Abstract
On large geographical scales, changes in animal population distribution and abundance are driven by environmental change due to climatic and anthropogenic processes. However, so far, little is known about population dynamics on a regional scale. We have investigated 92 archaeological horse remains from nine sites mainly adjacent to the Swiss Jura Mountains dating from c. 41,000–5,000 years BP. The time frame includes major environmental turning points such as the Last Glacial Maximum (LGM), followed by steppe vegetation, afforestation and initial re-opening of the landscape by human agricultural activities. To investigate matrilinear population dynamics, we assembled 240 base pairs of the mitochondrial d-loop. FST values indicate large genetic differentiation of the horse populations that were present during and directly after the LGM. After the retreat of the ice, a highly diverse population expanded as demonstrated by significantly negative results for Tajima’s D, Fu’s FS and mismatch analyses. At the same time, a different development took place in Asia where populations declined after the LGM. This first comprehensive investigation of wild horse remains on a regional scale reveals a discontinuous colonisation of succeeding populations, a pattern that diverges from the larger Eurasian trend.
Introduction
Population distributions and abundance patterns of species are driven by environmental change. Natural and anthropogenic processes impact the availability of food resources for large herbivores, but species respond individually to these challenges (e.g., [1, 2]). Generally, large animals are more likely to react to climate change due to longer generation intervals and smaller effective population size [3]. The Pleistocene-Holocene transition is characterised by profound climatic and thus environmental change, and species well adapted to the open steppe vegetation in Eurasia and North America were confronted with fragmentation and even loss of habitat, yet some were able to establish new niches and survived [1, 4]. A combination of human hunting pressure and habitat fragmentation caused by climate change seems to be the most appropriate explanation for most extinction events in the Old World [5–8] and is supported by both archaeological and palaeo-climatic evidence.
One of the key species of the Pleistocene steppe was the horse, Equus ferus caballus L. 1758. The palaeontological and archaeological record shows its abundance in an area ranging from North America to southern Europe. In the first genetic studies featuring Pleistocene horses from North America and Eurasia, it emerged that in addition to a mitochondrial clade consisting of North American horses, a second clade included both E. f. caballus from North America and Eurasia, and domestic horses [9, 10]. Cieslak et al. [11] further showed that while some matrilines were regionally confined to Alaska, the Eurasian steppe, and Iberia, others were extremely widely distributed suggesting a panmictic population. The approach of Lorenzen et al. [1] focused on (amongst others) horse population development in response to climate change, habitat distribution and human encroachment on a global scale, mainly featuring specimens from north-eastern Asia and north-western America. A positive correlation between available habitat size and genetic diversity supports their conclusion that climate had been the major driving force in population changes over the past 50 thousand years (ka). Horses were thriving particularly under cold and arid conditions. The authors found, however, that the drastic decline of genetic diversity in horses after the Last Glacial Maximum (LGM) could not be explained by habitat reduction alone and thus might reflect the impact of expanding human populations in Eurasia as indicated by the prevalence of horse remains in the archaeological (not palaeontological) record [1]. This scenario was supported by Orlando et al. [12], who found support for models indicating population reduction in the interglacial phases and expansion during the cold stages of Marine Isotope Stages (MIS) 4 and 3, followed by a 100-fold collapse after the LGM. In contrast to these large scale developments, little is as yet known about the effect of environmental change on regional horse populations, and it is probable that they responded more variably, depending on local conditions.
To investigate this issue, we focused on local horse population development through the course of c. 50 ka in the heterogeneous landscape between and including the Alps and the Jura Mountain Chain—present day Switzerland. This region was subjected to sometimes rapid environmental change. Both Alpine and Jura glaciers reached their maximal extent at c. 25 ka BP and started to retreat between 22 and 21 ka BP [13–15]. Deglaciation progressed quite rapidly; the northern Jura c. 50 km south of Basel (Rhine knee area) was ice-free by 19 ka BP [16], while around 18 ka BP, soil development had started in the Alpine foreland [17] when 80% of the LGM ice had melted [18]. Environmental conditions improved rapidly and human (temporary) settlements, first at the foot of the mountains and in caves, later towards the lakes [19], became more numerous [20]. Palynological data indicate herbaceous, heliophilous vegetation until c. 14.7 ka BP, forming grassland interspersed with dwarf shrubs [20–22]. At 14.7 ka BP, mean temperature rose by c. 5°C [21] and in the course of the following 2,000 years the landscape turned into open woodland; by 11 ka BP, forest canopy was probably closed [23]. Anthropogenic influence (agricultural activity) becomes traceable around 7 ka BP in the Jura [21] and in the Alpine foreland [24]. Since the Neolithic, large game species have lost significance in human diets [25], but since agriculture and domestic animal husbandry demanded open landscapes, wild species were increasingly displaced. Horses are absent from the Mesolithic archaeological record in Switzerland despite numerous known sites (yet less than from earlier and later periods) which include faunal assemblages [19]. Most likely, the last wild horses in Switzerland stem from Neolithic lakeshore settlements where horses are present in very low amounts; however, it cannot be ultimately ruled out that they represent first domestics [26]. It is assumed that from the Bronze Age onwards, all horse remains stem from domestic animals [27].
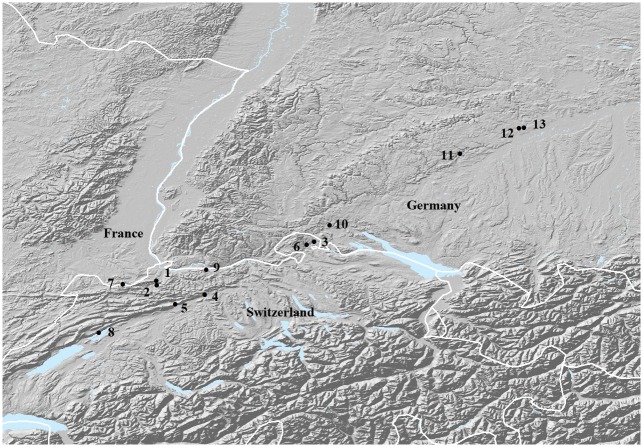
We have investigated horse teeth and bones from nine sites mainly adjacent to the Swiss Jura Mountains dating from c. 41 to 5 ka BP (Fig 1, Table 1, S1 Table). The dataset comprises all known Pleistocene sites with more than one horse remain. To address matrilinear population dynamics of Upper Pleistocene and Holocene wild horses, we have assembled 240 base pairs (bp) of the mitochondrial d-loop. We aim to contextualise population developments and natural and anthropogenic changes of the environment in a fringe area of the Eurasian steppe biota. These results are compared with published data from Northern Asia and the Ural region, the heartland of the steppe.
Fig 1. Map of investigated sites in Switzerland and added sites in Germany.
Site numbers according to Table 1.
Table 1. Details of the investigated sites, number of samples and haplogroups.
Archaeological cultures according to Leesch [28]. Mitochondrial haplogroup nomenclature follows Cieslak et al. [11]. Haplogroups identified from directly dated specimens are indicated in italics. For individual dates, skeletal elements and references see S1 Table.
| Name of site | 14C Age (cal ka BP) | Archaeological culture | Number of positive samples | Mitochondrial haplogroups | |
|---|---|---|---|---|---|
| 1 | Schalberghöhle | 41–37 | - | 3 | A; X3 |
| 12.5 | Azilian | 1 | B | ||
| 2 | Kohlerhöhle | 24–23 | Badegoulian | 11 | B; H; X3 |
| 15–13.5 | Magdalenian | 12 | A; B; D; K | ||
| 3 | Kesslerloch | 18–14 | Magdalenian | 43 | A; B; D; H; K; X3; X4b |
| 4 | Käsloch | 17–15 | Magdalenian | 3 | B; H |
| 5 | Rislisberghöhle | 15–12.5 | Magdalenian—Azilian | 4 | K |
| 6 | Schweizersbild | 15–14 | Magdalenian | 6 | A; B; X3 |
| 7 | Abri Neumühle | 14.5 | Magdalenian | 1 | C |
| 8 | Twann-Bahnhof | 5.5 | Neolithic | 6 | A; B; D |
| 9 | Mumpf | 5 | Neolithic | 2 | D |
| 10 | Petersfelsa, b | 15–14 | Magdalenian | 2 | A; B |
| 11 | Hohlefelsa | 14.8 | Magdalenian | 1 | C |
| 12 | Bocksteinhöhlea | 50 | - | 1 | A |
| 13 | Vogelherdhöhlea | 17 | Magdalenian | 1 | A |
| total = 97 | |||||
Materials and methods
Archaeological samples
A total of 202 horse (Equus sp.) teeth and bones were sampled either from Palaeolithic or Neolithic anthropogenic cultural layers, or from palaeontological contexts associated with hyena hunting activity and chance finds; 92 of them yielded amplifiable mtDNA [29]. All sites are located in or close to the Jura Mountains in Switzerland (Fig 1, Table 1, S1 Table). The samples had been stored in museums and archaeological collections since their excavations. To obtain direct dates from each layer at the respective sites, 31 samples were chosen for 14C dating using accelerator mass spectrometry (AMS) at ETH Zurich, Switzerland, and calibrated with CalPal [30]. Additional 14C dates were assembled from the literature. In some cases the age was projected from dendrochronology or typology (S1 Table). Five 14C dated sequences of Pleistocene horse remains (DQ007558/DQ007611: 14’751 cal BP, DQ007556/DQ007609: 14’752 cal BP, DQ007591: 16’928 cal BP, DQ007590: 50’735 cal BP; [10] FJ204352: 14’500 cal BP; [1] from the Swabian Jura were added to the dataset, resulting in 97 specimens to be analysed.
Processing of ancient samples
Preparation, extraction, amplification and Sanger sequencing of ancient samples were performed as described in Elsner et al. [29] in dedicated ancient DNA facilities following established standards for aDNA work [31], including multiple independent extractions and PCR, and routine cloning. Mitochondrial d-loop positions 15,492 to 15,669 and 15,696 to 15,758 [32] were targeted in seven partially overlapping fragments [33]. Contamination of ancient samples was never detected; all PCR amplification products in the extraction and PCR blank controls came from microorganisms or were unidentifiable according to GenBank Blast search and are most likely explained by the permissive PCR set up (low annealing temperature, up to 70 cycles).
Data analysis
Sequences were edited and aligned by eye with BioEdit [34]. A consensus sequence was built from at least three amplifications from a minimum of two independent extractions based on majority. To deal with sequences with missing nucleotides on the one hand, and to avoid specimens that stemmed from potentially mixed-up layers and an overrepresentation of Magdalenian samples, the analyses were done on three datasets (Table 2).
Table 2. Datasets used for analyses.
| Selection criterion | Number of sequences | |||||||
|---|---|---|---|---|---|---|---|---|
| Total | Palaeontological | Badegoulian | Madgalenian | Magd.+Azilian | Azilian | Neolithic | ||
| Dataset 1 | all samples | 97 | 4 | 11 | 70 | 74 | 4 | 8 |
| Dataset 2 | samples with > 40% missing nucleotides excluded | 78 | 4 | 11 | 53 | 57 | 4 | 6 |
| Dataset 3 | only 14C dated samples | 36 | 4 | 5 | 20 | - | 3 | 4 |
Within the datasets, samples were assembled into time bins according to similar environmental conditions. Magdalenian and Azilian samples were handled both in combination and separately because, on the one hand, the transition between the cultural horizons is marked by the temperature increase at c. 14.7 ka BP, but, on the other hand, possibly mixed up layers within the sites may have led to mis-assignment of individual specimens. The combined category could thus be dropped for dataset 3. Both Azilian and Neolithic date into the early Holocene, yet we assume different dynamics in the Neolithic due to anthropogenic interference to the landscape leading to a dilution of the interdependence of latitude/temperature and vegetation, and thus did not combine those samples.
Nucleotide diversity is defined as the average number of nucleotide differences per site between two DNA sequences in all possible pairs in the population studied, while haplotype diversity is a way to describe the uniqueness of a haplotype in a population. These indices as well as FST values and tests to detect recent population expansion (Tajima’s D, Fu’s FS, sum of squared deviations SSD, Harpending’s raggedness index) were computed with Arlequin 3.5 [35] with missing data coded as ‘?’ and allowed level of missing data set to 5%. Tajima’s D [36] uses the mean average number of pairwise nucleotide differences and the number of segregating sites, each scaled so that they are expected to be the same in a neutrally evolving population of constant size; Fu’s FS [37] is based on the number of alleles (haplotypes). It is generally assumed that Fu’s FS is more sensitive in detecting population expansion than Tajima’s D. The raggedness index [38, 39] is also used to detect recent population expansions, which is rejected by non-significant results. The SSD between observed and expected mismatch (distribution of the number of sequence differences) quantifies the smoothness of the observed mismatch.
To reject a statistical bias in the analyses introduced by uneven sample sizes in the respective time bins, directly compared time bins (e.g. Magdalenian and LGM) were randomized (10k permutations with replacement) using nucleotide and haplotype diversity estimated with the packages pegas [40] and seqinR [41] implemented in R [42] using the option ‘pairwise deletion of missing data’. This was done by pooling the sequences of two time bins, repeatedly creating two pseudo-groups of the same size of the original bins from the pool and collecting diversity parameters from them. In case the combined pseudo diversity deviated from the original diversity (threshold 0.05), sampling bias has to be assumed.
For the construction of Median Joining Networks (MJN) [43] with the program Network (fluxus-engineering.com) polymorphic nucleotide positions were down-weighted according to the number of polymorphisms (default 50) [33]. The transition: transversion weight was set to 1: 10 (S2 Table). Principal component analysis (PCA) was based on relative haplogroup frequencies within the Eurasian dataset and computed with PAST [44].
Results
Swiss and Swabian wild horse populations
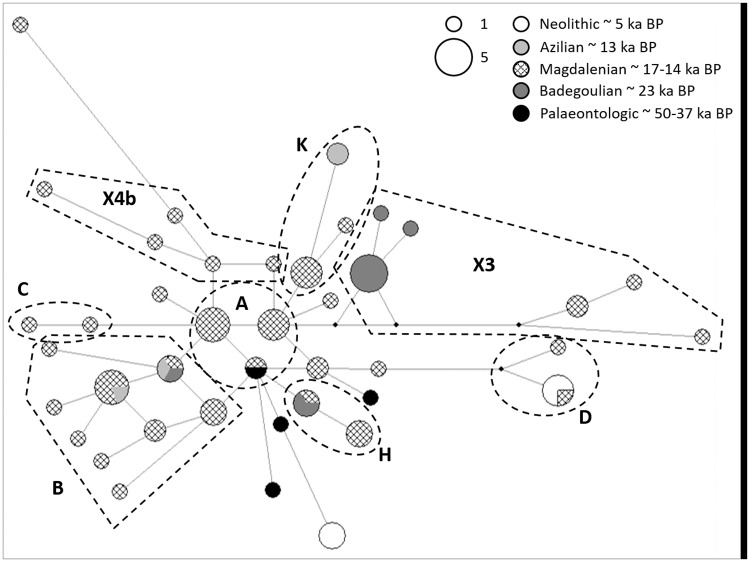
The maximal sequence length of 241 bp (without primers) could be assembled for 50 specimens, 231 bp for 15 samples. The remaining 27 specimens had missing data mostly between positions 15,564 and 15,669 relative to the horse reference mitogenome sequence [32]. Amongst the 92 Swiss and five Swabian horse samples, 36 polymorphic sites were present resulting in 41 haplotypes (ht), which can be summarized into eight haplogroups (hg) according to Cieslak et al. [11]: A, B, C, D; H, K and X3 (Fig 2; S3 Table). One previously unnamed haplogroup was labelled X4b following [11], distinguished by nucleotide positions 15495, 15540, 15602, (15718) and 15720 (defining nucleotide positions in bold, optional position in parenthesis).
Fig 2. Median Joining Network (MJN) of horse populations (97 samples, max. 241 bp).
Nodes are proportional to frequencies and branch length according to number of substitutions. Haplogroup nomenclature follows Cieslak et al. [11].
Nucleotide and haplotype diversities are shown in Table 3 (see also S4 Table). Both nucleotide and haplotype diversities are highest in the Magdalenian and lowest in the Badegoulian. Note that nucleotide diversity of the Neolithic deme is relatively high compared to the lower haplotype diversity, indicating population fragmentation.
Table 3. Nucleotide and haplotype diversities in horse populations from Switzerland and the Swabian Jura (dataset 2).
| Time period | Number of samples | Number of haplotypes | Nucleotide diversity | Haplotype diversity |
|---|---|---|---|---|
| Palaeontological | 4 | 4 | 0.0104 | 1 |
| Badegoulian | 11 | 4 | 0.0093 | 0.6 |
| Magdalenian | 53 | 28 | 0.016 | 0.95 |
| Magd. + Azilian | 57 | 29 | 0.0159 | 0.95 |
| Azilian | 4 | 3 | 0.0104 | 0.83 |
| Neolithic | 6 | 2 | 0.0149 | 0.6 |
The Median Joining Network (241 bp) shows only little haplotype continuity between the time bins. Two lineages (hgs B and H) are found in Switzerland during and after the LGM, and two lineages of hg B continue into the Azilian (Figs 2 and 3). All other haplotypes are restricted to single time bins or at least do not occur in succeeding time bins. For reasons of comparability, we tested whether a bias was introduced by uneven sample sizes within the time bins. This can be rejected for the comparison between Magdalenian (+ Azilian) and Badegoulian by permutation testing (S1 Fig). FST values for these time bins portend great genetic differentiation (0.17, p < 0.001) (S5 Table).
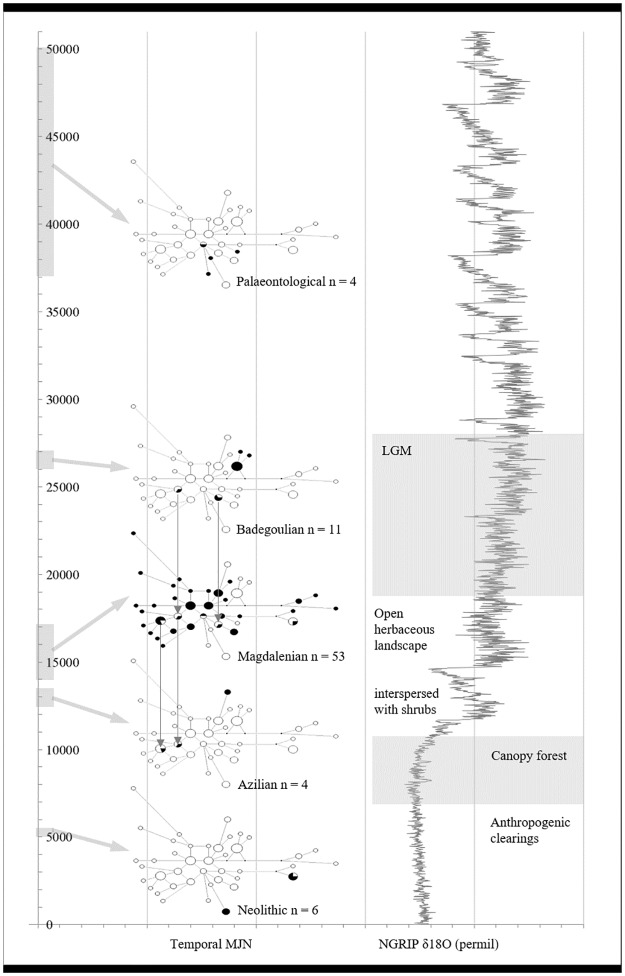
Fig 3. Temporal Median Joining Networks (MJN) of Swiss and Swabian horse populations (97 samples, max. 241 bp) in the context of environmental conditions and temperature.
Light grey boxes and arrows show the age of the sequences the particular MJN is based on. Empty nodes represent haplotypes absent from particular time bin. Vertical arrows indicate continuity of haplotypes from subsequent time bins. δ18O record of the North Greenland Ice Core Project (NGRIP) after [49–53], vegetation data from [21, 54].
Simulations according to Tajima [36] and Fu [37] indicate recent population expansion for the Magdalenian and the combined Magdalenian and Azilian sample sets (Table 4, S6 Table). This is supported by the distribution of the number of sequence differences between haplotypes (mismatch analysis, S2 Fig). Both the palaeontological and Magdalenian (+ Azilian) bins exhibit unimodal distributions. Equally, neither the sum of the squared deviations (SSD) nor Harpending’s raggedness index [38, 39] were statistically significant, further indicating population expansion (Table 4). This applies for the Badegoulian and Azilian time bins as well. However, an increased SSD and raggedness as well as multimodal distribution point to populations with secondary contact, i.e. populations that have received genetic input from further populations [45]. For the Neolithic time bin, population expansion can be rejected explicitly; the very high SSD and raggedness index might even indicate that individuals from the same site stemmed originally from independent populations.
Table 4. Tajima’s D, Fu’s FS, sum of squared deviations (SSD) and Harpending’s raggedness index results for horse populations from Switzerland and the Swabian Jura (dataset 2).
Significant results are in bold.
| Time period | Tajima’s D | p | Fu’s FS | p | SSD | p | Raggedness index | p |
|---|---|---|---|---|---|---|---|---|
| Palaeontological | -0.8 | .2 | -1.51 | .06 | 0.02 | 0.8 | 0.1 | 0.9 |
| Badegoulian | -0.09 | .5 | 0.77 | .7 | 0.13 | 0.1 | 0.3 | 0.1 |
| Magdalenian | -1.32 | .07 | -21.71 | 0 | 0.001 | 0.6 | 0.02 | 0.6 |
| Magd. + Azilian | -1.29 | .08 | -22.59 | 0 | 0.001 | 0.5 | 0.02 | 0.6 |
| Azilian | 1.37 | .9 | 0.46 | .5 | 0.09 | 0.3 | 0.25 | 0.7 |
| Neolithic | 2.12 | 1 | 4.51 | 1 | 0.38 | 0.02 | 0.88 | 0.03 |
In Fig 3, a temporal Median Joining Network (MJN) of 97 sequences (up to 241 bp) from the Swiss and Swabian Jura is put into context with the δ18O record of the North Greenland Ice Core Project (NGRIP) and prevailing regional environment. During the LGM, three distinct haplogroups are present (all from one site), two of which are also found in the Magdalenian. Most lineages present during the LGM did not reoccur in later time periods. The dominating haplotype X3 has as yet not been detected in Eurasian Pleistocene horses [1, 10–12] but is frequent in some modern breeds, both in Iberia (e.g. [46]) and Asia (e.g. [47, 48]). During the Magdalenian, when an open herbaceous landscape prevailed, the network topology shows a star-like expansion pattern. After c. 14.7 ka BP when global atmospheric temperature rose by c. 5°C and forestation began, only two genetically distant lineages are recovered from the Azilian. By c. 12 ka BP temperatures levelled off at present day conditions. Two distinct matrilines, one of which occurred in the Magdalenian, are present in the Neolithic when early human impact on the environment is observed. Generally, very little continuity existed through time.
Comparison with Pleistocene horse sequences from across Eurasia
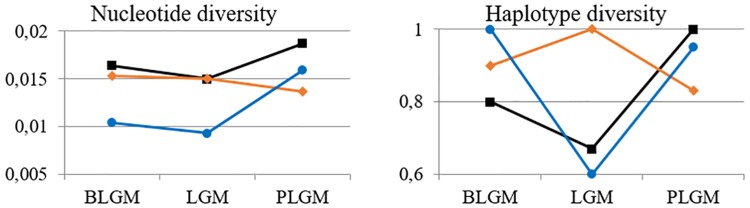
The Swiss and Swabian Jura sequences were put into context with published samples from Northern Asia and the Urals regions (S7 Table). They were sorted into time bins: before the LGM (BLGM, c. 50–27.5 ka BP), during the LGM (LGM, c. 25–22.5 ka BP), and after the LGM (PLGM, c. 18–12.5 ka BP). Nucleotide diversity is highest in Asia at all times, and is decreasing westwards (Fig 4). During the LGM, it drops slightly and rises to higher level afterwards in Asia and Switzerland, yet decreases in the Ural region. Haplotype diversity has to be regarded with caution due to low sample sizes (see Fig 5, S7 Table). Generally the trend seen from nucleotide diversity is repeated (Fig 4, S8 Table).
Fig 4. Nucleotide (left panel) and haplotype (right panel) diversity in Asia (black squares), Ural region (red diamonds) and the Swiss and Swabian Jura (blue circles) before (BLGM), during (LGM) and after (PLGM) the Last Glacial Maximum.
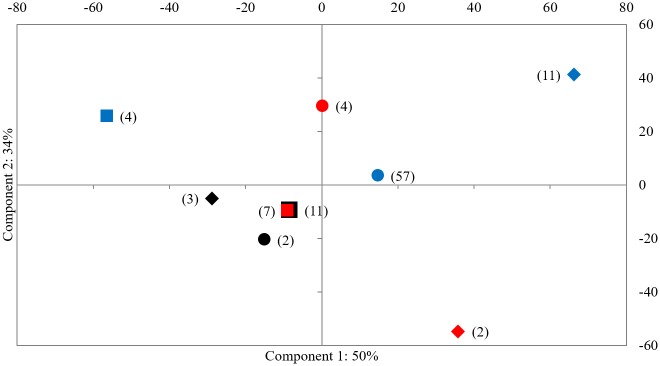
Fig 5. PCA graph based on relative haplogroup frequencies.
Asia (black), Ural region (red) and Swiss/Swabian (blue) samples divided in time bins: square = before the LGM, diamond = LGM, circle = after the LGM. Sample numbers are given in parenthesis. The first two components explain 84% of the variation (see S4 Fig for loadings).
Because the sample sizes in the Eurasian datasets are low compared to the Swiss/Swabian Jura, an unbiased comparison of FST values is only possible for the time bins Swiss/Swabian LGM and Swiss/Swabian PLGM (see above) and Swiss/Swabian LGM and Asian BLGM (FST 0.23, p < 0.001; S3 Fig, S9 Table). A principal component analysis (PCA) based on relative haplogroup frequencies demonstrates that while the Asian samples are genetically close to each other through time, and might, together with the Ural BLGM lineages, be addressed as panmictic, the Swiss/Swabian samples are more distinct. Moreover, the interruptive nature of the LGM concerning population continuity is apparent as the LGM demes from all regions stand apart from the preceding and succeeding populations (Fig 5, S4 Fig).
Discussion
The investigation of mt d-loop sequences from 97 horse teeth and bones dating from 50 to 5 ka BP from the region of the Swiss and Swabian Jura is the first regional study of Pleistocene to early Holocene horse population development. The investigated time frame includes the LGM as a major environmental change, which was followed by open steppe and later afforestation after the temperature rise c. 14.7 ka BP, leading to a densely forested landscape which was in turn partially re-opened for humans’ agricultural demands. The archaeological context of the horse remains studied here mirror hunter-gatherer colonisation, settlement and hunting strategies in the region. We have investigated all known E. f. caballus remains from Pleistocene Switzerland; open-air dry- and wetland sites did not contain specimens with amplifiable mtDNA [29].
In the region of the Swiss and Swabian Jura, we observe only little continuity of horse matrilines through time, particularly in the populations present during (Badegoulian) and directly after (Magdalenian) the LGM. Horses were most abundant when the landscape was open and comprised the typical characteristics of steppe biota like herbaceous grassland interspersed with shrub flora. Statistical analyses portend population expansion in the Magdalenian. Despite numerous archaeological sites from the Azilian, the Mesolithic and the Neolithic, horse finds from these horizons are extremely rare [19, 26]. Population fragmentation is indicated by comparatively high nucleotide and low haplotype diversity, and for the Neolithic deme, expansion can be rejected based on a significant raggedness index.
Throughout the investigated time frame, diversity patterns in Eurasia change. The general trend of decreasing nucleotide and haplotype diversity from east to west supports models of an initial population expansion of wild horses in eastern Asia [55]. However, the idea of a panmictic horse population across Eurasia during the last 50 ka (e.g. [1, 11]) has to be challenged, even for pre-LGM times, based on relative haplogroup frequencies. During the LGM, when large parts of the continent were covered by ice or un-inhabitable due to extremely cold and arid conditions [56], populations were isolated and fragmented as partially supported by FST values. Subsequently, this led to a diversification particularly in the Ural and Swiss/Swabian Jura regions. The finding of a regional population expansion in the Magdalenian contradicts previous studies of horse population development [1, 12] that proposed a rapid decline directly after the LGM.
Wild horses might have gone extinct in the region of present-day Switzerland in the Neolithic, yet due to low numbers of remains in the archaeological record from the Azilian onwards it is challenging to trace this development genetically. It seems likely that climate, and not human activity, was the major driving force behind abundance and diversity of horse populations in this region: the expansion time is contemporaneous with intensified human encroachment of the area after the LGM. However, Neolithic land use for farming and domestic animal husbandry presumably replaced the last remnants of the wild horse population in Switzerland; genetically, this remains to be proven.
Conclusion
In summary, the region of present-day Switzerland was inhabited by discontinuous horse populations and we cannot assume a panmictic deme over the investigated time period of 50 ka. Horse populations mostly replaced each other during and after the LGM, and only little continuity is observable. In the Magdalenian, diversity was highest as the population expanded into the newly accessible landscape. When the landscape transformed from open steppe into more and more densely forested woodland, the population probably shrunk and became fragmented.
Comparing horse matrilineages from Asia, the Ural, and the Swiss and Swabian Jura regions, population-specific developments are detectable. Wild horses possibly never formed a panmictic deme throughout their distribution range, and the LGM led to additional population fragmentation which subsequently persisted.
Besides the methodological challenges due to the discontinuous and unbalanced representation of equid sequences, this paper provides the first comprehensive investigation of wild horse remains from one restricted region. This approach has offered the opportunity to focus on aspects of horse population development that might be overlooked in the global picture by demonstrating specific reaction patterns to changing environmental conditions.
Supporting information
Unbiased and thus comparable pairs are framed. A: Dataset 1; B: Dataset 2; C: Dataset 3.
(DOCX)
X-axis: pairwise differences, y-axis: number of pairs. A: Dataset 1; B: Dataset 2; C: Dataset 3.
(DOCX)
Swiss/Swabian samples: dataset 2. Rejections of null hypothesis are framed blue (Swiss LGM vs. Asia BLGM 0.0208; Swiss LGM vs. Swiss PLGM 0.034).
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Haplogroup defining nucleotide positions relative to the horse reference mitogenome [1] are shown according to their position. All deviations from the reference sequence are given, with mandatory defining positions in bold and optional nucleotide positions in parenthesis. Note that transitions on nucleotide positions 15,585; 15,604 and 15,650 occur sporadically in all haplogroups; these positions are regarded as hotspots and therefore dismissed.
(DOCX)
(DOCX)
Lower triangle: FST values, upper triangle: p values. Comparable populations are boxed, significant FST values are in bold.
(DOCX)
Significant results are in bold. NaN = not a number because only one haplotype was present.
(DOCX)
Sequences marked with an ‘a’ are part of draft full genomes which are obtainable as SRA-Illumina runs on GenBank.
(DOCX)
(DOCX)
Lower triangle: FST values, upper triangle: p values. Comparable populations are boxed, significant FST values are in bold. Swiss/Swabian samples: dataset 2.
(DOCX)
Acknowledgments
Samples were kindly provided by Kantonsarchäologie Aargau (Mumpf), Archäologische Bodenforschung Basel-Stadt (Riehen-Ausserberg), Naturhistorisches Museum Basel (Allschwil-Ziegelei, Birseck-Ermitage, Münchenstein-Steinbruch, Schalberghöhle), Archäologie und Kantonsmuseum Basel-Landschaft (Brügglihöhle, Kohlerhöhle), Archäologischer Dienst Bern (Abri Neumühle, Twann-Bahnhof), Université de Neuchâtel (Hauterive-Champréveyres, Monruz), Kontansarchäologie Schaffhausen (Kesslerloch, Schweizersbild), Kantonsarchäologie Solothurn (Käsloch, Rislisberg) and by J. Sedlmeier. The authors thank L. Orlando and M. Muschick for help with statistical analysis. This study was funded by Swiss National Science Foundation (K-31K1_120528/1), Freiwillige Akademische Gesellschaft Basel, and Nikolaus and Bertha Burckhardt-Bürgin-Foundation.
Data Availability
Data are available from GenBank (ncbi.nlm.nih.gov/genbank) with accession numbers KC893753-KC893844.
Funding Statement
Julia Elsner was funded by Swiss National Science Foundation (K-31K1_120528/1), Freiwillige Akademische Gesellschaft Basel, and Nikolaus and Bertha Burckhardt-Bürgin-Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Lorenzen ED, Nogues-Bravo D, Orlando L, Weinstock J, Binladen J, Marske KA, et al. Species-specific responses of Late Quaternary megafauna to climate and humans. Nature. 2011;479(7373):359–64. 10.1038/nature10574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crees JJ, Carbone C, Sommer RS, Benecke N, Turvey ST. Millennial-scale faunal record reveals differential resilience of European large mammals to human impacts across the Holocene. P Roy Soc Lond B Bio. 2016;283(1827). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCain CM, King SB. Body sizes and activity times mediate mammalian responses to climate change. Global Change Biol. 2014. [DOI] [PubMed] [Google Scholar]
- 4.Campos PF, Willerslev E, Sher A, Orlando L, Axelsson E, Tikhonov A, et al. Ancient DNA analyses exclude humans as the driving force behind late Pleistocene musk ox (Ovibos moschatus) population dynamics. PNAS. 2010;107(12):5675–80. 10.1073/pnas.0907189107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnosky AD, Koch PL, Feranec RS, Wing SL, Shabel AB. Assessing the causes of late Pleistocene extinctions on the continents. Science. 2004;306(5693):70–5. 10.1126/science.1101476 [DOI] [PubMed] [Google Scholar]
- 6.Koch PL, Barnosky AD. Late quaternary extinctions: State of the debate. Annu Rev Ecol Evol S. 2006;37:215–50. [Google Scholar]
- 7.Hofreiter M, Stewart J. Ecological change, range fluctuations and population dynamics during the Pleistocene. Curr Biol. 2009;19(14):R584–R94. 10.1016/j.cub.2009.06.030 [DOI] [PubMed] [Google Scholar]
- 8.Stuart AJ. Late Quaternary megafaunal extinctions on the continents: a short review. Geol J. 2015;50(3):338–63. [Google Scholar]
- 9.Vila C, Leonard JA, Gotherstrom A, Marklund S, Sandberg K, Liden K, et al. Widespread origins of domestic horse lineages. Science. 2001;291(5503):474–7. 10.1126/science.291.5503.474 [DOI] [PubMed] [Google Scholar]
- 10.Weinstock J, Willerslev E, Sher A, Tong W, Ho SY, Rubenstein D, et al. Evolution, systematics, and phylogeography of Pleistocene horses in the new world: a molecular perspective. PLoS Biol. 2005;3(8):e241 10.1371/journal.pbio.0030241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cieslak M, Pruvost M, Benecke N, Hofreiter M, Morales A, Reissmann M, et al. Origin and history of mitochondrial DNA lineages in domestic horses. PLoS ONE. 2010;5(12):e15311 10.1371/journal.pone.0015311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orlando L, Ginolhac A, Zhang G, Froese D, Albrechtsen A, Stiller M, et al. Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature. 2013;499(7456):74–8. 10.1038/nature12323 [DOI] [PubMed] [Google Scholar]
- 13.Buoncristiani J-F, Campy M. The palaeogeography of the last two glacial episodes in France: the Alps and Jura In: Ehlers J, Gibbard PL, editors. Quaternary Glaciations Extent and Chronology Part I: Europe. Amsterdam: Elsevier; 2004. p. 101–10. [Google Scholar]
- 14.Ivy-Ochs S, Kerschner H, Reuther A, Maisch M, Sailer R, Schaefer J, et al. The timing of glacier advances in the northern European Alps based on surface exposure dating with cosmogenic 10Be, 26Al, 36Cl, and 21Ne. Geol S Am S. 2006;415:43–60. [Google Scholar]
- 15.Preusser F, Graf HR, Keller O, Krayss E, Schlüchter C. Quaternary glaciation history of northern Switzerland. Quatern Sci J. 2011;60(2–3):282–305. [Google Scholar]
- 16.Ivy-Ochs S, Schafer J, Kubik PW, Synal HA, Schluchter C. Timing of deglaciation on the northern Alpine foreland (Switzerland). Eclogae Geol Helv. 2004;97(1):47–55. [Google Scholar]
- 17.Jäger H, Achermann M, Waroszewski J, Kabała C, Malkiewicz M, Gärtner H, et al. Pre-alpine mire sediments as a mirror of erosion, soil formation and landscape evolution during the last 45 ka. CATENA. 2015;128(0):63–79. [Google Scholar]
- 18.Ivy-Ochs S, Kerschner H, Reuther A, Preusser F, Heine K, Maisch M, et al. Chronology of the last glacial cycle in the European Alps. J Quatern Sci. 2008;23(6–7):559–73. [Google Scholar]
- 19.Nielsen EH. Paläolithikum und Mesolithikum in der Zentralschweiz Mensch und Umwelt zwischen 17 000 und 5 500 v. Chr. Luzern: Archäologische Schriften; 2009. [Google Scholar]
- 20.Leesch D, Müller W, Nielsen E, Bullinger J. The Magdalenian in Switzerland: Re-colonization of a newly accessible landscape. Quatern Int. 2012;272(0):191–208. [Google Scholar]
- 21.Cupillard C, Magny M, Bocherens H, Bridault A, Bégeot C, Bichet V, et al. Changes in ecosystems, climate and societies in the Jura Mountains between 40 and 8 ka cal BP. Quatern Int. 2014(0). [Google Scholar]
- 22.Ammann B, von Grafenstein U, van Raden UJ. Biotic responses to rapid warming about 14,685yr BP: Introduction to a case study at Gerzensee (Switzerland). Palaeogeogr, Palaeoclimatol, Palaeoecol. 2013;391(0):3–12. [Google Scholar]
- 23.Ammann B, van Leeuwen JFN, van der Knaap WO, Lischke H, Heiri O, Tinner W. Vegetation responses to rapid warming and to minor climatic fluctuations during the Late-Glacial Interstadial (GI-1) at Gerzensee (Switzerland). Palaeogeogr, Palaeoclimatol, Palaeoecol. 2013;391(0):40–59. [Google Scholar]
- 24.Erny-Rodmann C, Gross-Klee E, Haas JN, Jacomet S, Zoller H. Früher "human impact" und Ackerbau im Uebergangsbereich Spätmesolitikum-Frühneolithikum im schweizerischen Mittelland. Jahrbuch der Schweizerischen Gesellschaft für Ur- und Frühgeschichte. 1997;80:27–56. [Google Scholar]
- 25.Schibler J. The economy and environment of the 4th and 3rd millennia BC in the northern Alpine foreland based on studies of animal bones. Environmental Archaeology. 2006;11(1):49–64. [Google Scholar]
- 26.Schibler J, Hüster-Plogmann H. Die neolithische Wildtierfauna und ihr Aussagegehalt betreffend Umwelt und Umweltveränderungen In: Stöckli WE, Niffeler U, Gross-Klee E, editors. SPM II: Neolithikum. Die Schweiz vom Paläolithikum bis zum frühen Mittelalter. Basel: Gesellschaft für Ur- und Frühgeschichte der Schweiz; 1995. p. 76–83. [Google Scholar]
- 27.Schibler J, Studer J. Haustierhaltung und Jagd während der Bronzezeit der Schweiz In: Hochuli S, Niffeler U, Rychener J, editors. SPM III: Bronzezeit. Die Schweiz vom Paläolithikum bis zum frühen Mittelalter. Basel: Gesellschaft für Ur- und Frühgeschichte der Schweiz; 1998. p. 171–92. [Google Scholar]
- 28.Leesch D. Das späte Jungpaläolithikum. Zeitlicher Rahmen und Fundinventare In: LeTensorer J-M, Niffeler U, editors. SPM I: Paläolithikum und Mesolithikum. Die Schweiz vom Paläolithikum bis zum frühen Mittelalter. Basel: Gesellschaft für Ur- und Frühgeschichte der Schweiz; 1993. p. 153–64. [Google Scholar]
- 29.Elsner J, Schibler J, Hofreiter M, Schlumbaum A. Burial condition is the most important factor for mtDNA PCR amplification success in Palaeolithic equid remains from the Alpine foreland. Archeol Anthropol Sci. 2014;7(4):505–15. [Google Scholar]
- 30.CalPal-2007 online [Internet]. 2012. http://www.calpal-online.de.
- 31.Shapiro B, Hofreiter M, editors. Ancient DNA—methods and protocols. New York: Springer; 2012. [Google Scholar]
- 32.Xu X, Arnason U. The complete mitochondrial DNA sequence of the horse, Equus caballus: extensive heteroplasmy of the control region. Gene. 1994;148(2):357–62. [DOI] [PubMed] [Google Scholar]
- 33.Elsner J, Deschler-Erb S, Stopp B, Hofreiter M, Schibler J, Schlumbaum A. Mitochondrial d-loop variation, coat colour and sex identification of Late Iron Age horses in Switzerland. J Archaeol Sci: Rep. 2016;6:386–96. [Google Scholar]
- 34.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acid S. 1999;41:95–8. [Google Scholar]
- 35.Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10(3):564–7. 10.1111/j.1755-0998.2010.02847.x [DOI] [PubMed] [Google Scholar]
- 36.Tajima F. Evolutionary relationship of DNA sequences in finite populations. Genetics. 1983;105(2):437–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fu YX. Estimating effective population-size or mutation-rate using the frequencies of mutations of various classes in a sample of DNA-sequences. Genetics. 1994;138(4):1375–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harpending HC, Sherry ST, Rogers AR, Stoneking M. The genetic structure of ancient human populations. Curr Anthrop. 1993;34(4):483–96. [Google Scholar]
- 39.Harpending HC. Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Hum Biol. 1994;66(4):591–600. [PubMed] [Google Scholar]
- 40.Paradis E. pegas: an R package for population genetics with an integrated–modular approach. Bioinformatics. 2010;26:419–20. 10.1093/bioinformatics/btp696 [DOI] [PubMed] [Google Scholar]
- 41.Charif D, Lobry JR. SeqinR 1.0–2: a contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis In: Bastolla U, Porto MJ, Roman HE, Vendruscolo M, editors. Structural approaches to sequence evolution: molecules, networks, populations. Biological and Medical Physics, Biomedical Engineering. New York: Springer; 2007. p. 207–32. [Google Scholar]
- 42.R Development Core Team RDCT. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2014. [Google Scholar]
- 43.Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16(1):37–48. [DOI] [PubMed] [Google Scholar]
- 44.Hammer O, Harper DAT, Ryan PD. PAST: PAleontological STatistics software package for education and data analysis. Palaeontol Electron. 2001;4(1):1–9. [Google Scholar]
- 45.Frankham R, Ballou JD, Briscoe DA. Introduction to conservation genetics. Cambridge: Cambridge University Press; 2002. 617 p. [Google Scholar]
- 46.Luís C, Bastos-Silveira C, Costa-Ferreira J, Cothran EG, Do Mar Oom M. A lost Sorraia maternal lineage found in the Lusitano horse breed. J Anim Breed Genet. 2006;123:399–402. 10.1111/j.1439-0388.2006.00612.x [DOI] [PubMed] [Google Scholar]
- 47.McGahern A, Bower MAM, Edwards CJ, Brophy PO, Sulimova G, Zakharov I, et al. Evidence for biogeographic pattering of mitochondrial DNA sequences in Eastern horse populations. Anim Genet. 2006;37(5):494–7. 10.1111/j.1365-2052.2006.01495.x [DOI] [PubMed] [Google Scholar]
- 48.Yang Y, Zhu Q, Liu S, Zhao C, Wu C. The origin of Chinese domestic horses revealed with novel mtDNA variants. Animal Science Journal. 2016. [DOI] [PubMed] [Google Scholar]
- 49.Vinther BM, Clausen HB, Johnsen SJ, Rasmussen SO, Andersen KK, Buchardt SL, et al. A synchronized dating of three Greenland ice cores throughout the Holocene. J Geophys Res-Atmos. 2006;111(D13):D13102. [Google Scholar]
- 50.Rasmussen SO, Andersen KK, Svensson AM, Steffensen JP, Vinther BM, Clausen HB, et al. A new Greenland ice core chronology for the last glacial termination. J Geophys Res-Atmos. 2006;111(D6):D06102. [Google Scholar]
- 51.Andersen KK, Svensson A, Johnsen SJ, Rasmussen SO, Bigler M, Röthlisberger R, et al. The Greenland Ice Core Chronology 2005, 15–42ka. Part 1: constructing the time scale. Quatern Sci Rev. 2006;25(23–24):3246–57. [Google Scholar]
- 52.Svensson A, Andersen KK, Bigler M, Clausen HB, Dahl-Jensen D, Davies SM, et al. A 60 000 year Greenland stratigraphic ice core chronology. Clim Past. 2008;4(1):47–57. [Google Scholar]
- 53.Wolff EW, Chappellaz J, Blunier T, Rasmussen SO, Svensson A. Millennial-scale variability during the last glacial: The ice core record. Quatern Sci Rev. 2010;29(21–22):2828–38. [Google Scholar]
- 54.Heiri O, Koinig KA, Spötl C, Barrett S, Brauer A, Drescher-Schneider R, et al. Palaeoclimate records 60–8 ka in the Austrian and Swiss Alps and their forelands. Quatern Sci Rev. 2014(0). [Google Scholar]
- 55.Warmuth V, Eriksson A, Bower MA, Barker G, Barrett E, Hanks BK, et al. Reconstructing the origin and spread of horse domestication in the Eurasian steppe. PNAS. 2012;109(21):8202–6. 10.1073/pnas.1111122109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hughes PD, Gibbard PL, Ehlers J. Timing of glaciation during the last glacial cycle: evaluating the concept of a global ‘Last Glacial Maximum’ (LGM). Earth-Sci Rev. 2013;125(0):171–98. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Unbiased and thus comparable pairs are framed. A: Dataset 1; B: Dataset 2; C: Dataset 3.
(DOCX)
X-axis: pairwise differences, y-axis: number of pairs. A: Dataset 1; B: Dataset 2; C: Dataset 3.
(DOCX)
Swiss/Swabian samples: dataset 2. Rejections of null hypothesis are framed blue (Swiss LGM vs. Asia BLGM 0.0208; Swiss LGM vs. Swiss PLGM 0.034).
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Haplogroup defining nucleotide positions relative to the horse reference mitogenome [1] are shown according to their position. All deviations from the reference sequence are given, with mandatory defining positions in bold and optional nucleotide positions in parenthesis. Note that transitions on nucleotide positions 15,585; 15,604 and 15,650 occur sporadically in all haplogroups; these positions are regarded as hotspots and therefore dismissed.
(DOCX)
(DOCX)
Lower triangle: FST values, upper triangle: p values. Comparable populations are boxed, significant FST values are in bold.
(DOCX)
Significant results are in bold. NaN = not a number because only one haplotype was present.
(DOCX)
Sequences marked with an ‘a’ are part of draft full genomes which are obtainable as SRA-Illumina runs on GenBank.
(DOCX)
(DOCX)
Lower triangle: FST values, upper triangle: p values. Comparable populations are boxed, significant FST values are in bold. Swiss/Swabian samples: dataset 2.
(DOCX)
Data Availability Statement
Data are available from GenBank (ncbi.nlm.nih.gov/genbank) with accession numbers KC893753-KC893844.