Abstract
A screen for the systematic identification of cis-regulatory elements within large (>100 kb) genomic domains containing Hox genes was performed by using the basal chordate Ciona intestinalis. Randomly generated DNA fragments from bacterial artificial chromosomes containing two clusters of Hox genes were inserted into a vector upstream of a minimal promoter and lacZ reporter gene. A total of 222 resultant fusion genes were separately electroporated into fertilized eggs, and their regulatory activities were monitored in larvae. In sum, 21 separable cis-regulatory elements were found. These include eight Hox linked domains that drive expression in nested anterior–posterior domains of ectodermally derived tissues. In addition to vertebrate-like CNS regulation, the discovery of cis-regulatory domains that drive epidermal transcription suggests that C. intestinalis has arthropod-like Hox patterning in the epidermis.
Keywords: cis-regulation, hox cluster, ascidian
Members of the Hox family of homeobox-containing genes encode DNA-binding proteins whose structures, genomic organizations, expression patterns, and biological functions are highly conserved throughout the higher metazoans. In vertebrates, axial Hox expression is seen in the neural tube and some paraxial mesodermal derivatives such as the somites (1, 2), whereas, in arthropods, Hox gene expression is found in the ventral nerve cord, visceral mesoderm, and epidermis (3). In all lineages examined thus far, Hox genes are expressed in nested anterior to posterior axial domains that are coincident with their linear positions within gene complexes. The pan-metazoan conservation of this colinearity suggests that there has been strong stabilizing selection on shared Hox cis-regulatory elements. Identifying these elements remains a primary goal of developmental genetics.
Because it is not yet possible to predict gene expression patterns based on sequence analysis alone, and cell culture assays fail to capture spatial or temporal information at high resolution, testing regulatory activity by transgenesis is the current method of choice for identifying and characterizing sequence-specific regulatory elements. However, the experimental difficulties involved in the generation of transgenic metazoan embryos limits the rate at which this work can proceed. These limitations are largely eliminated by the use of the protochordate Ciona intestinalis, a model system that can be used to characterize cis-regulatory DNAs by using high-throughput functional genomics techniques (4). Here, we adapt these methods to screen for cis-regulatory activity in >200 kb of genomic DNA containing C. intestinalis Hox genes. Regulatory elements flanking and internal to Hox genes are described.
Materials and Methods
Ascidians. Adult C. intestinalis were collected from Pillar Point Harbor in San Mateo County, CA, under scientific permit of the State of California Fish and Game Department. These animals were kept in recirculating artificial seawater at 16°C and used for artificial fertilization within 2 weeks of their removal from the wild.
Bacterial Artificial Chromosome (BAC) Clones. A BAC library made from C. intestinalis collected in Maizuru, Japan, was hybridized with probes prepared from putative C. intestinalis Hox2, Hox3, Hox4, and Hox11/12. Two Hox-containing BACs, designated XNI and XNE, were isolated. The BACs were sequenced and assembled by using the hierarchical shotgun sequencing strategy (5–7) with ABI3700 DNA sequencers. Gene models were predicted by using genscan (8) and by blast alignments to known genes and EST sequences (9, 10).
Cis-Screening Libraries. The screening libraries were built in the pCES vector (4) that contained a multiple cloning site and the C. intestinalis fkh basal promoter (11) fused to a lacZ reporter gene. The fkh core promoter sequence is essentially inactive in electroporated embryos but can be activated in every major tissue when linked to appropriate tissue-specific enhancers. The XNI and XNE BACs were separately sheared by Hydroshear (Genemachines, San Carlos, CA) to generate two random sets of DNA fragments ≈3.0 kb in size. These fragments were end-filled, then ligated into the blunted BamHI site of pCES to produce the xne and xni screening libraries. Colonies from these libraries were randomly picked and numbered, and both ends of the inserts were sequenced by using standard high-throughput sequencing protocols (ref. 12; see also www.jgi.doe.gov). The fkh basal promoter was chosen for this study, because it is known that the native fkh gene is expressed in a broad range of tissues and is therefore unlikely to contain tissue-specific silencers. This core promoter has been shown to produce faithful patterns of expression when combined with heterologous enhancers (13, 14).
Transgenics. Plasmids containing inserts were prepped by using QIAquick midi-kits (Qiagen, Valencia, CA). Electroporation, fixation, and 5-bromo-4-chloro-3-indolyl β-d-galactoside staining were performed as described (15). Aliquots containing 75 μg of a single experimental plasmid plus 25 μg of a control plasmid (-3.5CiBra>GFP) (16) were used in each electroporation. Batches of electroporated embryos were allowed to develop at 16°C for 18 h after fertilization. Because of the intracellular stability of the β-galactosidase protein, staining in a given cell is indicative of lacZ transcription at some time during that cell's ontogeny rather than active transcription at the time of fixation. Batches of electroporated embryos were prescreened for percentage of fully developed animals and expression of the control plasmid. Controls indicate that there are low levels of background expression in the mesenchyme (undifferentiated cells in the posterior trunk that contribute to postmetamorphosis mesodermal structures).
Whole-Mount in Situ Hybridization. In situ hybridizations were done with whole-mount staged embryos as described (15). Embryos were allowed to develop to the indicated stages at 16°C, fixed with paraformaldehyde, and stored in ethanol at -20°C. Digoxigenin-labeled RNA probes were synthesized by using T7 and T3 RNA polymerase (Promega).
Results
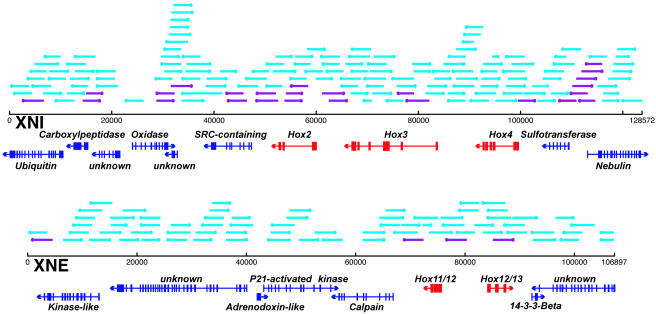
Six BACs that contain Hox DNA were isolated by low stringency hybridization. Sequence analysis revealed that these fell into two groups of three BACs with overlapping sequence. Two of the BACs (designated XNI and XNE) were sequenced by random shotgun methods and subsequently finished by primer walking. They were found to contain five putative Hox genes and predicted coding domains for 10 additional non-Hox genes. The orthologs of the vertebrate Hox2, Hox3, and Hox4 genes are linked on a BAC DNA clone (XNI) that is 12,873 bp in length, whereas the Hox11/12 and Hox12/13 genes are contained on a separate 106,897-bp BAC (XNE) (Fig. 1). Comparison to the draft genomic sequence (17) confirms these results. Additionally, both the current genomic draft and a recent cosmid walking effort (18) show that the C. intestinalis Hox genes are located in five separate genomic regions: [Hox1], [Hox2, Hox3, and Hox4], [Hox5 and Hox6], [Hox10], and [Hox11/12 and Hox12/13]. All of the putative Hox transcription units contained within the two BAC DNAs are shown in Fig. 1. As discussed in Spagnuolo et al. (18) and Wada et al. (19), analysis of the predicted amino acid sequences and genomic structures of Hox11/12 and Hox12/13 do not allow conclusive assignments to single parology groups for these two loci. Although the sequence of Hox5 is not completely conclusive, its position and orientation relative to the neighboring Hox6 locus is sufficient to assign it putative membership in the fifth Hox parology group (19).
Fig. 1.
Two Hox containing genomic BACs of C. intestinalis DNA and clones screened for cis-regulatory activity. Shown is the predicted structure of CiHox2, CiHox3, CiHox4, CiHox11/12, and CiHox 12/13, along with multiple non-Hox genes, in the XNI and XNE BACs. A total of 222 random library clones (≈3 kb) were randomly picked, and their positions on the BACs were identified. The sides of the inserts closest to the basal promoter in the vectors are indicated by dots. When individually electroporated into single-cell C. intestinalis embryos, 29 of the clones, shown in purple, were found to direct distinct, repeatable expression of the lacZ reporter gene. Full sequence information for these BACs can be retrieved from the GenBank database.
The C. intestinalis Hox genes examined here possess a number of genomic structure anomalies when compared with other metazoans. Two of the genes, Hox11/12 and Hox12/13, are divergently transcribed (Fig. 1). In virtually all other animals, Hox genes are transcribed in the same orientation (20). In addition, all mammalian Hox genes identified to date contain two exons, with the homeodomain present in the 3′ exon (21). In C. intestinalis, the Hox genes frequently contain three or more predicted exons (Fig. 1). The major peculiarity is the fact that the Hox genes are not tightly linked into a single cohesive complex. In most other animals, the Hox complexes are not interrupted by foreign genes or, at most, contain just one or two interruptions, as seen in Drosophila (22).
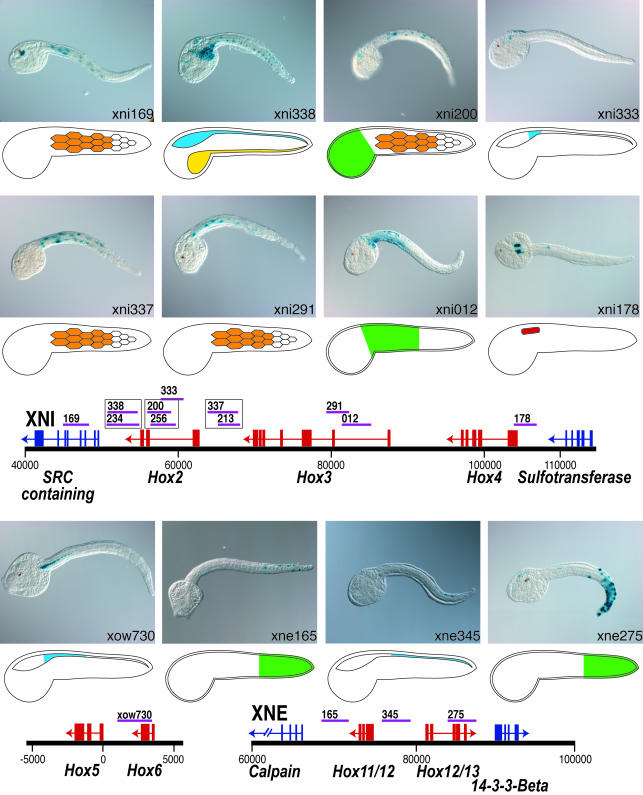
Cis-regulatory screening libraries were made by randomly cloning ≈3-kb fragments from each of the BACs into the pCES reporter vector. A total of 222 insert-containing clones from these libraries were individually electroporated into C. intestinalis embryos. Insert locations relative to the complete BACs XNI and XNE are shown in Fig. 1, with the subset that activated the lacZ reporter gene in one or more tissues shown in purple. Of these 222 constructs, 29 exhibited specific lacZ staining patterns (Table 1, which is published as supporting information on the PNAS web site). Taking overlapping fragments that drive identical expression patterns into account, 21 domains with distinct cis-regulatory activity were found in the BACs. Within the 100 kb that contain the two target Hox gene clusters, 14 fragments with cis-regulatory activity were identified (Fig. 2); 11 of these seem to be independent cis-regulatory domains. A 12th enhancer was identified associated with the Hox6 and Hox5 genes (Fig. 2).
Fig. 2.
Cis-regulatory activity from genomic fragments neighboring Hox genes. Fifteen fragments that were tightly linked to Hox loci drive distinct domains of LacZ activity, indicating that the cloned fragments contained cis-regulatory activity. Specific tissues and domains of expression are represented diagrammatically: CNS, blue; epidermis, green; gut, yellow; muscle, orange; trunk lateral cells, red.
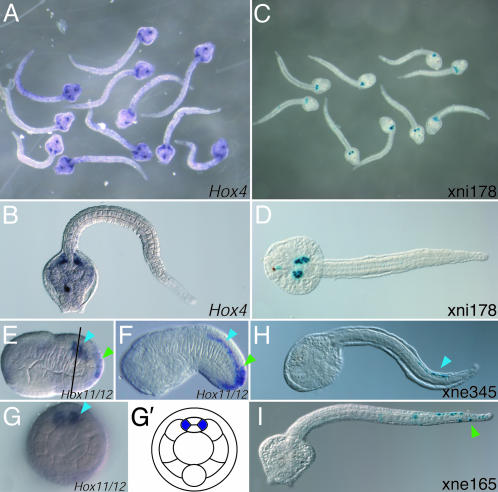
Endogenous expression patterns have been characterized by in situ hybridization for four of the C. intestinalis Hox genes addressed in the current study. Hox3 and Hox5 have been described (23, 24). The native expression patterns of Hox4 and Hox11/12 were characterized by in situ hybridization of RNA probes (Fig. 3). Enhancers identified in this screen drive expression consistent with in situ patterns for three of these genes: Hox4, Hox5, and Hox11/12.
Fig. 3.
Characterization of native CiHox4 and CiHox11/12 transcription by whole-mount in situ hybridization. Digoxigenin-labeled RNA antisense probes synthesized from exon regions of CiHox4 (A and B) and CiHox11/12 (E–G) were hybridized to C. intestinalis embryos. (A and B) In late tailbud embryos (18 h after fertilization at 16°C), CiHox4 transcription is detected in the trunk lateral cells, a domain which is identical to that driven by the xni178 construct (C and D). (E–G) In early tailbud embryos (E and G, 10 h after fertilization at 16°C; F, 12 h after fertilization at 15°C), CiHox12 transcript is detected in both the posterior neural tube (blue arrows) and the posterior tail epidermis (green arrows). (G and G′) Optical cross section and schematic representation at the approximate position of the line shown in E shows that the CiHox11/12 CNS expression is only detected in the paired lateral cells of the neural tube. The epidermis and neural tube patterns are a summation of the domains driven by the xne345 (H, posterior neural tube, blue arrow) and xne165 (I, posterior tail epidermis, green arrow).
At 18 h after fertilization, Hox4 is expressed in the trunk lateral cells (Fig. 3 A and B). These clusters of undifferentiated mesoderm cells flank the junction between the cerebral vesicle and the neural tube and give rise to multiple adult tissues including subsets of the blood, body wall muscle, and gill slits (25, 26). Expression of one other C. intestinalis Hox gene, Hox5, has also been described by in situ hybridization in these cells (23). The xni178 DNA, which overlaps the 5′ terminus of the Hox4 transcription unit, activates lacZ transcription in the same trunk lateral cell domain (Figs. 2 and 3 C and D), indicating that it probably contains the authentic enhancer for this gene.
In addition to the trunk lateral cells, Hox5 is expressed in the lateral cells of the anterior nerve cord (23). This expression domain starts at the trunk–tail boundary and extends through the anterior fourth of the tail. Although not part of the random screen of the two BACs, a DNA fragment, xow730, was found to activate an identical pattern of neural tube expression (Fig. 2). This fragment extends across much of the intragenic domain between Hox5 and Hox6, with a partial overlap of the 3′ of the Hox6 locus. The exact match between the xow730-driven expression and the neural tube expression of Hox5 suggests that it includes the authentic enhancer for this subdomain of Hox5 expression.
In early tailbud embryos, in situ hybridization with Hox11/12 probe detects expression in two different tissues in the posterior tail, the epidermis and the neural tube (Fig. 3 E and F). As with the Hox5 expression domain, the neural tube expression of Hox11/12 seems to be limited to the lateral cells (Fig. 3G). The Hox11/12 locus is flanked by two domains shown to have cis-regulatory activity. xne345, located to the 5′ of Hox11/12, drives marker gene expression in the posterior half of the neural tube (Figs. 2 and 3H). xne165, located to the 3′ of Hox11/12, drives marker gene expression in the epidermis of the posterior half of the tail (Figs. 2 and 3I). Thus, the full endogenous Hox11/12 expression pattern is recapitulated by these two separate enhancers.
Hox3 in situ hybridization studies in hatched tadpoles have shown that this gene is expressed in the posterior cerebral vesicle and anterior neural tube (24). The present survey identifies four DNA domains that exhibit cis-regulatory activity flanking or overlapping the Hox3 locus (Fig. 2). However, none of these direct expression in the CNS. Specifically, xni337 and xni213, which overlap and probably represent a single regulatory module, both produce expression in the tail muscles. xni291 and xni012 direct expression in the tail muscles and epidermis, respectively. Whereas there is no a priori reason to believe that the muscle expression represents authentic Hox enhancer activity, two other C. intestinalis Hox genes have been shown to be expressed in the epidermis in restricted anterior–posterior domains. In addition to the Hox11/12 posterior tail epidermis staining described here (Fig. 3), in situ studies using probes for Hox1 have shown that this gene is expressed in the epidermis at the trunk–tail boundary (27). The similarity between epidermal staining driven by xni012 and these known Hox expression domains suggests that the xni012 may be an authentic epidermal enhancer. Interestingly, DNA fragments that overlap xni291 and xni012 have been shown to direct expression in the CNS (24).
There are alternate potential explanations for the unexpected muscle expression driven by these constructs. First, this might be an experimentally induced artifact. The heterologous fkh promoter constructs might fail to mediate authentic patterns of expression when paired with certain enhancers, or the inserts, that were cloned from randomly sheared DNA, and might contain only portions of the full enhancer elements, leading to inappropriate regulation. Alternatively, the muscle expression might be the result of authentic enhancers that regulate neighboring non-Hox genes, such as Nebulin, whose vertebrate orthologs are known to express in muscle. Under this model, these muscle enhancers would normally be prevented from activating the Hox genes by repressor elements located elsewhere.
The remaining Hox domain DNAs that were found to encode cis-regulatory activity are linked to Hox2 or Hox12/13. Because there are currently no in situ localization data for these two genes, there is no direct reference for ascertaining the authenticity of these expression patterns, which include both expected CNS as well as other tissues. The overlapping xni338/xni234 fragments located at the 3′ end of the Hox2 gene drive expression throughout the CNS and endoderm. Although this CNS expression is similar to vertebrate Hox expression, homologous endodermal Hox expression has not been described in other chordates. The overlapping xni200/xni256 fragments, which are intronic to Hox2, direct expression in the head epidermis and tail muscles, but also overlap much of xni333, which drives CNS expression at the head–tail boundary in a pattern typical of orthologous vertebrate Hox genes. The overlapping xni337/xni213 fragments, which lie 5′ of Hox2 and 3′ of Hox3, also drive expression in the tail muscles. The xne275 fragment, located to the 3′ of Hox12/13, drives expression in a posterior tail epidermis domain similar to that driven by xne165. Although some of these expression patterns may be the result of experimental artifact or regulation of flanking genes (as described above), it is likely that, at minimum, the CNS expression is authentic.
Discussion
A typical metazoan Hox complex contains 9 or 10 linked genes, each with the same 5′ to 3′ orientation. The position of the genes within the complex relates in a colinear manner to the spatial, and in some cases temporal, sequence in which they are expressed along the anterior–posterior axis of the embryo (28). It is believed that complex interactions between shared regulatory elements are responsible for maintaining the genomic structures of these complexes. The genomic structure of the C. intestinalis Hox genes clearly does not exhibit this tight linkage. It is interesting to note that the C. intestinalis ParaHox genes are also unlinked (29). It is reasonable to propose that these lineage-specific losses of tight linkage have been associated with simplification of regulation involving the loss of cis-regulatory element sharing. The similar splintering of Hox linkage in Caenorhabditis elegans (30), another metazoan with a simplified body plan, suggests that the evolution of these lineages may have involved similar genomic changes.
Early stage cephalochordate (i.e., amphioxus) and vertebrate embryos exhibit localized patterns of Hox expression in the neural tube and derivatives of the mesoderm (usually paraxial mesoderm such as somites, which have no confirmed homologs in ascidians). Five of the C. intestinalis enhancers discovered here direct sequential patterns of expression in the neural tube or mesodermal trunk lateral cells. These findings fit well with expectations based on studies of vertebrates. Arthropods exhibit localized expression of Hox genes in the ventral nerve cord, visceral mesoderm, and epidermis (3). An unexpected finding of this study is the occurrence of Hox expression in the epidermis, as clearly confirmed by Hox11/12 in situ staining (Fig. 3E). An enhancer responsible for this epidermal expression was identified, as were three other fragments that drove epidermal expression. As with the CNS-associated enhancers, these elements, xni200, xni012, xne165, and xne275, drive expression in nested anterior to posterior domains. Combined with the observation that Hox1 is expressed in epidermis (27), this finding suggests that nested epidermal Hox expression is an authentic feature of C. intestinalis development. Although this may represent an ascidian-specific innovation, it also raises the possibility that the epidermal Hox expression of the arthropods represents the ancestral state of the bilaterian lineages. Recent work showing nested epidermal expression of Hox genes in the hemichordates, a basal deuterostome phylum, raises additional questions about whether the ancestral role of Hox genes is exclusive to nerve cord/neural tube patterning (31, 32). Further exploration of the relationship between CNS and epidermal Hox cis-regulation in the urochordates may be able to shed light on this issue.
This study represents the most intensive and comprehensive search for tissue-specific metazoan enhancers within large genomic intervals. Twenty-one separable enhancers were identified within the targeted regions, demonstrating the feasibility of conducting a targeted high-throughput screen for tissue-specific enhancers. C. intestinalis may use >10,000 such enhancers in the developing tadpole (17). As bioinformatic predictions of enhancers improve through modeling and comparative techniques, it will be possible to adapt the high throughput methods described here for the functional characterization of a significant percentage of the C. intestinalis enhancers. The end result of these efforts would be a regulatory atlas of the Ciona genome. This atlas could, in turn, provide a foundation for finding and decoding the cis-regulatory DNAs contained within the more complex vertebrate genomes.
Supplementary Material
Acknowledgments
This work was performed under the auspices of the U.S. Department of Energy, Office of Biological and Environmental Research, the University of California, Lawrence Berkeley National Laboratory under Contract DE-AC03-76SF00098, Lawrence Livermore National Laboratory under Contract W-7405-ENG-48, and Los Alamos National Laboratory under Contract W-7405-ENG-36.
Author contributions: D.N.K., B.-i.L., A.D.G., J.C.D., D.S.R., T.L.H., and P.M.R. designed research; D.N.K., B.-i.L., A.D.G., N.H., J.C.D., M.W., O.K., S.A., C.Z., S.A.D., N.S., Y.S., H.S., A.T.C., and P.M.R. performed research; D.N.K., A.D.G., N.H., N.S., Y.S., H.S., A.T.C., and P.M.R. contributed new reagents/analytic tools; D.N.K., B.-i.L., A.D.G., N.H., C.Z., S.A.D., M.L., and P.M.R. analyzed data; and D.N.K., B.-i.L., and P.M.R. wrote the paper.
Abbreviation: BAC, bacterial artificial chromosome.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AY827840 (XNI BAC) and AY851478 (XNE BAC)].
References
- 1.Altmann, C. R. & Brivanlou, A. H. (2001) Int. Rev. Cytol. 203, 447-482. [DOI] [PubMed] [Google Scholar]
- 2.Ruddle, F. H., Amemiya, C. T., Carr, J. L., Kim, C. B., Ledje, C., Shashikant, C. S. & Wagner, G. P. (1999) Ann. N.Y. Acad. Sci. 870, 238-248. [DOI] [PubMed] [Google Scholar]
- 3.Gindhart, J. G., King, A. N. & Kaufman, T. C. (1995) Genetics 139, 781-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harafuji, N., Keys, D. N. & Levine, M. (2002) Proc. Natl. Acad. Sci. USA 99, 6802-6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson, S. (1981) Nucleic Acids Res. 9, 3015-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gardner, R. C., Howarth, A. J., Hahn, P., Brown-Luedi, M., Shepherd, R. J. & Messing, J. (1981) Nucleic Acids Res. 9, 2871-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deininger, P. L. (1983) Anal. Biochem. 129, 216-233. [DOI] [PubMed] [Google Scholar]
- 8.Burge, C. & Karlin, S. (1997) J. Mol. Biol. 268, 78-94. [DOI] [PubMed] [Google Scholar]
- 9.Gish, W. & States, D. J. (1993) Nat. Genet. 3, 266-272. [DOI] [PubMed] [Google Scholar]
- 10.Satou, Y., Takatori, N., Fujiwara, S., Nishikata, T., Saiga, H., Kusakabe, T., Shin-I., T., Kohara, Y. & Satoh, N. (2002) Gene 287, 83-96. [DOI] [PubMed] [Google Scholar]
- 11.Di Gregorio, A., Corbo, J. C. & Levine, M. (2001) Dev. Biol. 229, 31-43. [DOI] [PubMed] [Google Scholar]
- 12.Detter, J. C., Jett, J. M., Lucas, S. M., Dalin, E., Arellano, A. R., Wang, M., Nelson, J. R., Chapman, J., Lou, Y., Rokhsar, D., et al. (2002) Genomics 80, 691-698. [DOI] [PubMed] [Google Scholar]
- 13.Erives, A., Corbo, J. C. & Levine, M. (1998) Dev. Biol. 194, 213-225. [DOI] [PubMed] [Google Scholar]
- 14.Di Gregorio, A. & Levine, M. (1999) Development (Cambridge, U.K.) 126, 5599-5609. [DOI] [PubMed] [Google Scholar]
- 15.Corbo, J. C., Erives, A., Di Gregorio, A., Chang, A. & Levine, M. (1997) Development (Cambridge, U.K.) 124, 2335-2344. [DOI] [PubMed] [Google Scholar]
- 16.Corbo, J. C., Levine, M. & Zeller, R. W. (1997) Development (Cambridge, U.K.) 124, 589-602. [DOI] [PubMed] [Google Scholar]
- 17.Dehal, P., Satou, Y., Campbell, R. K., Chapman, J., Degnan, B., De Tomaso, A., Davidson, B., Di Gregorio, A., Gelpke, M., Goodstein, D. M., et al. (2002) Science 298, 2157-2167. [DOI] [PubMed] [Google Scholar]
- 18.Spagnuolo, A., Ristoratore, F., Di Gregorio, A., Aniello, F., Branno, M. & Di Lauro, R. (2003) Gene 309, 71-79. [DOI] [PubMed] [Google Scholar]
- 19.Wada, S., Tokuoka, M., Shoguchi, E., Kobayashi, K., Di Gregorio, A., Spagnuolo, A., Branno, M., Kohara, Y., Rokhsar, D., Levine, M., et al. (2003) Dev. Genes Evol. 213, 222-234. [DOI] [PubMed] [Google Scholar]
- 20.Duboule, D. (1998) Curr. Opin. Genet. Dev. 8, 514-518. [DOI] [PubMed] [Google Scholar]
- 21.Hostikka, S. L. & Capecchi, M. R. (1998) Mech. Dev. 70, 133-145. [DOI] [PubMed] [Google Scholar]
- 22.Martin, C. H., Mayeda, C. A., Davis, C. A., Ericsson, C. L., Knafels, J. D., Mathog, D. R., Celniker, S. E., Lewis, E. B. & Palazzolo, M. J. (1995) Proc. Natl. Acad. Sci. USA 92, 8398-8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gionti, M., Ristoratore, F., Di Gregorio, A., Aniello, F., Branno, M. & Di Lauro, R. (1998) Dev. Genes Evol. 207, 515-523. [DOI] [PubMed] [Google Scholar]
- 24.Locascio, A., Aniello, F., Amoroso, A., Manzanares, M., Krumlauf, R. & Branno, M. (1999) Development (Cambridge, U.K.) 126, 4737-4748. [DOI] [PubMed] [Google Scholar]
- 25.Hirano, T. & Nishida, H. (1997) Dev. Biol. 192, 199-210. [DOI] [PubMed] [Google Scholar]
- 26.Nishide, K., Nishikata, T. & Satoh, N. (1989) Dev. Growth Differ. 31, 595-600. [DOI] [PubMed] [Google Scholar]
- 27.Nagatomo, K. & Fujiwara, S. (2003) Gene Expression Patterns 3, 273-277. [DOI] [PubMed] [Google Scholar]
- 28.McGinnis, W. & Krumlauf, R. (1992) Cell 68, 283-302. [DOI] [PubMed] [Google Scholar]
- 29.Ferrier, D. E. K. & Holland, P. W. H. (2002) Mol. Phylogenet. Evol. 24, 412-417. [DOI] [PubMed] [Google Scholar]
- 30.Van Auken, K., Weaver, D. C., Edgar, L. G. & Wood, W. B. (2000) Proc. Natl. Acad. Sci. USA 97, 4499-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lowe, C. J., Wu, M., Salic, A., Evans, L., Lander, E., Stange-Thomann, N., Gruber, C. E., Gerhart, J. & Kirschner, M. (2003) Cell 113, 853-865. [DOI] [PubMed] [Google Scholar]
- 32.Holland, N. D. (2003) Nat. Rev. Neurol. 4, 617-627. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.