Abstract
Human glucose-6-phosphatase plays a vital role in blood glucose homeostasis and holds promise as a therapeutic target for diabetes. Expression of its catalytic subunit gene 1 (G6PC1) is tightly regulated by metabolic-response transcription factors such as FoxO1 and CREB. Although at least three potential FoxO1 binding sites (insulin response elements, IREs) and one CREB binding site (cAMP response element, CRE) within the proximal region of the G6PC1 promoter have been identified, the interplay between FoxO1 and CREB and between FoxO1 bound at multiple IREs has not been well characterized. Here we present the crystal structures of the FoxO1 DNA binding domain in complex with the G6PC1 promoter. These complexes reveal the presence of a new non-consensus FoxO1 binding site that overlaps the CRE, suggesting a mutual exclusion mechanism for FoxO1 and CREB binding at the G6PC1 promoter. Additional findings include (i) non-canonical FoxO1 recognition sites, (ii) incomplete FoxO1 occupancies at the available IRE sites, and (iii) FoxO1 dimeric interactions that may play a role in stabilizing DNA looping. These findings provide insight into the regulation of G6PC1 gene transcription by FoxO1, and demonstrate a high versatility of target gene recognition by FoxO1 that correlates with its diverse roles in biology.
Keywords: Gluconeogenesis, glucose-6-phosphatase, FoxO1, CREB, gene regulation, protein-DNA interaction, crystal structure
1. Introduction
Glucose homeostasis is tightly regulated and mainly controlled by regulating endogenous glucose production in the liver and glucose uptake by peripheral tissues in response to various hormones and nutritional signals. In the liver, de novo glucose production or gluconeogenesis is a focal point of regulation, and is repressed during feeding conditions and reactivated upon fasting to meet the fuel requirement of peripheral tissues (Nordlie et al., 1999). The final step in the gluconeogenic pathway is catalyzed by the enzyme glucose-6-phosphatase (G6Pase).
G6Pase is a membrane-bound multi-component system, composed of a catalytic unit and transporter units for the substrate (glucose-6-phosphate) and the products (glucose and inorganic phosphate)(van Schaftingen and Gerin, 2002). The catalytic subunit of G6Pase is encoded by one of three genes, G6PC1, G6PC2 and G6PC3 whereas the antiporter that catalyzes transport of glucose-6-phosphate and inorganic phosphate is encoded by the SLC37A4 gene (Chou and Mansfield, 2014; Hutton and O’Brien, 2009). Hepatic gluconeogenesis is regulated in part by transcription mechanisms with both G6PC1 and SLC37A4 being targets for hormone- and nutrition-dependent modulation of expression (Mihaylova et al., 2011; Oh et al., 2013a). In particular, G6PC1 gene expression is directly regulated by insulin and glucagon. Insulin action is mediated through a multi-component insulin responsive unit that is composed of two regulatory regions in the proximal promoter, designated regions A and B. Region A binds HNF1α which serves as an accessory factor that is thought to enhance FoxO1 binding to two insulin response elements (IREs), designated IRE1 and IRE2 within Region B (Streeper et al., 2000; Vander Kooi et al., 2005). A third IRE, designated IRE3, is also present in Region B, but previous binding studies with a GST-FoxO1 fusion protein suggested that it does not bind FoxO1(Vander Kooi et al., 2003; Vander Kooi et al., 2005). On the other hand, glucagon action is mediated through CREB that recognizes cAMP response element (CRE) within Region B (Hornbuckle et al., 2004; Oh et al., 2013b) (Figure 1).
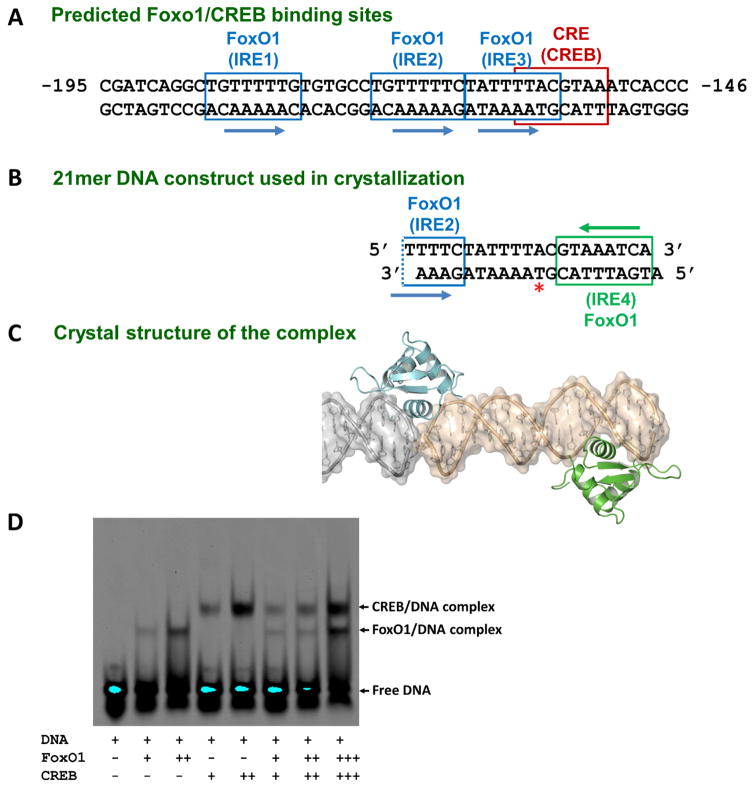
Figure 1. Sequence of the human G6PC1 promoter Region B and its composite binding site for FoxO1 and CREB.
(A) Region B contains the insulin-responsive elements (IRE: FoxO1 binding sites) and cAMP responsive element (CRE: CREB binding site). Multiple FoxO1 binding sites (IREs) and the adjacent palindromic CREB binding site (CRE) are shown. DNA sequence in this region is strictly conserved among vertebrates. (B) 21mer overhang dsDNA used in crystallization which contains the majority of IRE2 and the entire IRE3 and IRE4. Crystal packing-mediated pseudo-continuous helix formation creates a non-natural IRE2 site (IRE2′). The iodine-labeled thymidine nucleotide position (Thy11) is highlighted by a red asterisk. (C) Surface and ribbon representation of the FoxO1-DBD-21mer DNA complex structure. Two proteins (turquoise: IRE2′ site and green: IRE4 site) are bound to dsDNA, and one entire DNA construct and a portion of the neighboring DNA used in crystallization are shown in gold and gray, respectively. (D) Gel shift assay (EMSA) of FoxO1 and CREB binding to the same 21mer overhang dsDNA used in crystallization. FoxO1 and CREB can only bind individually and cannot bind at the same time for synergistic activation (lanes 6–8). The same amount of DNA was used for each lane while the protein amounts varied indicated by plus signs. Our newly determined crystal structures reveal considerable overlap of their binding sites, thus preventing co-occupancy, consistent with this biochemical data.
FoxO1, also known as FoxO1a (and originally as FKHR), is a member of the O subclass of the Forkhead transcription factor family (Huang and Tindall, 2007), while CREB is the founding member of a group of CRE binding factors that contain a basic leucine zipper DNA binding motif (De Cesare and Sassone-Corsi, 2000). FoxO1 is one of four FoxO proteins in mammals (FoxO1, 3, 4, and 6), and represents the predominant FoxO isoform (Kousteni, 2011; Wang et al., 2009) as it plays a wide range of physiological roles such as metabolic regulation, cell cycle control, and longevity (Calnan and Brunet, 2008; Huang and Tindall, 2007; Maiese et al., 2008). While FoxO proteins have overlapping expression profiles and activities and share significant sequence similarities in their DNA binding domains, their physiological roles are unique (Arden, 2008; Hosaka et al., 2004) and their functional modes of action are quite diverse (Lalmansingh et al., 2012).
FoxO1 and CREB play important roles in the regulation of hepatic glucose production (HGP) and elevated HGP is a key feature of both type and type 2 diabetes (Matsumoto et al., 2007; Oh et al., 2013b; Roden and Bernroider, 2003). A comparison of G6PC1 promoter sequences among vertebrates shows that the G6PC1 IREs and CRE are highly conserved. While multiple potential IREs and one CRE have been identified in the human G6PC1 promoter (Figure 1A), the exact mechanisms by which FoxO factors and CREB regulate G6PC1 gene expression and the central question of how overall enhanceosome assembly and mutual and/or specific interactions are made by these proteins have not been well characterized.
In this work, we present the crystal structures of FoxO1 DNA binding domain (DBD) bound to the G6PC1 promoter, which suggest that (i) the binding of FoxO1 and CREB is mutually exclusive, (ii) FoxO1 binding sites at the G6PC1 promoter involves multiple binding states due to different binding affinities, (iii) FoxO1 displays loose DNA binding selectivity towards target gene recognition, and (iv) FoxO1 dimeric interactions could induce DNA bending and mediate DNA looping. These findings and ensuing analytical studies have important implications for the regulatory mechanisms underlying HGP during fasting and in diabetes and provide novel insights into human gene regulation.
2. Materials and methods
2.1. Protein expression, DNA oligo preparation, and complex formation
A fragment of human FoxO1 cDNA corresponding to the DNA binding domain (amino acids 154–262) was subcloned by standard PCR into a pET41a vector (GE Healthcare). The pET41a vector was modified to replace the existing thrombin cleavage site with a more specific TEV (Tobacco etch virus) protease cleavage site. FoxO1-DBD was over-expressed in Escherichia coli BL21(DE3)-Rosetta2 (Novagen) with induction of 0.5 mM IPTG at an OD600 of 0.8–1.0 at 37°C and harvested after culturing for additional 3–4 hours at 30°C. The cells were lysed by sonication and the expressed GST-fusion proteins were isolated in the presence of 0.6 M NaCl to prevent nonspecific binding to bacterial DNA. FoxO1 was released by TEV protease digestion from glutathione-agarose beads (Invitrogen) after overnight incubation at 4°C and further purified by ion-exchange chromatography (Mono-S FPLC). The purified protein was estimated to be at least 98% pure judged by staining with Coomassie on 8–25% gradient SDS-PAGE gel. Fractions were pooled and stored at −80°C as a 10% (v/v) glycerol stock.
Tritylated oligonucleotides corresponding to the G6PC1 promoter sequence (20–42mers), including the iodine-labeled DNAs, were purchased from the Midland Certified Reagent Company (Midland, TX) and further purified by reverse phase HPLC on a C8 XTerra prep column (Waters), using a linear 5% to 50% (v/v) acetonitrile gradient in 50mM triethyl amine acetate buffer (pH 7.0). Excess mobile phase containing acetonitrile was removed using HiTrapQ (GE Healthcare), and the trityl groups were removed with 80% (v/v) acetic acid. The deprotected oligonucleotides were precipitated with 75% (v/v) ethanol, dissolved in water for concentration measurement by A260 and lyophilized before storage at −80°C. Double-stranded DNAs were generated for crystallization by heating equimolar amounts of complementary oligonucleotides to 95°C for 10 min and slowly cooling to 4°C. The annealing buffer was 20mM Tris (pH 8.0), 200mM NaCl, and 1mM EDTA.
2.2. Crystallization and data collection
The initial crystallization trials were carried out at 22°C in 96-well plates with the sparse matrix (Jancarik and Kim, 1991) by the hanging-drop vapor diffusion method, utilizing the Phenix nano-drop crystallization robot system (Art Robbins Instruments). Drops consisting of 0.1μl protein-DNA solution (after mixing the protein and DNA at a molar ratio of 2:1.2 for 21mer and 4:1.2 for 40mer DNA) were mixed with an equal volume of reservoir solution and equilibrated against 500μl of reservoir solution. Conditions yielding small crystals were further optimized by variations of crystallization parameters and additives in 24-well plates. Although many different DNA constructs were used for screenings, diffraction-quality crystals were reproducibly obtained only using the over-hang 21mer (nucleotide positions −169 to −149 of the G6PC1 promoter sequence) and the blunt-end 40mer (nucleotide positions −188 to −149). The optimized 21mer crystals were grown with well solution containing 14–18% (v/v) PEG8000, 100mM ammonium sulfate, 20mM magnesium chloride, 50mM MES pH5.6 and 10% glycerol, and transferred into mother liquor containing an additional 15% (v/v) glycerol as cryoprotectant before being directly plunged into liquid nitrogen and stored for data collection. The crystals with iodine-labeled DNAs were grown under the same condition as the wild type. The 40mer crystals were grown under the similar condition, but with 22–26% (v/v) PEG8000. All the native data were collected at 100K at APS (SER-CAT 22ID) using a MAR-300 CCD detector and processed using HKL2000 (Otwinowski and Minor, 1997). Typical crystals and the resulting diffraction patterns are shown in supplementary Figure S1.
2.3. Structure Determination and refinement
The 21mer crystals belong to the space group P21 with unit cell dimensions a = 46.61 Å, b = 79.03 Å, c = 48.27 Å and β = 90.79 °, and diffract to 2.3 Å resolution while the 40mer crystals belong to the space group C2 with unit cell dimensions a = 104.36 Å, b = 251.99 Å, c = 124.37 Å and β = 89.78 °, and diffract to 5.0 Å resolution. There is one complex in the asymmetric unit (46.0% solvent content) for 21mer crystals and four complexes in the asymmetric unit (59.0% solvent content) for 40mer crystals. The structures were solved by the molecular replacement method by use of MOLREP (Vagin and Teplyakov, 1997). As a search model, we used a previously determined structure of the FoxO1-DNA complex (PDB accession code 3CO6). For 21mer crystals, the best solution with two Foxo1-DNA pairs had a correlation coefficient of 38.5%, 11.4% above the second best solution. The Rcryst value after molecular replacement was 0.514. After one round of rigid-body refinement, Rcryst and Rfree dropped to 0.452 and 0.408, respectively. For 40mer crystals, a similar repetitive molecular replacement approach using the partial solution from previous job was applied until all thirteen Foxo1-DNA pairs were found and there were no more solutions. Further refinement was carried out with Refmacs as run by PHENIX (Adams et al., 2010) alternating with manual fitting in COOT (Emsley and Cowtan, 2004) until convergence. Individual atomic coordinates, group B-factors and NCS restraints were utilized for initial rounds of refinement. For nucleotides, Watson-Crick base pairing and global restraints were placed on bond lengths, bond angles, nonbonded contacts, and temperature factors of neighboring atoms. Towards the end of refinement, individual B-factors and TLS refinement parameters were employed, with geometry and B-factor restraint weightings set to their default values. Solvent molecules were added with phenix.refine and manually inspected in COOT. The final model was validated with MolProbity (Chen et al., 2010) prior to deposition in the PDB. Data and refinement statistics for 21mer crystals are provided in Table 1. Data and refinement statistics for 40mer crystals are not provided due to low resolution and no-confidence in individual atom parameters. However, the overall structure and the positions/orientations of individual protein/DNAs are so apparent. Strict NCS constraints were applied during refinement. Figures were prepared with PyMOL (The PyMOL Molecular Graphics System).
Table 1.
Statistics of 21mer complex crystallographic analysis
| Data collection | |
|---|---|
| Wavelength (Å) | 1.000 |
| Space group | P21 |
| Unit-cell parameters | |
| a, b, c (Å) | 46.61, 79.03, 48.27 |
| α, β, γ (°) | 90.00, 90.79, 90.00 |
| Resolution (Å) | 40.15–2.30 (2.34–2.30)a |
| Completeness (%) | 94.5 (85.5)a |
| Average multiplicity | 4.5 (2.6)a |
| <I/σ(I)> | 16.5 (1.84)a |
| Rmerge (%) | 8.0 (46.5)a |
|
| |
| Refinement | |
|
| |
| Resolution (Å) | 40.15–2.30 |
| Number of reflections | 12191 |
| Rwork | 0.208 |
| Rfree§ | 0.245 |
| Number of atoms | |
| Protein | 1368 |
| DNA | 855 |
| Solvent | 51 |
| Average B-factor (Å2) | |
| Protein atoms | 68.1 |
| DNA | 73.0 |
| Solvent | 45.2 |
| R.M.S.D. from ideal geometry | |
| Bond lengths (Å) | 0.002 |
| Bond angles (°) | 0.758 |
| Ramchandran Plot | |
| Favored (%) | 94.6 |
| Allowed (%) | 5.4 |
| Outliers (%) | 0 |
| Rotamer outliers (%) | 1.34 |
Values in parentheses are for the highest-resolution shell
5% of the reflection data excluded from refinement
2.3.1. Accession number
Protein Data Bank: coordinates and structure factors for the FoxO1–21mer DNA complex have been deposited under the accession code 5DUI.
2.4. Electrophoretic Mobility Shift Assay (EMSA)
Synthetic oligonucleotides labeled with an infrared dye (IRDye 700 phosphoramidite) were purchased from Integrated DNA Technology (IDT), quantified and annealed to form dsDNAs. Binding reactions of different protein/DNA molar ratios were assembled at 21°C in a total volume of 5 μl in 25mM Tris pH 7.5, 50mM NaCl, 2mM MgCl2, 1mM DTT, 100 μg/ml BSA and 10% (v/v) glycerol. Binding reactants were loaded onto an actively running 5% non-denaturing polyacrylamide gel in 0.5X TBE (45mM Tris, 45mM borate, 1mM EDTA, pH 8.3) that had been pre-electrophoresed for 30 min at 4°C. Electrophoresis continued for 60 min at 4°C before the gel was analyzed with the Odyssey Infrared Imaging System (LI-COR Biosciences). For non-labeled DNAs, ethidium bromide was used to stain the samples.
2.5. Site-directed mutagenesis analysis
The ‘Quick change Multi site-directed mutagenesis’ kit (Stratagene) was used to generate the constructs with each point mutation of the human G6PC1 reporter gene construct according to the manufacturer’s instructions. The plasmid template used in the mutations was pGL3-G6PC1 (see below). Single mutants were generated by substituting A to T, G to C, and vice versa. All of the generated constructs with the mutated sequences were verified with DNA sequencing (Supplementary Figure S3).
2.6. Transient transfection and transcription assays (Luciferase reporter assays)
The full length cDNA of human FoxO1, wild type or the mutant, were subcloned into the pCMV Sport6 vector (Life Technologies), and the reporter vector pGL3-G6PC1 containing the region of the human G6PC1 promoter encompassing the dominant region B was constructed and used for luciferase assays. HeLa cells were cultured in DMEM medium supplemented with 10% fetal bovine serum, 50 units/ml penicillin G, 50 μg/ml streptomycin (Sigma), and 0.1 mM non-essential amino acids (Invitrogen). HeLa cells were transfected using Opti-MEM and recommendations. LipofectAMINE 2000 reagent (Invitrogen) according to the manufacturer’s Briefly, a total of 30 ng of pCMV FoxO1 and 50 ng of pGL3-G6PC1 and 10 ng of pRL-TK (control renilla luciferase vector) were used for transfection of 1 × 105 cells seeded on a 24-well plate one day before transfection. After transfection and incubation, cells were washed with 1X PBS and lysed with luciferase lysis buffer supplied with the Luciferase assay kit (Promega). Luciferase activity was measured using the Dual Luciferase assay system (Promega) and Lmax Luminometer (Molecular Devices). All values were normalized by the relative ratio of firefly luciferase activity and renilla luciferase activity. At least four independent transfections were performed in duplicate.
3. Results
3.1. Lack of evidence for synergistic activation of the G6PC1 promoter by FoxO1 and CREB
Transcription factors often function in a combinatorial manner to regulate cellular gene expression by recruiting the general transcriptional apparatus cooperatively to the promoter (Panne, 2008). Based on the proximity of their binding sites, we hypothesized that FoxO1 and CREB may bind synergistically to the human G6PC1 promoter (Figures 1A–C). Thus, the initial goal of these studies was to examine the structure of FoxO1 and CREB bound simultaneously to the G6PC1 promoter. However, in gel mobility shift assays, CREB and FoxO1 proteins appeared to bind independently to the G6PC1 promoter, with no evidence for CREB/FoxO1/DNA ternary complex formation (Figure 1D).
Indirect evidence for the absence of synergistic binding was also apparent from our initial crystallization trials. Despite the presence of all three components (CREB, FoxO1, and 21mer DNA containing the IRE3 and CRE elements) in crystallization drops, crystals containing only FoxO1 and DNA appeared. Although the crystallization process selects only packable species for a given crystal and the resulting crystal may not represent all of the available species in the drops, this outcome indirectly supports the same observation made from the gel-shift assays. Based on these findings, we subsequently set up crystal trays with the samples containing only FoxO1 and DNA and determined the structures.
3.2. Structure determination of a FoxO1-DNA complex and the discovery of a new FoxO1 binding site on the G6PC1 promoter
The methods of crystallization and structure determination are described in the Methods section. Typical crystals and the diffraction patterns are shown in supplementary Figure S1 and the final refinement statistics for the high resolution structure are summarized in Table 1.
For crystallization studies we initially used a double stranded DNA 21mer containing two of the three IREs (the entire IRE3 and the majority of IRE2) as well as the CRE in the G6PC1 promoter (Figures 1A–B). Under the crystallization conditions, this self-complementary over-hang DNA construct (Figure 1B) forms pseudo-continuous helices in the crystal packing through end-to-end Watson Crick base-pairs and base-stacking interactions. While the FoxO1 proteins bound to the predicted IRE sites on the G6PC1 promoter should have a head-to-tail orientation (Figures 1A–B), the two proteins found in our crystal structure showed a head-to-head orientation (Figure 1C and Figure 2A), which hinted an unsuspected new binding site with an opposite orientation. FoxO1 binding sites (IREs) are known to have the consensus sequence ‘T[G/A]TTTAC’ or ‘T[G/A]TTTTG’ (Brent et al., 2008). The only potential FoxO1 binding site on the opposite strand with a consecutive ‘TTT’ sequence is the region 3′ of IRE3, which we refer to as IRE4 (Figures 1B–C). This potential FoxO1 binding site might have been overlooked in the past due to its weak correlation with the consensus and its considerable overlap with the existing CREB binding site. Based on this observation, we initially assigned the two protein-occupied sites as IRE3 and IRE4, rather than IRE1 and IRE4 or IRE2 and IRE4 because the spatial separation between each binding site matches reasonably well with the estimated value from the sequence between IRE3 and IRE4 (Figure 2A gray dotted circle). However, the FoxO1 binding modes in this model would be completely reversed from the ones observed in previous FoxO1/DNA complex structures (Brent et al., 2008; Obsil and Obsilova, 2011). This contrary finding prompted us to reexamine our initial DNA sequence assignment.
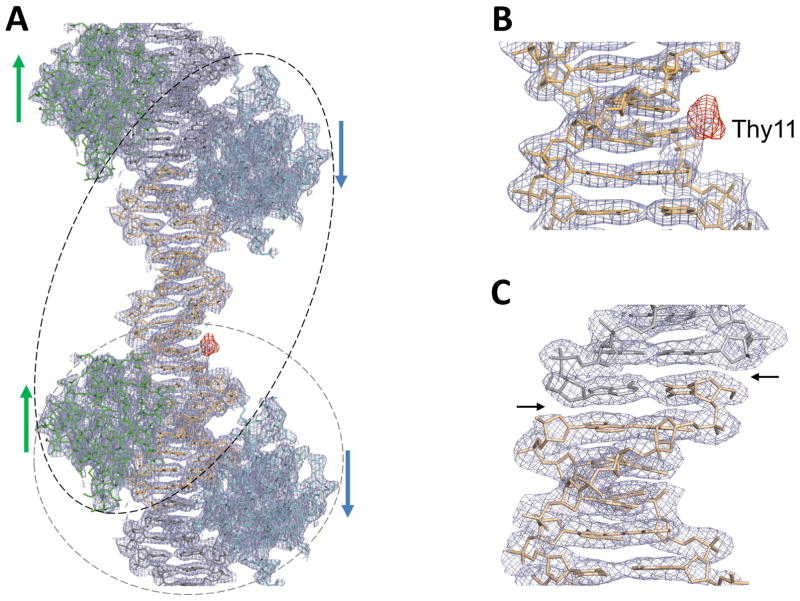
Figure 2. Overall structures of the FoxO1-DBD-21mer DNA complex and verification of DNA sequence assignment of the pseudo-continuous helix within the crystal lattice.
(A) Overall electron density maps of the 21mer structure at 2.3 Å resolution (the final 2Fo-Fc map (gray mesh) contoured at 1σ calculated with the phases from the final model and the difference map (red mesh) contoured at 3σ calculated with iodine-labeled crystals using the coefficient Fiodine-Fnative and the phases from the final native model). (B) A zoomed-up view of the difference map, together with the final 2Fo-Fc map, clearly displaying a strong peak at the expected position of CH3 group of Thy11 nucleotide. This difference map unequivocally highlights the correct position of iodine. (C) A zoomed-up view of the electron density breaks at the ends of the overhang DNA construct used in crystallization. Break points are indicated by black arrows.
To verify the correct register of the DNA sequence, we utilized iodine-labeled DNAs during crystallization and solved the crystal structure of the FoxO1/labeled-DNA complex. Iodine substitution was made at the methyl group of the Thy11 base (Figure 1B) and the crystals were grown under the same conditions as wild type. By employing the electron density maps for the position of the iodine peak and for the presence of electron density breaks at both ends of the overhang 21mer DNA (Figures 2), we assigned the correct DNA sequence of the pseudo-continuous helix within the crystal lattice. A strong iodine peak clearly showed up in the Fiodine-Fnative difference map (Figure 2A–B) and DNA backbone break points were clearly visible in the final 2Fo-Fc map (Figure 2C). The data show that the asymmetric unit contains IRE2 and IRE4 (black dotted circle in Figure 2A) as opposed to IRE3 and IRE4 with non-canonical DNA binding modes as initially assigned (gray dotted circle). With the correct register of the DNA sequence, both proteins display previously known binding modes with a canonical orientation. The IRE3 site is vacant in this structure due to crystal packing constraints, as a result of weaker binding (see below).
3.3. Supporting evidence for a new FoxO1 binding site and a crystal packing-induced non-native binding site on the G6PC1 promoter
We also performed point-mutagenesis studies to evaluate the importance of this newly discovered IRE on the G6PC1 promoter. Point mutations in IRE4 as well as IRE3 resulted in considerably reduced FoxO1-stimulated G6PC1-luciferase fusion gene expression in transient transfection assays (Figure 3A). The mutations within IRE4 that affected fusion gene expression correlate very well with the base-specific interaction patterns observed in the crystal structure (see below Detailed Protein-DNA interactions). These data strongly support our structural findings and suggest that IRE4 has the potential to play an important role in induction of G6PC1 transcription by FoxO1.
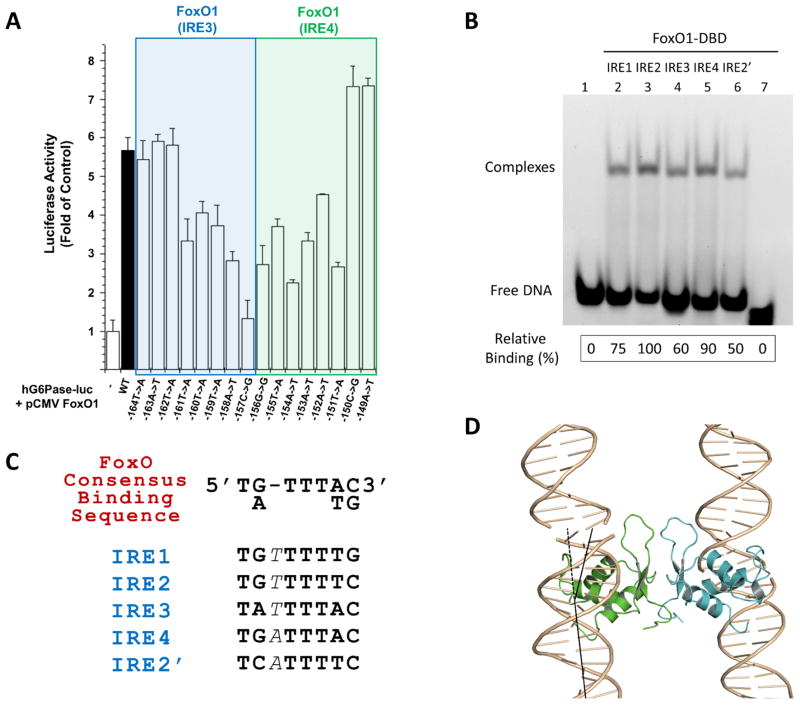
Figure 3. Functional features of IRE3/IRE4 point mutations within the G6PC1 promoter.
(A) Overall transcriptional activity measured by standard luciferase-based transcriptional reporter assays with the G6PC1 promoter mutants. The first lane (control) refers to an empty vector without FoxO1 transfection, and all data have been normalized against firefly Renilla luciferase activity. Data represent the mean±SE (n=4). Each nucleotide position is indicated by the position numbers shown in Fig. 1A. The sequence of individual mutations have been confirmed (supplementary Fig. S3). (B) Gel shift assays with individual IRE sites (14mer dsDNA containing single IRE site in the middle with natural flanking sequences). Lane assignments are shown on top. The first lane refers to IRE1 without FoxO1-DBD protein loading while the last lane refers to a 14mer dsDNA of unrelated sequence with FoxO1-DBD protein loading. The same amount of protein loading was ensured for each lane and the 1:1 protein-DNA molar ratio was used. DNA binding affinity of FoxO1 was estimated by quantification of each shifted band (complex) using Gel Logic 212 Pro (Carestream Molecular Imaging) and its software, which is shown at the bottom by relative binding (%) values. (C) The consensus FoxO recognition sequence and the novel IRE sites on the G6PC1 promoter. One extra base in each IRE site is indicated by italicized font. (D) FoxO1 dimeric interactions that mediates crystal packing in a horizontal direction. Vertical direction crystal contacts are solely made by the end-to-end interactions of neighboring DNA strands. The same FoxO1 dimeric interactions are observed for both proteins present in the 21mer crystal lattice as well as 40mer crystals. FoxO1 binding induces about 25.84° bending of individual dsDNA molecules in the crystal, shown by black lines.
We next evaluated FoxO1 binding to IRE4 by gel-shift assay using oligos containing single binding sites (Figure 3B). Consistent with the reporter studies, binding of FoxO1 to IRE4 was compatible in affinity to one of the known binding sites (IRE2). FoxO1 displays the strongest binding to IRE2 followed by IRE4, IRE1, and IRE3. Previously, FoxO1 binding to IRE3 was not detected in gel retardation (Vander Kooi et al., 2003) or footprinting experiments (Vander Kooi et al., 2005) whereas we observe FoxO1 binding to IRE3 in both gel retardation (Figure 3B) and crystallographic analysis with a low resolution structure (Fig. 4A) (see below 40mer Complex Structure description). We hypothesize that these differences might be due to the nature of the proteins and DNA samples used in the studies (cellular extract containing full-length fusion protein vs. purified DBD-only protein and/or small oligos vs. larger PCR products, respectively), and/or concentration effects during analysis. Nevertheless, our crystal structures and mutational analyses suggest that IRE3 is also a functional FoxO1 binding site.
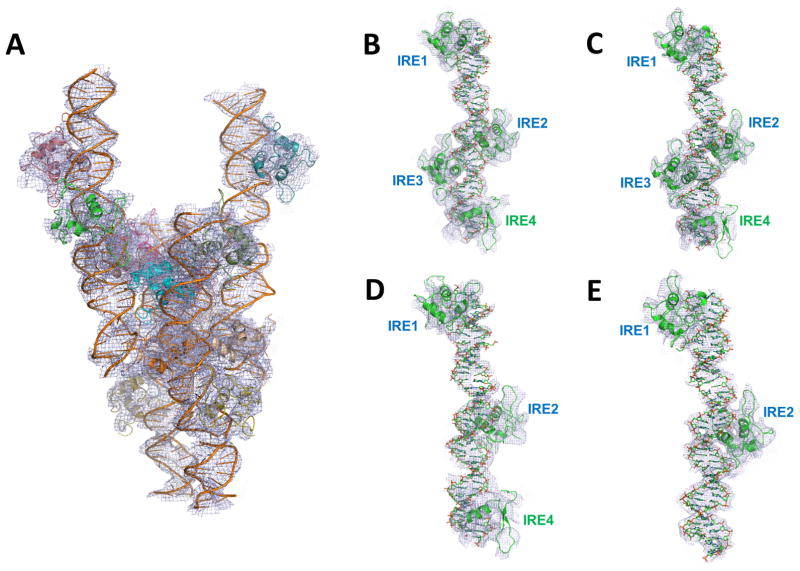
Figure 4. Low resolution crystal structure of the FoxO1-DBD-40mer DNA and the makeup of each complex within the asymmetric unit.
(A) Low resolution (5.0 Ǻ) crystal structure of the 40mer complex. The entire asymmetric unit content with four independent complexes is shown in which each complex contains a different number of FoxO1 proteins bound to DNA containing all four IRE sites. (B–E) Makeup of each complex. Two complexes had all IRE sites occupied (B–C) while two other complexes had partially occupied IRE sites (D: 3 sites and E: 2 sites). Foxo1-bound IRE sites in each complex are labeled. Electron density maps were contoured at 0.8σ.
More importantly, to our surprise, FoxO1 was also bound to the IRE2 site although the 21mer DNA construct used in our crystallization did not contain the full IRE2 sequence (Figure 1B). Apparently, a non-natural IRE2 site (referred to as IRE2′) was created by crystal packing interactions between the neighboring DNA molecules, thus completing the non-natural binding site within a pseudo-continuous helices (Figures 1C, 2A and 2C). This newly created IRE2′ site is almost identical to the native sequence except that the 5′ end sequences were altered from TGT to TCA (Figures 1A–B and 3C). A synthetic oligo containing this IRE2′ sequence is indeed recognized by FoxO1 (with ~50% reduced binding affinity compared to the natural IRE2 construct) as demonstrated by a gel shift assay (Figure 3B, lane 6). On the other hand, although the IRE3 site was included in our DNA construct, this site was vacant in the 21mer complex structure although its binding was observed in two of the four complexes in the 40mer complex structure (see below). This is consistent with the previous DNA binding studies indicating that the IRE3 site has the weakest binding affinity for FoxO1 (Vander Kooi et al., 2003; Vander Kooi et al., 2005), although slightly stronger than the non-native IRE2′ site when tested by gel shift assays (Figure 3B lane 4 and 6).
3.4. Additional supporting evidence from a 40mer complex structure and multiple variations in FoxO1 occupancy on G6PC1 promoter
Under similar crystallization conditions, we also generated a FoxO1-DBD-DNA complex with a 40mer blunt end DNA containing all four potential FoxO1 binding sites. Crystals weakly diffracted (supplementary Figure S1) and produced the best synchrotron data set at 5 Å resolution (C2 space group with a=104.36, b=251.99, c=124.37 Å, and β=89.78°). Molecular replacement solution and the subsequent rigid-body refinement indicate that each asymmetric unit contains four protein/DNA complexes with different FoxO1 protein occupancies (Figure 4A). Although some of the protein electron densities are poorly defined due to low resolution and partial occupancy at each binding site (when a protein is missing at a certain position in this crystal packing, the crystal lattice can still be maintained by additional packing interactions at the neighboring sites), the currently refined structure (Rwork=27% and Rfree=36% after reference structure-based and strictly restricted refinement) reveals that the asymmetric unit contains two complexes with FoxO1 bound to all four IREs, one with FoxO1 bound to IRE1, IRE2, and IRE4, and one with FoxO1 bound to only IRE1 and IRE2. The makeup of each complex and its electron density map are shown in Figures 4B–E. Although correct DNA sequence assignment for pseudo-continuous helices in this crystal form could have been difficult, if not impossible at this resolution, recognition of relative spacing between each binding site enabled correct identification of each binding site. These findings suggest that there exits an inhomogeneous population of FoxO1 binding to this promoter region and that the delicate balance between FoxO1 binding at specific sites in conjunction with other site-specific DNA-binding proteins may contribute to combinatorial gene regulation (Voss and Hager, 2014; Weingarten-Gabbay and Segal, 2014). These findings are also in good agreement with the relative binding affinity estimations for each IRE site (Fig. 3B). Although there are no protein-protein interactions between the adjacent FoxO1 DNA binding domains on the DNA duplex, the FoxO1 dimeric interactions between the neighboring DNA strands for crystal packing observed in the 21mer structure are also observed in the 40mer structure (see below Foxo1 Dimeric Interactions).
3.5. Overall description of the high resolution FoxO1-DBD-DNA complex structure
FoxO family members possess a conserved winged-helix DBD known as the ‘Forkhead box’ or Fox (Kaestner et al., 2000; Wang et al., 2009). Several crystal structures of FoxO family members have been determined, including FoxO1-DBD in complex with two variants of DNA target sequences (Obsil and Obsilova, 2008; Obsil and Obsilova, 2011). They share similar overall folding and DNA binding modes.
The FoxO1-DBD presented herein adopts the expected forkhead winged-helix fold made of three α-helices (H1, H2 and H3), three β-strands (S1, S2 and S3) and two wing-like loops (W1 and W2) (Fig 1C and supplementary Fig. S3). The helix H3 is positioned into the major groove roughly perpendicular to the DNA axis and provides the majority of protein-DNA interactions. However, as seen in the previous FoxO1-DBD-DNA complex structures (Brent et al., 2008; Obsil and Obsilova, 2011), the lack of electron density at the C-terminal second wing (W2) indicates its flexibility and disorderness in FoxO1. Although we used the entire FoxO1-DBD (amino acids 154–262) in crystallization, the final electron density was only visible for the regions between amino acid 160 and 245 for both monomers. The current structure superimposes very well with the previous FoxO1-DNA complex structures, with an overall root mean square deviation of 0.442 Å or less for Cα atoms (supplementary Fig. S2). One noticeable difference at the DNA binding interface among structures is the slightly altered conformation of the loop between helices H2 and H3 in our crystal structure, which provides additional non-specific backbone interactions with the major groove through Asn204 and Ser205.
3.6. Detailed protein-DNA interactions and loose FoxO1 DNA target selectivity
Although FoxO family members share similar overall folding and DNA binding modes, each FoxO member and its target sites pose unique recognition modes and regulatory mechanisms (Obsil and Obsilova, 2011). The mechanism by which different classes of FoxO proteins recognize diverse DNA sequences adjacent to the core sequence is still not fully understood (Obsil and Obsilova, 2011), although all FoxO proteins are known to recognize two consensus sequences: 5′-T[G/A]TTTTG-3′, known as the insulin response element (IRE); and 5′-T[G/A]TTTAC-3′, known as the Daf-16 family member-binding element (DBE) (Brent et al., 2008; Furuyama et al., 2000) (Figure 3C). Brent and colleagues have previously reported FoxO1 structures with two distinctive DNA target sequences (IRE and DBE), revealing a substantial degree of plasticity and different binding affinities (Brent et al., 2008). A comparison of these structures with our structures in complex with non-canonical recognition sites in the G6PC1 promoter further highlights the intriguing plasticity of molecular recognition by FoxO1.
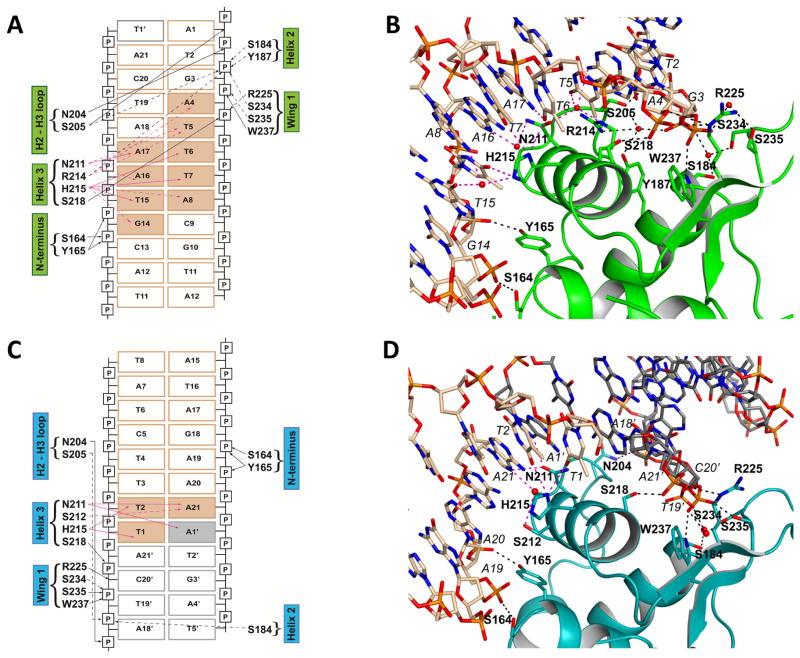
For each monomer, schematic drawings of FoxO1-DNA interactions observed in our high resolution crystal structure are shown in Figure 5. At each binding site, the FoxO1-DBD binds to the DNA duplex in a similar manner as observed in other FoxO-DBD-DNA complex structures, with the recognition helix H3 docked into the major groove of the DNA providing all of the base-specific interactions. Additional DNA backbone interactions are made by the wing W1 and a few other residues from helix H2, the H2-H3 loop and the N-terminus. These interactions are made by a combination of direct and water-mediated interactions (Figures 5). Base-specific interactions are made by the invariant residues N211 and H215 at both binding sites, and additionally R214 for IRE4 and S212 for the IRE2′ binding site. Additional weak van der Waals contacts are observed between helix H3 side chains (R214, H215, S218, and L219) and the DNA base methyl groups of the core recognition sequence for base-specific interactions.
Figure 5. Schematic diagram and detailed protein-DNA interactions between FoxO1-DBD at the downstream binding site (IRE4) (A and B) and at the upstream non-native binding site (IRE2′) (C and D).
Direct interactions are indicated by solid arrows while water-mediated interactions are indicated by dotted lines. Water molecules are omitted in schematic diagram for clarity (A and C), but they are shown in actual structure presentations (B and D). Base-specific interactions are shown with pink arrows (solid or dotted) and color-filled rectangles for interacting DNAs. Non-specific DNA backbone interactions are shown with black arrows (solid or dotted). Van der Waals contacts are not included in the figures. Gray boxed-DNAs come from the neighboring molecule in the crystal lattice. Individual amino acids and their secondary structure elements are indicated.
Surprisingly, these new FoxO1 structures reveal loose selectivity and plasticity in DNA target sequence recognition. While extensive base-specific interactions are made at the IRE4 site, only minimal base-specific interactions are made at the IRE2′ site. Base-specific recognition occurs through only 2 base pairs at IRE2′ (Figures 5C–D). Moreover, the non-canonical IRE sites in the G6PC1 promoter that are physically recognized by FoxO1 all contain one extra base pair at the core of the recognition sequence compared to the known FoxO consensus binding sequences (Figure 3C). Interestingly, the IRE3 and IRE4 sites contain the 3′ end sequence defined as DBE rather than IRE (AC instead of TG). These findings suggest that perhaps the FoxO recognition sequence needs to be expanded and that many more FoxO target genes may exist that are yet to be identified in human genome.
3.7. FoxO1 dimeric interactions and potential DNA looping
While the vertical crystal packing interactions in our crystals are provided by DNA stacking interactions that result in pseudo-continuous helices, the horizontal crystal packing interactions are solely provided by identical symmetric protein-protein interactions throughout the crystal lattice (Figure 3D). The same dimeric interactions are observed in both 21mer and 40mer crystals. Moreover, similar FoxO homo-dimers have been observed in previous FoxO1-DNA(Brent et al., 2008) and FoxO3a-DNA complex structures (Tsai et al., 2007). Although no biological functions have been ascribed for such a dimer, these interactions could be involved in physiological long-range interactions within gene promoters, such as stabilization of DNA-loops (Nolis et al., 2009; Sanyal et al., 2012). The individual dsDNA molecules in the crystals are bent about 25.84° due to FoxO1 binding at IRE4 as calculated with the program CURVES+ (Lavery et al., 2009), with a slight increase in the width of the major groove in the core-sequence region. Although the observed bending is compensated for by FoxO1 binding at the adjacent IRE site, resulting in a relatively straight orientation of DNAs in the crystal packings of both crystal forms, a considerable degree of DNA bending upon FoxO1 binding suggests that potential interactions between FoxO1 monomers may contribute to long-range DNA looping.
4. Discussion
Controlled gene expression is a central element for all living systems and is often combinatorial in nature. A typical gene promoter contains multiple transcription factor binding sites whose synergistic activities in a time- and signal-dependent manner account for well-coordinated programs of cellular responses (Merika and Thanos, 2001; Weingarten-Gabbay and Segal, 2014). Moreover, clustering of multiple binding sites for the same transcription factors (homotypic clustering) is widespread in vertebrate genomes and predominantly found in promoter and enhancer regions (Gotea et al., 2010; He et al., 2012; Kazemian et al., 2013). However, the exact roles of such homotypic clustering and the interplay among various transcription factors have not been well characterized. We have investigated the molecular basis of target recognition by FoxO1 on the human G6PC1 promoter whose gene product performs a key role in the control of blood glucose homeostasis. Here we report several novel and unexpected findings which include (i) a discovery of a new FoxO1 binding site that suggests mutual exclusion of FoxO1 and CREB binding to the G6PC1 promoter, (ii) a demonstration that homotypic clustering of multiple FoxO1 binding sites at the G6PC1 promoter involves FoxO1 binding with widely different binding affinities, (iii) a discovery of loose DNA selectivity by FoxO1, and (iv) potential DNA looping mediated by FoxO1 dimeric interactions.
Multiple transcription factors including FoxO1, CREB, and hepatocyte nuclear factors (HNFs) are involved in G6PC1 gene regulation and distinctive roles for these transcription factors have been reported (Hirota et al., 2008; Onuma et al., 2009; Vander Kooi et al., 2005). Current evidence suggests considerable cross-talk between FoxO1 and CREB pathways on hormone-responsive gluconeogenic genes including G6PC1 (Gross et al., 2008; Herzig et al., 2001; Oh et al., 2013b). Although FoxO1 and CREB are regulated by independent signal-transduction pathways and by multiple mechanisms involving change in phosphorylation/acetylation/ubiquitination, these two pathways coordinately regulate gluconeogenesis (Gross et al., 2008; Herzig et al., 2001; Oh et al., 2013b; Ravnskjaer et al., 2016). However, whether G6PC1 gene expression is regulated independently by these two pathways upon binding of FoxO1 and CREB to their respective binding sites or whether FoxO1 and CREB interact synergistically on the G6PC1 promoter had not been studied.
Our data suggest that although the conserved IRE4/CRE elements on the G6PC1 promoter accommodates both FoxO1 and CREB, their binding is likely mutually exclusive such that they cannot occupy the G6PC1 promoter at the same time. Our crystal structures reveal a new FoxO1 binding site and this newly discovered IRE site (IRE4) exhibits considerable overlap with the CREB binding site, which is in good agreement with the results of gel-shift assay using CREB and FoxO1 proteins in combination (Figure 1D). This conclusion was further supported by the results from transcriptional assays using mutant G6PC1 fusion genes and binding assays using oligos containing individual IRE elements within the G6PC1 promoter. While FoxO1 and CREB cannot occupy the IRE4/CRE site at the same time we think it is unlikely that they compete for binding to this overlapping site. Instead, previous discoveries that CREB and FoxO1 are sequentially involved in gluconeogenic gene expression via activator/coactivator exchange during fasting suggest that FoxO1 and CREB share this site (Liu et al., 2008; Oh et al., 2013b). Thus, we think FoxO1 and CREB stimulate G6PC1 gene expression during fasting by sequentially occupying the IRE4/CRE site, in addition to FoxO1 binding the other three IREs. In the fasted state, increases in circulating glucagon first stimulate hepatic gluconeogenesis via the PKA-mediated phosphorylation of CREB and the promotion of the CREB-CRTC2 axis (Liu et al., 2008), which is repressed by AMPK(Lee et al., 2010), while decreases in circulating insulin then increase gluconeogenic gene expression via the de-phosphorylation of the forkhead transcription factor FoxO1 reversing the repression of FoxO1 by Akt upon insulin activation(Gross et al., 2009; Park et al., 2010). Our findings therefore support this sequential utilization of the CREB and FoxO1 pathways during the early (short-term) and late (prolonged) fasting stages, illustrating how the exchange of two gluconeogenic regulators during fasting maintains energy balance (Liu et al., 2008; Oh et al., 2013b). It is worth noting that the IRE4 site also constitutes one of the two half-sites of a glucocorticoid response element within the G6PC1 promoter (Vander Kooi et al., 2005), making this region a potential hotspot for integration of hormonal responses.
Our crystal structures also represent a new class of FoxO-DNA interactions. Although FoxO proteins display a conserved mode of DNA docking, their sequence recognition modes vary considerably (Obsil and Obsilova, 2008). The G6PC1 promoter contains multiple non-canonical FoxO binding sites, and surprisingly, they all contain one additional base pair within the core recognition sequence. The presence of novel FoxO binding sites on the G6PC1 promoter suggests the existence of versatile DNA recognition by FoxO1, and our structures provide new insights into how non-canonical DNA target sites are recognized. Our high resolution crystal structure also reveals a non-native binding site created by DNA stacking within crystal lattice. Despite weaker interactions, the IRE2′-FoxO1 interactions were captured in our crystal structure. Surprisingly, this recognition requires only two base-pair specific interactions and DNA backbone interactions using the same protein structural elements and residues. Our structure unexpectedly reveals loose selectivity and high versatility of DNA recognition by a FoxO family member. These finding, therefore, expand the repertoire of the FoxO recognition sequences and suggest that many more FoxO target genes will be found in the human genome.
FoxO1 has been known to bind cooperatively to G6PC1 IREs (Onuma et al., 2006). However, no physical protein-protein interactions are observed between the FoxO1 DNA binding domains although interactions between the full-length proteins cannot be ruled out. Transcriptional activity of FoxO proteins are also tightly controlled by various post-translational modifications such as phosphorylation, acetylation, methylation and ubiquitination, as well as subcellular localization and direct protein-protein interaction (Calnan and Brunet, 2008; Huang and Tindall, 2007). These modification sites include S249, S256, R251, R253, K245, K248, and K262 within the FoxO1-DBD and their potential molecular effects are well described in earlier publications (Brent et al., 2008; Obsil and Obsilova, 2008; Obsil and Obsilova, 2011; Tsai et al., 2007). Also, additional FoxO family members are known to act on the G6PC1 promoter (Haeusler et al., 2010; Kim et al., 2011; Onuma et al., 2006). FoxO1 orthologs, such as FoxO3 and FoxO4, are known to bind to the same IRE sites and the integrated control of insulin-responsive G6PC1 gene expression by FoxO family members has been reported (Haeusler et al., 2014; Onuma et al., 2006). Furthermore, interdependent binding of Fox family members, i.e. FoxO1 and FoxA1/A2, to insulin-sensitive genes including G6PC1 is known to nucleate transcriptional events in chromatin in response to signaling events leading to regulation of hepatic glucose metabolism (Yalley et al., 2016). Therefore, whether or not other members of the FoxO family bind to all IRE sites and how these multiple IRE sites on the G6PC1 promoter are synergistically utilized for coactivator recruitment and chromatin remodeling warrant further investigation.
Finally, there is a growing interest in developing agents that specifically modulate hepatic glucose production by targeting FoxO1 and/or G6PC1. Since excessive hepatic glucose production is a contributing factor to fasting hyperglycemia in diabetes, G6PC1/FoxO1 continues to be an attractive therapeutic target for the treatment of type 2 diabetes mellitus and has proven very effective (Nagashima et al., 2010; Pandey et al., 2016; Samuel et al., 2006). Even though the structures of individual FoxO family proteins in complex with DNA have been already determined, our new crystal structures and the molecular basis of novel DNA recognition of the G6PC1 promoter by FoxO1 provide additional insights into the molecular functions of FoxO1 which can potentially be specifically targeted for modulating G6PC1 expression and glucose production as a potential diabetes therapy.
Supplementary Material
Acknowledgments
We wish to thank the staff at SER-CAT beamlines 22-ID and 22-BM for data collection. We are also grateful to Guangteng Wu and Geun Bae Rha, for their assistance in sample preps during the early stage of the project. This work was supported by the American Diabetes Association (7-08-CD-03) and the Commonwealth of Kentucky Diabetes Research Trust Fund (KDR-PP09-09) to Y.C., and the American Diabetes Association (7-13-BS-119) and the National Institute of Health (R01DK92589) to R.M.O.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://....
Footnotes
Conflict of Interest
The authors declare that thru have no conflicts of interest with the contents of this article.
Author contributions
Y.C. and R.M.O. conceived the experiments; Y.C. and P.S. purified the proteins and nucleic acids and crystallized the complexes; P.S. and Y.C. collected and analyzed the structural data; Y.C., P.S. and J.A.E. solved and refined the structures; E.H. performed the G6PC1 promoter mutagenesis and luciferase assays; P.S. performed the EMSA experiment; Y.C. and R.M.O. wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallogr D Struc Biol. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arden KC. FOXO animal models reveal a variety of diverse roles for FOXO transcription factors. Oncogene. 2008;27:2345–2350. doi: 10.1038/onc.2008.27. [DOI] [PubMed] [Google Scholar]
- Brent MM, Anand R, Marmorstein R. Structural basis for DNA recognition by FoxO1 and its regulation by posttranslational modification. Structure. 2008;16:1407–1416. doi: 10.1016/j.str.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calnan DR, Brunet A. The FoxO code. Oncogene. 2008;27:2276–2288. doi: 10.1038/onc.2008.21. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta crystallogr D Struc Biol. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou JY, Mansfield BC. The SLC37 family of sugar-phosphate/phosphate exchangers. Curr Top Membr. 2014;73:357–382. doi: 10.1016/B978-0-12-800223-0.00010-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cesare D, Sassone-Corsi P. Transcriptional regulation by cyclic AMP-responsive factors. Prog Nucleic Acid Res Mol Biol. 2000;64:343–369. doi: 10.1016/s0079-6603(00)64009-6. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Struc Biol. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Furuyama T, Nakazawa T, Nakano I, Mori N. Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem J. 2000;349:629–634. doi: 10.1042/0264-6021:3490629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotea V, Visel A, Westlund JM, Nobrega MA, Pennacchio LA, Ovcharenko I. Homotypic clusters of transcription factor binding sites are a key component of human promoters and enhancers. Genome Res. 2010;20:565–577. doi: 10.1101/gr.104471.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross DN, van den Heuvel AP, Birnbaum MJ. The role of FoxO in the regulation of metabolism. Oncogene. 2008;27:2320–2336. doi: 10.1038/onc.2008.25. [DOI] [PubMed] [Google Scholar]
- Gross DN, Wan M, Birnbaum MJ. The role of FOXO in the regulation of metabolism. Curr Diab Rep. 2009;9:208–214. doi: 10.1007/s11892-009-0034-5. [DOI] [PubMed] [Google Scholar]
- Haeusler RA, Kaestner KH, Accili D. FoxOs function synergistically to promote glucose production. J Biol Chem. 2010;285:35245–35248. doi: 10.1074/jbc.C110.175851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler RA, Hartil K, Vaitheesvaran B, Arrieta-Cruz I, Knight CM, Cook JR, Kammoun HL, Febbraio MA, Gutierrez-Juarez R, Kurland IJ, Accili D. Integrated control of hepatic lipogenesis versus glucose production requires FoxO transcription factors. Nat Commun. 2014;5:5190. doi: 10.1038/ncomms6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Duque TS, Sinha S. Evolutionary origins of transcription factor binding site clusters. Mol Biol Evol. 2012;29:1059–1070. doi: 10.1093/molbev/msr277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzig S, Long F, Jhala US, Hedrick S, Quinn R, Bauer A, Rudolph D, Schutz G, Yoon C, Puigserver P, Spiegelman B, Montminy M. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- Hirota K, Sakamaki J, Ishida J, Shimamoto Y, Nishihara S, Kodama N, Ohta K, Yamamoto M, Tanimoto K, Fukamizu A. A combination of HNF-4 and Foxo1 is required for reciprocal transcriptional regulation of glucokinase and glucose-6-phosphatase genes in response to fasting and feeding. J Biol Chem. 2008;283:32432–32441. doi: 10.1074/jbc.M806179200. [DOI] [PubMed] [Google Scholar]
- Hornbuckle LA, Everett CA, Martin CC, Gustavson SS, Svitek CA, Oeser JK, Neal DW, Cherrington AD, O’Brien RM. Selective stimulation of G-6-Pase catalytic subunit but not G-6-P transporter gene expression by glucagon in vivo and cAMP in situ. Am J Physiol Endocrinol Metab. 2004;286:E795–808. doi: 10.1152/ajpendo.00455.2003. [DOI] [PubMed] [Google Scholar]
- Hosaka T, Biggs WH, 3rd, Tieu D, Boyer AD, Varki NM, Cavenee WK, Arden KC. Disruption of forkhead transcription factor (FOXO) family members in mice reveals their functional diversification. Proc Natl Acad Sci U S A. 2004;101:2975–2980. doi: 10.1073/pnas.0400093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Tindall DJ. Dynamic FoxO transcription factors. J Cell Sci. 2007;120:2479–2487. doi: 10.1242/jcs.001222. [DOI] [PubMed] [Google Scholar]
- Hutton JC, O’Brien RM. Glucose-6-phosphatase catalytic subunit gene family. J Biol Chem. 2009;284:29241–29245. doi: 10.1074/jbc.R109.025544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jancarik J, Kim SH. Sparse matrix sampling: a screening method for crystallization of proteins. J Appl Cryst. 1991;24:409–411. [Google Scholar]
- Kaestner KH, Knochel W, Martinez DE. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 2000;14:142–146. [PubMed] [Google Scholar]
- Kazemian M, Pham H, Wolfe SA, Brodsky MH, Sinha S. Widespread evidence of cooperative DNA binding by transcription factors in Drosophila development. Nucleic Acids Res. 2013;41:8237–8252. doi: 10.1093/nar/gkt598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Perdomo G, Zhang T, Slusher S, Lee S, Phillips BE, Fan Y, Giannoukakis N, Gramignoli R, Strom S, Ringquist S, Dong HH. FoxO6 integrates insulin signaling with gluconeogenesis in the liver. Diabetes. 2011;60:2763–2774. doi: 10.2337/db11-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kousteni S. FoxO1: a molecule for all seasons. J Bone Miner Res. 2011;26:912–917. doi: 10.1002/jbmr.306. [DOI] [PubMed] [Google Scholar]
- Lalmansingh AS, Karmakar S, Jin Y, Nagaich AK. Multiple modes of chromatin remodeling by Forkhead box proteins. Biochim Biophys Acta. 2012;1819:707–715. doi: 10.1016/j.bbagrm.2012.02.018. [DOI] [PubMed] [Google Scholar]
- Lavery R, Moakher M, Maddocks JH, Petkeviciute D, Zakrzewska K. Conformational analysis of nucleic acids revisited: Curves+ Nucleic Acids Res. 2009;37:5917–5929. doi: 10.1093/nar/gkp608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Seo WY, Song KH, Chanda D, Kim YD, Kim DK, Lee MW, Ryu D, Kim YH, Noh JR, Lee CH, Chiang JY, Koo SH, Choi HS. AMPK-dependent repression of hepatic gluconeogenesis via disruption of CREB.CRTC2 complex by orphan nuclear receptor small heterodimer partner. J Biol Chem. 2010;285:32182–32191. doi: 10.1074/jbc.M110.134890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Dentin R, Chen D, Hedrick S, Ravnskjaer K, Schenk S, Milne J, Meyers DJ, Cole P, Yates J, 3rd, Olefsky J, Guarente L, Montminy M. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature. 2008;456:269–273. doi: 10.1038/nature07349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiese K, Chong ZZ, Shang YC. OutFOXOing disease and disability: the therapeutic potential of targeting FoxO proteins. Trends Mol Med. 2008;14:219–227. doi: 10.1016/j.molmed.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M, Pocai A, Rossetti L, Depinho RA, Accili D. Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor Foxo1 in liver. Cell Metab. 2007;6:208–216. doi: 10.1016/j.cmet.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Merika M, Thanos D. Enhanceosomes. Curr Opin Genet Dev. 2001;11:205–208. doi: 10.1016/s0959-437x(00)00180-5. [DOI] [PubMed] [Google Scholar]
- Mihaylova MM, Vasquez DS, Ravnskjaer K, Denechaud PD, Yu RT, Alvarez JG, Downes M, Evans RM, Montminy M, Shaw RJ. Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell. 2011;145:607–621. doi: 10.1016/j.cell.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima T, Shigematsu N, Maruki R, Urano Y, Tanaka H, Shimaya A, Shimokawa T, Shibasaki M. Discovery of novel forkhead box O1 inhibitors for treating type 2 diabetes: improvement of fasting glycemia in diabetic db/db mice. Mol Pharmacol. 2010;78:961–970. doi: 10.1124/mol.110.065714. [DOI] [PubMed] [Google Scholar]
- Nolis IK, McKay DJ, Mantouvalou E, Lomvardas S, Merika M, Thanos D. Transcription factors mediate long-range enhancer-promoter interactions. Proc Natl Acad Sci U S A. 2009;106:20222–20227. doi: 10.1073/pnas.0902454106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordlie RC, Foster JD, Lange AJ. Regulation of glucose production by the liver. Annu Rev Nutr. 1999;19:379–406. doi: 10.1146/annurev.nutr.19.1.379. [DOI] [PubMed] [Google Scholar]
- Obsil T, Obsilova V. Structure/function relationships underlying regulation of FOXO transcription factors. Oncogene. 2008;27:2263–2275. doi: 10.1038/onc.2008.20. [DOI] [PubMed] [Google Scholar]
- Obsil T, Obsilova V. Structural basis for DNA recognition by FOXO proteins. Biochim Biophys Acta. 2011;1813:1946–1953. doi: 10.1016/j.bbamcr.2010.11.025. [DOI] [PubMed] [Google Scholar]
- Oh KJ, Han HS, Kim MJ, Koo SH. Transcriptional regulators of hepatic gluconeogenesis. Arch Pharm Res. 2013a;36:189–200. doi: 10.1007/s12272-013-0018-5. [DOI] [PubMed] [Google Scholar]
- Oh KJ, Han HS, Kim MJ, Koo SH. CREB and FoxO1: two transcription factors for the regulation of hepatic gluconeogenesis. BMB Rep. 2013b;46:567–574. doi: 10.5483/BMBRep.2013.46.12.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onuma H, Vander Kooi BT, Boustead JN, Oeser JK, O’Brien RM. Correlation between FOXO1a (FKHR) and FOXO3a (FKHRL1) binding and the inhibition of basal glucose-6-phosphatase catalytic subunit gene transcription by insulin. Mol Endocrinol. 2006;20:2831–2847. doi: 10.1210/me.2006-0085. [DOI] [PubMed] [Google Scholar]
- Onuma H, Oeser JK, Nelson BA, Wang Y, Flemming BP, Scheving LA, Russell WE, O’Brien RM. Insulin and epidermal growth factor suppress basal glucose-6-phosphatase catalytic subunit gene transcription through overlapping but distinct mechanisms. Biochem J. 2009;417:611–620. doi: 10.1042/BJ20080999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pandey A, Kumar GS, Kadakol A, Malek V, Gaikwad AB. FoxO1 Inhibitors: The Future Medicine for Metabolic Disorders? Curr Diabetes Rev. 2016;12:223–230. doi: 10.2174/1573399811666150804104858. [DOI] [PubMed] [Google Scholar]
- Panne D. The enhanceosome. Curr Opin Struct Biol. 2008;18:236–242. doi: 10.1016/j.sbi.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Park JM, Kim TH, Bae JS, Kim MY, Kim KS, Ahn YH. Role of resveratrol in FOXO1-mediated gluconeogenic gene expression in the liver. Biochem Biophys Res Commun. 2010;403:329–334. doi: 10.1016/j.bbrc.2010.11.028. [DOI] [PubMed] [Google Scholar]
- Ravnskjaer K, Madiraju A, Montminy M. Role of the cAMP Pathway in Glucose and Lipid Metabolism. Handb Exp Pharmacol. 2016;233:29–49. doi: 10.1007/164_2015_32. [DOI] [PubMed] [Google Scholar]
- Roden M, Bernroider E. Hepatic glucose metabolism in humans--its role in health and disease. Best Pract Res Clin Endocrinol Metab. 2003;17:365–383. doi: 10.1016/s1521-690x(03)00031-9. [DOI] [PubMed] [Google Scholar]
- Samuel VT, Choi CS, Phillips TG, Romanelli AJ, Geisler JG, Bhanot S, McKay R, Monia B, Shutter JR, Lindberg RA, Shulman GI, Veniant MM. Targeting foxo1 in mice using antisense oligonucleotide improves hepatic and peripheral insulin action. Diabetes. 2006;55:2042–2050. doi: 10.2337/db05-0705. [DOI] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streeper RS, Svitek CA, Goldman JK, O’Brien RM. Differential role of hepatocyte nuclear factor-1 in the regulation of glucose-6-phosphatase catalytic subunit gene transcription by cAMP in liver- and kidney-derived cell lines. J Biol Chem. 2000;275:12108–12118. doi: 10.1074/jbc.275.16.12108. [DOI] [PubMed] [Google Scholar]
- Tsai KL, Sun YJ, Huang CY, Yang JY, Hung MC, Hsiao CD. Crystal structure of the human FOXO3a-DBD/DNA complex suggests the effects of post-translational modification. Nucleic Acids Res. 2007;35:6984–6994. doi: 10.1093/nar/gkm703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Cryst. 1997;30:1002–1025. [Google Scholar]
- van Schaftingen E, Gerin I. The glucose-6-phosphatase system. J Biol Chem. 2002;362:513–532. doi: 10.1042/0264-6021:3620513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Kooi BT, Streeper RS, Svitek CA, Oeser JK, Powell DR, O’Brien RM. The three insulin response sequences in the glucose-6-phosphatase catalytic subunit gene promoter are functionally distinct. J Biol Chem. 2003;278:11782–11793. doi: 10.1074/jbc.M212570200. [DOI] [PubMed] [Google Scholar]
- Vander Kooi BT, Onuma H, Oeser JK, Svitek CA, Allen SR, Vander Kooi CW, Chazin WJ, O’Brien RM. The glucose-6-phosphatase catalytic subunit gene promoter contains both positive and negative glucocorticoid response elements. Mol Endocrinol. 2005;19:3001–3022. doi: 10.1210/me.2004-0497. [DOI] [PubMed] [Google Scholar]
- Voss TC, Hager GL. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat Rev Genet. 2014;15:69–81. doi: 10.1038/nrg3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Zhang X, Zhao H, Wang Q, Pan Y. FoxO gene family evolution in vertebrates. BMC Evol Biol. 2009;9:222. doi: 10.1186/1471-2148-9-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingarten-Gabbay S, Segal E. The grammar of transcriptional regulation. Hum Genet. 2014;133:701–711. doi: 10.1007/s00439-013-1413-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalley A, Schill D, Hatta M, Johnson N, Cirillo LA. Loss of Interdependent Binding by the FoxO1 and FoxA1/A2 Forkhead Transcription Factors Culminates in Perturbation of Active Chromatin Marks and Binding of Transcriptional Regulators at Insulin-sensitive Genes. J Biol Chem. 2016;291:8848–8861. doi: 10.1074/jbc.M115.677583. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.