Abstract
Immune cell cosignaling receptors are important modulators of immune cell function. For T cells, cosignaling receptors supply necessary secondary signals supporting activation or attenuation after engagement of antigen-presenting cells. The primary cosignaling receptors belong to either the Ig (CD28-like) or TNF receptor (TNFR) superfamilies. The CD28 family is comprised of coinhibitory and costimulatory receptors. The three coinhibitory receptors are cytotoxic T lymphocyte antigen 4, programmed death-1, and B and T lymphocyte attenuator (BTLA). Although cytotoxic T lymphocyte antigen 4 and programmed death-1 interact with B7-Ig family counter receptors, the ligand for BTLA is less clear. From a protein–protein interaction screen, we identified the TNFR family member herpesvirus entry mediator (HVEM) as a counter receptor for BTLA. Here we show that HVEM binds BTLA with high affinity and can form a ternary complex with its known ligands homologous to lymphotoxin, showing inducible expression, and competing with herpes simplex virus glycoprotein D for HVEM, a receptor expressed by T lymphocytes (LIGHT) or lymphotoxin α and BTLA. In addition, binding of HVEM to BTLA attenuates T cell activation, identifying HVEM/BTLA as a coinhibitory receptor pair. This study is a demonstration of a direct interaction between the primary T cell cosignaling receptors of the CD28 and TNFR families.
Keywords: T cells, B7-Ig family, cosignaling receptors
Naive T lymphocytes are stimulated via T cell receptor (TCR) engagement with peptide-MHC on antigen-presenting cells. The fate of T cells is also dependent on a secondary signal through the ligation of coreceptors expressed on T cells with their ligands on antigen-presenting cells. Although costimulatory signal is essential for clonal expansion of T cells and protective immune responses, the coinhibitory signal maintains T cells self tolerance and prevents autoimmunity (1). The balance of the stimulatory and inhibitory signals dictates the outcomes of immune responses.
The primary cosignaling receptors belong to either the Ig CD28-like or TNF receptor (TNFR) superfamilies (1–5). Currently, the CD28 family consists of five lymphoid-specific coreceptors [CD28, inducible T cell costimulator (ICOS), cytotoxic T lymphocyte antigen 4 (CTLA-4), programmed death-1 (PD-1), and B and T lymphocyte attenuator (BTLA)] (1–3). CD28 and ICOS are single Ig-variable (IgV) domain glycoproteins that promote T cell activation, whereas the structurally related CTLA-4, PD-1, and BTLA receptors function to attenuate T cell activation. To date, all of the ligands that have been described for the CD28-like family members belong to the B7 superfamily (1–3, 6–9). Six B7 family members have been described, all of which have conserved extracellular IgV and Ig-constant domains (3). In the TNFR-TNF superfamily, five receptor–ligand interactions have been described that act as positive regulators. These include OX40–OX40L, 4-1BB–4-1BBL, CD27–CD70, CD30–CD30L, and herpesvirus entry mediator (HVEM)–homologous to lymphotoxin, which shows inducible expression and competes with herpes simplex virus (HSV) glycoprotein D (gD) for HVEM, a receptor expressed by T lymphocytes (HVEM–LIGHT) (4).
The most recently identified CD28 family member is the inhibitory coreceptor, BTLA (10–12). BTLA is expressed on developing B and T cells, all mature lymphocytes, splenic macrophages, and mature marrow-derived dendritic cells (10, 11). BTLA contains a single IgV domain and two intracellular immunoreceptor tyrosine-based inhibitory motifs that are phosphorylated after BTLA coligation to antigen receptors, resulting in recruitment of protein tyrosine phosphatases SHP-1 and SHP-2 (13). Because of this, and that coligation of BTLA to the TCR inhibits T cell activation, BTLA is implicated as a negative regulator of T cell activation (10, 11). This finding is further supported by the observation that BTLA-deficient T cells show increased proliferation and that BTLA–/– mice have increased Ab response and show increased incidence and severity to an autoimmune disorder (9, 10).
Initially, BTLA was proposed to interact with a B7 family member called B7x (10, 14). This conclusion, however, was based on an indirect binding study testing the interaction of B7x-Ig fusion to spleen and lymph node cells from either WT or BTLA-deficient mice. Furthermore, we have been unable to detect any specific binding of BTLA-Fc to B7x-transfected cells (data not shown). Spurred by the inability to confirm the BTLA–B7x interaction, we screened a secreted protein library (15) by using surface plasmon resonance (SPR) and identified HVEM as a coreceptor for human BTLA.
Here we show that BTLA and HVEM interact with high affinity and can form a trimeric complex with TNF ligands LIGHT or lymphotoxin α (LTα). Our binding studies suggest that BTLA interacts with the outer surface of the HVEM/TNF complex, suggesting structural models for how HVEM might engage BTLA on the cell surface. Finally, we demonstrate that binding of HVEM to BTLA results in the inhibition of T cell proliferation, suggesting that HVEM and BTLA form an inhibitory coreceptor pair.
Materials and Methods
Protein Reagents. Human BTLA (Met-1 to Leu-155) and follistatin (Met-1 to Trp-344), or murine HVEM (Met-1-Val-207) and CTLA-4 (Met-1-Ser-160) were cloned into the expression vector pRK5 as fusions to the Fc portion of human or murine (mHVEM) IgG1. Proteins were produced by transient transfection of Chinese hamster ovary cells by using DMRIE-C (Invitrogen). Human HVEM (Met-1 to Ser-199) was cloned into expression vector pSVI7 (16) as an Fc fusion. HVEM-Fc was produced from a stable expressing Chinese hamster ovary cell line. All Fc-tagged proteins were purified to >90% purity by affinity chromatography by using protein-A Sepharose (Amersham Pharmacia). Aggregates were separated from dimers with an S-300 (Pharmacia) gel-filtration column. The follistatin-Fc protein was used as a control Fc-fusion protein. Human LTα (17) and BP-2 peptide (18) were generated as described. gD (Δ290–299) (19) was a generous gift from Gary Cohen (University of Pennsylvania, Philadelphia).
SPR Screening. A protein library consisting of frozen aliquots of >2,000 individual secreted proteins (15), purified through standard chromatographic techniques by using either Fc- or His-tags, was used to conduct the protein interaction screen. BTLA-Fc or control Fc-fusion protein were amine-coupled to a Biacore CM5 sensor chip and the reference flow cells blocked with ethanolamine. Proteins were individually tested at a concentration of 2 μg/ml in HBS-P buffer (0.01 M Hepes, pH 7.4/0.15 M NaCl/0.005% Surfactant P20) supplemented with 0.15 M NaCl. All SPR analyses were performed at 25°C by using a flow rate of 5 μl/min. Recombinant human CD28-Fc, CTLA4-Fc, ICOS-Fc, and PD-1-Fc (R & D Systems) were amine-coupled to individual flow cells of a CM5 sensor chip. The activity of each immobilized CD28 family member was confirmed by testing the binding of the appropriate B7 ligand (R & D Systems). Binding responses were measured after a 1-min injection of 2 μg/ml HVEM-Fc in HBS-P buffer. Binding of individual TNFR family members to immobilized BTLA-Fc was tested in an identical manner. For competition and cobinding studies, LIGHT (Alexis Biochemicals), LTα, BP-2, or gD (Δ290–299) were preincubated with HVEM-Fc (10 nM) for >1 h and injected over an amine-coupled BTLA-Fc CM5 sensor chip. Sensorgrams were run in random order and in duplicate. Response levels were recorded 15 sec before the end of a 2-min injection. All sensor chips were regenerated, without loss of activity, by using 10 mM glycine, pH 2.5. The SPR cobinding experiment was conducted on a CM5 sensor chip with HVEM-Fc amine-coupled at 150 response units. Low-level immobilization allowed near saturation binding for LIGHT and BTLA-Fc.
Flow Cytometry Reagents. Phycoerythrin-conjugated goat anti-human IgG (Fc specific) F(Ab′)2 fragment (Caltag); biotinylated mouse anti-FLAG M2 Ab (Sigma); FITC-conjugated mouse-anti-human HVEM (clone 122, MBL); and mouse-anti-human BTLA (clone 5F5 raised against BTLA; Genentech) were used for flow cytometry. FLAG and BTLA Abs were biotinylated according to the manufacturer's instructions by using Sulfo-NHS-LC-biotin (Pierce) and detected with Tri-color-conjugated streptavidin (BD Biosciences or Caltag). Anti-BTLA and anti-HVEM Abs were used to select for BTLA- or HVEM-expressing cells, respectively.
Transient Transfection and Stable Cell Lines. AD-293 cells (Stratagene) were transiently transfected with Polyfect (Qiagen) by using 16 μg of DNA per 15-cm Petri dish for 48–72 h. Empty pRK5 vector was used as a control. Cells were removed from the plate by using PBS cell dissociation solution (Sigma), washed with PBS, and then resuspended in human cell buffer [PBS with 2% FBS (HyClone)/2 mM EDTA, pH 8] for flow cytometry binding studies or RPMI medium 1680 with 2% FBS and 10 mM Hepes (pH 7.2) for radiolabeled protein-binding studies. A stable human BTLA 3T3 cell line was generated by using retroviral infection with a MSCV-hBTLA construct (BD Biosciences).
Cell-Binding Assays. BTLA and LIGHT were iodinated by the lactoperoxidase (Biotrend, Cologne, Germany) method and HVEM by using Bolton & Hunter reagent (Amersham Pharmacia). Displacement binding studies were done in triplicate with 0.5 nM or 0.05 nM of labeled protein. The iodinated protein was incubated with 125,000 AD-293 cells in the presence of varying concentrations of unlabeled protein for 4 h at 4°C. Cells were harvested on Millipore filter plates and washed five times with PBS. Dried filters were counted, and Scatchard analysis was performed with newligand 1.05 software (Genentech). For flow cytometry assays, proteins were incubated with 1 × 106 AD-293 transfected or mock-transfected cells for 15–20 min at room temperature in human cell buffer. Washes and all further incubations were done at 4°C in PBS containing 2% FBS and 0.1% sodium azide. Analysis was performed either immediately or after overnight fixation at 4°C with 1% paraformaldehyde. At least 10,000 events were acquired on a Becton Dickinson FAC-Scan cytometer. Binding of BTLA-Fc or LIGHT-FLAG was detected by flow cytometry by using an anti-Fc or anti-FLAG. Neither HVEM-Fc nor BTLA-Fc bound vector-transfected control cells, compared with Fc isotype control protein, and neither Fc nor FLAG isotype control proteins bound to HVEM- or BTLA-expressing cells. For blocking assays, 1 million stable transfected human BTLA 3T3 cells were first pretreated with 1 μg of the specified anti-human BTLA or control Ab for 15 min at 4°C. After washing with PBS, cells were incubated with 1 μg of human HVEM-Fc for 15 min at 4°C. Cells were then washed twice with PBS and stained with goat F(ab)2 anti-mouse IgG (Fc specific) (Caltag Laboratories). Cells were then analyzed by flow cytometry.
Human T Cell Proliferation Assays. Human peripheral blood mononuclear cells were isolated from whole blood by Ficoll gradient and activated at 2 million cells per ml in 96-well plates with different concentrations of phytohemagglutin. Human CD4+ T cells were purified from the whole blood of healthy donors by using the RosetteSep CD4+ enrichment mixture (StemCell Technologies, Vancouver). Cell purity by FACS was >90%. Flat-bottom 96-well plates (Costar) were coated with mouse anti-human CD3 clone UCHT1 (BD Pharmingen) at various concentrations and mouse anti-human IgG1-Fc clone MH1015 (Caltag Laboratories) at 5 μg/ml overnight at 4°C. Plates were washed twice with PBS. Follistatin-Fc, HVEM-Fc, or decoy receptor 3 (DCR3)-Fc (R & D Systems) was added at 10 μg/ml and incubated for 2 h at 37°C. After aspiration, 200,000 cells per well were added with or without BTLA mAbs (clone 3B1 and 5F5 raised against human BTLA; Genentech) or isotype control Ab at various concentrations. The cells were incubated for 72 h at 37°C, with 7% CO2, and pulsed with [3H]thymidine for the last 18 h of culture. [3H]thymidine incorporation was measured by using a microplate scintillation counter (TopCount, PerkinElmer).
Murine T Cell Proliferation Assays. BALB/c splenocytes were isolated over a gradient (Ficoll-Paque 1119, Sigma), and CD4+ T cells were then isolated by using MACS CD4+ T cell isolation kit. For ConA activation, splenocytes were plated at 4 × 105 cells per 200 μl in 96-well plates and activated by ConA (Sigma Aldrich) at 1 μg/ml. For anti-CD3 activation, plates were coated overnight at 4°C with 100 μl of PBS containing 5 μg of anti-mouse IgG (clone A85-3, BD Pharmingen). Plates were subsequently washed twice with PBS and then coated with 1 μg/ml anti-mouse-CD3 (clone 145-2C11, BD Pharmingen) and 10 μg/ml of each fusion protein in 100 μl of PBS for 4 h at 37°C. To the coated plates, 4 × 105 CD4+ cells in 200 μl of DMEM media plus 10% FCS (HyClone), glutamine, nonessential amino acids, and anti-mouse CD28 (clone 37.51, BD Pharmingen) at 1 μg/ml were added. Changes in proliferation were determined by incubating the cells for 60 h at 37°C with 7% CO2 and pulsing with [3H]thymidine for the last 12 h of culture. [3H]thymidine incorporation was measured by using a microplate scintillation counter (TopCount, PerkinElmer).
Results
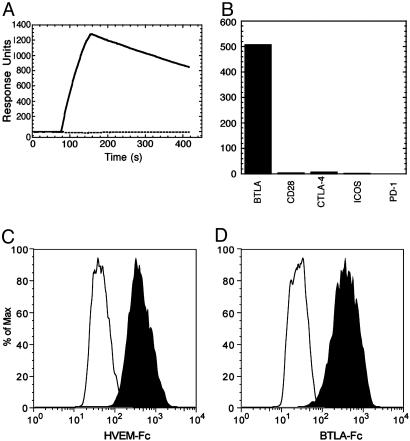
BTLA Binds HVEM Specifically. We screened a secreted protein library by using SPR to identify a coreceptor for human BTLA (15). A single specific hit was identified to HVEM (Fig. 1A). HVEM belongs to the TNFR superfamily, is one of several characterized T cell costimulatory TNFRs (2, 4), and mediates herpesvirus entry into host cells (20–22). To determine whether BTLA-Fc binding was specific to HVEM, we tested binding to other T cell costimulatory TNFRs, including 4-1BB, OX40, CD27, CD30, CD40, and DcR3 and did not detect any interaction (data not shown). Conversely, CD28, ICOS, PD-1, and CTLA-4 did not bind HVEM (Fig. 1B). Binding of BTLA-Fc to HVEM-transfected AD-293 cells and, reciprocally, binding of HVEM-Fc to BTLA-transfected AD-293 cells show that the interaction between these proteins can occur on the cell surface (Fig. 1 C and D). Binding analyses to BTLA or HVEM expressing AD-293 cells by using either radiolabeled HVEM-Fc or BTLA-Fc revealed high-affinity interactions with equilibrium dissociation constants (Kd) of 5.5 and 25 nM, respectively.
Fig. 1.
HVEM binds BTLA. (A) The sensorgrams represent binding of HVEM-Fc to immobilized BTLA-Fc (solid line) or control Fc-fusion protein (dashed line). (B) SPR-binding responses of HVEM-Fc to immobilized CD28 family members. Binding of HVEM-Fc (C), BTLA-Fc (D) (both shaded black), or Fc-fusion control protein (all at 500 nM) to BTLA-expressing (C) or HVEM-expressing (D) AD-293 cells, detected by flow cytometry.
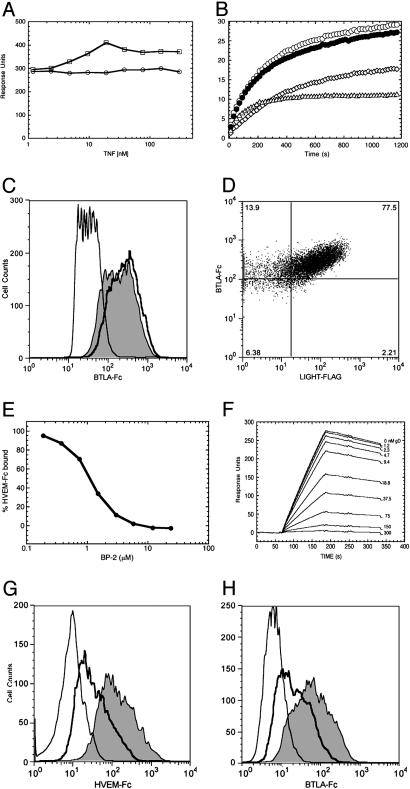
Formation of a Ternary Complex Between BTLA/HVEM and TNF Ligand LIGHT. HVEM binds the TNF ligands LIGHT and LTα (23). To determine whether either of these ligands could block HVEM binding to BTLA, we conducted an in solution SPR competition experiment. We found that a high molar excess of LIGHT or LTα could not block HVEM binding to BTLA (Fig. 2A). LIGHT or LTα did not bind to BTLA in the absence of HVEM (data not shown). Under conditions of saturation binding to immobilized HVEM, a mixture of BTLA-Fc and LIGHT resulted in a nearly additive SPR signal, compared with the individually bound samples (Fig. 2B), suggesting that LIGHT and BTLA can bind simultaneously to HVEM. This finding is supported by cell-binding assays, which show BTLA-Fc binding is not inhibited by excess LIGHT and that both LIGHT and BTLA-Fc, at saturating concentrations, can simultaneously bind HVEM expressed on AD-293 cells (Fig. 2 C and D). In addition, HVEM-Fc binds to AD-293 cells that coexpress LIGHT and BTLA with higher apparent affinity (500 pM) than to BTLA (5.5 nM) or LIGHT (7 nM) individually. This increased affinity may be explained by an avidity effect from binding LIGHT and BTLA simultaneously or by enhanced individual binding resulting from an induced allosteric change. Finally, to confirm that the ternary interaction occurs in solution, we conducted a pull-down assay using BTLA-Fc, an untagged HVEM construct, and LIGHT (data not shown). We find that LIGHT is pulled down by protein A-Sepharose only in the presence of HVEM. Taken together, these data indicate that BTLA binds a site on HVEM that is distinct from LIGHT, and that BTLA/HVEM/LIGHT can form a ternary complex.
Fig. 2.
Competition studies among BTLA, LIGHT, and gD for HVEM binding. (A) HVEM-Fc (10 nM) preincubated with LIGHT (○) or LTα (□) at the indicated concentrations, injected over immobilized BTLA-Fc and monitored by SPR. (B) Sensorgrams of LIGHT (25 nM) (⋄) or BTLA-Fc (17 nM) (▵) and binding of a mixture of LIGHT (25 nM) and BTLA-Fc (17 nM) (○) to immobilized HVEM-Fc. Calculated sum of individual LIGHT and BTLA-Fc sensorgrams (•). (C) Binding of 21 nM BTLA-Fc in the presence of 210 nM LIGHT to HVEM-expressing human embryonic kidney AD-293 (solid line) compared with binding of 21 nM BTLA-Fc (shaded gray) or control protein (thin line). (D) Simultaneous binding of BTLA-Fc and LIGHT at 500 nM to HVEM-expressing 293 cells (percentage positive cells indicated in each quadrant). (E) Samples containing increasing concentrations of BP-2 peptide preincubated with HVEM-Fc (10 nM), injected over immobilized BTLA-Fc, and analyzed by SPR. (F) Sensorgrams of HVEM-Fc (10 nM) binding to immobilized BTLA-Fc in the presence of increasing amounts of gD (Δ290–299). (G and H) Binding of 10 nM HVEM-Fc or BTLA-Fc in the presence (solid line) or absence (shaded) of 600 nM gD (Δ290–299) to BTLA or HVEM AD-293 cells, respectively. Fc-control protein (thin line).
HSV-1 gD Competes with BTLA for Binding to HVEM. HVEM was initially identified as a cellular mediator of HSV-1 entry and shown to bind the HSV-1 gD (19, 21). HVEM contains three of the characteristic cysteine-rich domains (CRDs) common to all of the TNFR family members. A crystal structure of herpesvirus gD protein complexed with HVEM and mutagenesis experiments show that the first CRD of HVEM contains the required binding site, whereas the CRD2 domain of HVEM provides structural support for the binding site in CRD1 (24–26). In addition, gD- and the LTα/LIGHT-binding sites appear to be on opposite sides of HVEM and likely do not overlap (24). Although LIGHT has been shown to inhibit gD binding, this inhibition may be due to a locally induced allosteric change in HVEM (24, 27). These structural studies and the fact that LIGHT and BTLA do not compete for HVEM binding (Fig. 2 A–D) raise the possibility that BTLA, like gD, interacts with the outer surface of the HVEM/LIGHT complex. To test this hypothesis, we used a phage-derived peptide (BP-2) that is capable of blocking HVEM binding to gD, but not to LIGHT (24, 27). At concentrations that inhibit gD binding, BP-2 inhibited the binding of HVEM to BTLA (Fig. 2E). More directly, recombinant gD (Δ290–299) (19) also inhibited HVEM binding to BTLA (Fig. 2F). This observation was repeated in cell-binding assays that show that the recombinant gD (Δ290–299) inhibits the binding of soluble BTLA-Fc and HVEM-Fc to AD-293 cells expressing HVEM or BTLA, respectively (Fig. 2 G and H). Together, these results indicate that BTLA likely interacts with the first CRD of HVEM on a site distinct from the LIGHT/LTα-binding sites.
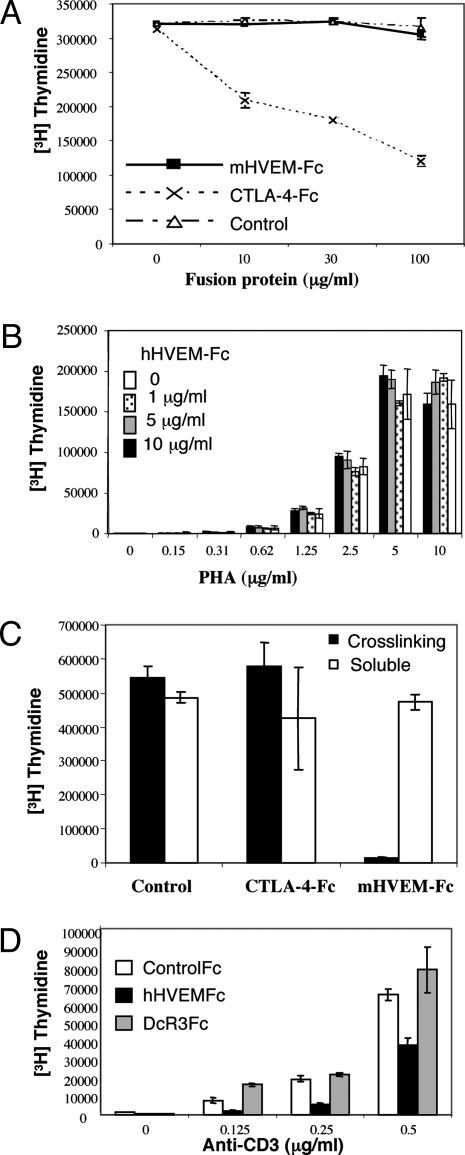
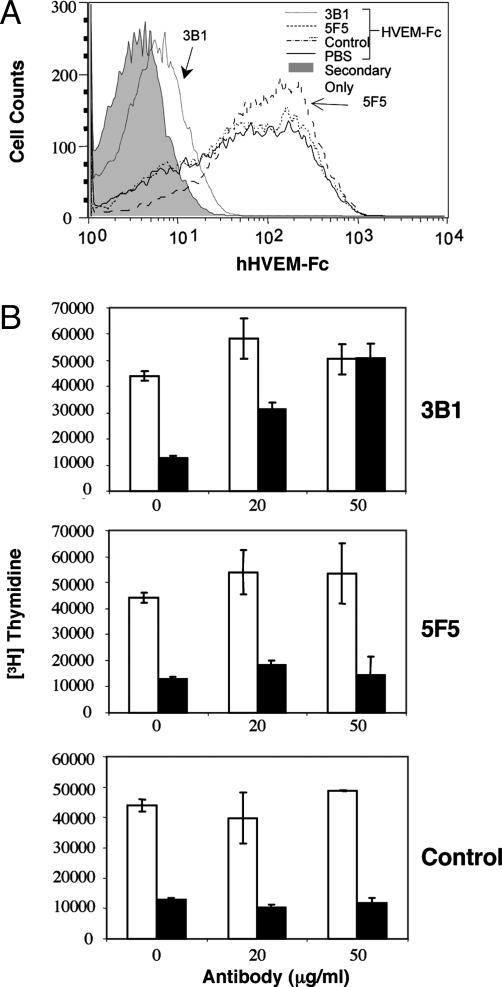
HVEM Inhibits T Cell Activation Through BTLA. The intracellular domain of BTLA contains dual immunoreceptor tyrosine-based inhibitory motif domains that are inducibly phosphorylated resulting in the recruitment of SHP-1 and SHP-2, consistent with an inhibitory role (10, 11, 13). This finding is supported by the observation that crosslinking of BTLA with the TCR inhibits activation (10, 11). We hypothesized that HVEM could potentially deliver inhibitory signal to T cells through the interaction with BTLA. Previous studies have shown that soluble murine HVEM-Fc fusion protein significantly inhibited T cell proliferation in vitro without being crosslinked to the TCR (28, 29). However, our purified murine HVEM-Fc dimer failed to suppress ConA induced proliferation of T cells from BALB/c mice (Fig. 3A), whereas addition of CTLA-4-Fc, which binds B7-1 and B7-2 expressed on antigen-presenting cells and prevents their interaction with CD28, resulted in the inhibition of proliferation as shown (29, 30). We further confirmed this result by using human HVEM-Fc. No inhibition was observed when soluble HVEM-Fc was added to the media of phytohemagglutin-activated human peripheral blood mononuclear cells (Fig. 3B) or anti-CD3 activated human CD4+ T cells (not shown). The reason for the discrepancy between our results and the previously published work on the effect of soluble HVEM-Fc on T cell activation is unclear at this time. However, we have found that, if we do not remove high-molecular-weight aggregates from our murine HVEM-Fc, it is a potent inhibitor of T cell proliferation in vitro. Importantly, the aggregates showed significantly reduced affinity to BTLA when analyzed by Biacore (data not shown). Nevertheless, consistent with the notion that immunoreceptor tyrosine-based inhibitory motifs containing coinhibitory receptors exert their role when coligated with the TCR, crosslinking murine HVEM-Fc significantly inhibits anti-CD3 induced activation of purified murine CD4+ T cells (Fig. 3C). Consistent with previous data (29, 31), there was no inhibitory effect observed with CTLA-4-Fc under the same conditions. Similarly, we also show here that crosslinking human HVEM-Fc with antigen receptors attenuates human T cell activation (Fig. 3D). This inhibition was at least, in part, mediated by the HVEM/BTLA interaction, because crosslinking DcR3-Fc, which interacts with LIGHT but not BTLA, failed to suppress T cell activation (Fig. 3D). To further demonstrate that the interaction between HVEM and BTLA accounted for this inhibitory function, we screened anti-human BTLA Abs for their ability to block the HVEM–BTLA interaction. One Ab, 3B1, both specifically blocked HVEM-Fc binding to cells expressing human BTLA (Fig. 4A) and reversed the inhibitory effect of HVEM-Fc on T cell proliferation whereas a nonblocking anti-BTLA Ab did not (Fig. 4B). None of our anti-BTLA Abs affected T cell activation and proliferation when added to the media (data not shown).
Fig. 3.
HVEM-Fc inhibits T cell activation when crosslinked. (A) Splenocytes from BALB/c mice were activated by either 1 μg/ml ConA plus different concentration of murine HVEM-Fc CTLA-4-Fc or control protein as indicated. Cells were incubated for 60 h with [3H]thymidine incorporated for the last 12 h. Data represented average value from triplicate wells. (B) Human peripheral blood mononuclear cells were activated by different concentration of phytohemagglutin. Human HVEM was added to the media at the indicated concentration. Proliferation was measured 72 h later. The results represent the average of triplicate wells. (C) CD4+ T cells were purified from BALB/c spleens and activated by plate-bound anti-CD3, or anti-CD3 and murine HVEM-Fc. Proliferation was measured 60 h later. Data represent average of triplicate wells. Similar data were obtained in three independent experiments. (D) Primary human CD4+ T cells were stimulated with plate-bound anti-CD3. A control Fc-fusion protein (open bar), HVEM-Fc (filled bar), or DcR3-Fc (gray bar) also was crosslinked on the plate by precoated anti-Fc Ab. The proliferation of T cells was measured 72 h later. The results represent the average of triplicate wells. Similar results were obtained from seven independent healthy donors.
Fig. 4.
HVEM-Fc inhibits T cell activation through the interaction with BTLA. (A) The 3T3 cells expressing human BTLA were blocked with anti-human BTLA Abs or isotype control, incubated with HVEM-Fc, followed by anti-Fc Ab, and analyzed by FACS scan. Data represent one out three independent results. (B) As indicated in Fig. 3D, CD4+ T cells were stimulated by plate-bound anti-CD3 (0.25 μg/ml) with (filled bar) or without HVEM-Fc (open bar), in the presence of anti-BTLA monoclonal 3B1 (Top), 5F5 Ab (Middle), or isotype control (Bottom). The data represent the average from triplicate wells. Similar results were obtained from four independent healthy donors.
Discussion
In this report, we demonstrate a direct and specific interaction between the CD28 family member BTLA and the TNFR family member HVEM. These two cosignaling receptor families and their B7 and TNF ligands have been well characterized with regard to amplifying or attenuating T cell responses (1–5). HVEM was initially thought to function as positive regulator of T cell activation through its interaction with LIGHT (22), whereas CD28 family member, BTLA, acts as a negative regulator of T cell responses (10, 11). Our data suggest that HVEM also may act as a negative regulator through binding to BTLA and that HVEM/BTLA represent a new coinhibitory receptor pair.
Previously, it has been shown that crosslinking BTLA to the TCR was necessary for association of SHP-1 and SHP-2 and inhibition of T cell activation (13). Similarly, we show here that HVEM-Fc fusion protein inhibited T cell proliferation in vitro when crosslinked and coimmobilized with anti-CD3. It is possible that the immobilized HVEM-Fc inhibited T cell activation by preventing the interaction of LIGHT with cellular HVEM. However, this mechanism is unlikely because DcR3, which binds LIGHT, did not inhibit T cell activation. Furthermore, in the presence of an anti-BTLA Ab that blocks HVEM binding to BTLA, HVEM-Fc did not inhibit T cell proliferation. This finding indicates that the inhibitory effect of HVEM-Fc fusion protein is mediated through its interaction with BTLA rather than simply blocking the HVEM–LIGHT interaction. In contrast with previous reports (28), we were unable to show inhibition of T cell activation by HVEM without coimmobilizing with anti-CD3 in either murine or human T cell proliferation assays. However, we did find that, if aggregated material was not removed, murine HVEM-Fc preparations were inhibitory without coimmobilizing with anti-CD3 and that this inhibition was BTLA-independent.
Whether the binding of BTLA to HVEM or the formation of the HVEM/BTLA/LIGHT ternary complex results in a stimulatory effect that is mediated through HVEM is unclear at this time. However, we have found that coimmobilizing BTLA-Fc fusion protein with anti-CD3 has no effect on T cell activation in vitro (data not shown), suggesting that binding of BTLA to HVEM on T cells does not trigger signaling through HVEM. In addition, the phenotype of BTLA- and LIGHT-deficient mice supports an inhibitory role for the HVEM–BTLA interaction. T cells from BTLA-deficient mice exhibit hyperproliferation, whereas T cell activation is minimally, if at all, affected by deletion of LIGHT (10, 11, 32, 33).
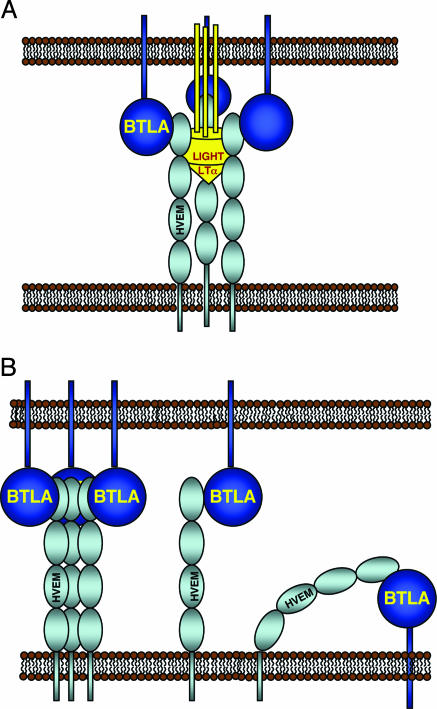
Because the binding of BTLA is inhibited by HSV-1 gD, we propose that BTLA interacts with the first CRD of HVEM. In comparison with other TNFR/TNF crystal structures, engagement of LIGHT or LTα by HVEM is predicted to impose a rigid 3-fold symmetrical structure spanning a length of ≈80–100 Å (34). The extracellular IgV domain of BTLA, which is ≈120 aa long, likely adopts a structure similar to CTLA-4, which spans ≈50 Å on its longest dimension (35–38). This distance in addition to the number of linker residues (<10 aa) between the IgV domain and the transmembrane domain of BTLA is unlikely to provide enough distance to accommodate a complex where HVEM/LIGHT/LTα and BTLA interact on the same membrane. Instead, in the presence of LIGHT or LTα, we propose that HVEM and BTLA interact from apposed membranes during periods of cell–cell contact (Fig. 5A). Receptor-ligand pairs, such as CTLA-4-B7, MHC-TCR, and CD2-CD58, that span the central zone of the immunological synapse generally range from 100 to 140 Å (36). Our model of BTLA interacting with HVEM on CRD1 would fit within this central zone. In the absence of LIGHT or LTα, both an inter- or intracellular interaction may be possible (Fig. 5B). If HVEM exists as a preformed trimer, as has been described for other TNFRs (34, 36), HVEM could serve to recruit three BTLA molecules into close proximity and trigger downstream signaling events (Fig. 5B). Alternatively, an intercellular dimeric complex could form between monomeric HVEM and BTLA, or HVEM might have sufficient conformational freedom to tilt over and engage BTLA on the same membrane (Fig. 5B). Determining which of these models represent active signaling complexes will require further experimentation. It is interesting to speculate that, if BTLA dimerization occurs in the context of the immunological synapse, it could provide a mechanism for superclustering of the HVEM/LIGHT/BTLA. This hypothesis is similar to what has been proposed for the CTLA-4/B7-1 or B7-2 complex (35–37).
Fig. 5.
Receptor interaction models. (A) Proposed model for the BTLA, HVEM, and LIGHT/LTα complex. (B) Model of a pretrimerized HVEM interacting with BTLA (Left) and monomeric intercellular (Center) and intracellular (Right) BTLA/HVEM complexes.
Thus, our data suggest that the HVEM–BTLA interaction represents a new coinhibitory pathway for T cells. That BTLA and HVEM are both expressed on T cells and antigen-presenting cells (10, 11, 22) further suggests that this interaction may serve as a regulator of T cell activation.
Acknowledgments
We thank Hilary Clark, Dylan Daniel, Austin Gurney, and Sarah Hymowitz for helpful discussions; Gary Cohen for the generous gift of gD protein; and Makia Nakamura and Phil Hass, both of Genentech, for protein reagents.
Author contributions: L.C.G., K.M.L., W.O., and D.L.E. designed research; L.C.G., K.M.L., J.C.-F., and V.C. performed research; B.W. contributed new reagents/analytic tools; L.C.G., K.M.L., W.O., and D.L.E. analyzed data; and L.C.G., K.M.L., W.O., and D.L.E. wrote the paper.
Abbreviations: BTLA, B and T lymphocyte attenuator; CRD, cysteine-rich domain; CTLA-4, cytotoxic T lymphocyte antigen 4; DcR3, decoy receptor 3; gD, glycoprotein D; HSV, herpes simplex virus; HVEM, herpesvirus entry mediator; LTα, lymphotoxin α; PD-1, programmed death-1; SPR, surface plasmon resonance; TCR, T cell receptor; TNFR, TNF receptor; ICOS, inducible T cell costimulator; IgV, Ig-variable.
References
- 1.Sharpe, A. H. & Freeman, G. J. (2002) Nat. Rev. Immunol. 2, 116–126. [DOI] [PubMed] [Google Scholar]
- 2.Rothstein, D. M. & Sayegh, M. H. (2003) Immunol. Rev. 196, 85–108. [DOI] [PubMed] [Google Scholar]
- 3.Coyle, A. J. & Gutierrez-Ramos, J. C. (2001) Nat. Immunol. 2, 203–209. [DOI] [PubMed] [Google Scholar]
- 4.Croft, M. (2003) Nat. Rev. Immunol. 3, 609–620. [DOI] [PubMed] [Google Scholar]
- 5.June, C. H. & Riley, J. L. (2005) Blood 105, 13–21. [DOI] [PubMed] [Google Scholar]
- 6.Chen, L. (2004) Nat. Rev. Immunol. 4, 336–347. [DOI] [PubMed] [Google Scholar]
- 7.Khoury, S. J. & Sayegh, M. H. (2004) Immunity 20, 529–538. [DOI] [PubMed] [Google Scholar]
- 8.Greenwald, R. J., Latchman, Y. E. & Sharpe, A. H. (2002) Curr. Opin. Immunol. 14, 391–396. [DOI] [PubMed] [Google Scholar]
- 9.Liang, L. & Sha, W. C. (2002) Curr. Opin. Immunol. 14, 384–390. [DOI] [PubMed] [Google Scholar]
- 10.Watanabe, N., Gavrieli, M., Sedy, J. R., Yang, J., Fallarino, F., Loftin, S. K., Hurchla, M. A., Zimmerman, N., Sim, J., Zang, X., et al. (2003) Nat. Immunol. 4, 670–679. [DOI] [PubMed] [Google Scholar]
- 11.Han, P., Goularte, O. D., Rufner, K., Wilkinson, B. & Kaye, J. (2004) J. Immunol. 172, 5931–5939. [DOI] [PubMed] [Google Scholar]
- 12.Carreno, B. M. & Collins, M. (2003) Trends Immunol. 24, 524–527. [DOI] [PubMed] [Google Scholar]
- 13.Gavrieli, M., Watanabe, N., Loftin, S. K., Murphy, T. L. & Murphy, K. M. (2003) Biochem. Biophys. Res. Commun. 312, 1236–1243. [DOI] [PubMed] [Google Scholar]
- 14.Zang, X., Loke, P., Kim, J., Murphy, K., Waitz, R. & Allison, J. P. (2003) Proc. Natl. Acad. Sci. USA 100, 10388–10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clark, H. F., Gurney, A. L., Abaya, E., Baker, K., Baldwin, D., Brush, J., Chen, J., Chow, B., Chui, C., Crowley, C., et al. (2003) Genome Res. 13, 2265–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirchhofer, D., Peek, M., Li, W., Stamos, J., Eigenbrot, C., Kadkhodayan, S., Elliott, J. M., Corpuz, R. T., Lazarus, R. A. & Moran, P. (2003) J. Biol. Chem. 278, 36341–36349. [DOI] [PubMed] [Google Scholar]
- 17.Gray, P. W., Aggarwal, B. B., Benton, C. V., Bringman, T. S., Henzel, W. J., Jarrett, J. A., Leung, D. W., Moffat, B., Ng, P., Svedersky, L. P., et al. (1984) Nature 312, 721–724. [DOI] [PubMed] [Google Scholar]
- 18.Sarrias, M. R., Whitbeck, J. C., Rooney, I., Spruce, L., Kay, B. K., Montgomery, R. I., Spear, P. G., Ware, C. F., Eisenberg, R. J., Cohen, G. H., et al. (1999) J. Virol. 73, 5681–5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitbeck, J. C., Peng, C., Lou, H., Xu, R., Willis, S. H., Ponce de Leon, M., Peng, T., Nicola, A. V., Montgomery, R. I., Warner, M. S., et al. (1997) J. Virol. 71, 6083–6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwon, B. S., Tan, K. B., Ni, J., Oh, K. O., Lee, Z. H., Kim, K. K., Kim, Y. J., Wang, S., Gentz, R., Yu, G. L., et al. (1997) J. Biol. Chem. 272, 14272–14276. [DOI] [PubMed] [Google Scholar]
- 21.Montgomery, R. I., Warner, M. S., Lum, B. J. & Spear, P. G. (1996) Cell 87, 427–436. [DOI] [PubMed] [Google Scholar]
- 22.Granger, S. W. & Rickert, S. (2003) Cytokine Growth Factor Rev. 14, 289–296. [DOI] [PubMed] [Google Scholar]
- 23.Mauri, D. N., Ebner, R., Montgomery, R. I., Kochel, K. D., Cheung, T. C., Yu, G. L., Ruben, S., Murphy, M., Eisenberg, R. J., Cohen, G. H., et al. (1998) Immunity 8, 21–30. [DOI] [PubMed] [Google Scholar]
- 24.Carfi, A., Willis, S. H., Whitbeck, J. C., Krummenacher, C., Cohen, G. H., Eisenberg, R. J. & Wiley, D. C. (2001) Mol. Cell 8, 169–179. [DOI] [PubMed] [Google Scholar]
- 25.Connolly, S. A., Landsburg, D. J., Carfi, A., Wiley, D. C., Eisenberg, R. J. & Cohen, G. H. (2002) J. Virol. 76, 10894–10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whitbeck, J. C., Connolly, S. A., Willis, S. H., Hou, W., Krummenacher, C., Ponce de Leon, M., Lou, H., Baribaud, I., Eisenberg, R. J. & Cohen, G. H. (2001) J. Virol. 75, 171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarrias, M. R., Whitbeck, J. C., Rooney, I., Ware, C. F., Eisenberg, R. J., Cohen, G. H. & Lambris, J. D. (2000) Mol. Immunol. 37, 665–673. [DOI] [PubMed] [Google Scholar]
- 28.Tamada, K., Shimozaki, K., Chapoval, A. I., Zhai, Y. F., Su, J., Chen, S. F., Hsieh, S. L., Nagata, S., Ni, J. & Chen, L. P. (2000) J. Immunol. 164, 4105–4110. [DOI] [PubMed] [Google Scholar]
- 29.Wang, J., Lo, J. C., Foster, A., Yu, P., Chen, H. M., Wang, Y., Tamada, K., Chen, L. & Fu, Y.-X. (2001) J. Clin. Invest. 108, 1771–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shahinian, A., Pfeffer, K., Lee, K. P., Kundig, T. M., Kishihara, K., Wakeham, A., Kawai, K., Ohashi, P. S., Thompson, C. B. & Mak, T. W. (1993) Science 261, 609–612. [DOI] [PubMed] [Google Scholar]
- 31.Green, J. M., Noel, P. J., Sperling, A. I., Walunas, T. L., Gray, G. S., Bluestone, J. A. & Thompson, C. B. (1994) Immunity 1, 501–508. [DOI] [PubMed] [Google Scholar]
- 32.Scheu, S., Alferink, J., Potzel, T., Barchet, W., Kalinke, U. & Pfeffer, K. (2002) J. Exp. Med. 195, 1613–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamada, K., Ni, J., Zhu, G. F., Fiscella, M., Teng, B. Q., van Deursen, J. M. A. & Chen, L. P. (2002) J. Immunol. 168, 4832–4835. [DOI] [PubMed] [Google Scholar]
- 34.Bodmer, J. L., Schneider, P. & Tschopp, J. (2002) Trends Biochem. Sci. 27, 19–26. [DOI] [PubMed] [Google Scholar]
- 35.Schwartz, J. C., Zhang, X., Fedorov, A. A., Nathenson, S. G. & Almo, S. C. (2001) Nature 410, 604–608. [DOI] [PubMed] [Google Scholar]
- 36.Schwartz, J. C., Zhang, X., Nathenson, S. G. & Almo, S. C. (2002) Nat. Immunol. 3, 427–434. [DOI] [PubMed] [Google Scholar]
- 37.Stamper, C. C., Zhang, Y., Tobin, J. F., Erbe, D. V., Ikemizu, S., Davis, S. J., Stahl, M. L., Seehra, J., Somers, W. S. & Mosyak, L. (2001) Nature 410, 608–611. [DOI] [PubMed] [Google Scholar]
- 38.Zhang, X., Schwartz, J. C., Guo, X., Bhatia, S., Cao, E., Lorenz, M., Cammer, M., Chen, L., Zhang, Z. Y., Edidin, M. A., et al. (2004) Immunity 20, 337–347. [DOI] [PubMed] [Google Scholar]