detected isochromosome Xq and low-level mosaicism as low as 5%.
Conclusion:
We present an accurate method of diagnosing TS using WES, including cases with low-level mosaicism, isochromosome Xq, and cryptic Y-chromosome material. Given the potential use of next-generation sequencing for NBS in many different diseases and syndromes, we propose WES can be used as a screening test for TS in newborns.
Whole-exome sequencing is an effective tool for diagnosis of Turner syndrome. Incorporation of this technology in newborn screening may lead to earlier diagnosis and improved patient outcomes.
Turner syndrome (TS) is complete or partial loss of 1 X chromosome in phenotypic female patients. Common characteristics include short stature, infertility, hypothyroidism, autoimmune diseases, hearing loss, and specific cognitive deficits. The gold standard of diagnosis is karyotype (1). Some infants are diagnosed in utero or at birth based on ultrasound findings, lymphedema, or congenital heart malformations such as coarctation of the aorta. Prompt diagnosis allows for therapies such as growth hormone (GH) therapy as early as 4 years of age (2, 3), age-appropriate puberty induction with hormone therapy, and evaluation for congenital heart disease (1). Many girls with TS are diagnosed later and miss the opportunity for life-changing therapies and comorbidity screenings (4–6). Even with advances in prenatal testing, some studies have shown that >20% of patients are diagnosed after the age of 12 years (7), and that the average delay in diagnosis after girls with TS fall below the fifth percentile in height is 5 years (4).
There is currently no newborn screening (NBS) test for TS, even though such a test would most likely be beneficial (1, 8). Karyotype would be too labor and time intensive and costly to be a practicable test. A few groups have proposed diagnostic genetic testing by sequencing small portions of the X chromosome or real-time polymerase chain reaction gene quantification; however, these techniques are specific to TS and do not have applicability to other genetic conditions (8, 9). Chromosomal microarray has been shown to be both sensitive and specific for detection of TS but is limited to detection of copy number variations (CNVs) only (10). In contrast, next-generation sequencing-based techniques (e.g., genome, exome, gene-panel sequencing) have the potential to identify a much larger number of diseases than are now screened for using NBS and other methods (11–13).
In this study, we show that whole-exome sequencing (WES) is both sensitive and specific for diagnosing TS, using two orthogonal analyses of exome sequencing: predicting genomic CNV using differences in depth of sequencing and B allele frequency (BAF) calculation from informative single nucleotide polymorphisms (SNPs). Because next-generation sequencing technology has the potential to diagnose the majority of conditions on current newborn screens in addition to other genetic conditions not screened (11–13), we show that our study is a portal into the future of genomic screening.
Methods
Patients
Patients gave consent for and were enrolled in the National Human Genome Research Institute protocol 11-HG-0093 (Personalized Genomic Research). Inclusion criteria were a 50-metaphase karyotype or chromosomal microarray diagnosing a chromosome X deficiency and either complete or partial X-chromosome deletion or mosaic (Table 1). Control subjects were healthy female and male children and adults without a disorder of sex development. In total, 27 individuals with TS, 37 46,XX female control subjects, and 27 46,XY control male subjects were evaluated in this study.
Table 1.
Description of Patients With TS Who Participated in This Study
| Patient No. | Age, y | Karyotype, No. of Metaphase Cells | Missing X (if Known) | Congenital Heart Disease | Height, cma | Gynecologic History | Otherb | Received GH | Received Estrogen Therapy |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 29 | Mosaic 45,X/46,XX | BAV | 149 | Primary amenorrhea | None | No | Yes | |
| 2 | 27 | 45,X | Paternal | None | 150 | Primary amenorrhea | Osteoporosis, hearing loss | Yes | Yes |
| 3 | 40 | 45,X | None | 138 | Primary amenorrhea | Hypothyroidism, osteopenia, hearing loss, renal anomaly | Yes | Yes | |
| 4 | 60 | 45,X | Maternal | None | 143 | Primary amenorrhea | Hypothyroidism, osteopenia, hearing loss | No | No |
| 5 | 37 | 45,X | Paternal | BAV; aortic dilatation | 143 | Primary amenorrhea | Hearing loss | No | Yes |
| 6 | 34 | 45,X | Paternal | BAV; aortic dilatation | 152 | Primary amenorrhea | Osteoporosis, hearing loss | No | Yes |
| 7 | 53 | 45,X | None | 136 | Primary amenorrhea | Osteoporosis, hearing loss | No | No | |
| 8 | 19 | 45,X | Maternal | None | 157 | Primary amenorrhea | Hypothyroidism, hearing loss | Yes | Yes |
| 9 | 32 | 45,X | Maternal | BAV; aorta coarctation (repaired); aorta dilatation | 133 | Primary amenorrhea | Hearing loss, osteopenia, absent left kidney | No | Yes |
| 10 | 51 | 45,X | Paternal | aorta dilatation | 142 | Primary amenorrhea | Hearing loss, cleft palate | No | Yes |
| 11 | 31 | 45,X | Paternal | BAV | 152 | Primary amenorrhea | Multiple nevi | No | Yes |
| 12 | 35 | 45,X | Paternal | BAV; aorta coarctation (repaired); aorta dilatation | 152 | Primary amenorrhea | Hypothyroidism | No | Yes |
| 13 | 33 | 45,X | Paternal | BAV; history of aorta dissection and repair | 150 | Primary amenorrhea | Depression | Yes | Yes |
| 14 | 35 | 45,X | Paternal | BAV; aorta coarctation (repaired) | 146 | Primary amenorrhea | Hearing loss | No | Yes |
| 15 | 70 | 45,X | none | 128 | History of 2 unassisted pregnancies | Hypothyroidism, osteoporosis, CAD, hearing loss, depression | No | No | |
| 16 | 61 | 45,X | Paternal | BAV; aorta coarctation (repaired); aorta dilatation | 139 | Primary amenorrhea | Osteoporosis, hearing loss, single kidney | No | No |
| 17 | 47 | 45,X | Paternal | None | 142 | Primary amenorrhea | Hypothyroidism, osteoporosis, fatty liver, hearing loss | No | Yes |
| 18 | 32 | 45,X[49]/47,XXX[1] | Paternal | None | 143 | History of 2 unassisted pregnancies | None | No | No |
| 19 | 67 | 45,X | None | 142 | Primary amenorrhea | Osteoporosis | No | Yes | |
| 20 | 61 | 45,X | None | 145 | Primary amenorrhea | None | No | No | |
| 21 | 55 | 45,X | Paternal | None | 137 | Primary amenorrhea | Hypothyroidism | No | Yes |
| 22 | 42 | 45,X | None | 139 | Primary amenorrhea | Hypothyroidism, osteoporosis | No | Yes | |
| 23 | 39 | 45,X | None | 145 | Primary amenorrhea | Osteoporosis, hearing loss | No | Yes | |
| 24 | 40 | 45,X | Paternal | Aorta dilatation | 152 | Primary amenorrhea | Hypothyroidism, osteopenia | No | Yes |
| 25 | 38 | 45,X | BAV | 139 | History of unassisted pregnancy | Hypothyroidism, hearing loss | Yes | No | |
| 26 | 24 | 45,X | Paternal | None | 157 | Primary amenorrhea | Depression | Yes | Yes |
| 27 | 44 | 45,X | None | 145 | Primary amenorrhea | Hypothyroidism, hearing loss | No | Yes |
Abbreviations: BAV, bicuspid aortic valve; CAD, coronary artery disease.
Mean height of women in the United States is 162 cm when all ethnicities are included; fifth percentile for height is 151 cm (14).
Phenotypes associated with TS.
DNA extraction
Genomic DNA was isolated from whole blood of patients with TS and of control subjects, using the QIAamp DNA Blood Maxi Kit (QIAGEN, Valencia, CA) and following the manufacturer’s instructions.
Chromosomal microarray
The Illumina HumanExome BeadChip-12v1_A (Illumina Inc., San Diego, CA) SNP chip (~250,000 markers) was used for chromosomal microarray analysis. Samples with calls below Illumina’s expected 99% SNP call rates were excluded.
WES
Exome-enriched libraries were prepared from genomic DNA using the NimbleGen SeqCap EZ version 3.0 + UTR capture kit (Roche, Madison, WI). Libraries were sequenced at the National Institutes of Health Intramural Sequencing Center on the Illumina HiSEquation 2500 platform using 125-bp paired-end reads. Subsequent processing used the computational resources of the National Institutes of Health HPC Biowulf cluster (http://hpc.nih.gov). Binary alignment map (BAM) files were produced according to GATK (version 3.6) best practices (15–17) by mapping FASTQ files to the GRCh37 human reference assembly using the Burrows-Wheeler Aligner (18) (version 0.7.15) and processing with Picard tools (v.2.1.1; http://broadinstitute.github.io/picard).
Data analysis
EXCAVATOR2
CNV prediction from exome sequence data was done using EXCAVATOR2 (19) (version 1.1). This recently released tool uses reads aligning outside targeted regions to improve detection of CNVs as compared with other exome-based CNV callers such as XHMM and CoNIFER (19). We ran each of our BAM files (i.e., of 27 TS, 37 female control subjects, 27 male control subjects) through the EXCAVATOR2 pipeline using the standard settings, but we adjusted the algorithm to also analyze chromosome Y. All TS samples were compared with those of female and male control subjects separately. Each female control subject was also processed individually to confirm a 46,XX genotype. The final list of predicted CNVs for each sample was then annotated for genes, presence in the Database of Genomic Variants (20), and cytogenetic regions, using ANNOVAR (21). Similar to microarrays, EXCAVATOR2 generates log R ratio (LRR) values across chromosomal regions, where LRR is defined as log2(RCcase/RCcontrol) and the read count (RC) is the mean RC over a specified interval. Deviation of LRR from 0 is evidence of copy number change. We estimated the level of mosaicism for 45,X using the following formula:
Polymorphic variant calling
We downloaded from the University of California, Santa Cruz, Genome Browser (22) a list of coding variants outside the pseudoautosomal regions on chromosomes X and Y with minor allele frequency ≥1% in the SNP database (23) build 146. In total, there were 2,068 and 31 such SNPs on chromosome X and Y, respectively. We then determined the sequencing depth for the reference and any alternate alleles at each of these locations using the SAMtools (24) (version 1.3.1) mpileup command on each of our processed BAM files. For each SNP with a minimum depth of sequencing ≥10 reads, BAF was calculated as:
Heterozygous calls were defined as BAF between 0.4 and 0.6, and were compared between the TS cohort, female control subjects, and male control subjects. Mosaicism was calculated from BAF using the formula:
Statistical analysis
Microsoft Excel for Mac (version 15.27) was used to tabulate data and calculate odds ratios, confidence intervals, and perform Student t tests.
Mosaic and 46,X,iso(Xq) simulation
To simulate various states of mosaicism, we selected BAM files of a patient with nonmosaic TS and a female control subject with comparable nonchromosome X sequencing coverage. As expected, the chromosome X coverage in the TS sample was approximately half that of the control subject. We then used the Picard DownsampleSam function to reduce the average coverage of the BAM files by various fractions commensurate with predetermined levels of mosaicism. After merging the appropriate downsampled BAM files, using SAMtools, we then ran each simulated mosaic alignment through the EXCAVATOR2 pipeline. To simulate a case with isochromosome Xq, we extracted the Xq alignment from a TS BAM file using SAMtools. We then merged the Xq and TS alignments, simulating 1 normal X chromosome and 1 long-arm isochromosome X derivative. Finally, we ran this simulated 46,X,iso(Xq) BAM file through the EXCAVATOR2 pipeline.
Results
CNV analysis
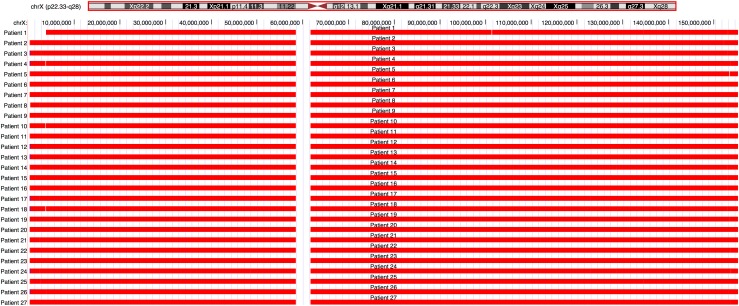
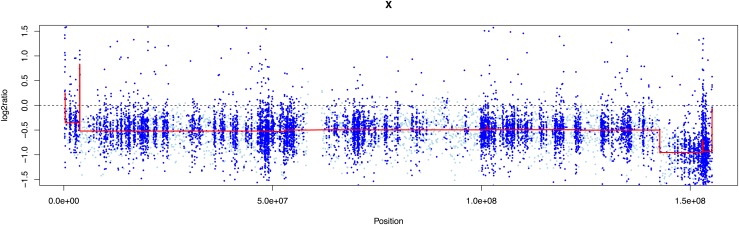
Using CNV prediction, we were able to demonstrate monosomy of chromosome X in all 27 TS samples with no false-negative results (Fig. 1). There were also no false-positive results, because all 37 female control subjects were correctly assigned a 46,XX genotype (Supplemental Fig. 1 (178.2KB, pdf) ). Thus, the sensitivity and specificity were both 100% for this assay. In 1 case, we confirmed an individual who was mosaic for 45,X and 46,XX cell lines on karyotype. The mean LRR on chromosome X in this sample (patient 1) was −0.56 (Fig. 2), corresponding to approximately a 64% level of mosaicism for 45,X, consistent with the 70% value obtained on microarray. In another case (patient 14), we were able to detect the presence of Y-chromosome material [Fig. 3(a)], a finding confirmed with microarray but not previously seen on karyotype. Specifically, EXCAVATOR2 predicted the presence of a 4.6-Mb stretch on Yp11.2 containing 8 coding genes. The reported size on microarray was slightly smaller at 3.4 Mb, a difference likely due to a limited number of SNP probes in this region on array. Analysis of the LRR plot for chromosome Y in this sample was suggestive of a mosaic state (Supplemental Fig. 2 (178.2KB, pdf) ), which may explain why it was not seen on karyotype. Using our simulated data, we detected mosaicism for 45,X to a level as low as 5% (Table 2). We were also able to easily identify isochromosome Xq [Fig. 3(b)], further demonstrating EXCAVATOR2’ s ability to detect subtle differences in read depth.
Figure 1.
University of California, Santa Cruz, Genome Browser screenshot demonstrating EXCAVATOR2-based CNV prediction on chromosome X for individuals with TS (red = deletion). The first line represents patient 1, known to be mosaic for 45,X/46,XX.
Figure 2.
LRR plot for patient 1 who had TS with known 45,X/46,XX mosaicism. The mean LRR (red line) was −0.56, corresponding to approximately 64% 45,X and 36% 46,XX. Note full 100% monosomy for distal Xq, confirmed on microarray.
Figure 3.
University of California, Santa Cruz, Genome Browser screenshots demonstrating EXCAVATOR2-based CNV detection. (a) Y-chromosome material present in patient 14, who had TS. Black bars and blue bars indicate microarray and exome data predictions, respectively. (b) 46,X, iso(Xq) detection using simulated data. Monosomy of Xp and trisomy of Xq is indicated by red bars and blue bars, respectively.
Table 2.
EXCAVATOR2-Based Predictions of Percent Mosaicism From Simulated Data
| Simulated Percent Mosaicism | Predicted Percent Mosaicism |
|---|---|
| 5 | 3.5 |
| 10 | 8.9 |
| 15 | 14.5 |
| 20 | 19.9 |
| 25 | 24.9 |
| 30 | 30 |
| 35 | 34.8 |
| 40 | 40.2 |
| 45 | 45.2 |
| 50 | 50.3 |
| 55 | 55.5 |
| 60 | 60.5 |
| 65 | 65.6 |
| 70 | 70.4 |
| 75 | 75.1 |
| 80 | 80.2 |
| 85 | 84.8 |
| 90 | 89.5 |
| 95 | 94.1 |
Informative SNPs
The mean number of SNP-database informative X-chromosome calls per sample captured by exome sequencing for the TS, and female and male control subjects was 1818, 1842, and 1743, respectively (Table 3). Consistent with a hemizygous state, the mean number of heterozygous X-chromosome calls per sample for TS was 4, representing 0.22% of the total calls. In contrast, the average number of heterozygous X-chromosome calls per sample for female control subjects was 178, representing 9.6% of the total calls. As expected, the female control subjects averaged a 45-fold higher heterozygous count on chromosome X than the TS group (Student t test P = 9.0 × 10−25) because of their second X chromosome. The mean male control subject heterozygous count on chromosome X was 4.8 per sample, or 0.27% of the total calls. As anticipated, this was not significantly different from the TS group (P = 0.2), given that the male subjects also only had a single X chromosome.
Table 3.
Informative SNP Analysis
| Patients With TS | 46,XX Female Control Subjects | 46,XY Male Control Subjects | |
|---|---|---|---|
| Total calls,a no. (95% CI) | 1818 (1807–1830) | 1842 (1828–1856) | 1743 (1726–1761) |
| Heterozygous calls,a no. [% total] (95% CI) | 4 [0.22] (3.0–5.1) | 178 [9.6] (164–191) | 4.8 [0.27] (3.8–5.8) |
| Individuals with Y-chromosome calls,b absolute no. | 1/27 | 0/37 | 27/27 |
Abbreviation: CI, confidence interval.
Rows 1 and 2 describe the average number of calls per sample in each group.
Line 3 describes the number of individuals in each group with SNP calls on chromosome Y.
Patient 1 was known to be mosaic for 45,X as previously described. Using BAF data from informative SNPs, we calculated a 71% level of mosaicism for 45,X. This was very close to the 64% value calculated using LRR data from EXCAVATOR2 and the 70% value from microarray. We also confirmed Y-chromosome material in the other case (patient 14) by this method, in that informative SNPs were found at 5 loci on the TSPY2 and TSPY8 genes.
Discussion
We used 2 different techniques based on WES data, CNV analysis with EXCAVATOR2 and informative SNP calling, to diagnose TS with high sensitivity and specificity. With the possibility of next-generation sequencing as part of future NBS analysis (11–13), screening for TS in newborns with next-generation sequencing is possible.
Diagnosis of TS has traditionally been done using karyotype or microarray. Although accurate, these tests are costly ($400 vs $1200, respectively) and usually conducted as part of an evaluation for short stature or delayed puberty (25). Though there is already a rapid pyrosequencing-based method for TS diagnosis based on genotyping of SNPs (8), we believe a standardized comprehensive screening test is necessary. Bodian et al. (11) recently demonstrated the advantages of incorporating next-generation sequencing into NBS by evaluating for single-nucleotide variants and small insertions/deletions in 163 NBS genes. Our work shows that such data can also be used to assess for large genomic CNVs like those in TS. The cost of exome sequencing continues to fall and many providers offer it for <$400 (26). Furthermore, data analysis is becoming increasingly inexpensive through automation. The time is quickly approaching when cost is no longer the major obstacle in implementing such an effort.
The motivation for diagnosing TS early is related to its multisystemic nature and increased mortality and morbidity compared with the general population (6). One review of TS described a threefold increase in premature death and >10-year reduction in life expectancy in women with TS, mainly because of cardiovascular complications (27). Bicuspid aortic valve occurs in 15% to 30% of female patients with TS and is associated with development of aortic valve dysfunction and ascending aortic dilatation later in life (27). Aortic coarctation is found in 17% of women with TS and may present in infancy with congestive heart failure necessitating urgent surgical intervention (25, 27, 28). Aortic dissection is 6 times more common in TS than the general population, with increased incidence even in girls and adolescents (29). One study described several cases of aortic dissection in the first decade of life, including a 4-year-old girl (30). Current TS guidelines recommend a thorough cardiovascular evaluation including echocardiography and blood pressure measurement in all extremities at the time of diagnosis, regardless of age (1). Thus, the early detection of cardiovascular system abnormalities in TS allows for close surveillance and intervention, should it be needed.
Haploinsufficiency of the SHOX gene located in the pseudoautosomal region of the X chromosome leads to short stature in 95% to 99% of female patients with TS (27), explaining the rationale for GH treatment. Initiating GH by 4 years of age significantly increases height in girls with TS, with 80% to 93% reaching a normal height after several years of treatment (2, 3). Notably, patients who start GH at a younger age and continue treatment of longer periods generally experience greater height gains (31). In fact, 1 group recommends initiating GH in patients with TS in the first year of life (32).
There are other benefits to early diagnosis of TS. Hearing loss is common among female patients with TS and can necessitate the use of hearing aids during childhood (1). Evidence suggests some individuals with TS have cognitive and neurodevelopmental challenges that may benefit from early educational intervention (1). Furthermore, the ability to perform ovarian biopsy in young girls with TS while they still have viable follicles may be a realistic way to prevent infertility (33). Despite these and other benefits to early identification, the diagnosis of TS is often delayed. In 1 study, one-half of girls were diagnosed after infancy, with an average delay of 5 years between diagnosis and when they had fallen below the fifth percentile in height (4). Other studies showed that 22% of women were not diagnosed until after the age of 12 years and until adulthood in up to 10% of cases. Our technique would diagnose infants at birth and permit early interventions to maximize overall quality of life.
With our protocol, we were able to detect 27 of 27 cases of karyotype-confirmed TS with a sensitivity and specificity of 100%. According to the American College of Medical Genetics and Genomics, nonmosaic monosomy X is found in only 45% of patients with TS (34). The remaining 55% are either mosaic for 45,X and another cell line or possess a structural chromosome abnormality. Thus, it is necessary for any diagnostic test to detect all such mechanisms. We successfully confirmed a true 45,X/46,XX mosaic case in our cohort. And our simulated data analysis showed this method can detect isochromosome Xq and low-level mosaicism. Mosaic TS karyotypes are associated with a variable and generally less severe phenotype (35). In 1 recent study, low-level 45,X/46,XX mosaicism up to 10% was not associated with congenital heart disease or thoracic aorta dilatation (36). Our automated technique can detect mosaicism as low as 5%. However, we could easily set a mosaicism threshold below which the algorithm does not generate an abnormal result. This would alleviate the uncertainty for parents whose newborn has low-level mosaicism but may exhibit few or none of the TS signs. Further work is needed to determine this precise level of mosaicism at which we begin to monitor for features of TS. We also identified another patient in our cohort with previously unreported Y-chromosome material containing 8 coding genes. Approximately 6% to 8% of individuals with TS have Y-chromosome material detectable by karyotype or molecular techniques (37). This is notable because of the associated 10% to 30% risk of gonadoblastoma in such cases (37). This gonadal tumor can develop even in the first decade of life and may transform into a malignant germ-cell neoplasm (37). Thus, current guidelines recommend prophylactic gonadectomy in patients with TS with Y-chromosome material (1, 37). Testing for cryptic Y-chromosome material is currently only done if a marker chromosome is seen on karyotype or signs of virilization are present (1). In contrast, our technique permits detection of Y-chromosome material in the newborn period and early screening for gonadoblastoma. In addition, whereas previous assays evaluating for cryptic Y-chromosome material used only a few centromeric or SRY markers (37–39), our method provides precise information about which genes on the Y chromosome are present. This may lead to a better understanding of the mechanism of gonadoblastoma in women with TS.
The issue of “secondary” or “incidental” findings discovered through genome-scale sequencing is important. The American College of Medical Genetics and Genomics in 2013 recommended that known pathogenic or expected pathogenic variants in 56 (now 59) genes should be disclosed to patients undergoing such testing (40). Although many of the disorders associated with these genes are adult onset, others, like Marfan syndrome (FBN1 gene) or familial adenomatous polyposis (APC gene), have important manifestations during childhood that must be monitored. This was a proof-of-principle research effort and our patients agreed not to be informed of secondary findings. Nevertheless, if this technique is someday incorporated into NBS, we feel there is an obligation to disclose these findings, given the opportunity to reduce overall mortality and morbidity.
The main limitation of this paper is the small patient cohort comprising mainly classic 45,X women. We addressed this shortcoming by using simulated data to test different scenarios, such as low-level mosaicism and structural X-chromosome abnormalities. Our results suggest this technique is at least comparable to array-based diagnostic testing for TS. By extension, this method could also be used to assess for other aneuploidies such as trisomy 21 or microdeletions like 22q11.2.
Obviously, there are a number of considerations, including ethical issues, costs, and clinical validity, involving next-generation sequencing and NBS, and these issues are beyond the scope of this text. But these challenges should not dissuade us from using this powerful tool given the important potential benefit it offers. In conclusion, we feel our results demonstrate the ability of WES-based techniques to detect TS, which strengthens the argument for the incorporation of next-generation sequencing into standard NBS.
Acknowledgments
Acknowledgments
This research is part of National Human Genome Research Institute Study 11-HG-0093.
Clinical trial registry: ClinicalTrials.gov no. NCT01294345 (registered 10 February 2011).
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- BAF
- B allele frequency
- BAM
- binary alignment map
- GH
- growth hormone
- LRR
- log R ratio
- NBS
- newborn screening
- RC
- read count
- SNP
- single nucleotide polymorphism
- TS
- Turner syndrome
- WES
- whole-exome sequencing.
References
- 1.Bondy CA; Turner Syndrome Study Group . Care of girls and women with Turner syndrome: a guideline of the Turner Syndrome Study Group. J Clin Endocrinol Metab. 2007;92(1):10–25. [DOI] [PubMed] [Google Scholar]
- 2.Davenport ML, Crowe BJ, Travers SH, Rubin K, Ross JL, Fechner PY, Gunther DF, Liu C, Geffner ME, Thrailkill K, Huseman C, Zagar AJ, Quigley CA. Growth hormone treatment of early growth failure in toddlers with Turner syndrome: a randomized, controlled, multicenter trial. J Clin Endocrinol Metab. 2007;92(9):3406–3416. [DOI] [PubMed] [Google Scholar]
- 3.Linglart A, Cabrol S, Berlier P, Stuckens C, Wagner K, de Kerdanet M, Limoni C, Carel JC, Chaussain JL; French Collaborative Young Turner Study Group . Growth hormone treatment before the age of 4 years prevents short stature in young girls with Turner syndrome. Eur J Endocrinol. 2011;164(6):891–897. [DOI] [PubMed] [Google Scholar]
- 4.Sävendahl L, Davenport ML. Delayed diagnoses of Turner’s syndrome: proposed guidelines for change. J Pediatr. 2000;137(4):455–459. [DOI] [PubMed] [Google Scholar]
- 5.Mohamed S, Roche EF, Hoey HMCV. Mode of initial presentation and chromosomal abnormalities in Irish patients with Turner syndrome: a single-centre experience. J Pediatr Endocrinol Metab. 2015;28(11-12):1215–1218. [DOI] [PubMed] [Google Scholar]
- 6.Stochholm K, Juul S, Juel K, Naeraa RW, Gravholt CH. Prevalence, incidence, diagnostic delay, and mortality in Turner syndrome. J Clin Endocrinol Metab. 2006;91(10):3897–3902. [DOI] [PubMed] [Google Scholar]
- 7.Massa G, Verlinde F, De Schepper J, Thomas M, Bourguignon JP, Craen M, de Zegher F, François I, Du Caju M, Maes M, Heinrichs C; Belgian Study Group for Paediatric Endocrinology . Trends in age at diagnosis of Turner syndrome. Arch Dis Child. 2005;90(3):267–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rivkees SA, Hager K, Hosono S, Wise A, Li P, Rinder HM, Gruen JR. A highly sensitive, high-throughput assay for the detection of Turner syndrome. J Clin Endocrinol Metab. 2011;96(3):699–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corrêa SC, Rocha MN, Richeti F, Kochi C, Silva E Lima LA, Magalhães M, Longui CA. Neonatal detection of Turner syndrome by real-time PCR gene quantification of the ARSE and MAGEH1 genes. Genet Mol Res. 2014;13(4):9068–9076. [DOI] [PubMed] [Google Scholar]
- 10.Prakash S, Guo D, Maslen CL, Silberbach M, Milewicz D, Bondy CA; GenTAC Investigators . Single-nucleotide polymorphism array genotyping is equivalent to metaphase cytogenetics for diagnosis of Turner syndrome. Genet Med. 2014;16(1):53–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bodian DL, Klein E, Iyer RK, Wong WS, Kothiyal P, Stauffer D, Huddleston KC, Gaither AD, Remsburg I, Khromykh A, Baker RL, Maxwell GL, Vockley JG, Niederhuber JE, Solomon BD. Utility of whole-genome sequencing for detection of newborn screening disorders in a population cohort of 1,696 neonates. Genet Med. 2016;18(3):221–230. [DOI] [PubMed] [Google Scholar]
- 12.Berg JS, Powell CM. Potential uses and inherent challenges of using genome-scale sequencing to augment current newborn screening. Cold Spring Harb Perspect Med. 2015;5(12):a023150 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Landau YE, Lichter-Konecki U, Levy HL. Genomics in newborn screening. J Pediatr. 2014;164(1):14–19. [DOI] [PubMed] [Google Scholar]
- 14.Fryar CD, Gu Q, Ogden CL. Anthropometric reference data for children and adults: United States, 2007-2010. Vital Heal Stat 11. 2012;(252):1–40. [PubMed] [Google Scholar]
- 15.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van der Auwera GA, Carneiro MO, Hartl C, et al. . From fastQ data to high-confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:1110.1–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D’Aurizio R, Pippucci T, Tattini L, Giusti B, Pellegrini M, Magi A. Enhanced copy number variants detection from whole-exome sequencing data using EXCAVATOR2. Nucleic Acids Res. 2016;44(20):e154 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacDonald JR, Ziman R, Yuen RKC, Feuk L, Scherer SW. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 2014; 42(Database issue, D1):D986–D992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuhn RM, Haussler D, Kent WJ. The UCSC genome browser and associated tools. Brief Bioinform. 2013;14(2):144–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup . The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eckhauser A, South ST, Meyers L, Bleyl SB, Botto LD. Turner syndrome in girls presenting with coarctation of the aorta. J Pediatr. 2015;167(5):1062–1066. [DOI] [PubMed] [Google Scholar]
- 26.Science Exchange. Science Exchange homepage. Available at: www.scienceexchange.com. Accessed 24 January 2017.
- 27.Mortensen KH, Andersen NH, Gravholt CH. Cardiovascular phenotype in Turner syndrome--integrating cardiology, genetics, and endocrinology. Endocr Rev. 2012;33(5):677–714. [DOI] [PubMed] [Google Scholar]
- 28.Turtle EJ, Sule AA, Webb DJ, Bath LE. Aortic dissection in children and adolescents with Turner syndrome: risk factors and management recommendations. Arch Dis Child. 2015;100(7):662–666. [DOI] [PubMed] [Google Scholar]
- 29.Gravholt CH, Landin-Wilhelmsen K, Stochholm K, Hjerrild BE, Ledet T, Djurhuus CB, Sylvén L, Baandrup U, Kristensen BØ, Christiansen JS. Clinical and epidemiological description of aortic dissection in Turner’s syndrome. Cardiol Young. 2006;16(5):430–436. [DOI] [PubMed] [Google Scholar]
- 30.Sybert VP. Cardiovascular malformations and complications in Turner syndrome. Pediatrics. 1998;101(1):E11. [DOI] [PubMed] [Google Scholar]
- 31.Ross J, Lee PA, Gut R, Germak J. Impact of age and duration of growth hormone therapy in children with Turner syndrome. Horm Res Paediatr. 2011;76(6):392–399. [DOI] [PubMed] [Google Scholar]
- 32.Hughes IP, Choong CS, Harris M, Ambler GR, Cutfield WS, Hofman PL, Cowell CT, Werther G, Cotterill A, Davies PS; Australasian Paediatric Endocrine Group (APEG) . Growth hormone treatment for Turner syndrome in Australia reveals that younger age and increased dose interact to improve response. Clin Endocrinol (Oxf). 2011;74(4):473–480. [DOI] [PubMed] [Google Scholar]
- 33.Borgström B, Hreinsson J, Rasmussen C, Sheikhi M, Fried G, Keros V, Fridström M, Hovatta O. Fertility preservation in girls with turner syndrome: prognostic signs of the presence of ovarian follicles [published correction appears in J Clin Endocrinol Metab. 2009;94(4):1478]. J Clin Endocrinol Metab. 2009;94(1):74–80. [DOI] [PubMed] [Google Scholar]
- 34.Wolff DJ, Van Dyke DL, Powell CM; Working Group of the ACMG Laboratory Quality Assurance Committee . Laboratory guideline for Turner syndrome. Genet Med. 2010;12(1):52–55. [DOI] [PubMed] [Google Scholar]
- 35.El-Mansoury M, Barrenäs ML, Bryman I, Hanson C, Larsson C, Wilhelmsen L, Landin-Wilhelmsen K. Chromosomal mosaicism mitigates stigmata and cardiovascular risk factors in Turner syndrome. Clin Endocrinol (Oxf). 2007;66(5):744–751. [DOI] [PubMed] [Google Scholar]
- 36.Klásková E, Tüdös Z, Sobek A, Zapletalová J, Dostál J, Zbořilová B, Sobek A Jr, Adamová K, Lattová V, Dostálová Z, Procházka M. Low-level 45,X/46,XX mosaicism is not associated with congenital heart disease and thoracic aorta dilatation:prospective magnetic resonance imaging and ultrasound study. Ultrasound Obstet Gynecol. 2015;45(6):722–727. [DOI] [PubMed] [Google Scholar]
- 37.Mazzanti L, Cicognani A, Baldazzi L, et al. . Gonadoblastoma in Turner syndrome and Y-chromosome-derived material. Am J Med Genet A. 2005;135(2):150–154. [DOI] [PubMed] [Google Scholar]
- 38.Álvarez-Nava F, Soto M, Sánchez MA, Fernández E, Lanes R. Molecular analysis in Turner syndrome. J Pediatr. 2003;142(3):336–340. [DOI] [PubMed] [Google Scholar]
- 39.Bianco B, Lipay M, Guedes A, Oliveira K, Verreschi ITN. SRY gene increases the risk of developing gonadoblastoma and/or nontumoral gonadal lesions in Turner syndrome. Int J Gynecol Pathol. 2009;28(2):197–202. [DOI] [PubMed] [Google Scholar]
- 40.Green RC, Berg JS, Grody WW, Kalia SS, Korf BR, Martin CL, McGuire AL, Nussbaum RL, O’Daniel JM, Ormond KE, Rehm HL, Watson MS, Williams MS, Biesecker LG; American College of Medical Genetics and Genomics . ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15(7):565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]