Abstract
Context:
Central precocious puberty (CPP) results from premature activation of the hypothalamic–pituitary–gonadal axis. Few genetic causes of CPP have been identified, with the most common being mutations in the paternally expressed imprinted gene MKRN3.
Objective:
To identify the genetic etiology of CPP in a large multigenerational family.
Design:
Linkage analysis followed by whole-genome sequencing was performed in a family with five female members with nonsyndromic CPP. Detailed phenotyping was performed at the time of initial diagnosis and long-term follow-up, and circulating levels of Delta-like 1 homolog (DLK1) were measured in affected individuals. Expression of DLK1 was measured in mouse hypothalamus and in kisspeptin-secreting neuronal cell lines in vitro.
Setting:
Endocrine clinic of an academic medical center.
Patients:
Patients with familial CPP were studied.
Results:
A complex defect of DLK1 (∼14-kb deletion and 269-bp duplication) was identified in this family. This deletion included the 5′ untranslated region and the first exon of DLK1, including the translational start site. Only family members who inherited the defect from their father have precocious puberty, consistent with the known imprinting of DLK1. The patients did not demonstrate additional features of the imprinted disorder Temple syndrome except for increased fat mass. Serum DLK1 levels were undetectable in all affected individuals. Dlk1 was expressed in mouse hypothalamus and in kisspeptin neuron-derived cell lines.
Conclusion:
We identified a genomic defect in DLK1 associated with isolated familial CPP. MKRN3 and DLK1 are both paternally expressed imprinted genes. These findings suggest a role of genomic imprinting in regulating the timing of human puberty.
Through a combination of linkage analysis and whole genome-sequencing, a mutation in the paternally expressed imprinted gene DLK1 in a family with central precocious puberty is identified.
Pubertal development is an essential biological process marking the transition from childhood to adulthood and ultimately resulting in the ability of an individual to reproduce. In humans, puberty is initiated through activation of the hypothalamic–pituitary–gonadal axis, marked by an increase in pulsatile gonadotropin-releasing hormone (GnRH) release leading to pituitary secretion of luteinizing hormone (LH) and follicle-stimulating hormone. Notably, early activation of the reproductive axis results in central precocious puberty (CPP), marked by thelarche prior to age 8 years in girls or testicular enlargement prior to 9 years in boys. CPP is an abnormal pediatric condition that can be attributable to cerebral congenital malformations, acquired insults, or genetic alterations (1, 2).
Activating mutations in KISS1 and KISS1R have been reported in association with CPP, but mutations of these genes are very rare in sporadic cases of CPP (3, 4). In 2013, we described the first human mutations in MKRN3, a paternally expressed imprinted gene, in a cohort of families with CPP (5). Whole-exome sequencing revealed loss-of-function mutations in MKRN3 in five of 15 families with CPP. The mutations were inherited from the father in all 15 affected individuals, consistent with the maternal imprinting of MKRN3. Other loss-of-function mutations of MKRN3 have since been reported in familial CPP (6–9). The current frequency of MKRN3 mutations in familial CPP is up to 46% (10). MKRN3 encodes makorin RING-finger protein 3, a protein with domains implicated in E3 ubiquitin ligase and RNA binding activity.
The important role of MKRN3 in human puberty initiation was reinforced by large genome-wide studies involving women of European descent from 57 studies (11). In this comprehensive study, menarche signals were found for paternal inheritance at the imprinted MKRN3 and Delta-like 1 homolog (DLK1) loci (11). DLK1, also known as preadipocyte factor 1, is a transmembrane protein containing epidermal growth factor–like repeats in its extracellular domain (12). It is a noncanonical ligand in the Delta–Notch signaling pathway and plays a role in inhibiting adipocyte differentiation (12, 13). DLK1 is widely expressed in embryonic tissue, but, in humans, postnatal expression is highest in adrenal, pituitary, and ovarian tissue (http://www.gtexportal.org/home/gene/DLK1). A neuroendocrine function of DLK1 was suggested by evidence of postnatal DLK1 expression in several hypothalamic nuclei (14). Loss of DLK1 has been shown to affect pituitary hormone content (15), but currently there is no definitive mechanistic link between DLK1 function and pubertal development. In this study, we describe a novel genomic defect characterized by a deletion of the first exon of DLK1 in a family with CPP.
Case Report
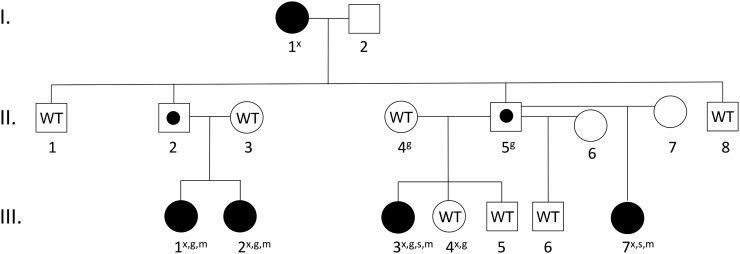
We report an Afro-descendant Brazilian family in which four female members (two sisters and two paternal half-sisters) had progressive CPP (Fig. 1). Their paternal grandmother reported her age of menarche as 9 to 10 years and is presumed to have had CPP, as this timing is 2 standard deviations (SD) below the average age of menarche (16, 17). Biochemical investigations were not performed on the grandmother when she was a child. In the four affected girls, breast development occurred at age 4.6 to 5.9 years, accompanied by growth acceleration (>6 cm/y) and advanced bone age (Table 1). Precocious pubarche was also observed in two of them. The hormonal profile confirmed early activation of the central axis, as demonstrated by their pubertal LH levels under basal conditions (>0.6 U/L) and after GnRH stimulation (>6.9 U/L for girls). All of them had normal findings on magnetic resonance imaging of the hypothalamic–pituitary region, excluding potential hypothalamic lesions. Pelvic ultrasound scans demonstrated increased uterine and ovarian volumes, consistent with CPP. They were treated with long-acting GnRH agonists with adequate clinical and hormonal control. Interestingly, the children’s fathers had normal pubertal timing, suggesting either incomplete penetrance or an imprinted disorder, similar to MKRN3 affected families (5). Additional clinical details at the time of initial diagnosis and long-term follow-up are presented in Table 1 and in the Supplemental Data (616.8KB, docx) (Supplemental Clinical Histories (616.8KB, docx) and Supplemental Table 1 (616.8KB, docx) ).
Figure 1.
Pedigree of family 1. Affected individuals are filled in black. All affected individuals carry the DLK1 mutation. Unaffected carriers, due to the imprinted pattern of inheritance, are marked with a black dot (II.2 and II.5). Individuals whose DNA was tested and did not carry the deletion are marked with the letters WT (wild-type). All wild-type individuals had normal pubertal timing. Individuals marked by empty symbols (I.2, II.6, II.7) were not tested. An “x” denotes that the subject underwent exome sequencing; a “g” denotes that the subject underwent whole-genome sequencing; and “s” denotes that the subject underwent genome-wide SNP microarray genotyping; and an “m” denotes that the subject underwent regional MLPA analysis.
Table 1.
Clinical and Hormonal Features of Affected Girls With Familial CPP Associated With Deletion of Exon 1 of DLK1 at Initial Diagnosis and Last Visit
| Patient III.3 | Patient III.2 | Patient III.1 | Patient III.7 | |
|---|---|---|---|---|
| Initial diagnosis | ||||
| Clinical manifestation, y | Thelarche (5.5) Pubarche (6.0) | Thelarche (5.0) | Thelarche (4.6) | Thelarche (5.9) Pubarche (6.0) |
| Age at first visit, y | 6.7 | 8.0 | 6.4 | 6.9 |
| Height, cm (SD) | 129.5 (2.5) | 136.3 (1.2) | 125.5 (1.7) | 127.5 (1.6) |
| BMI, kg/m2 (percentile) | 17.9 (87%) | 19.5 (95%) | 16.8 (83%) | 17.3 (78%) |
| Breast development (Tanner stage) | 4 | 4 | 3 | 4 |
| Pubic hair (Tanner stage) | 2 | 1 | 1 | 3 |
| Bone age, y | 11 | 11 | 7 | 10.6 |
| Basal and stimulated LH levels, UI/La | 1.9/31.5 | 1.7/26.9 | <0.6/11.4 | 1.6/23.5 |
| Estradiol, pg/mlb | 53.3 | 34.9 | <13 | 29 |
| Last visit | ||||
| Age, y | 22 | 21 | 18 | 16 |
| Duration of GnRHa treatment, y | 4.2 | 3.0 | 4.2 | 3.6 |
| Weight, kg | 71.6 | 55.5 | 84 | 56.1 |
| Target height, cm (SD) | 166 (0.6) | 158.5 (−0.6) | 158.5 (−0.6) | 166.5 (0.7) |
| Height, cm | 156.5 | 159.7 | 159.3 | 160.5 |
| BMI, kg/m2 | 29.2 | 22 | 33.1 | 22.1 |
| Age of menarche, y | 12 | 12 | 12 | 10.8 |
| Basal LH/FSH, UI/La | 3.3/5.1 | 2.9/2.5 | 5.5/7.0 | 0.2/0.6 |
| Estradiol, pg/mLb | 83 | 170.1 | <15 | 26.5 |
Breast and pubic hair Tanner stage and bone age were assessed at the time of first visit.
Abbreviations: BMI: body mass index; FSH, follicle-stimulating hormone.
Immunofluorometric assay.
Fluoroimmunoassay.
Materials and Methods
The research protocol was approved by the Ethics Committee of São Paulo University. Written informed consent was obtained from all participants.
Linkage analysis
Linkage analysis was performed using data extracted from six individuals who previously underwent exome sequencing (Fig. 1). To model the potentially imprinted disorder, an autosomal dominant pattern of inheritance was used and the fathers of the affected subjects were marked as obligate carriers. Details of the linkage analysis can be found in the Supplemental Methods (616.8KB, docx) .
Multiplex ligation-dependent probe amplification analysis
Four affected subjects underwent methylation-specific multiplex ligation-dependent probe amplification (MLPA) analysis using SALSA MS–MLPA probemix ME032-A1 UPD7/UPD14 (MRC-Holland). The ME032-A1 probemix contains probes for the genes PLAGL1, GRB10, MEST, DLK1, MEG3, RTL1, and MIR380. Two probes for exons 3 and 4 of the DLK1 were used for copy number variation analysis. These probes were not located in imprinted regions and did not generate information on the methylation status of DLK1. Coffalyser was used for analysis using the standard.
Whole-genome sequencing
Whole-genome sequencing of six individuals from this family was performed at the Broad Institute. All samples had >30-fold mean coverage across the genome. Variants were called using the Genome Analysis Toolkit following best practices as implemented at the Broad Institute. All coordinates are given in the Genome Reference Consortium build 37. Each sample had an average of 3,838,185 single nucleotide polymorphism (SNPs; 99,137 novel) and 709,114 indels (127,593 novel). The 1000 control genomes included samples sequenced on the same platform at the Broad Institute for other indications. The Brazilian genomes were obtained from EPIGEN-Brasil Initiative (https://epigen.grude.ufmg.br). For the copy number analysis, raw bam files of four affected and five unaffected individuals (including three additional control genomes from a separate family) were sorted and indexed using Samtools version 1.3. The end of chromosome 14 from position 99,000,000 was extracted. The average read depth was computed on nonsliding windows of 1000 bases as a computationally efficient data reduction approach. The ratio of averages of read depths of affected over nonaffected sequences was then plotted using R version 3.1.3.
Polymerase chain reaction analysis of deletion
Polymerase chain reaction (PCR) primers were designed that spanned the deletion. The forward primer was located at chromosome 14:101179960–101179979 and the reverse primer at chromosome 14:101194449–101194468. This should result in a predicted product size of 14,509 bases. The primers use included: forward, 5′-ATGGAGAGGTTCATGCTGGG-3′, reverse, 5′-CCTCGGACTCCCAAAAGCAA-3′.
DLK1 serum measurements
Serum DLK1 levels were measured in all affected family members and controls using a soluble DLK1 enzyme-linked immunosorbent assay (ELISA; IBL-America, Minneapolis, MN) per the manufacturer’s instructions. Per the manufacturer’s data, the ELISA is specific for measurement of DLK1 and recovers an average of 92% of DLK1 when serum samples are spiked with known concentrations of human DLK1. DLK1 levels range in human, healthy control serum from 0.4 to >2.5 ng/mL. The lower limit of detection for the assay is ∼0.4 ng/mL with a mean intra-assay variability of 5% and mean interassay variability of 8.1%. This assay has not been validated in a large control population. Using two different ELISAs, mean DLK1 serum levels were found to be 0.69 ± 0.036 and 1.39 ± 1.37 ng/mL in 114 and 30 healthy controls, respectively (18, 19).
Dlk1 expression
Total RNA was harvested using the RNeasy mini kit with RNase-free DNase treatment (Qiagen, Valencia, CA) from 10-cm plates (n = 3 per group) of human embryonic kidney (HEK)293 cells and two immortalized mouse cell lines derived from the arcuate (KTaR-1) and anteroventral periventricular (AVPV; KTaV-3) nuclei of adult Kiss1–green fluorescent protein female mice (20). Mediobasal hypothalamic tissue, which includes the arcuate nucleus, was collected from juvenile (postnatal day 12) wild-type C57/B6 male mice (n = 3).
One microgram of total RNA from each of these sources was reverse transcribed using iScript reverse transcription supermix (Bio-Rad Laboratories, Hercules, CA). Real-time quantitative PCR was performed using an ABI Prism 7000 detection system (Applied Biosystems, Foster City, CA) using iQ SYBR Green Supermix (Bio-Rad Laboratories). The primers were designed from the mouse Dlk1 sequence (NM_0100524; forward, 5′-AGTGCGAAACCTGGGTGTC-3′, reverse, 5′-GCCTCCTTGTTGAAAGTGGTCA-3′). PCR conditions were as follows: 50°C for 2 minutes, 95°C for 10 minutes, 95°C for 15 seconds then 60°C for 40 seconds for 40 cycles, 95°C for 15 seconds, 60°C for 20 seconds then 95°C for 15 seconds. Results were analyzed using ABI Prism 7000 SDS software (Applied Biosystems). Levels of Dlk1 messenger RNA (mRNA) were normalized to Rpl19 mRNA levels as an internal control. Data were analyzed using Prism statistical software (GraphPad Software, San Diego, CA). Multiple comparisons were analyzed by one-way analysis of variance followed by a Tukey honest significant difference test. All data are presented as the mean ± standard error of the mean. Differences were considered significant when P < 0.05.
Results
Identification of a DLK1 rearrangement
As part of a prior study (5), six members from this family (Fig. 1) underwent exome sequencing. No novel nonsynonymous variants were found to segregate with the CPP phenotype in this family. Using the common SNPs identified within the exome data, we reduced the search space via linkage analysis, defining those regions of the genome segregating perfectly with disease status. As expected, because the maximum logarithm of odds score achievable in the analyzed individuals is 1.8 (Supplemental Methods (616.8KB, docx) ), discovered chromosome segments covered roughly 10−1.8 of the genome (Supplemental Table 2 (616.8KB, docx) ), and these segments defined our target for the mutation search. Interestingly, these segments did not include the genomic region containing MKRN3, which excluded that gene as causative in this family. Of the five identified regions, we were particularly interested in the linkage region at chromosome 14q32 for a number of reasons. First, it harbors a cluster of imprinted genes. None of the other linkage regions is known to contain any imprinted genes. Second, maternal uniparental disomy of chromosome 14 or paternal deletions of this region result in Temple syndrome, a complex syndrome that includes CPP (21). Third, a large genome-wide association study of age of menarche identified two variants within this locus that influence timing of menarche but only when inherited from the father (11). Specifically, one of these variants was found to correlate with expression of the imprinted gene DLK1 in adipose tissue, making DLK1 a leading candidate to explain this association. Taken together, all of these data suggest that there exists a paternally expressed imprinted gene in this region that affects pubertal timing.
Genome-wide SNP genotyping was performed using the Infinium CytoSNP-850K BeadChip platform in two affected individuals to assess for rare copy number variants in this region, but none was detected. Additionally, a specific regional MLPA kit (MRC-Holland) that includes 12 probes within the 14q32.2 region was performed in four affected individuals and did not detect any copy number variants or aberrant DNA methylation. We then performed whole-genome sequencing in six individuals from this family and focused the analysis on the region of linkage on 14q32. Owing to the rarity of familial CPP, we filtered out all variants present in any public database (1000 Genomes, Exome Aggregation Consortium, dbSNP) or in ∼1000 control genomes sequenced on the same platform. We also excluded any variants found in 37 Brazilian genomes (EPIGENE-Brasil Initiative project) (22). This resulted in 87 novel variants within the region of linkage. Of these, there was a single nonsynonymous variant, a novel frameshift in AHNAK2. This variant was not thought to be pathogenic, as this is a highly mutable gene with >130 loss-of-function variants reported in the Exome Aggregation Consortium database (http://exac.broadinstitute.org). None of the 86 noncoding variants was found within a well-defined gene promoter region or within a known or predicted puberty-related transcription factor binding site.
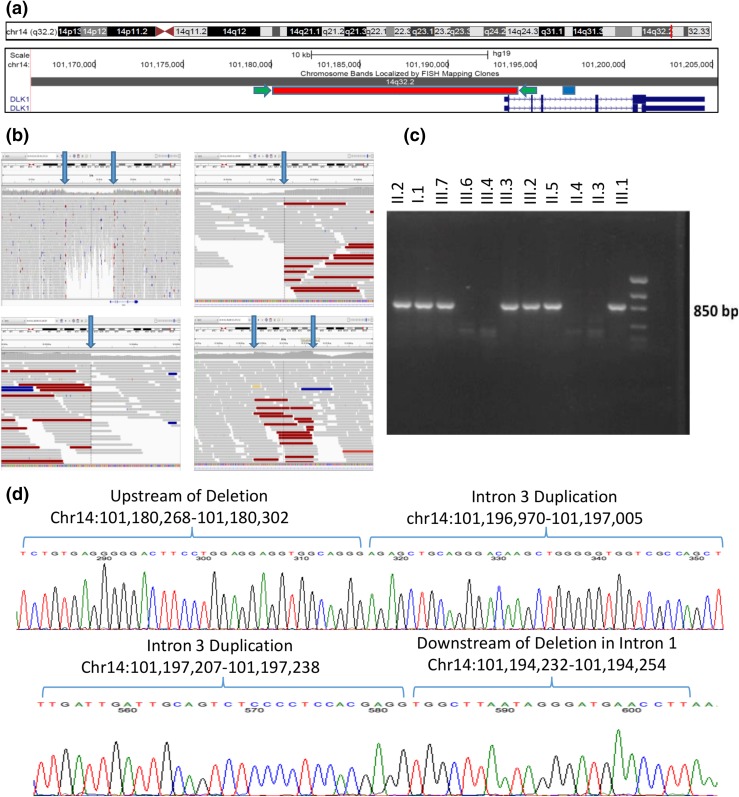
To assess for potential copy number variants, we calculated the whole-genome sequencing read depth across the linkage region in bins of 1000 bases and compared the average read depth in affected vs unaffected individuals (including an additional three control genomes). Using this approach, we identified a novel ∼14-kb heterozygous deletion that encompassed the entirety of the first exon of DLK1, including the translational start site [Fig. 2(a)]. DLK1 is known to be an imprinted paternally expressed gene (23–25). No other known genes including noncoding RNAs are present within the deletion. Upon manual review of the whole-genome sequencing data, we were able to map the precise breakpoints of the deletion to chromosome 14:101180303–101194231 [Fig. 2(b)]. This deletion was not present in any of ∼300 control genomes. PCR amplification using primers designed to span the deletion demonstrated perfect segregation of the deletion with the phenotype (or obligate carrier status) within the family [Fig. 2(c)], including additional family members who were not included in the linkage or whole-genome study (Fig. 1). The probability of this deletion segregating perfectly with the phenotype in this family is 5 × 10−4. Surprisingly, the PCR product was ∼270 bases longer than predicted based on the length of the deletion. Sanger sequencing was performed, which demonstrated that a 269-nucleotide segment from intron 3 of DLK1 had been duplicated and inserted between the ends of the genomic deletion (Fig. 2(d)]. On further review of the whole-genome data, increased read depth is evident in the area of this intronic duplication [Fig. 2(b), bottom right panel]. The deletion and duplication were not evident on the prior SNP arrays, as their lengths were below the detection limit of that array, and even on retrospective analysis of the exome data a difference in read depth was not evident, presumably due to the variability in efficiency of exon capture. Additionally, the commercial regional MLPA kit only includes probes in exons 3 and 4 of DLK1 as well as probes in additional downstream imprinted genes in the region, and thus could not detect the specific deletion or duplication found in this family. In summary, whole-genome sequencing revealed a novel complex chromosomal copy number variant resulting in deletion of the first exon of the imprinted gene DLK1 associated with CPP in this family. Segregation analysis of the DLK1 rearrangement followed an imprinted pattern with complete penetrance on paternal transmission consistent with the pubertal phenotype data in this family.
Figure 2.
Complex genomic rearrangement resulting in deletion of exon 1 of DLK1. (a) Screenshot of DLK1 region from the University of California Santa Cruz Genome Browser including two RefSeq DLK1 transcripts in dark blue. The region of the genomic deletion is highlighted in red, and the region of the duplication is highlighted in blue. The location of the PCR primers used to amplify the sequence of the deletion (c and d) are indicated by green arrows. (b) Screenshots from the Integrative Genomics Viewer of whole-genome data from subject III.2. In each panel, the top track lists the genomic coordinates shown on the screen. The middle section shows individual sequencing reads as gray bars with a histogram of individual base coverage above the reads. The colored reads represent individual paired-end sequence reads whose paired read is not located at the expected distance. This occurs due to the deletion/genomic rearrangement. Top left panel: overview of genomic region including DLK1 and its upstream region. The heterozygous deletion is clearly visible in the middle of the panel as a region of decreased sequencing coverage. The location of the DLK1 gene is noted at the bottom of this panel. Bottom left panel: zoomed-in view of the beginning of the deletion demonstrating a precipitous drop in sequencing coverage along with an abundance of reads with pairs at an unexpected distance. Top right panel: zoomed-in view of the end of the deletion. Bottom right panel: area of duplication in intron 3 of DLK1 showing increased coverage and abundance of reads with pairs at an unexpected distance. Blue arrows in all panels indicate the boundaries of the deletion or duplication. (c) PCR products demonstrating segregation of the deletion in the family. PCR primers span the deletion, and only individuals with the deletion demonstrate a PCR product of ∼850 bp. Subjects II.1, II.8, and III.5 were also tested and do not carry the deletion (data not shown). (d) Sanger sequencing of the PCR products in a subject with the complex genomic rearrangement. The sequence from intron 3 has been duplicated and inserted between the two ends of the large deletion. This results in a sequence that starts upstream of the deletion and then includes the whole duplicated region of intron 3 and concludes downstream of the deletion. The top panel’s sequence begins upstream of the deletion and shows the breakpoint where the upstream region joins the beginning of the duplicated region from intron 3 of DLK1. The bottom panel’s sequence starts with the end of the intron 3 duplicated region and joins to the region immediately downstream of the deletion in intron 1 of DLK1. All coordinates are given in the Genome Reference Consortium build 37.
Circulating DLK1 levels
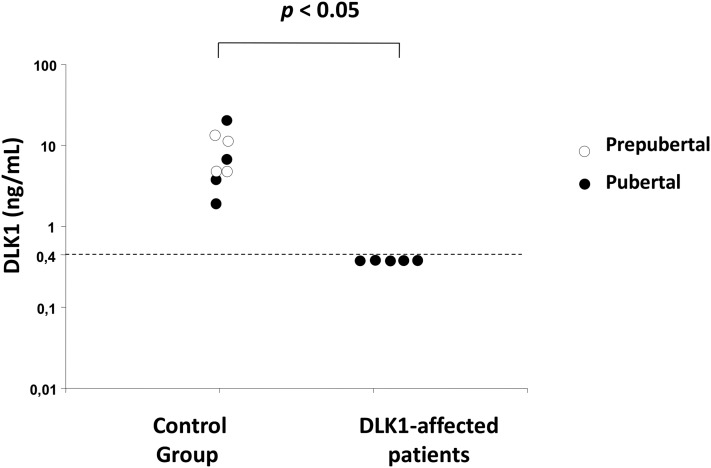
To investigate the effect of the genomic deletion on DLK1 production, serum DLK1 levels were measured in the four affected girls as well as in their affected grandmother and compared with eight controls, including four prepubertal individuals and four individuals who reported normal pubertal timing. DLK1 serum levels ranged from 1.9 to 20 ng/mL in the control group. In contrast, all five affected individuals had undetectable DLK1 levels (<0.4 ng/mL), supporting the notion that the genomic deletion leads to complete lack of DLK1 production in these individuals (Fig. 3).
Figure 3.
DLK1 serum levels in controls and in five patients with CPP due to DLK1 deletion. Affected patients had undetectable DLK1 serum levels (below the assay limit of detection of 0.4 ng/mL).
Dlk1 expression in the hypothalamus
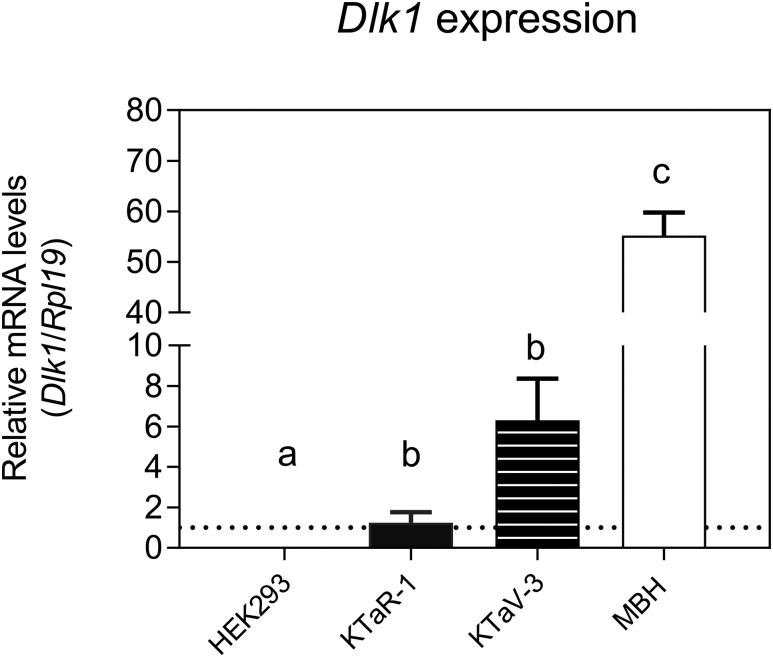
It has been shown previously that Dlk1 is expressed postnatally in the hypothalamus, in vasopressin and oxytocin neurons (14). The hypothalamus is a key locus for neuronal networks intimately involved in the neuroendocrine control of puberty, harboring GnRH neurons themselves as well as the upstream kisspeptin neurons, an essential component of the complex regulatory network that regulates GnRH secretion to control the timing of puberty onset (2). We measured Dlk1 expression in the mediobasal hypothalamus (MBH) of juvenile wild-type male mice. The MBH encompasses the hypothalamic arcuate nucleus, a key site for the control of GnRH secretion and the locus of a major population of kisspeptin neurons in males and females. Furthermore, we measured Dlk1 expression in two immortalized mouse cell lines, KTaR-1 and KTaV-3, derived from kisspeptin neurons in the arcuate and AVPV nuclei, respectively (20). The AVPV has been shown to harbor a second, sexually dimorphic population of kisspeptin neurons more abundant in females, implicated in mediating the ovulatory LH surge (26). Dlk1 expression was detected in the mouse MBH and in both cell lines, with significantly higher (P < 0.05) levels of expression than in HEK293 cells, in which no Dlk1 mRNA expression was observed (Fig. 4). Expression levels were significantly higher in the MBH than in the cell lines (P < 0.05). There was a trend toward higher expression in KTaV-3 cells compared with KTaR-1 cells, although this did not reach statistical significance (P = 0.072). Taken together, these data provide further supportive evidence that DLK1 may play a role in regulating pubertal timing, possibly by influencing kisspeptin signaling.
Figure 4.
Dlk1 mRNA expression in mouse MBH and in two kisspeptin-expressing mouse cell lines (KTaR-1 and KTaV-3). Dlk1 mRNA levels were measured by real-time quantitative PCR. HEK293 cells were used as a negative control. Results are shown as mean ± standard error of the mean for each sample (n = 3). Means with different letters are significantly different from one another (Tukey honest significant difference test, P < 0.05). Specifically, expression levels in HEK293 cells are significantly different than in KTaR-1 and KTaV-3 cells, which are also significantly different from MBH. There is no significant difference between KTaR-1 and KTaV-3. KTaR-1 is an arcuate-derived immortalized mouse cell line; KTaV-3 is an anteroventral periventricular-derived immortalized mouse cell line.
Clinical features of patients with DLK1 rearrangement
Abnormalities in a cluster of imprinted genes at chromosome 14q32.2, including the DLK1 locus, have been associated with Temple syndrome (21, 27). Potential clinical features of this complex imprinted syndrome were evaluated in the four female patients with the DLK1 deletion (Table 2). Typical central precocious puberty was the primary phenotype found in these four female patients. Prenatal and postnatal growth failure was excluded in these patients, considering their normal birth weight and childhood heights. Additionally, three of them achieved a normal final adult height (within their midparental target height range) after GnRH analog treatment. Other classical clinical features of Temple syndrome, such as history of abnormal feeding, facial dysmorphism (Supplemental Fig. 1 (616.8KB, docx) ), relative macrocephaly, small hands/feet, and neurologic abnormalities were also excluded in these CPP patients with DLK1 deletion, suggesting a nonsyndromic form of CPP. However, overweight or obesity was diagnosed in three patients at initial diagnosis. Furthermore, metabolic abnormalities were present in patient III.1 (mild obesity and abnormal hemoglobin A1c) and patient III.3 (overweight) as well as in her affected paternal grandmother (obesity and type 2 diabetes mellitus) during adulthood. All of them had increased body fat percentage by bioelectrical impedance analysis, with predominance of visceral abdominal fat. No other metabolic and hormonal defects were diagnosed in these patients.
Table 2.
Clinical Features of Temple Syndrome and CPP Patients With Deletion of Exon 1 of DLK1
| Clinical Features | Temple Syndrome (frequency %) | Patient III.3 | Patient III.2 | Patient III.1 | Patient III.7 |
|---|---|---|---|---|---|
| Gestational age and birth | Premature delivery (30%) | ∼40 wk vaginal | 39 wk vaginal | 39 wk vaginal | 39 wk vaginal |
| Birth length, cm (SD) | Intrauterine growth retardation (75%) | Not available | 51 (0.99) | 53 (2.07) | 48 (−0.62) |
| Birth weight, kg (SD) | Low birth weight (87%) | ∼3.0 kg (−0.52) | 3.9 (1.37) | 3.2 (−0.07) | 3.7 (0.98) |
| Feeding behavior | Feeding problems (43%) | Normal | Normal | Normal | Normal |
| Facial characteristics | Facial dysmorphism | Not detected | Not detected | Not detected | Not detected |
| Head circumference, cm (percentile %)a | Relative macrocephaly (56%) | 55.5 (50) | 54.5 (25–50) | 54.5 (25–50) | 56.5 (50–75) |
| Hands (finger × palm), cm (percentile %)b | Small hands (87%) | 8.2 × 11.3 (50) | 7.5 × 11 (50) | 7.5 × 11 (50) | 7.8 × 11.2 (50) |
| Shoe size (US) | Small feet (96%) | Polydactyly 7.5 | Short IV metatarsal 7.0 | 5.5 | 6.5 |
| Neurologic features | Hypotonia (93%) | Not present | Not present | Not present | Not present |
| Motor development delay (83%) | |||||
| Speech delay (59%), | |||||
| Mental retardation (39%) | |||||
| Puberty development | Early onset puberty (86%) | + | + | + | + |
| Metabolic alterations | Maturity-onset diabetes of the young (rare) Early-onset type 2 diabetes | Acanthosis nigricans | — | Acanthosis nigricans Prediabetes | — |
| Total cholesterol | Hypercholesterolemia (10%) | 182 | 122 | 196 | 171 |
| Low-density lipoprotein | 141 | 47 | 135 | 114 | |
| Triglycerides, mg/dL | 84 | 39 | 100 | 66 | |
| Body mass index, kg/m2 | Overweight and obesity | Overweight | Normal weight | Obese | Normal weight |
| Abdominal | Truncal obesity | 94 | 69.5 | 94.5 | 69 |
| Circumference (cm) and fat mass (%) | 43.6 | 35.4 | 50.5 | 34.2 |
Normal total cholesterol values are <200 mg/dL; normal low-density lipoprotein values are <100 mg/dL; high-density lipoprotein values are 160–189 mg/dL. Normal percentage of fat mass in bioelectrical impedance analysis is 18%–28%.
Adult head circumference was determined as in Nellhaus (37).
Middle finger length and palm length were determined as in Feingold and Bossert (38).
DLK1 analysis in other families with CPP
The DLK1 gene was analyzed in an additional 19 unrelated patients (18 girls and one boy) with idiopathic CPP. A family history of one or more affected relatives with precocious puberty was reported in 13 cases (Supplemental Table 3 (616.8KB, docx) ). Among these 19 patients, only two reported a history of CPP in their paternal relatives, whereas five were reported in their maternal relatives and 12 were considered undetermined (six unknown family history and six with an affected sibling only). Loss-of-function mutations of MKRN3 were previously excluded in all cases. Sanger sequencing of the five exons of the DLK1 gene, including the 5′ untranslated region, revealed only known polymorphisms in these 19 unrelated patients with familial CPP. An MLPA assay, containing probes for exons 3 and 4 of the DLK1, was also performed in 16 of these patients and no deletion was detected.
Discussion
Through a combination of linkage analysis and whole-genome sequencing, we have identified a complex genomic defect including a deletion of the first exon of DLK1 in a family with CPP. The inheritance pattern follows that of a paternally expressed imprinted gene consistent with the known imprinting of DLK1. There have been no prior reports of monogenic defects in DLK1 leading to isolated CPP.
Human DLK1 is encoded by a paternally expressed gene located on the long arm of chromosome 14, within a locus associated with Temple syndrome. This rare genetic condition is an imprinting disorder first described in 1991 (28) in a male with maternal uniparental disomy of chromosome 14. Since then, a total of 51 cases have been reported, most of whom have maternal uniparental disomy but a small number have paternal deletions including DLK1 and its surrounding cluster of imprinted genes (GTL2/MEG3, RTL1, MEG8, and DIO3). Characteristic features of Temple syndrome include intrauterine growth retardation, postnatal short stature, truncal hypotonia, small hands, mild facial dysmorphisms, and precocious puberty (21). The female patients described in the present study did not demonstrate any of these features outside of the precocious puberty (Table 2) distinguishing them from patients with classic Temple syndrome. Our patients had a deletion of the first exon of DLK1, which would presumably lead to a complete loss of DLK1, supported by their undetectable serum DLK1 levels. Attempts to delineate which genes within this region contribute to specific aspects of the Temple syndrome phenotype have not been definitive. A mouse knockout model of Dlk1 deficiency resulted in increased neonatal mortality and decreased growth and rib abnormalities in the surviving mice (29). Additionally, the knockout mice had increased adiposity and elevated levels of cholesterol and triglycerides, supporting the role of Dlk1 in regulating adipocyte generation in mice. Details of pubertal timing were not reported but there was no apparent difference. Interestingly, although our patients do not demonstrate the growth failure seen in the knockout mice, all four patients had elevated fat mass based on bioelectrical impedance despite two of them being normal weight. Lipid levels were not grossly abnormal except in the one obese patient (patient III.1). Taken together with the known loss of DLK1 expression in Temple syndrome as well as the knockout mouse model, our data suggest that loss of DLK1 function is the cause of precocious puberty in patients with Temple syndrome and may be the cause of those patients’ obesity but is not causal of any of the other Temple syndrome features.
Kagami et al. (30) reported a series of patients with deletions in the 14q.32 region, including two families in whom paternally transmitted deletions were found. In the first family, two individuals (one male and one female) inherited a 109-kb deletion encompassing DLK1 and MEG3. These two individuals had modest short stature with adult heights of −2.9 and −2.2 SD. The female had early onset puberty with menarche at age 10 years 3 months, and pubertal timing was unknown in the male. This report further supports the notion that loss of DLK1 expression leads to precocious puberty, as it was the only paternally expressed gene in their deleted region. The male subject was noted to be overweight, and both subjects had mild frontal bossing and small hands, in contrast to our patients. In the second family, a single female had a much larger 411-kb deletion involving multiple genes (WDR25, BEGAIN, DLK1, MEG3, RTL1, RTL1as, and MEG8). That patient had severe short stature with a height of −4.4 SD and a normal body mass index. She also had frontal bossing and small hands but was reported to have a normal age of menarche.
DLK1 is a noncanonical ligand in the Delta–Notch signaling pathway and plays a role in inhibiting adipocyte differentiation (12, 13). The connection between this known DLK1 function and control of pubertal timing is currently unknown. As noted earlier, a neuroendocrine function of DLK1 was suggested by evidence of postnatal Dlk1 expression in several hypothalamic nuclei (14). Interestingly, Villanueva et al. (14) found that Dlk1 expression increased progressively in mice postnatally, from postnatal day P6 to P20, correlating with increases in kisspeptin expression. This is in contrast to our previous findings with Mkrn3 where expression decreased with pubertal onset, consistent with MKRN3 acting as a negative inhibitor of pubertal timing (5). Intriguingly, the DLK1 intracellular domain has been shown to be a negative regulator of Notch signaling by disrupting the Notch1/RBP-Jk transcriptional complex (31). It was recently demonstrated that Rbpj-k–mediated Notch signaling is critical for kisspeptin neuronal development within the mouse arcuate nucleus and for subsequent Kiss1 expression (32). Paradoxically, persistent Notch signaling during development also interfered with the development of kisspeptin and other arcuate neurons, suggesting a complex control mechanism in which Notch signaling must be properly titrated for formation of kisspeptin neurons. Our data confirm prior findings that Dlk1 is expressed in the mouse MBH, which is essential for the neuroendocrine control of GnRH secretion and the timing of puberty onset, in large part through kisspeptin neurons. Furthermore, our data demonstrate Dlk1 expression in cell lines derived from kisspeptin neurons in the arcuate and AVPV hypothalamic nuclei. Although it is premature to speculate on the mechanism by which loss-of-function mutations in Dlk1 result in precocious puberty, these data, when taken together, suggest that it may involve regulation of kisspeptin neuron formation, maturation, and/or secretion of kisspeptin, likely involving the Notch signaling pathway. Interestingly, the opposing directions of regulation of Mkrn3 and Dlk1 expression prior to pubertal onset suggest that there may be competition or interactions between MKRN3 and DLK1 to control puberty onset. Indeed, this possibility is supported by a recent report that abnormalities of the Prader–Willi locus on chromosome 15 (which includes MKRN3) can modify the expression of genes of the DLK1-DIO3 locus on chromosome 14, supporting a complex trans-regulation between imprinted genes in these regions (33).
It is striking that potential loss-of-function mutations in two paternally expressed imprinted genes, MKRN3 and DLK1, are associated with central precocious puberty. Moreover, common SNPs near both of these genes affect timing of menarche in the general population when paternally inherited (11). According to kinship theory (34), imprinted genes have evolved within an evolutionary context in which maternal alleles are silenced for genes that increase demands on an individual’s mother. Therefore, it has been posited that maternally expressed genes should favor earlier puberty, thus reducing maternal demands, whereas paternally expressed imprinted genes delay puberty and thus prolong the maternally dependent stage of childhood (35, 36). The current findings that potential loss-of-function mutations in two paternally expressed imprinted genes result in CPP support this hypothesis.
In summary, using whole-genome sequencing, we have identified a complex chromosomal copy number variant leading to deletion of the first exon of DLK1 associated with CPP in a large Brazilian family. Loss of DLK1 function is the likely cause of precocious puberty in individuals with the imprinting disorder Temple syndrome. This gene potentially represents the second paternally expressed imprinted gene in which loss-of-function mutations cause familial CPP, supporting a role for imprinted genes in regulating the timing of pubertal development.
Acknowledgments
The authors thank Michael Guo for assistance in the genome analysis, Mark Daly for insightful comments, Alexandre Pereira for providing the genomic Brazilian data (EPIGEN-Brasil Initiative), and Ana Krepischi for support on the SNPs array.
Acknowledgments
This work was supported by Grant K23HD07335 (to A.D.) and Grant R01HD082314 (to U.B.K.) from the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health; Grant 302849/2015-7 (to A.C.L) from Conselho Nacional de Desenvolvimento Científico e Tecnológico; Grants 13/03236-5 (to A.C.L.) and 2013/06391-1 (to D.B.M.) from the Fundação de Amparo à Pesquisa do Estado de São Paulo; and by a grant from the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (to M.C.). The Brazilian EPIGEN Project was supported by the Brazilian Ministry of Health and Ministry of Science and Technology.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- AVPV
- anteroventral periventricular
- CPP
- central precocious puberty
- DLK1
- Delta-like 1 homolog
- ELISA
- enzyme-linked immunosorbent assay
- GnRH
- gonadotropin-releasing hormone
- HEK
- human embryonic kidney
- LH
- luteinizing hormone
- MBH
- mediobasal hypothalamus
- MLPA
- multiplex ligation-dependent probe amplification
- mRNA
- messenger RNA
- PCR
- polymerase chain reaction
- SD
- standard deviation
- SNP
- single nucleotide polymorphism.
References
- 1.Latronico AC, Brito VN, Carel JC. Causes, diagnosis, and treatment of central precocious puberty. Lancet Diabetes Endocrinol. 2016;4(3):265–274. [DOI] [PubMed] [Google Scholar]
- 2.Abreu AP, Kaiser UB. Pubertal development and regulation. Lancet Diabetes Endocrinol. 2016;4(3):254–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teles MG, Bianco SD, Brito VN, Trarbach EB, Kuohung W, Xu S, Seminara SB, Mendonca BB, Kaiser UB, Latronico ACA. A GPR54-activating mutation in a patient with central precocious puberty. N Engl J Med. 2008;358(7):709–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silveira LG, Noel SD, Silveira-Neto AP, Abreu AP, Brito VN, Santos MG, Bianco SD, Kuohung W, Xu S, Gryngarten M, Escobar ME, Arnhold IJ, Mendonca BB, Kaiser UB, Latronico AC. Mutations of the KISS1 gene in disorders of puberty. J Clin Endocrinol Metab. 2010;95(5):2276–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abreu AP, Dauber A, Macedo DB, Noel SD, Brito VN, Gill JC, Cukier P, Thompson IR, Navarro VM, Gagliardi PC, Rodrigues T, Kochi C, Longui CA, Beckers D, de Zegher F, Montenegro LR, Mendonca BB, Carroll RS, Hirschhorn JN, Latronico AC, Kaiser UB. Central precocious puberty caused by mutations in the imprinted gene MKRN3. N Engl J Med. 2013;368(26):2467–2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Settas N, Dacou-Voutetakis C, Karantza M, Kanaka-Gantenbein C, Chrousos GP, Voutetakis A. Central precocious puberty in a girl and early puberty in her brother caused by a novel mutation in the MKRN3 gene. J Clin Endocrinol Metab. 2014;99(4):E647–E651. [DOI] [PubMed] [Google Scholar]
- 7.Schreiner F, Gohlke B, Hamm M, Korsch E, Woelfle J. MKRN3 mutations in familial central precocious puberty. Horm Res Paediatr. 2014;82(2):122–126. [DOI] [PubMed] [Google Scholar]
- 8.Macedo DB, Abreu AP, Reis AC, Montenegro LR, Dauber A, Beneduzzi D, Cukier P, Silveira LF, Teles MG, Carroll RS, Junior GG, Filho GG, Gucev Z, Arnhold IJ, de Castro M, Moreira AC, Martinelli CE Jr, Hirschhorn JN, Mendonca BB, Brito VN, Antonini SR, Kaiser UB, Latronico AC. Central precocious puberty that appears to be sporadic caused by paternally inherited mutations in the imprinted gene makorin ring finger 3. J Clin Endocrinol Metab. 2014;99(6):E1097–E1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Vries L, Gat-Yablonski G, Dror N, Singer A, Phillip M. A novel MKRN3 missense mutation causing familial precocious puberty. Hum Reprod. 2014;29(12):2838–2843. [DOI] [PubMed] [Google Scholar]
- 10.Simon D, Ba I, Mekhail N, Ecosse E, Paulsen A, Zenaty D, Houang M, Jesuran Perelroizen M, de Filippo GP, Salerno M, Simonin G, Reynaud R, Carel JC, Léger J, de Roux N. Mutations in the maternally imprinted gene MKRN3 are common in familial central precocious puberty. Eur J Endocrinol. 2016;174(1):1–8. [DOI] [PubMed] [Google Scholar]
- 11.Perry JR, Day F, Elks CE, Sulem P, Thompson DJ, Ferreira T, He C, Chasman DI, Esko T, Thorleifsson G, Albrecht E, Ang WQ, Corre T, Cousminer DL, Feenstra B, Franceschini N, Ganna A, Johnson AD, Kjellqvist S, Lunetta KL, McMahon G, Nolte IM, Paternoster L, Porcu E, Smith AV, Stolk L, Teumer A, Tšernikova N, Tikkanen E, Ulivi S, Wagner EK, Amin N, Bierut LJ, Byrne EM, Hottenga JJ, Koller DL, Mangino M, Pers TH, Yerges-Armstrong LM, Hua Zhao J, Andrulis IL, Anton-Culver H, Atsma F, Bandinelli S, Beckmann MW, Benitez J, Blomqvist C, Bojesen SE, Bolla MK, Bonanni B, Brauch H, Brenner H, Buring JE, Chang-Claude J, Chanock S, Chen J, Chenevix-Trench G, Collée JM, Couch FJ, Couper D, Coviello AD, Cox A, Czene K, D’adamo AP, Davey Smith G, De Vivo I, Demerath EW, Dennis J, Devilee P, Dieffenbach AK, Dunning AM, Eiriksdottir G, Eriksson JG, Fasching PA, Ferrucci L, Flesch-Janys D, Flyger H, Foroud T, Franke L, Garcia ME, García-Closas M, Geller F, de Geus EE, Giles GG, Gudbjartsson DF, Gudnason V, Guénel P, Guo S, Hall P, Hamann U, Haring R, Hartman CA, Heath AC, Hofman A, Hooning MJ, Hopper JL, Hu FB, Hunter DJ, Karasik D, Kiel DP, Knight JA, Kosma VM, Kutalik Z, Lai S, Lambrechts D, Lindblom A, Mägi R, Magnusson PK, Mannermaa A, Martin NG, Masson G, McArdle PF, McArdle WL, Melbye M, Michailidou K, Mihailov E, Milani L, Milne RL, Nevanlinna H, Neven P, Nohr EA, Oldehinkel AJ, Oostra BA, Palotie A, Peacock M, Pedersen NL, Peterlongo P, Peto J, Pharoah PD, Postma DS, Pouta A, Pylkäs K, Radice P, Ring S, Rivadeneira F, Robino A, Rose LM, Rudolph A, Salomaa V, Sanna S, Schlessinger D, Schmidt MK, Southey MC, Sovio U, Stampfer MJ, Stöckl D, Storniolo AM, Timpson NJ, Tyrer J, Visser JA, Vollenweider P, Völzke H, Waeber G, Waldenberger M, Wallaschofski H, Wang Q, Willemsen G, Winqvist R, Wolffenbuttel BH, Wright MJ, Boomsma DI, Econs MJ, Khaw KT, Loos RJ, McCarthy MI, Montgomery GW, Rice JP, Streeten EA, Thorsteinsdottir U, van Duijn CM, Alizadeh BZ, Bergmann S, Boerwinkle E, Boyd HA, Crisponi L, Gasparini P, Gieger C, Harris TB, Ingelsson E, Järvelin MR, Kraft P, Lawlor D, Metspalu A, Pennell CE, Ridker PM, Snieder H, Sørensen TI, Spector TD, Strachan DP, Uitterlinden AG, Wareham NJ, Widen E, Zygmunt M, Murray A, Easton DF, Stefansson K, Murabito JM, Ong KK; Australian Ovarian Cancer Study; GENICA Network; kConFab; LifeLines Cohort Study; InterAct Consortium; Early Growth Genetics (EGG) Consortium . Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature. 2014;514(7520):92–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smas CM, Sul HS. Pref-1, a protein containing EGF-like repeats, inhibits adipocyte differentiation. Cell. 1993;73(4):725–734. [DOI] [PubMed] [Google Scholar]
- 13.Laborda J, Sausville EA, Hoffman T, Notario V. dlk, a putative mammalian homeotic gene differentially expressed in small cell lung carcinoma and neuroendocrine tumor cell line. J Biol Chem. 1993;268(6):3817–3820. [PubMed] [Google Scholar]
- 14.Villanueva C, Jacquier S, de Roux N. DLK1 is a somato-dendritic protein expressed in hypothalamic arginine-vasopressin and oxytocin neurons. PLoS One. 2012;7(4):e36134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheung LY, Rizzoti K, Lovell-Badge R, Le Tissier PR. Pituitary phenotypes of mice lacking the notch signalling ligand delta-like 1 homologue. J Neuroendocrinol. 2013;25(4):391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Vries L, Kauschansky A, Shohat M, Phillip M. Familial central precocious puberty suggests autosomal dominant inheritance. J Clin Endocrinol Metab. 2004;89(4):1794–1800. [DOI] [PubMed] [Google Scholar]
- 17.Partsch CJ, Heger S, Sippell WG. Management and outcome of central precocious puberty. Clin Endocrinol (Oxf). 2002;56(2):129–148. [DOI] [PubMed] [Google Scholar]
- 18.Li H, Cui ML, Chen TY, Xie HY, Cui Y, Tu H, Chen FH, Ge C, Li JJ. Serum DLK1 is a potential prognostic biomarker in patients with hepatocellular carcinoma. Tumour Biol. 2015;36(11):8399–8404. [DOI] [PubMed] [Google Scholar]
- 19.Peng T, Zhou Y, Li J, Li J, Wan W, Jia Y. Detection of Delta-like 1 ligand for the diagnosis of tuberculous meningitis: an effective and rapid diagnostic method. J Int Med Res. 2014;42(3):728–736. [DOI] [PubMed] [Google Scholar]
- 20.Jacobs DC, Veitch RE, Chappell PE. Evaluation of immortalized AVPV- and arcuate-specific neuronal kisspeptin cell lines to elucidate potential mechanisms of estrogen responsiveness and temporal gene expression in females. Endocrinology. 2016;157(9):3410–3419. [DOI] [PubMed] [Google Scholar]
- 21.Ioannides Y, Lokulo-Sodipe K, Mackay DJ, Davies JH, Temple IK. Temple syndrome: improving the recognition of an underdiagnosed chromosome 14 imprinting disorder: an analysis of 51 published cases. J Med Genet. 2014;51(8):495–501. [DOI] [PubMed] [Google Scholar]
- 22.Kehdy FS, Gouveia MH, Machado M, Magalhães WC, Horimoto AR, Horta BL, Moreira RG, Leal TP, Scliar MO, Soares-Souza GB, Rodrigues-Soares F, Araújo GS, Zamudio R, Sant Anna HP, Santos HC, Duarte NE, Fiaccone RL, Figueiredo CA, Silva TM, Costa GN, Beleza S, Berg DE, Cabrera L, Debortoli G, Duarte D, Ghirotto S, Gilman RH, Gonçalves VF, Marrero AR, Muniz YC, Weissensteiner H, Yeager M, Rodrigues LC, Barreto ML, Lima-Costa MF, Pereira AC, Rodrigues MR, Tarazona-Santos E; Brazilian EPIGEN Project Consortium . Origin and dynamics of admixture in Brazilians and its effect on the pattern of deleterious mutations. Proc Natl Acad Sci USA. 2015;112(28):8696–8701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmidt JV, Matteson PG, Jones BK, Guan XJ, Tilghman SM. The Dlk1 and Gtl2 genes are linked and reciprocally imprinted. Genes Dev. 2000;14(16):1997–2002. [PMC free article] [PubMed] [Google Scholar]
- 24.Takada S, Tevendale M, Baker J, Georgiades P, Campbell E, Freeman T, Johnson MH, Paulsen M, Ferguson-Smith AC. Delta-like and Gtl2 are reciprocally expressed, differentially methylated linked imprinted genes on mouse chromosome 12. Curr Biol. 2000;10(18):1135–1138. [DOI] [PubMed] [Google Scholar]
- 25.Wylie AA, Murphy SK, Orton TC, Jirtle RL. Novel imprinted DLK1/GTL2 domain on human chromosome 14 contains motifs that mimic those implicated in IGF2/H19 regulation. Genome Res. 2000;10(11):1711–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adachi S, Yamada S, Takatsu Y, Matsui H, Kinoshita M, Takase K, Sugiura H, Ohtaki T, Matsumoto H, Uenoyama Y, Tsukamura H, Inoue K, Maeda K. Involvement of anteroventral periventricular metastin/kisspeptin neurons in estrogen positive feedback action on luteinizing hormone release in female rats. J Reprod Dev. 2007;53(2):367–378. [DOI] [PubMed] [Google Scholar]
- 27.Briggs TA, Lokulo-Sodipe K, Chandler KE, Mackay DJ, Temple IK. Temple syndrome as a result of isolated hypomethylation of the 14q32 imprinted DLK1/MEG3 region. Am J Med Genet A. 2016;170A(1):170–175. [DOI] [PubMed] [Google Scholar]
- 28.Temple IK, Cockwell A, Hassold T, Pettay D, Jacobs P. Maternal uniparental disomy for chromosome 14. J Med Genet. 1991;28(8):511–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moon YS, Smas CM, Lee K, Villena JA, Kim KH, Yun EJ, Sul HS. Mice lacking paternally expressed Pref-1/Dlk1 display growth retardation and accelerated adiposity. Mol Cell Biol. 2002;22(15):5585–5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kagami M, Sekita Y, Nishimura G, Irie M, Kato F, Okada M, Yamamori S, Kishimoto H, Nakayama M, Tanaka Y, Matsuoka K, Takahashi T, Noguchi M, Tanaka Y, Masumoto K, Utsunomiya T, Kouzan H, Komatsu Y, Ohashi H, Kurosawa K, Kosaki K, Ferguson-Smith AC, Ishino F, Ogata T. Deletions and epimutations affecting the human 14q32.2 imprinted region in individuals with paternal and maternal upd(14)-like phenotypes. Nat Genet. 2008;40(2):237–242. [DOI] [PubMed] [Google Scholar]
- 31.Jung J, Mo JS, Kim MY, Ann EJ, Yoon JH, Park HS. Regulation of Notch1 signaling by Delta-like ligand 1 intracellular domain through physical interaction. Mol Cells. 2011;32(2):161–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biehl MJ, Raetzman LT. Rbpj-κ mediated Notch signaling plays a critical role in development of hypothalamic Kisspeptin neurons. Dev Biol. 2015;406(2):235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stelzer Y, Sagi I, Yanuka O, Eiges R, Benvenisty N. The noncoding RNA IPW regulates the imprinted DLK1-DIO3 locus in an induced pluripotent stem cell model of Prader-Willi syndrome. Nat Genet. 2014;46(6):551–557. [DOI] [PubMed] [Google Scholar]
- 34.Haig D. Genomic imprinting and kinship: how good is the evidence? Annu Rev Genet. 2004;38:553–585. [DOI] [PubMed] [Google Scholar]
- 35.Crespi B. The evolutionary biology of child health. Proc Biol Sci. 2011;278(1711):1441–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haig D. Colloquium papers: transfers and transitions: parent-offspring conflict, genomic imprinting, and the evolution of human life history. Proc Natl Acad Sci USA. 2010;107(Suppl 1):1731–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nellhaus G. Head circumference in children with idiopathic hypopituitarism. Pediatrics. 1968;42(1):210–211. [PubMed] [Google Scholar]
- 38.Feingold M, Bossert WH. Normal values for selected physical parameters: an aid to syndrome delineation. Birth Defects Orig Artic Ser. 1974;10(13):1–16. [PubMed] [Google Scholar]