Abstract
Several species of Brucella are known to be zoonotic, but B. neotomae infection has been thought to be limited to wood rats. In 2008 and 2011, however, B. neotomae was isolated from cerebrospinal fluid of 2 men with neurobrucellosis. The nonzoonotic status of B. neotomae should be reassessed.
Keywords: Brucella, Brucella neotomae, zoonoses, neurobrucellosis, bacteria, Costa Rica, brucellosis, neurological infections
Members of the genus Brucella are the infectious agents of brucellosis, a neglected disease responsible for economic losses resulting from abortion and low performance in production animals (1). The 4 species mainly responsible for this widespread bacterial zoonosis are B. melitensis, B. abortus, B. suis, and to a lesser extent B. canis. Underdiagnosis and limited awareness of the disease among healthcare practitioners is common in many countries (1).
B. neotomae, isolated in 1957 from wood rats (Neotoma lepida) in North America (2), has been considered nonzoonotic (3). It has been isolated from target organs of experimentally infected mice and guinea pigs (4,5). We report the isolation of B. neotomae from cerebrospinal fluid samples from 2 human patients with neurobrucellosis.
The Study
In 2008, a Brucella sp. isolate was submitted to the Tropical Diseases Research Center at the Universidad de Costa Rica. This isolate (denoted strain bneohCR2) was cultured from a cerebrospinal fluid sample obtained from a 64-year-old male patient at one of the main hospitals in San José, Costa Rica. In 2011, another isolate (denoted strain bneohCR1) was recovered from a cerebrospinal fluid sample from a 51-year-old male patient at a regional hospital in Costa Rica. As is common for other patients with brucellosis, the blood leukocyte count for each patient was almost within the reference range, and C-reactive protein level was within reference range. Both patients showed clinical signs compatible with neurobrucellosis (6), had positive Rose Bengal test results, and recovered after receiving 1 month of streptomycin (750 mg/d) and 3 months of doxycycline (100 mg/12 h).
Further bacteriologic analysis (7,8) confirmed that the isolates were a Brucella sp. (Table). Real-time PCR high-resolution melting analysis (9) confirmed genus designation but was inconclusive regarding species designation. Bruce-ladder multiplex PCR (10) and multiple loci variable number of tandem repeats–16-loci panel analysis (http://mlva.u-psud.fr/brucella/; Figure 1) indicated that the profiles for both DNA isolates corresponded to profiles for B. neotomae. Analysis of bneohCR2 by multiplex single-nucleotide polymorphism (SNP) primer extension assay (11) and by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry of protein extracts (12) (Technical Appendix Table 1) confirmed that the isolate was B. neotomae.
Table. Differential biochemical profile of Brucella isolates from 2 men with neurobrucellosis, Costa Rica, 2008 and 2011.
| Analysis |
bneohCR1 |
bneohCR2 |
| Biochemical tests | ||
| Oxidase | - | – |
| Citrate utilization | – | – |
| Nitrate reduction | + | + |
| CO2 required | – | – |
| H2S production | + | + |
| Urease activity, h |
0–0.5+ |
0–0.5+ |
| Growth in presence of dyes | ||
| Thionin 20 μg/mL |
– |
– |
| Basic fuchsin 20 μg/mL | – | – |
| A | + | + |
| M | – | – |
Figure 1.
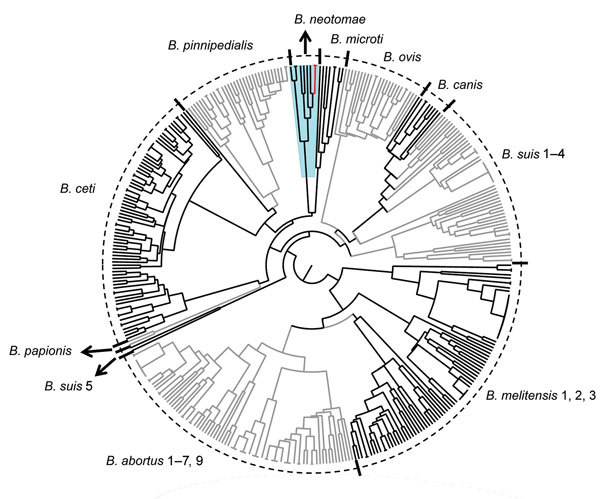
Dendrogram based on multiple locus variable number of tandem repeats–16-loci panel analysis of Brucella spp. (http://mlva.u-psud.fr/brucella/) and clinical isolates from human cerebrospinal fluid samples from 2 patients with brucellosis. The isolates bneohCR1 and bneohCR2 (red branches) showed a pattern consistent with previously reported profiles for Brucella neotomae (blue shading). Black, gray, and tic marks are used to differentiate between adjacent species. Arrows separate small neighboring clusters and indicate the B. neotomae cluster.
We performed whole-genome sequencing of bneohCR1 and bneohCR2 and resequencing of reference strain B. neotomae 5K33. Sequencing data were deposited at the European Nucleotide Archive (http://www.ebi.ac.uk/ena/) under accession codes ERS1563929 (bneohCR1), ERS1563928 (bneohCR2), and ERS162447 (5K33). To place the bneohCR1 and bneohCR2 in a phylogenetic context, publicly available reads from 51 Brucella whole-genome sequences (Technical Appendix Table 2) were aligned and then mapped to B. suis 1330 by using SMALT version 0.5.8 (ftp://ftp.sanger.ac.uk/pub/resources/software/smalt/). Reads from bneohCR1 and bneohCR2 genomes mapped to 98.6% of the B. suis 1330 genome. SNPs were called from the alignment by use of Samtools (http://samtools.sourceforge.net/), and 34,307 variable sites across all isolates were extracted by using SNP sites (13). The resulting alignment of SNPs was used for maximum-likelihood phylogenetic reconstruction by use of RAxML version 7.0.4 (https://github.com/stamatak/standard-RAxML). The generated phylogenetic tree confirmed that the bneohCR isolates clustered together with B. neotomae reference strain 5K33 (ENA accession no. JMSC01, assembly accession no. GCA_00742255.1) (Figure 2). Isolates bneohCR1 and bneohCR2 differed from the reference genome by 174 and 160 SNPs, respectively. This number of SNPs is smaller than that between B. abortus 9–941 and B. abortus 2308 (214 SNPs), which are 2 well-recognized strains of the same Brucella species.
Figure 2.
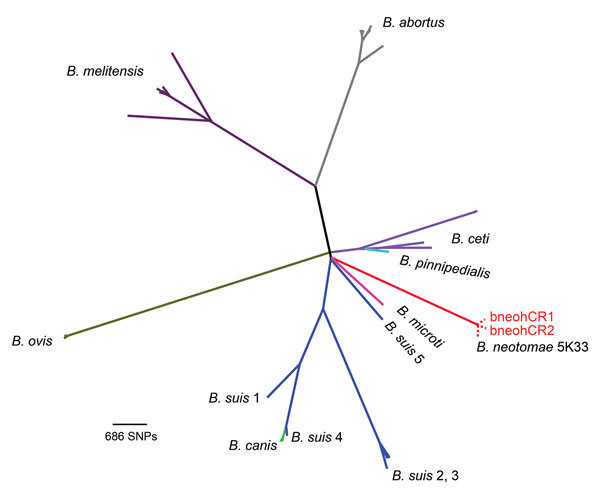
Phylogenetic tree based on 34,307 single-nucleotide polymorphisms (SNPs) found among 51 Brucella genome sequences. The clinical isolates bneohCR1 and bneohCR2 cluster with B. neotomae 5K33 and differ by 164 SNPs. A different color is used to represent each Brucella species. Dotted red lines denote the 3 B. neotomae isolates, which overlap at the tip of the branch because of the high identity among them.
Analysis of 23 previously reported genomic islands or anomalous genomic loci (14) was performed for both bneohCR genomes. For this analysis, a “genomic-island pseudo-molecule” was constructed by concatenation of 23 genomic regions obtained from different Brucella genomes. BLAST (https://github.com/sanger-pathogens/Farm_blast) comparison of this pseudo-molecule and the bneohCR draft genomes, generated by assembly with Velvet (15), showed that the genomic loci known as 26.5 kb, 12 kb, and GI-6 that are absent in B. neotomae (14) are also absent in the queried genomes.
Conclusions
This report of B. neotomae as a cause of zoonotic disease raises questions about possible underrepresentation of reported cases. This study also has implications for brucellosis diagnosis. Specifically, the differences among B. neotomae and the other Brucella species at the biochemical level are subtle. The major difference between B. neotomae and B. abortus, the main cause of human brucellosis in Costa Rica, is the presence of oxidase activity in B. abortus, which is assessed subjectively (7,8). Because B. neotomae has not, until now, been considered zoonotic, some cases of brucellosis reported as being caused by atypical zoonotic classical Brucella might have been misdiagnosed cases of B. neotomae infection. The introduction of whole-genome sequencing into the clinical field will thus improve diagnosis.
A lack of epidemiologic information with regard to the 2 patients reported here precluded the investigation of exposure or contact with hosts known to harbor B. neotomae. Further studies are needed to establish which animals may act as reservoirs for B. neotomae in Costa Rica.
In summary, B. neotomae should be considered a zoonosis risk for infection in humans. Incorporation of molecular techniques for diagnosis will help resolve the Brucella genus homogeneity obtained when only biochemical assays are used.
Protein mass values of Brucella reference strains and bneohCR2, and general information and accession numbers of the genomes included in the phylogenetic analysis.
Acknowledgments
We are grateful to Heilyn Estrada-Murillo for isolate information, Daphnne Garita for technical assistance, Gabriela Hernandez-Mora for provision of serotype-specific antiserum, and Gordon Dougan for helpful discussions.
This work was funded by Fondos del Sistema del Consejo Nacional de Rectores, Costa Rica, and Wellcome Trust. N.R.-V. and C.J.-R. were partially sponsored by University of Costa Rica scholarships. K.S.B. is funded by Wellcome Trust Fellowship 106690/A/14/Z. K.S.B. and N.R.T. were funded by Wellcome Trust grant no. 098051.
All procedures involving live Brucella were carried out according to the “Reglamento de Bioseguridad de la CCSS39975-0,” year 2012, after “Decreto Ejecutivo no. 0965-S,” year 2002 and protocol approved by SIA0434-14 Universidad Nacional, Costa Rica. These genetic resources were accessed in Costa Rica according to the Biodiversity Law no. 7788 and the Convention on Biological Diversity, under the terms of respect to equal and fair distribution of benefits among those who provided such resources.
Biography
Miss Suárez-Esquivel is a PhD student at Programa de Doctorado en Ciencias as Universidad de Costa Rica, working at Programa de Investigación en Enfermedades Tropicales at Universidad Nacional. Her primary research interest is the evolution of Brucella species and their host specificity.
Footnotes
Suggested citation for this article: Suárez-Esquivel M, Ruiz-Villalobos N, Jiménez-Rojas C, Barquero-Calvo E, Chacón-Díaz C, Víquez-Ruiz E, et al. Brucella neotomae infection in humans, Costa Rica. Emerg Infect Dis. 2017 June [date cited]. http://dx.doi.org/10.3201/eid2306.162018
Current affiliation: University of Liverpool, Liverpool, UK
References
- 1.World Health Organization. The control of neglected zoonotic diseases. In: NZD4 Organising Committee, editor. WHO conference report. Geneva: WHO Press; 2014. p. 23–35. [Google Scholar]
- 2.Stoenner HG, Lackman DB. A new species of Brucella isolated from the desert wood rat, Neotoma lepida Thomas. Am J Vet Res. 1957;18:947–51. [PubMed] [Google Scholar]
- 3.Moreno E. Retrospective and prospective perspectives on zoonotic brucellosis. Front Microbiol. 2014;5:213. 10.3389/fmicb.2014.00213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stoenner HG. The behavior of Brucella neotomae and Brucella suis in reciprocal superinfection experiments in mice and guinea pigs. Am J Vet Res. 1963;24:376–80. [PubMed] [Google Scholar]
- 5.Gibby IW, Gibby AM. Host–parasite relationships with Brucella neotomae. J Bacteriol. 1965;89:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ceran N, Turkoglu R, Erdem I, Inan A, Engin D, Tireli H, et al. Neurobrucellosis: clinical, diagnostic, therapeutic features and outcome. Unusual clinical presentations in an endemic region. Braz J Infect Dis. 2011;15:52–9. 10.1016/S1413-8670(11)70140-4 [DOI] [PubMed] [Google Scholar]
- 7.Alton GG, Jones LM, Angus RD, Verger JM. Techniques for the brucellosis laboratory. Paris: Institut National de la Recherche Agronomique; 1988. p. 39–41. [Google Scholar]
- 8.Weyant R, Popovic T, Bragg S. Basic laboratory protocols for the presumptive identification of Brucella species. Atlanta: Centers for Disease Control and Prevention; 2001. p. 1–16. [Google Scholar]
- 9.Winchell JM, Wolff BJ, Tiller R, Bowen MD, Hoffmaster AR. Rapid identification and discrimination of Brucella isolates by use of real-time PCR and high-resolution melt analysis. J Clin Microbiol. 2010;48:697–702. 10.1128/JCM.02021-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.García-Yoldi D, Marín CM, de Miguel M-J, Muñoz PM, Vizmanos JL, López-Goñi I. Multiplex PCR assay for the identification and differentiation of all Brucella species and the vaccine strains Brucella abortus S19 and RB51 and Brucella melitensis Rev1. Clin Chem. 2006;52:779–81. 10.1373/clinchem.2005.062596 [DOI] [PubMed] [Google Scholar]
- 11.Scott JC, Koylass MS, Stubberfield MR, Whatmore AM. Multiplex assay based on single-nucleotide polymorphisms for rapid identification of Brucella isolates at the species level. Appl Environ Microbiol. 2007;73:7331–7. 10.1128/AEM.00976-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Isidoro-Ayza M, Ruiz-Villalobos N, Pérez L, Guzmán-Verri C, Muñoz PM, Alegre F, et al. Brucella ceti infection in dolphins from the Western Mediterranean sea. BMC Vet Res. 2014;10:206. 10.1186/s12917-014-0206-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Page AJ, Taylor B, Delaney AJ, Soares J, Seemann T, Keane JA, et al. SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb Genomics. 2016. Apr 29. 10.1099/mgen.0.000056 [DOI] [PMC free article] [PubMed]
- 14.Mancilla M. The Brucella genomic islands. In: López-Goñi I, O’Callaghan D, editors. Brucella: Molecular Microbiology and Genomics. Poole (UK): Caister Academic Press; 2012. p. 36–57. [Google Scholar]
- 15.Page AJ, De Silva N, Hunt M, Quail MA, Parkhill J, Harris SR, et al. Robust high-throughput prokaryote de novo assembly and improvement pipeline for Illumina data. Microb Genomics. Microbiology Society; 2016. Aug 25. 10.1099/mgen.0.000083 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Protein mass values of Brucella reference strains and bneohCR2, and general information and accession numbers of the genomes included in the phylogenetic analysis.


