Abstract
Chloroplast development is an integral part of plant survival and growth, and occurs in parallel with chlorophyll biosynthesis. However, little is known about the mechanisms underlying chloroplast development in hexaploid wheat. Here, we obtained a spaceflight-induced wheat albino mutant mta. Chloroplast ultra-structural observation showed that chloroplasts of mta exhibit abnormal morphology and distribution compared to wild type. Photosynthetic pigments content was also significantly decreased in mta. Transcriptome and chloroplast proteome profiling of mta and wild type were done to identify differentially expressed genes (DEGs) and proteins (DEPs), respectively. In total 4,588 DEGs including 1,980 up- and 2,608 down-regulated, and 48 chloroplast DEPs including 15 up- and 33 down-regulated were identified in mta. Classification of DEGs revealed that most were involved in chloroplast development, chlorophyll biosynthesis, or photosynthesis. Besides, transcription factors such as PIF3, GLK and MYB which might participate in those pathways were also identified. The correlation analysis between DEGs and DEPs revealed that the transcript-to-protein in abundance was functioned into photosynthesis and chloroplast relevant groups. Real time qPCR analysis validated that the expression level of genes encoding photosynthetic proteins was significantly decreased in mta. Together, our results suggest that the molecular mechanism for albino leaf color formation in mta is a thoroughly regulated and complicated process. The combined analysis of transcriptome and proteome afford comprehensive information for further research on chloroplast development mechanism in wheat. And spaceflight provides a potential means for mutagenesis in crop breeding.
Introduction
Chloroplasts originated from an endosymbiotic event between a photosynthetic cyanobacterium and a eukaryotic host [1, 2]. They are not only essential for photosynthesis, but are also responsible for the production of many important metabolites, such as amino acids, lipids, starch, hormones, vitamins and isoprenoids in higher plants [3–5]. These functions make the chloroplast an indispensable organelle for plants survival and growth. Functional and photosynthetically active chloroplasts develop from undeveloped proplastids. During this differentiation proplastids enlarge and then thylakoids are formed and stacked into defined grana [6]. Chloroplast differentiation requires the participation of many proteins. Most of these proteins are nuclear-encoded and imported into the developing chloroplast. Therefore, the nucleus regulates essential aspects of chloroplast development [7]. In addition, retrograde signaling from the developing chloroplast to the nucleus also ensures the production of appropriate levels of protein complexes involved in chloroplast maturation [3, 8].
Chlorophyll (Chl) biosynthesis occurs in parallel with chloroplast development during leaf color formation. Chl is distributed in chloroplast thylakoid membrane and, as the main part of light harvesting complex, plays an important role in photosynthesis [9]. The Chl biosynthetic pathway and Chl metabolism have been extensively studied. The enzymes and their encoded nuclear genes for all 15 biosynthetic steps have been identified [10, 11]. The leaf color and photosynthetic efficiency are directly affected by chloroplast development, the number and size of chloroplasts, and Chl biosynthesis and content [12, 13]. In addition, leaf color is also affected by temperature; relatively low temperatures lead to temporary leaf color variation in some chlorophyll-deficient mutants [14]. The ratio of chlorophylls and carotenoids alters in low temperature is most likely the reason for this. Chl includes two different forms Chl a and Chl b, which occur in an approximate ratio of 3:1, influence the leaf green color. Carotenoids are the pigments responsible for the leaf orange color [15].
Leaf color is directly related to the production of photosynthetic pigments and chloroplast development. Thus, leaf color mutants are widely used to reveal the mechanisms for the regulation of chloroplast development and function, Chl biosynthesis, and photosynthesis [12, 16–18]. Thus far, many genes and transcription factors (TFs) affecting chloroplast development and division have been identified. The phytochrome-interacting factor 3 (PIF3) functions in mediating the initial phases of light-induced chloroplast development after the first exposure of green-faded seedlings to light, through the regulation of a subset of rapidly photoresponsive nuclear genes encoding plastid and photosynthesis-related components [19]. In addition, PIF3 is a repressor negatively regulating chloroplast development in Arabidopsis [18]. The Golden2-like (GLK) gene family includes a pair of partially redundant nuclear TFs, GLK1 and GLK2, that are required for the expression of nuclear-encoded photosynthetic genes and chloroplast development in diverse plant species, including maize (Zea mays), rice (Oryza sativa), and Arabidopsis thaliana [2, 20–22]. As in rice, GLK1 and GLK2 are expressed in partially overlapping domains in photosynthetic tissue, the glk1glk2 double mutant is pale green in all photosynthetic tissues and there is a reduction in grana thylakoids in the chloroplasts [20]. FtsZ is a key structural component of chloroplast division machinery in perhaps all photosynthetic eukaryotes. The chloroplast forms of FtsZ assemble into inner membrane-associated rings in the stromal compartments [23]. Members of the Accumulation and Replication of Chloroplast (ARC) gene family cooperate with FtsZ to regulate chloroplast division [24]. Besides, The three isoforms of NADPH: protochlorophyllide oxidoreductase (POR), PORA, PORB and PORC, are the key enzyme catalyzing Chl biosynthesis. PORA is expressed primarily early in development including etiolation, germination and greening. PORB is expressed throughout leaf. PORC expression pattern is similar to PORB when treated by high light [17, 25].
Despite knowledge of the genes involved in model plant species, the molecular mechanisms for chloroplast development and Chl biosynthesis are not well-understood in hexaploid wheat due to its large genome. In species such as wheat large-scale transcriptome profiling and chloroplast proteome analysis (2D-DIGE and MALDI-TOF /TOF) will be helpful in obtaining a global view of gene expression patterns and provide insight into the potential molecular mechanisms of mutagenesis generating [13, 26, 27]. Transcriptome database are valuable resources for genetic and genomic studies of multiple difference genes and pathways. For example, genes involved in chloroplast development and division, Chl biosynthesis, and pigment biosynthesis and transport can be identified via transcriptome analysis [13]. And wheat proteomic studies with a special focus on chloroplasts provide a better understanding of the proteins involved in photosynthesis [27].
We have obtained a novel chlorophyll-deficient mutant mta from the progeny of Mt6172 (MT), generating from the wild type (WT) Triticum aestivum L. H6172, which was exposed to spaceflight induction [14]. To explore the genetic alterations under space environment and uncover the mechanisms underlie the albino phenotype of mta, we performed chloroplast ultra-structural observation and photosynthetic pigments assays of mta and WT. We found that chloroplasts in mta exhibited distorted morphology and were less widely distributed than that of WT. The photosynthetic pigment content was significantly decreased in mta. Leaf transcriptome sequencing and chloroplast proteomic analyses of mta and WT were also carried out. The DEGs and DEPs associated with chloroplast components and development, Chl biosynthesis, and photosynthesis were identified. Omics data mining revealed new metabolic pathways and TFs involved in leaf color formation. Importantly, we found that the DEPs were significantly functioned in photosynthesis including PSⅠ, PSⅡ, cytochrome b6f complex and F-type ATPase. The expression levels of selected DEGs and genes encoding DEPs were validated by qRT-PCR. Integrated omics analyses revealed the main molecular mechanisms regulating leaf color formation, involving chloroplast development, chlorophyll biosynthesis, and photosynthesis. These findings provide new clues to binding omics means for discovering multiple differences in mutagenesis on polyploidy plant species. In addition, spaceflight provides an approach to generating mutants that can be used in crop breeding research.
Materials and methods
Plant materials
All wheat seeds were kindly selected and provided by Space Breeding Research Center, Chinese Academy of Agricultural Sciences. The WT and MT wheat seeds were grown in an experimental field at the Chinese Academy of Agricultural Sciences, Beijing for three weeks. At their three leaf stage, the whole leaves of WT and mta plants were separately collected for transcriptome sequencing analysis. The same plants leaves of WT and mta were used for chloroplast protein extraction and identification. For qRT-PCR validation, the leaves powder for RNA-seq were collected in tubes, frozen in liquid nitrogen, and then stored at -80°C until analysis.
Transmission electron microscopy (TEM)
The three-week-old seedling leaves of WT and mta were cut into 1×1 mm pieces and fixed in 2.5% glutaraldehyde for 24 hours at 4°C. After washing with 0.1 mol/L phosphate buffer (pH 7.2), leaf samples were fixed in 1% osmium tetroxide (OSO4) for 2 hours and washed with phosphate buffer again. Tissues were dehydrated through an ethanol series, 30, 50, 70, 80, 90, 95, and 100%, successively. Then embedded in Epon 812 and sectioned using a Leica ultramicrotome (Leica Microsystems, Ltd, Germany). After staining in 0.2% lead citrate, the ultrastructure of leaf cells was observed under a transmission electron microscope (HT-7700, Hitachi, Japan).
Contents of chlorophyll and carotenoid assays
A total of 0.2 g fresh leaves from WT and mta were cut into 2×2 mm pieces and submerged in 10 ml 95% alcohol for 24 hours in the dark with five replicates, respectively. Leaf extractions were made after swirled and oscillated the samples for 6–8 times until leaves turned white. The specific light absorption of leaf extracts containing chloroplast pigments were measured at 665, 649, and 470 nm using a SPECORD 200 spectrophotometer (AnalytikJena, Germany) according to a previously published method [28, 29]. Chlorophyll and carotenoid content was calculated as following formulas: Chl a = 13.95 × A665 − 6.88 × A649, Chl b = 24.96 × A649 − 7.32 × A665, Car = (1000A470 − 2.05Chl a − 114.8Chl b)/245.
RNA isolation and library preparation
All leaves of 80 WT and 70 mta plants were collected and a total of four libraries with two biological replicates were prepared. RNA was extracted using TRIzol-A+ reagent (TIANGEN BIOTECH, Beijing) and treated with RNase-free DNase I (TaKaRa). RNA quantity was measured using a Nanodrop Quibt 2.0 Flurometer (Life Technologies, CA, USA). RNA quality was evaluated with an Agilent Bioanalyzer Model 2100 (Agilent Technologies, Palo Alto, CA). Samples with an RNA Integrity Number (RIN) value greater than 6.4 were deemed acceptable according to the Illumina transcriptome sequencing protocol. All sequencing reads from the four libraries have been submitted to the Sequence Read Archive (SRA), National Center for Biotechnology Information (NCBI) with an accession of SRP106763 (BioProject ID of PRJNA386075; Four run ID of SRR5520556, SRR5527113, SRR5527114 and SRR5527116 represent mta_R1, mta_R2, WT_R1 and WT_R2 sample reads, respectively).
Transcriptome sequencing and assembly
WT and mta poly (A) mRNA were enriched using oligo dT magnetic beads, then fragmented using fragmentation buffer. The fragmented mRNA was used as template for cDNA synthesis. First strand cDNA was synthesized using reverse transcriptase and random primers, followed by second strand cDNA synthesis using DNA PolymeraseI and RNaseH. Samples were cleaned using the QIAquick PCR kit, and were eluted by eluent buffer for end repairing and sequencing adapter joining. Then the cDNA fragments were separated by agarose gel electrophoresis and fragments of 100–300 bp were enriched by PCR amplification to create cDNA libraries. The well-constructed cDNA libraries were then sequenced on the Illumina HiSeq 2500 platform. The fastq format raw reads were first processed using in-house perl scripts. In this step, clean reads were obtained by removing reads containing adapter, ploy-N, and low quality reads from raw reads. At the same time, Q30 and GC-content of the clean data were calculated. Assembly of the clean reads was performed using Trinity [30], according to the reference genome Chinese Spring wheat in the public databases.
Functional annotation and DEG identification
Gene function was annotated based on the following databases: NCBI non-redundant protein sequences (NR), Protein family (Pfam), UniProt/Swiss-Prot, Gene Ontology (GO), Clusters of Orthologous Groups of proteins (KOG/COG), and Kyoto Encyclopedia of Genes and Genomes (KEGG). Unigene expression levels were expressed as fragments per kilobase of transcript per million fragments mapped (FPKM). FPKM values were calculated using RSEM [31]. Differential expression analysis and identification of DEGs between WT and mta were performed using the DEGseq R package, which provides a statistical method for determining differential expression using a model based on the negative binomial distribution [32]. The resulting p-values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate (FDR). Unigenes with an adjusted p-value ≤ 0.01 and |log2 fold changes| ≥ 1 between mta and WT were designated DEGs. These DEGs were also annotated with GO, COG and KEGG assignments to obtain significantly enriched pathways based on DEG-enrichment by the right sided fisher exact test [33].
Chloroplast protein extraction
Chloroplasts were isolated from WT and mta following a modified protocol [34]. Each step was performed at 4°C or on ice. Briefly, 20 g fresh seedling leaves for each sample were harvested and homogenized with a mortar and pestle in extraction buffer. The homogenate was filtered through 6 layers of muslin, and this step was repeated twice. After that, the suspension containing chloroplasts was subsided by centrifugation, and the pellets containing the chloroplasts were resuspended in isolation buffer. The above resuspended material was then loaded on top of a Percoll step gradient. After centrifugation, the chloroplasts were isolated and from washed twice with isolation buffer and then directly used for protein extraction after visualization under a fluorescence microscope (OLYMPUS, U-RFL-T, Germany) to assess quality. Total chloroplast protein was extracted using TCA/acetone precipitation [35], and chloroplast pellets were resuspended in suspension buffer [36]. Protein concentration was measured with the Bio-Rad Protein Assay kit, which was based on the Bradford method using Bovine Serum Albumin (BSA) as a standard. Proteins were purified using a 2D Clean-up kit (GE Healthcare, USA) with all steps performed on ice, and all chloroplast proteins were dissolved in DIGE lysis-buffer (labeling buffer). Next, the Bradford method was again used to measure the protein concentration for quantitative analysis of the whole leave chloroplast proteins.
2D-DIGE and image analyses
Two-Dimensional Difference Gel Electrophoresis (2D-DIGE) was carried out after analytical gels labeling WT and mta chloroplast proteins with Cy2 (blue), Cy3 (green) and Cy5 (red) fluorescent dyes (5 nmol Cyanine Dye DIGE Fluor mimimal Dye labeling kit, GE, USA). Preparative gels staining with coomassie blue G-250. All steps were done according to the manufacturer’s instructions. The experimental strategy is shown in S1 Fig. Three biological replicate WT samples (50 μg each) were labeled with Cy3 (one sample) and Cy5 (two samples), and the mta samples (50 μg each) were labeled with Cy3 (two samples) and Cy5 (one sample). The labeled samples were combined and separated on 2-DE gels together with the internal standard (IS), which was prepared by mixing 25 μg WT and 25 μg mta samples and labeling with Cy2. Labeling reactions were carried out according to the manufacturer’s instructions. Isoelectric focusing (IEF) was performed using Ettan IPGphor according to GE Healthcare operating manual and a previously described method [37]. All gels were scanned using a scanner (GE Healthcare, USA) according to the manufacturer's protocol. The abundance of each protein spot in the scanned images was quantified using Image Master Platinum 7.0 software (GE Healthcare, USA).
Protein identification by MALDI-TOF MS
All selected spots were manually excised from the WT and mta chloroplast proteins 2D-DIGE gels. The normalized volume of each spot was assumed to represent the abundance of the detected protein. A criterion of ≥ 1.5-fold change (1.5-fold increase / decrease, p-value ≤ 0.05) was used to define significant differences when comparing spot sizes between groups. In-gel digestion and MS acquisition were performed as described [38]. TOF mass spectra (TOF-MS) were searched against the NCBInr (http://www.ncbi.nlm.nih.gov/) protein database using the MASCOT search engine (http://www.matrixscience.com, Matrix Science). Identification was based on the combination of a MASCOT score, maximum peptide coverage, and additional experimental pI and MW of the protein spots in the gels. The NCBI and TAIR (http://www.arabidopsis.org/) database were used to obtain information about protein functional annotation, encoding genes and subcellular localization.
Bisulfite sequencing PCR (BSP)
BSP was utilized to determine the methylation status at single CpG resolution of TaPORA promoter fragments. Genomic DNA was isolated from the leaves of WT and mta using a DNA-quick Plant System kit (Tiangen Biotech, Beijing, China) according to the manufacturer’s instructions. Approximately 500 ng of genomic DNA were treated with bisulfate CT conversion reagent following the protocols in the EZ DNA Methylation-Gold TM kit (ZYMO research corp, USA). TaPORA promoter primers for BSP (S5 Table) were designed using the online tool MethPrimer (http://www.urogene.org/methprimer/). BSP reaction with 25 μl per tube was done using high fidelity PrimeSTAR GXL DNA polymerase (Takara Biotech, Dalian, China) and BSP primers. The -PCR protocols were as follows: 98°C for 2 min; 35 cycles of 98°C for 10 s, 57°C for 15 s, and 68°C for 20 s; and a final extension 68°C for 6 min. The PCR products were separated by electrophoresis on 2% agarose gels and were then purified using a gel extraction kit (Axygen Biotech, Hangzhou, China) following standard protocols. The purified fragments were cloned into the pLB vector (Tiangen Biotech) and verified by sequencing in Sangon Company (Shanghai, China). At least five clones were sequenced for each BSP reaction.
Quantitative real-time PCR (qRT-PCR) validation
qRT-PCR analysis using the Bio-Rad CFX Manager (Bio-Rad, USA) with SsoFastTM EvaGreen Supermix (Bio-Rad) was employed to verify the DEG and DEP expression results. Primers for specific genes encoding photosynthetic proteins and photosynthesis DEGs (S6 Table) were designed using Beacon Designer 7 (Bio-Rad, USA). qRT-PCR assays were performed in triplicate (technical repeats) with two independent biological replicates, and ACTIN (GenBank: AAW78915) was used as theIS [39]. Relative expression levels were determined using a relative quantitative method (2−ΔΔCT) [40].
Statistical analysis
Statistical analysis was performed by SAS8.01 software (SAS Institute, Cary, NC, USA). The data of Chl and carotenoid contents were expressed as the mean ± standard deviation. All data were analyzed using one-way ANOVA. The p-value 0.05 and 0.01 represent different significant level with confidence of 95% and 99%, respectively.
Results
Phenotypic properties of mta: Leaf color and chloroplasts
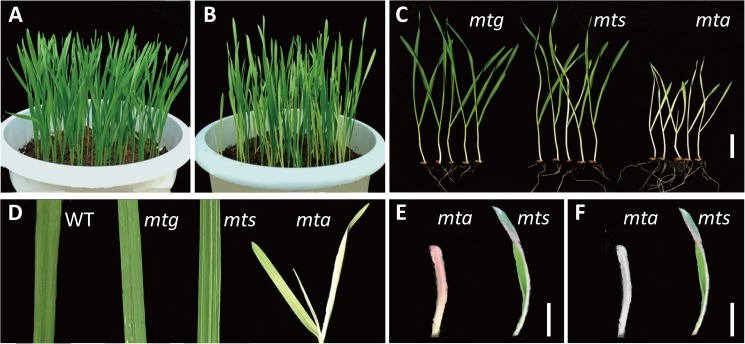
Mt6172 (MT) was a spaceflight-induced mutant originating from a winter wheat cultivar, H6172 (WT) (Fig 1A). The self-fertilized progenies of MT segregated three types of leaf color: green, narrow-white striped and albino. These mutants were named mtg, mts and mta, respectively (Fig 1B and 1C). The mta mutant had completely albino leaves and died after 25–28 days. The white leaf tissue in mts and mta turned pink or purple when temperature decreased to 4–10°C, but returned to albino as temperature increased (Fig 1D–1F). Although transiently affected by an environmental factor, mta leaf color of mta was mainly controlled by genetic regulation.
Fig 1. Phenotype of wild type and leaf color mutants of Triticum aestivum L.
(A) Wild type; (B) Leaf color mutants; (C) Type of leaf color mutants: mtg, green leaf; mts, narrow-white striped leaf; mta, albino leaf. (D) Leaf of wild type and mutants. (E) Phenotype of mta and mts white tissue in low temperature; (F) Phenotype of mta and mts white tissue as temperature increased. (All white bar = 2 cm).
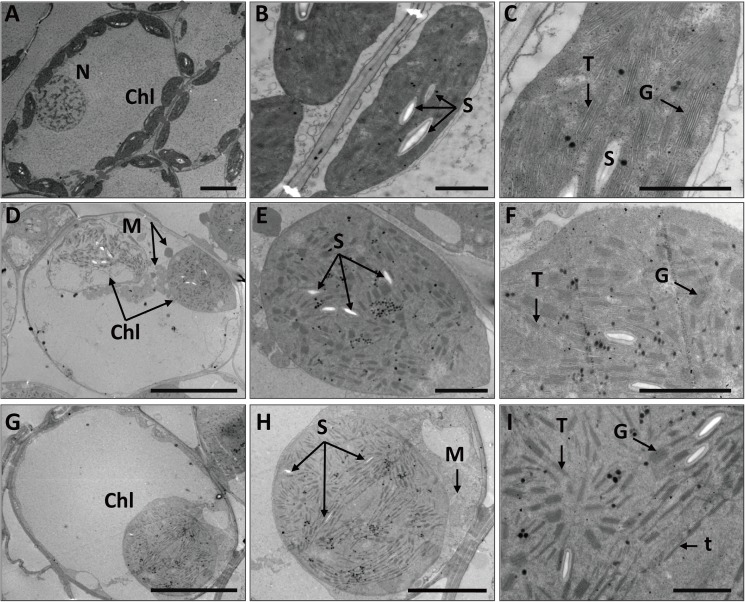
To investigate chloroplast morphology and distribution in mta, we further analyzed the chloroplast ultrastructure of mta and WT. As shown in Fig 2, WT chloroplasts exhibited typical structures and had a highly organized inner membrane system, numerous granum and inter-granum thylakoids and several starch granules (Fig 2A–2C). In mta, mesophyll cells contained rarely or none chloroplasts, and the chloroplasts were abnormal appearing inflated (Fig 2D–2I). The inner structure of inflated chloroplasts almost disappeared and contained disordered granum lamellae. These chloroplasts had smaller and more abundant starch granules than those in WT (Fig 2D–2F). Another large-sized chloroplasts in mta had no intact membrane structures, and contained many single thylakoid sacs and small starch granules (Fig 2G–2I). These results confirm that chloroplast development in mta is impaired.
Fig 2. Chloroplast ultrastructures of WT and mta.
(A-C) Chloroplast ultrastructure of WT (A, bar = 10 μm; B, bar = 2 μm; C, bar = 1 μm); (D-I) Chloroplast ultrastructure of mta (D, G bar = 10 μm; E, bar = 2 μm; F, bar = 1 μm; H, bar = 5 μm; I, bar = 1 μm). In these pictures, Chl, denotes chloroplast; N, denotes nucleus; M, denotes mitochondria; S, denotes starch; T, denotes thylakoid grana; G, denotes grana; t, denotes single thylakoid sac.
Chl and carotenoid content were also significantly different between mta and WT (Table 1). Total Chl, Chl a, Chl b, and carotenoid content in mta were much lower than in WT. The ratio of Chl a/Chl b in mta was very close to WT, but the ratio of carotenoid/Chl in mta was slightly higher than that of WT, indicating that the total Chl content decreased more than the carotenoid content in mta. These data suggest that albino leaf phenotype of mta is directly affected by decreased pigment content.
Table 1. Chlorophyll and carotenoid content in WT and mta.
| Sample | Chl a (mg·g-1) | Chl b (mg·g-1) | Chl a + b (mg·g-1) | Chl a/b | Carotenoid (mg·g-1) | Carotenoid/Chl |
|---|---|---|---|---|---|---|
| WT | 1.19 ± 0.06 | 0.29 ± 0.02 | 1.48 ± 0.07 | 4.06 ± 0.15 | 0.30 ± 0.02 | 0.20 ± 0.01 |
| mta | 0.19 ± 0.17a | 0.05 ± 0.01b | 0.24 ± 0.03a | 4.00 ± 0.26a | 0.06 ± 0.01a | 0.24 ± 0.01a |
a Significant difference (P≤0.05)
b Significant difference (P≤0.01).
Transcriptome analysis
Illumina sequencing and unigenes assembly
To gain insight into the mechanism of abnormal chloroplast development in mta, we used transcriptome analysis to identify chloroplast-related genes that are differentially expressed between WT and mta. Four cDNA libraries yielded 30,608,728 to 78,882,496 pair-end clean reads (S1 Table). The total reads were mapped to the reference Chinese Spring genome (Triticum aestivum IWGSC1_popseq.31). Using Trinity, the clean reads of WT and mta were assembled into 105,265 transcripts (Table 2), which comprised 64,999 unigenes. The length of 44.85% of the transcripts ranged from 300 bp to 1000 bp, 21.2% were shorter than 300 bp and 33.92% were longer than 1000 bp. These results indicated that clean data was in good quality. We therefore used these data for the following analyses.
Table 2. Summary of functional annotations and length distributions of unigenes and DEGs between WT and mta.
| Total | GO | COG | KEGG | NR | Swiss-Prot | |
|---|---|---|---|---|---|---|
| All unigenes | ||||||
| Unigenes | 105,256 | 89,470 | 36,855 | 37,883 | 105,214 | 77,679 |
| Length < 300 | 22,339 | 18,420 | 4,356 | 7,346 | 22,339 | 13,698 |
| 300 ≤ Length < 1000 | 47,211 | 39,777 | 16,797 | 17,393 | 47,185 | 34,732 |
| 1000 ≤ Length | 35,706 | 31,273 | 15,702 | 13,144 | 35,700 | 29,249 |
| DEG Set | ||||||
| mta / WT | 4,588 | 4,065 | 2,583 | 1,703 | 4,498 | 3,844 |
| Mta_R1 / WT_R1 | 6,191 | 5,468 | 2,443 | 2,251 | 6,060 | 5,143 |
| Mta_R2 / WT_R2 | 6,421 | 5,673 | 2,497 | 2,501 | 6,417 | 5,143 |
Unigenes functional annotation and DEG classification
To obtain transcript functional annotations, all assembled genes were searched against five public databases (GO, COG, KEGG, Swiss-Prot, and NR). The correlation coefficients for both the WT and mta biological replicates reached significant level and were higher than 0.96 (S2 Fig). This result demonstrated that the unigenes were suitable for further analysis. Among these unigenes, a total of 4,588 DEGs were identified between WT and mta. The summary of DEGs functional annotation and classification were provided in Table 2.
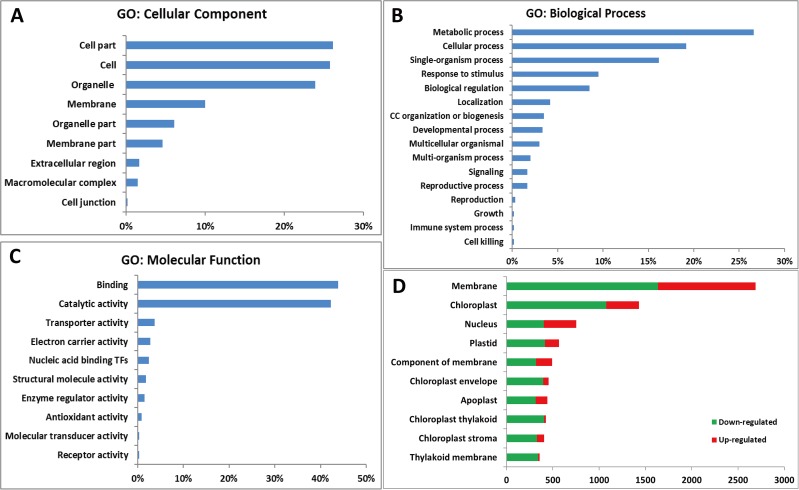
GO annotation of all 4,588 DEGs was performed using the WEGO software [41] (http://www.geneontology.org/), and 4,065 DEGs were categorized into cellular component, biological process and molecular function using blast topGO bio-conductor (http://www.blast2go.org/) (Fig 3). The cellular components category mainly included DEGs related to cell part, cell, organelle part, and membrane (Fig 3A). Biological processes category was classified as metabolic term (27%), cellular term (19%), single-organism process (16%) and other processes (Fig 3B). Molecular functions contained 44% DEGs in binding function and 42% in catalytic activity (Fig 3C). Further analysis of different cellular components showed that DEGs were significantly enriched in membrane, chloroplast and nucleus, and most DEGs were down-regulated in mta. Specially, almost all the DEGs involved in chloroplast thylakoid and thylakoid membrane were down-regulated (Fig 3D).
Fig 3. Functional categorization of DEGs between WT and mta based on GO annotation.
The proportion of each category is displayed based on (A) cellular component, (B) biological process, or (C) molecular function. (D) The number of DEGs in different cellular component categories, green color represents down-regulated in mta while red color represents up-regulated in mta.
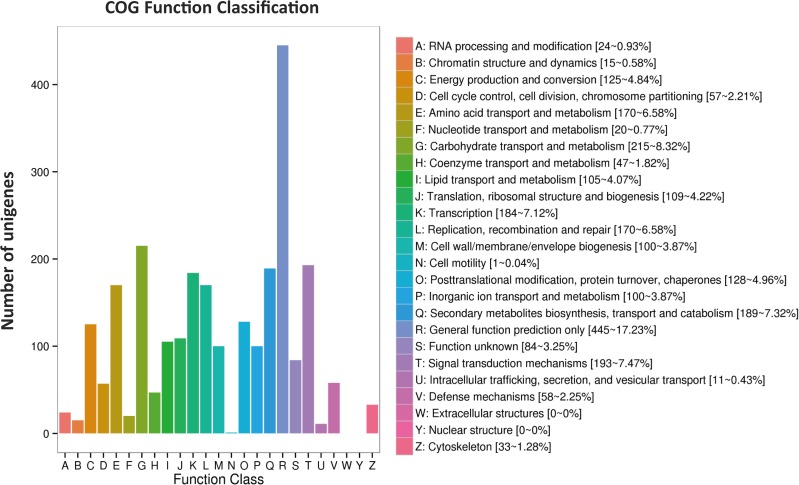
All 4,588 DEGs were further evaluated based on COG annotation. A total of 2,583 (56.30%) were clustered into 24 COG categories (Fig 4). The “general functional prediction only” cluster (445, 17.23%) represented the largest group, followed by “carbohydrate transport and metabolism” (215, 8.32%), “signal transduction mechanisms” (193, 7.47%), “amino acid transport and metabolism” (170, 6.58%), “posttranslational modification, protein turnover, chaperones” (128, 4.96%), “energy production and conversion” (162, 4.57%), “cell wall/ membrane/ envelope biogenesis” (100, 3.87%), “cytoskeleton” (33, 1.28%), and “RNA processing and modification” (24, 0.93%). No genes were assigned to “extracellular structures” and “nuclear structure”.
Fig 4. Histogram of COG classifications.
2,583 DEGs were grouped into 24 COG categories.
In order to reconstruct the metabolic pathways that might be involved in chloroplast development, Chl biosynthesis and photosynthesis, all of the DEGs were searched against KEGG database (http://www.genome.jp/kegg/) and mapped onto 111 KEGG pathways. The top 20 enriched pathways were selected (Table 3). Among these pathways, “phenylalanine metabolism”, “carbon fixation in photosynthetic organisms”, “porphyrin and chlorophyll metabolism”, “photosynthesis”, and “photosynthesis- antenna proteins” were significantly enriched with a q-value cut-off of 2.20×E-49.
Table 3. The top 20 significantly enriched pathways identified by KEGG analysis.
| Rank | KEGG pathway term | Gene number | Rich factor a | q-value b | Pathway ID |
|---|---|---|---|---|---|
| 1 | Phenylalanine metabolism | 111 | 2.029766 | 0.00E+00 | ko00360 |
| 2 | Carbon fixation in photosynthetic organisms | 88 | 2.958113 | 0.00E+00 | ko00710 |
| 3 | Porphyrin and chlorophyll metabolism | 62 | 4.840904 | 0.00E+00 | ko00860 |
| 4 | Photosynthesis | 75 | 8.138023 | 2.20E-49 | ko00195 |
| Photosynthesis—antenna proteins | ko00196 | ||||
| 5 | Glyoxylate and dicarboxylate metabolism | 61 | 2.736091 | 1.30E-10 | ko00630 |
| 6 | Carbon metabolism | 137 | 1.866761 | 4.04E-10 | ko01200 |
| 7 | Phenylpropanoid biosynthesis | 149 | 1.957957 | 1.19E-09 | ko00940 |
| 8 | Monoterpenoid biosynthesis | 20 | 4.838344 | 1.72E-07 | ko00902 |
| 9 | Cutin, suberine and wax biosynthesis | 29 | 3.291935 | 1.03E-06 | ko00073 |
| 10 | Starch and sucrose metabolism | 104 | 1.697702 | 7.79E-06 | ko00500 |
| 11 | Pentose phosphate pathway | 35 | 2.402294 | 1.32E-04 | ko00030 |
| 12 | Glycolysis / Gluconeogenesis | 69 | 1.749535 | 4.53E-04 | ko00010 |
| 13 | Fatty acid elongation | 32 | 2.372977 | 4.86E-04 | ko00062 |
| 14 | Glutathione metabolism | 57 | 1.848673 | 5.96E-04 | ko00480 |
| 15 | Thiamine metabolism | 14 | 3.562022 | 2.68E-03 | ko00730 |
| 16 | Fatty acid metabolism | 43 | 1.787461 | 1.75E-02 | ko01212 |
| 17 | alpha-Linolenic acid metabolism | 34 | 1.900516 | 2.62E-02 | ko00592 |
| 18 | Limonene and pinene degradation | 10 | 3.689237 | 2.83E-02 | ko00903 |
| 19 | Nitrogen metabolism | 26 | 1.937781 | 1.01E-01 | ko00910 |
| 20 | Plant hormone signal transduction | 76 | 1.430521 | 1.14E-01 | ko04075 |
a Reflect the enrichment level of DEGs on pathway, the higher number shows more significant
b Multiply hypothesis testing calibration of p-value, the top 20 significantly enriched pathways were selected based on q-value.
Identification of DEGs related to chloroplast development and chlorophyll biosynthesis
Based on the DEGs annotation and classification above, we found genes involving in the chloroplast development and division, Chl and pigment biosynthesis, and other TFs were all included in the transcriptome database. These gene expression levels in mta were significantly different from those in WT (Table 4 and S2 Table). Many genes and TFs regulated chloroplast development were found in mta, three genes Triticum aestivumLinn_newGene_8077, Traes5AS_049A22D30, and Traes5AS_EBCEA6601 annotated as transcription factor PIF3 were up-regulated. The GLK family including TRAES3BF062500050CFD_g, Traes_7AL_6279FCE8B and Traes_7DL_58DDB90E3 was down-regulated. Chloroplast division gene FtsZ (Traes2AL_6EE01D4C2) was also down-regulated in mta. DEGs participating in porphyrin and chlorophyll metabolism (S3 Fig), such as HEMA1, CHLD, GUN4, CHLI1, CRD1, PORA, and PORB were all down-regulated, except HEME1, which was slightly up regulated in mta. Among all those DEGs, which were identified as involving in chlorophyll biosynthesis, the PORA gene expression level was much lower than other genes (S2 Table). Methylated CpG sites in the TaPORA promoter were predicted in CpGisland (http://www.urogene.org/). The methylation level of TaPORA promoter was further detected, and result showed that the methylation level in mta was much higher compared with WT (S4 Fig). In addition, a total of 261 TFs including MYB, bHLH and other proteins, such as MATE, WRKY, NAC, and ethylene-responsive, auxin-responsive, light-inducible proteins, and fatty acyl-CoA reductase (FAR), ribulose bisphosphate carboxylase/oxygenase activase A, were identified as participating in chloroplast biosynthesis and movement, pigment biosynthesis, and stress response (S3 Table). The transcriptome analysis was consistent with the phenotypes of mta and WT. These results suggest that differential expression of genes might affect chloroplast development and chlorophyll biosynthesis, and thus contributed to the albino leaf phenotype of mta.
Table 4. DEGs and TFs involved in chloroplast development, and chlorophyll biosynthesis in mta transcriptome.
| Function | Gene or TF family | Expression level in mta | Annotation |
|---|---|---|---|
| Chloroplast development | PIF3 | Up-regulated | Transcription factor PIF3 |
| GLK | Down-regulated | Transcription factor GLK1 | |
| Chloroplast division | FtsZ | Down-regulated | Cell division protein, chloroplastic |
|
Chlorophyll biosynthesis |
HEMA1 | Down-regulated | Glutamyl-tRNA reductase 1, chloroplastic |
| HEME1 | Up-regulated | Heme oxygenase 1, chloroplastic | |
| CHLD | Down-regulated | Magnesium-chelatase subunit ChlD, chloroplastic | |
| GUN4 | Down-regulated | Tetrapyrrole-binding protein, chloroplastic | |
| CHLI1 | Down-regulated | Magnesium-chelatase subunit ChlI, chloroplastic | |
| CRD1 | Down-regulated | Magnesium-protoporphyrin IX monomethyl ester | |
| PORA | Down-regulated | Protochlorophyllide reductase A, chloroplastic | |
| PORB | Down-regulated | Protochlorophyllide reductase B, chloroplastic | |
| Pigment biosynthesis | ARR2 | Up-regulated | Two-component response regulator ARR2 |
| CHS | Up-regulated | Chalcone synthase | |
| DFR | Up-regulated | Dihydroflavonol-4-reductase | |
| Transcription factors | MYB | Down-regulated | Myb-related protein |
| bHLH | Down-regulated | Transcription factor bHLH | |
| MATE | Up-regulated | MATE efflux family protein 5 | |
| WRKY | Up-regulated | WRKY transcription factor | |
| NAC | Up-regulated | NAC transcription factor | |
| IAA | Down-regulated | Auxin-responsive protein |
Proteomic analysis
Chloroplast protein preparation and 2D-DIGE analysis
Protein sample preparation is a critical step in proteomic analysis. Chloroplast organelles were isolated and subsequently observed by a fluorescence microscope. Consistent with the chloroplast ultrastructure observation (Fig 2), chloroplasts in WT had a spindle shape, and the chloroplasts in mta were inflated and fewer in number (S5 Fig). The Bradford method was used to quantify analysis whole leaves chloroplast proteins. 2D-DIGE was used to identify DEPs. Coomassie blue G-250 stained proteins were directly matched to CyeDye images of analytical gels (S6 and S7 Figs). Approximately 1,645 spots were selected and 100 differential spots were excised for MS analysis. A total of 48 proteins including 33 down-regulated and 15 up-regulated in mta, were identified from chloroplast DEP spots. The size of the chloroplast proteins ranged from 14.4 to 94 kDa, and the isoelectric points (pI) of most of the proteins ranged from 5.08 to 9.4. Proteins with adjusted fold change (average volume ratio) values > 0 were up regulation, otherwise down regulation in mta (S4 Table).
DEP identification and functional classification
The normalized spot volume for each spot was calculated relative to the internal standard using DeCyder software and reflects protein abundance. Protein identification by MALDI-TOF/TOF MS revealed that a number of DEPs were found in multiple spots. And the 48 DEPs were divided into eleven categories based on functional groups. These categories mainly included photosynthesis, antioxidant/ hydrogen peroxide enzyme, photosynthesis-antenna proteins, component of chloroplast ribosome, chloroplast movement, stress response, oxidation-reduction reaction, photorespiration, protein phosphatase, phospholipid metabolism and other unknown function (Table 5). Most of the DEPs are nuclear-encoded and imported into the chloroplast and thylakoid membrane. These results indicate that the nucleus regulates essential aspects of albino leaf color formation in mta.
Table 5. Summary of chloroplast proteins and functional annotations.
| Functional group | Protein function | Protein name | Coding genome | Subcellular localization |
|---|---|---|---|---|
| Ⅰ | Photosynthesis/photosystemⅠ/ATP synthase | ATPB, ATPE,ATPA | Chloroplast | T |
| Photosynthesis/photosystemⅠ | PsaC | Chloroplast | T | |
| Photosynthesis/photosystemⅡ | Lhca1, PsbP, PsbO, HCF136, CYP38, PPL2 | Nuclear | T | |
| Photosynthesis/cytochrome b6f complex | PETC, RISP | Nuclear | T | |
| Photosynthesis/NADPH dehydrogenase complex | NDF4, NDF1, CYP20-2 | Nuclear | T | |
| Photosynthesis/Calvin-Benson cycle | GGT1, RBCS1A, RCA | Nuclear | C | |
| Photosynthesis/Calvin-Benson cycle | RBCL | Chloroplast | C | |
| Ⅱ | Antioxidant/Hydrogen peroxide enzyme | CAT2 | Nuclear | C |
| Antioxidant/Dimethylglycine dehydrogenase | GLDP2 | Nuclear | C | |
| Ⅲ | Cation channels | VDAC | Nuclear | C |
| Ⅳ | Embryonic development | EMB140 | Nuclear | C |
| Ⅴ | Component of chloroplast ribosome | RPL12 | Nuclear | C |
| Chloroplast movement | CHUP1 | Nuclear | C | |
| Ⅵ | Stress response | NDPK1 | Nuclear | C |
| Ⅶ | Oxidation-reduction reaction | MOA2.2 | Nuclear | C |
| Ⅷ | Photorespiration | CI76 | Nuclear | C |
| Ⅸ | Protein phosphatase | PAP27 | Nuclear | C |
| Ⅹ | Phospholipid metabolism | ACBP | Nuclear | Unknown |
| Ⅺ | Unknown | F13M7.10,T7F6.22, T21B4 |
Nuclear | Unknown |
T: Thylakoid membrane; C: Chloroplast.
Transcriptome and proteome data mining
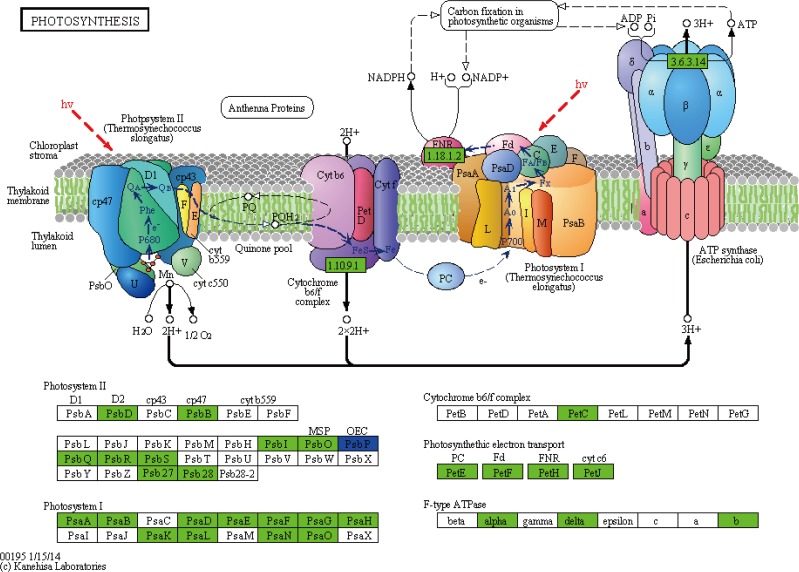
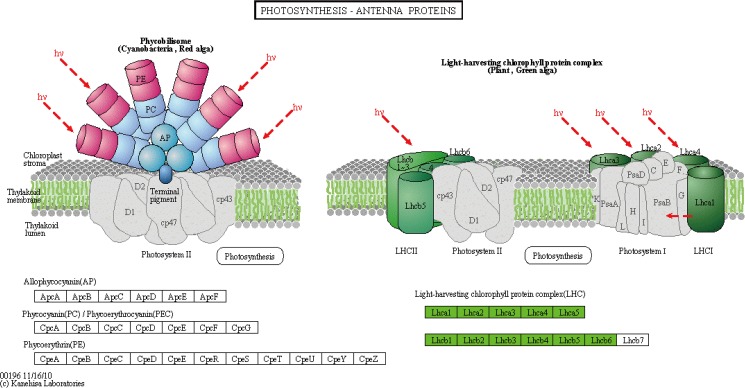
Integrative analysis of the transcriptome and the chloroplast proteome data provided an important tool for verifying the expression of key genes in mta. The distribution of DEGs and DEPs among species was counted based on NCBInr (S8 Fig). The close matches were from Triticum aestivum, Aegilliops tauschii, Triticum urartu, Hordeum vulgare, Oryza sativa, Zea mays, and Arabidopsis thaliana. To reveal the correlation between DEGs and DEPs precisely, the genes and proteins coded by them were combined analyzed. The photosynthesis pathway contained 78 DEGs, 24 genes of them were in photosystem I (PSI), 28 genes were in PSII, two genes were in the cytochrome b6/f complex, 18 genes were in the photosynthetic electron transport (Fig 5), and six genes were in the light-harvesting chlorophyll protein complex (LHC) (Fig 6). The DEPs were also functioned in those items, including one in PSI, six in PSII, two in the cytochrome b6/f complex, three in the F-type ATPase, four in LHC, and three in NADH dehydrogenase complex. The expression levels of all these photosynthetic proteins were lower in mta compared to WT. The result was in accordance with the DEGs expression level (Figs 5 and 6, S4 Table). The genes and corresponding proteins participated in photosynthesis may be associated with the leaf color variation in mta.
Fig 5. Differentially expressed genes and proteins mapped to photosynthesis pathway.
The known pathways were obtained from KEGG database. Green color denotes lower expression in mta compared with WT, while red color denotes higher expression. Blue color denotes both up- and down-regulated genes in mta compared to WT.
Fig 6. Differentially expressed genes and proteins mapped to photosynthesis-antenna proteins pathway.
Green color denotes lower expression in mta compared with WT.
Despite those, proteins related to chloroplast, stress response, and other metabolic pathways were identified (Table 5). The chloroplast protein-coding genes were located in the nucleus and were functioned as involving in the same pathways. Generally, the relationship between DEGs and chloroplast DEPs expression level was highly consistent with the transcriptome and proteomic analyses. These data further demonstrate that the leaf color formation of spaceflight-induced mutant mta is likely to be caused by gene mutation or epigenetic modification, which directly regulate chloroplast development and photosynthesis.
Verification of selected DEPs and DEGs
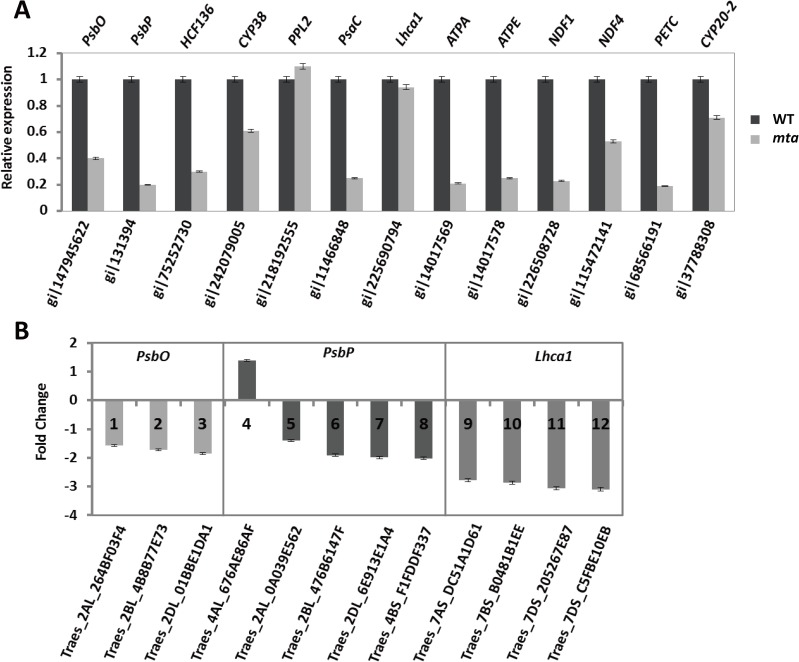
To validate the transcriptome and proteomic analyses, qRT-PCR was performed for a few key photosynthetic genes selected based on DEP expressed sequence tags (EST) and DEGs. Only one proteomic PPL2 expression levels slight varied compared with the transcript levels determined from qRT-PCR analysis. The down-regulation pattern of 12 chloroplast DEPs in mta, including PsbO, PsbP, HCF136, CYP38, Lhca, PsaC, ATPA, ATPE, NDF1, NDF4, PETC and CYP20-2 was consistent with the corresponding transcripts based on qRT-PCR (Fig 7A, S5 Table). The fold-change values of the 12 photosynthesis-related DEGs, including PsbO, PsbP and Lhca1 were largely keeping the same expression levels in mta (Fig 7B). The verification results demonstrated that the photosynthetic genes showed similar expression patterns between DEPs and DEGs from omics analysis.
Fig 7. Quantitative real-time PCR analysis of 13 DEP-coding genes.
The relative gene expression levels of selected target genes were normalized to a BestKeeper composed of the endogenous reference gene TaActin. (A) The black and gray bars represent WT and mta, respectively. Low values indicate down-regulation in mta, while high values indicate up-regulation. (B) The different colors represent 12 DEGs, including PsbO (1–3), PsbP (4–8), Lhca1 (9–12). Positive values indicate higher expression in mta and negative values denote higher expression in WT. Error bars represent the standard deviation.
Discussion
During spaceflight, various factors including cosmic irradiation, microgravity and space magnetic fields possibly interact to produce a unique environment that imposes both mutagenic and non-mutagenic stress on plant seeds. Thus, spaceflight has been an effective method for inducing a mutation [42]. The albino-lethal mutant mta used in this study and other leaf color mutants were obtained from spaceflight mutagenic induction. We speculated all changes in mta were caused by gene mutation or genetic modification. However, the molecular mechanisms leading to leaf color variation exposed to spaceflight in hexaploid wheat chlorophyll deficiency mutants are still not-well known.
The chloroplasts development and photosynthetic pigments are the primary factors affecting leaf color formation in higher plants. To date, leaf color mutants and genes regulating chloroplast development and Chl biosynthesis have been reported in many plant species including Arabidopsis, rice, maize and tomato [43–46]. Mutants with dysfunctional chloroplasts usually have leaves that lose their green color. In addition, abnormal plastid number and development may lead to impaired formation of the thylakoid membranes and reduced accumulation of Chl a/ b- binding proteins of the light-harvesting complexes I and II [46]. Thus, changes in leaf color could reflect the abnormal development and function of the plastid. In this study, the structure and quantity of chloroplasts in WT and mta were observed using TEM and fluorescence microscopy, respectively. Our results strongly suggest a relationship between chloroplast morphology and leaf color. The number and shape and inner structure of chloroplasts in mta were distinct from those of WT (Fig 2 and S5 Fig). Furthermore, the Chl and carotenoid content in mta were significantly lower than that in WT. Together, our results show that leaf color variation was directly determined by chloroplast number and development, and Chl biosynthesis. In addition, low temperature stress made a temporary influence on leaf color variation. This might be caused by the effects on the construction of chloroplast thylakoid membranes and changes in carotenoid/Chl content.
Integrating transcriptome and proteome profiling data is generally believed to provide a better understanding of the differences in gene regulation and complex biological processes between wild type and variations [47]. As transcriptome analyses provide information about quantitative changes in gene expression, these data can be used to obtain fundamental insights into specific pathways and genes associated with certain species [13]. In addition, proteomic analysis using approaches such as 2D-DIGE and MALDI-TOF/TOF are helpful in identifying the differences and categories of abundant proteins [26].
Here, we cataloged differences in mRNA and protein abundances that might be associated with leaf color formation in WT and mta through integrated profiling of gene expression using RNA-Seq and chloroplast proteomic analyses. We obtained 4,588 DEGs and 48 chloroplast DEPs. Many DEGs annotations were related to chloroplast, chloroplast envelope, thylakoid membrane and stroma. These genes were enriched in KEGG pathways, such as “photosynthesis”, “photosynthesis-antenna proteins” and “porphyrin and chlorophyll metabolism” (Table 3). Meanwhile, most of chloroplast DEPs was involved in photosynthesis, followed by component of chloroplast activity and other metabolic pathways (Table 5). As in photosynthetic pathway, the DEGs and DEPs were significantly involved in PSI, PSII, cytochrome b6/f complex, F-type ATPase, NADH dehydrogenase complex and LHC. The DEGs and corresponding DEPs showed the same expression level (Figs 5 and 6). These results were consistent with our original prediction that leaf color formation is greatly affected by chloroplast development, and photosynthesis. Importantly, this is the first study to provide transcriptome information for a leaf color mutant in hexaploid wheat. Our mta chloroplast proteome results are consistent with those of a previously reported wheat chloroplastic proteome [27]. Thus, the unigene from omics data will aid researchers in identifying the specific genes relevant to leaf color formation in Triticum aestivum L.
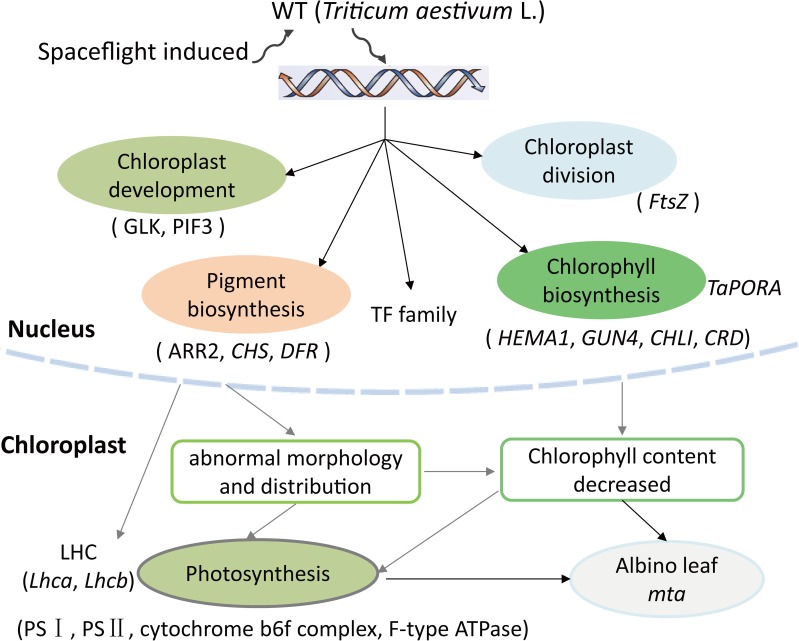
Previous studies have shown that chloroplast development is a complex and highly regulated process that includes light signaling during photo-morphogenesis, the transition from proplastids to chloroplasts, import of nuclear-encoded chloroplast proteins, thylakoid biosynthesis, chloroplast division, and retrograde signaling from the chloroplast to the nucleus [7, 8, 48]. PIF3 and GLK had been identified as important TFs regulating chloroplast development. PIF3 is a repressor that negatively regulates chloroplast development in Arabidopsis [18]. PIF3 mediates the initial phases of light-induced chloroplast development through regulation of a subset of rapidly photoresponsive nuclear genes encoding plastid and photosynthesis-related components [19]. GLK is required for the expression of nuclear photosynthetic genes and chloroplast development in diverse chlorophyll deficiency plant species, including maize, rice, and Arabidopsis [2, 20–22]. FtsZ is a key structural component of the chloroplast division machinery in perhaps all photosynthetic eukaryotes [23]. The mechanism of Chl biosynthesis and most the genes involved have been identified in Arabidopsis [10]. In rice, OsPORB is essential for maintaining light-dependent Chl synthesis throughout leaf development, whereas OsPORA mainly functions in the early stages of leaf development [17]. Despite those, many other transcription factors may be potentially regulated the leaf color formation or plant growth and development. For example, auxin-responsive factor acts indirectly to regulate chloroplast movements [49]. MYB family is responsible for the biosynthesis of phenylpropanoids, including anthocyanin and phlobaphene pigments [50]. Consistent with what is known about these genes and TF family, in our study, we found several genes and TFs regulating chloroplast development and division, Chl biosynthesis, and pigment biosynthesis (Table 4 and S2 Table). The TF family contained GLK, repressor PIF3, FtsZ, ARR2 and other transcription factors, such as MYB, bHLH, WRKY, NAC, and auxin-responsive factor. The related genes included HEMA1, GUN4, PORA, PORB, CHS, and DFR. The expression level and functional annotations of all these genes provide support for our hypothesis that leaf color variation is determined by combined regulation of multiple genes. We also suppose that TaGLK, TaPIF3, and TaFtsZ may regulate chloroplast development and division, while TaHEMA, TaCHLD, TaPORB, and TaPORA participate in chlorophyll biosynthesis in wheat. Changes in these genes expression may affect photosynthesis and all together lead to the albino phenotype in mta (Fig 8).
Fig 8. Possible leaf color formation pathways of albino mutant mta in wheat (Triticum aestivum L.).
To find the cause of decreased chlorophyll content in mta, we chose one gene TaPORA and detected the promoter methylation level. We found TaPORA promoter methylation level was much higher in mta compared to WT (S4 Fig). Previous studies have shown that plant genomes are characterized by a relatively high degree of nuclear DNA methylation [51], and methylation in promoters is responsible for low gene expression or silencing [52, 53]. In addition, other research indicates that spaceflight induces both transient and heritable alterations in DNA methylation and gene expression in rice [54]. We speculate that spaceflight environment might induce heritable alteration in TaPORA promoter methylation in mta. And the promoter methylation might play important role in the TaPORA lower expression. However, we found many SNPs existed between WT and mta. These SNPs uniformly distributed on the 21 wheat chromosomes within gene or Intergenic regions (S9 Fig). We also surmise that gene mutation in the nucleus is the main factor controlling the expression of albino trait in mta, which should be investigated in the future study.
Conclusions
In conclusion, the chloroplast structure and photosynthetic pigments content in the spaceflight-induced mutant mta were significantly different from those in WT H6172 (Triticum aestivum L.). Transcriptome and chloroplast proteomic analyses of WT and mta revealed that DEGs and DEPs were mainly involved in chloroplast activities, and photosynthesis pathways. In addition, these DEGs and corresponding DEPs showed similar expression pattern. In addition, genes participated in chloroplast development and division, chlorophyll and pigment biosynthesis were identified from albino mutant mta and WT transcriptome database. qRT-PCR verified that those photosynthetic DEGs and DEPs were differentially expressed in mta. Based on these results, we speculate that changes in chloroplast development, Chl biosynthesis, and the protein levels of photosynthetic proteins all contribute to the differences in leaf color formation in the mta (Fig 8). We also show that spaceflight might be used of mutagenesis in crop breeding.
Supporting information
(PDF)
(A) wild type samples (WT_R1 and WT_R2) and (B) albino mutant samples (mta_R1 and mta_R2).
(PDF)
(PDF)
(A) CpG island distribution in the TaPORA promoter methylation, BSP primer design regions and sequencing data. (B) TaPORA promoter methylation level. (C) TaPORA expression determined level by qRT-PCR analysis.
(PDF)
(a, c) Chloroplasts visualized under white light, (b, d) chloroplast visualized under a fluorescence from High Pressure Mercury (HPM) lamp. (1) large quantity of chloroplasts; (2) broken chloroplasts; (3) the size of intact chloroplasts in mta is bigger than in WT. Magnification bar = 20 μm.
(PDF)
(PDF)
(a, b, c) Three replicated experiments of the internal standard; (d, e, f) three replicated experiments of WT chloroplast proteins; (g, h, i) three replicated experiments of mta chloroplast proteins. The blue, red and green gel images were labeled with fluorescent Cy2, Cy5 and Cy3 dyes, respectively.
(PDF)
(PDF)
Blue color represents number of SNPs, and red color represents number of genes with SNP.
(PDF)
Q30 (base content of clean reads with a quality no less than 30).
(XLSX)
(XLSX)
(XLSX)
a Spot no are given in S6 Fig. b Fold change: average volume ratio (mta/ WT). c Mascot score: more than 20 is significant (P≤0.05) from MS data. d SC: sequence coverage. e Molecular weights (MW, kDa) and isoelectric point (pI) of spots from the gel.
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We would like to thank all the members who were involved in the experiment in this study. The authors deeply appreciate all the technicians for their assistance in experiments. We particularly thank the reviewers for giving us the constructive suggestions about the manuscript.
Data Availability
All sequencing reads from the four libraries have been submitted to the Sequence Read Archive (SRA), National Center for Biotechnology Information (NCBI) with an accession of SRP106763 (BioProject ID of PRJNA386075; Four run ID of SRR5520556, SRR5527113, SRR5527114 and SRR5527116 represent mta_R1, mta_R2, WT_R1 and WT_R2 sample reads, respectively).
Funding Statement
This work was supported by the National Key Research and Development Program of China: grant no. 2016YFD0102101, the National 973 Program: grant no. 2014CB138101, and Core Research Budget of the Non-profit Governmental Research Institutions (ICS, CAAS).
References
- 1.Reyes-Prieto A, Weber APM, Bhattacharya D. The Origin and Establishment of the Plastid in Algae and Plants. Annual Review of Genetics. 2007;41(1):147–68. [DOI] [PubMed] [Google Scholar]
- 2.Waters MT, Moylan EC, Langdale JA. GLK transcription factors regulate chloroplast development in a cell-autonomous manner. Plant J. 2008;56(3):432–44. doi: 10.1111/j.1365-313X.2008.03616.x [DOI] [PubMed] [Google Scholar]
- 3.Chan KX, Phua SY, Crisp P, McQuinn R, Pogson BJ. Learning the Languages of the Chloroplast: Retrograde Signaling and Beyond. Annual review of plant biology. 2016;67:25–53. doi: 10.1146/annurev-arplant-043015-111854 [DOI] [PubMed] [Google Scholar]
- 4.Chiang YH, Zubo YO, Tapken W, Kim HJ, Lavanway AM, Howard L, et al. Functional characterization of the GATA transcription factors GNC and CGA1 reveals their key role in chloroplast development, growth, and division in Arabidopsis. Plant physiology. 2012;160(1):332–48. PubMed Central PMCID: PMCPMC3440210. doi: 10.1104/pp.112.198705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luna-Valdez LAd, Martinez-Batallar AG, Hernandez-Ortiz M, Encarnacion-Guevara S, Ramos-Vega M, Lopez-Bucio JS, et al. Proteomic analysis of chloroplast biogenesis (clb) mutants uncovers novel proteins potentially involved in the development of Arabidopsis thaliana chloroplasts. J Proteomics. 2014;111:148–64. doi: 10.1016/j.jprot.2014.07.003 [DOI] [PubMed] [Google Scholar]
- 6.Pogson BJ, Ganguly D, Albrecht-Borth V. Insights into chloroplast biogenesis and development. Biochimica et biophysica acta. 2015;1847(9):1017–24. doi: 10.1016/j.bbabio.2015.02.003 [DOI] [PubMed] [Google Scholar]
- 7.Avendaño-Vázquez A-O, Cordoba E, Llamas E., San Román C, Nisar N., De la Torre S, … León P. An Uncharacterized Apocarotenoid-Derived Signal Generated in ζ-Carotene Desaturase Mutants Regulates Leaf Development and the Expression of Chloroplast and Nuclear Genes in Arabidopsis. The Plant Cell. 2014;26 (6):2524–37. doi: 10.1105/tpc.114.123349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pogson BJ, Woo NS, Forster B, Small ID. Plastid signalling to the nucleus and beyond. Trends in plant science. 2008;13(11):602–9. doi: 10.1016/j.tplants.2008.08.008 [DOI] [PubMed] [Google Scholar]
- 9.Sandhu D, Atkinson T, Noll A, Johnson C, Espinosa K, Boelter J, et al. Soybean proteins GmTic110 and GmPsbP are crucial for chloroplastdevelopment and function. Plant Science. 2016;252:76–87. doi: 10.1016/j.plantsci.2016.07.006 [DOI] [PubMed] [Google Scholar]
- 10.Beale SI. Green genes gleaned. Trends in plant science. 2005;10(7):309–12. doi: 10.1016/j.tplants.2005.05.005 [DOI] [PubMed] [Google Scholar]
- 11.Masuda T, Fujita Y. Regulation and evolution of chlorophyll metabolism. Photochemical & Photobiological Sciences. 2008;7(10):1131. [DOI] [PubMed] [Google Scholar]
- 12.Ma C, Cao J, Li J, Zhou B, Tang J, Miao A. Phenotypic, histological and proteomic analyses reveal multiple differences associated with chloroplast development in yellow and variegated variants from Camellia sinensis. Scientific reports. 2016;6:33369 PubMed Central PMCID: PMC5025893. doi: 10.1038/srep33369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Y, Chen X, Xu B, Li Y, Ma Y, Wang G. Phenotype and transcriptome analysis reveals chloroplast development and pigment biosynthesis together influenced the leaf color formation in mutants of Anthurium andraeanum 'Sonate'. Frontiers in plant science. 2015;6:139 PubMed Central PMCID: PMCPMC4356079. doi: 10.3389/fpls.2015.00139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo H-j, Zhao H-b, Zhao L-s, Gu J-y, Zhao S-r, Li J-h, et al. Characterization of a Novel Chlorophyll-Deficient Mutant Mt6172 in Wheat. Journal of Integrative Agriculture. 2012;11(6):888–97. [Google Scholar]
- 15.Sanchez C, Baranda AB, Martinez de Maranon I. The effect of High Pressure and High Temperature processing on carotenoids and chlorophylls content in some vegetables. Food Chem. 2014;163:37–45. doi: 10.1016/j.foodchem.2014.04.041 [DOI] [PubMed] [Google Scholar]
- 16.Deng XJ, Zhang HQ, Wang Y, He F, Liu JL, Xiao X, et al. Mapped clone and functional analysis of leaf-color gene Ygl7 in a rice hybrid (Oryza sativa L. ssp. indica). PloS one. 2014;9(6):e99564 PubMed Central PMCID: PMC4059691. doi: 10.1371/journal.pone.0099564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakuraba Y, Rahman ML, Cho S-H, Kim Y-S, Koh H-J, Yoo S-C, et al. The ricefaded green leaflocus encodes protochlorophyllide oxidoreductase B and is essential for chlorophyll synthesis under high light conditions. The Plant Journal. 2013;74(1):122–33. doi: 10.1111/tpj.12110 [DOI] [PubMed] [Google Scholar]
- 18.Stephenson PG, Fankhauser C, Terry MJ. PIF3 is a repressor of chloroplast development. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(18):7654–9. PubMed Central PMCID: PMCPMC2678601. doi: 10.1073/pnas.0811684106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monte E, Tepperman JM, Al-Sady B, Kaczorowski KA, Alonso JM, Ecker JR, et al. The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(46):16091–8. PubMed Central PMCID: PMCPMC528976. doi: 10.1073/pnas.0407107101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fitter DW, Martin DJ, Copley MJ, Scotland RW, Langdale JA. GLK gene pairs regulate chloroplast development in diverse plant species. The Plant Journal. 2002;31(6):713–27. [DOI] [PubMed] [Google Scholar]
- 21.Rossini L, Cribb L, Martin DJ, Langdale JA. The Maize Golden2 Gene Defines a Novel Class of Transcriptional Regulators in Plants. The Plant Cell. 2001;13:1231–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yasumura Y, Moylan EC, Langdale JA. A conserved transcription factor mediates nuclear control of organelle biogenesis in anciently diverged land plants. Plant Cell. 2005;17(7):1894–907. PubMed Central PMCID: PMCPMC1167540. doi: 10.1105/tpc.105.033191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osteryoung KW, Nunnari J. The Division of Endosymbiotic Organelles. Science. 2003;302:1698–704. doi: 10.1126/science.1082192 [DOI] [PubMed] [Google Scholar]
- 24.Maple J, Moller SG. Plastid Division: Evolution, Mechanism and Complexity. Annals of Botany. 2006;99(4):565–79. doi: 10.1093/aob/mcl249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gabruk M, Stecka A, Strzalka W, Kruk J, Strzalka K, Mysliwa-Kurdziel B. Photoactive protochlorophyllide-enzyme complexes reconstituted with PORA, PORB and PORC proteins of A. thaliana: fluorescence and catalytic properties. PloS one. 2015;10(2):e0116990 PubMed Central PMCID: PMC4319759. doi: 10.1371/journal.pone.0116990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hellinger R, Koehbach J, Soltis DE, Carpenter EJ, Wong GK, Gruber CW. Peptidomics of Circular Cysteine-Rich Plant Peptides: Analysis of the Diversity of Cyclotides from Viola tricolor by Transcriptome and Proteome Mining. J Proteome Res. 2015;14(11):4851–62. PubMed Central PMCID: PMCPMC4642221. doi: 10.1021/acs.jproteome.5b00681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamal AH, Cho K, Choi JS, Bae KH, Komatsu S, Uozumi N, et al. The wheat chloroplastic proteome. J Proteomics. 2013;93:326–42. doi: 10.1016/j.jprot.2013.03.009 [DOI] [PubMed] [Google Scholar]
- 28.Bao W, Neng L. Determination methods for photosynthetic pigment content of Bryophytew ITH special relation of extracting solvents. China J Appl Environ Biol. 2005;11(2):235–7. [Google Scholar]
- 29.Arnon DI. Copper enzymes in isolated chloroplasts, polyphenol oxidase in Beta vulgris. Plant physiology. 1949;24:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnology. 2011;29(7):644–52. doi: 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323 PubMed Central PMCID: PMCPMC3163565. doi: 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26(1):136–8. doi: 10.1093/bioinformatics/btp612 [DOI] [PubMed] [Google Scholar]
- 33.Jung S-H. Stratified Fisher's exact test and its sample size calculation. Biometrical Journal. 2014;56(1):129–40. doi: 10.1002/bimj.201300048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan P, Wang X, Kuang T, Li Y. An efficient method for the extraction of chloroplast proteins compatible for 2-DE and MS analysis. Electrophoresis. 2009;30(17):3024–33. doi: 10.1002/elps.200900172 [DOI] [PubMed] [Google Scholar]
- 35.Zienkiewicz A, Rejon JD, de Dios Alche J, Rodriguez-Garcia MI, Castro AJ. A protocol for protein extraction from lipid-rich plant tissues suitable for electrophoresis. Methods in molecular biology. 2014;1072:85–91. doi: 10.1007/978-1-62703-631-3_7 [DOI] [PubMed] [Google Scholar]
- 36.Hou DY, Du GY, Lin JT, Duan M, Guo AG. Proteome analysis of chloroplast proteins in stage albinism line of winter wheat (triticum aestivum) FA85. BMB reports. 2009. [DOI] [PubMed] [Google Scholar]
- 37.Andrew Alban SOD, Bjorkesten Lennart, Andersson Christian, Sloge Erik, Lewis Steve, Currie Ian. A novel experimental design for comparative two-dimensional gel analysis: Two-dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics. 2003;3:36–44. doi: 10.1002/pmic.200390006 [DOI] [PubMed] [Google Scholar]
- 38.Wang X, Li X, Deng X, Han H, Shi W, Li Y. A protein extraction method compatible with proteomic analysis for the euhalophyte Salicornia europaea. Electrophoresis. 2007;28(21):3976–87. doi: 10.1002/elps.200600805 [DOI] [PubMed] [Google Scholar]
- 39.Gu J, Wang Q, Cui M, Han B, Guo H, Zhao L, et al. Cloning and characterization of Ku70 and Ku80 homologues involved in DNA repair process in wheat (Triticum aestivum L.). Plant Genetic Resources. 2014;12(S1):S99–S103. [Google Scholar]
- 40.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001;29(900). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, et al. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 2006;34(Web Server issue):W293–7. PubMed Central PMCID: PMCPMC1538768. doi: 10.1093/nar/gkl031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang J, Shen S, Zhang T, Chen GD, Liu H, Ma XB, et al. Morphological variation of mutant sunflowers (Helianthus annuus) induced by space flight and their genetic background detection by SSR primers. Genet Mol Res. 2012;11(3):3379–88. doi: 10.4238/2012.September.25.6 [DOI] [PubMed] [Google Scholar]
- 43.Lin YP, Lee TY, Tanaka A, Charng YY. Analysis of an Arabidopsis heat-sensitive mutant reveals that chlorophyll synthase is involved in reutilization of chlorophyllide during chlorophyll turnover. Plant J. 2014;80(1):14–26. doi: 10.1111/tpj.12611 [DOI] [PubMed] [Google Scholar]
- 44.Zhou K, Ren Y, Lv J, Wang Y, Liu F, Zhou F, et al. Young Leaf Chlorosis 1, a chloroplast-localized gene required for chlorophyll and lutein accumulation during early leaf development in rice. Planta. 2013;237:279–92. doi: 10.1007/s00425-012-1756-1 [DOI] [PubMed] [Google Scholar]
- 45.Shi D, Zheng X, Li L, Lin W, Xie W, Yang J, et al. Chlorophyll deficiency in the maize elongated mesocotyl2 mutant is caused by a defective heme oxygenase and delaying grana stacking. PloS one. 2013;8(11):e80107 PubMed Central PMCID: PMCPMC3823864. doi: 10.1371/journal.pone.0080107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barry CS, Aldridge GM, Herzog G, Ma Q, McQuinn RP, Hirschberg J, et al. Altered chloroplast development and delayed fruit ripening caused by mutations in a zinc metalloprotease at the lutescent2 locus of tomato. Plant physiology. 2012;159(3):1086–98. PubMed Central PMCID: PMCPMC3387696. doi: 10.1104/pp.112.197483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumar D, Bansal G, Narang A, Basak T, Abbas T, Dash D. Integrating transcriptome and proteome profiling: Strategies and applications. Proteomics. 2016;16(19):2533–44. doi: 10.1002/pmic.201600140 [DOI] [PubMed] [Google Scholar]
- 48.Waters MT, Langdale JA. The making of a chloroplast. The EMBO Journal. 2009;28(19):2861–73. doi: 10.1038/emboj.2009.264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eckstein A, Krzeszowiec W, Waligorski P, Gabrys H. Auxin and chloroplast movements. Physiol Plant. 2016;156(3):351–66. doi: 10.1111/ppl.12396 [DOI] [PubMed] [Google Scholar]
- 50.Uimari A, Strommer J. Myb26: a MYB-like protein of pea flowers with affinity for promoters of phenylpropanoid genes. The Plant Journal. 1997;12(6):1273–84. [DOI] [PubMed] [Google Scholar]
- 51.Vanyushin BF, Ashapkin VV. DNA methylation in higher plants: past, present and future. Biochimica et biophysica acta. 2011;1809(8):360–8. doi: 10.1016/j.bbagrm.2011.04.006 [DOI] [PubMed] [Google Scholar]
- 52.Vincent A, Omura N, Hong SM, Jaffe A, Eshleman J, Goggins M. Genome-wide analysis of promoter methylation associated with gene expression profile in pancreatic adenocarcinoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2011;17(13):4341–54. PubMed Central PMCID: PMC3131423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao Y, Sun J, Zhang H, Guo S, Gu J, Wang W, et al. High-frequency aberrantly methylated targets in pancreatic adenocarcinoma identified via global DNA methylation analysis using methylCap-seq. Clin Epigenetics. 2014;6(1):18 PubMed Central PMCID: PMCPMC4177372. doi: 10.1186/1868-7083-6-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ou X, Long L, Zhang Y, Xue Y, Liu J, Lin X, et al. Spaceflight induces both transient and heritable alterations in DNA methylation and gene expression in rice (Oryza sativa L.). Mutation research. 2009;662(1–2):44–53. doi: 10.1016/j.mrfmmm.2008.12.004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(A) wild type samples (WT_R1 and WT_R2) and (B) albino mutant samples (mta_R1 and mta_R2).
(PDF)
(PDF)
(A) CpG island distribution in the TaPORA promoter methylation, BSP primer design regions and sequencing data. (B) TaPORA promoter methylation level. (C) TaPORA expression determined level by qRT-PCR analysis.
(PDF)
(a, c) Chloroplasts visualized under white light, (b, d) chloroplast visualized under a fluorescence from High Pressure Mercury (HPM) lamp. (1) large quantity of chloroplasts; (2) broken chloroplasts; (3) the size of intact chloroplasts in mta is bigger than in WT. Magnification bar = 20 μm.
(PDF)
(PDF)
(a, b, c) Three replicated experiments of the internal standard; (d, e, f) three replicated experiments of WT chloroplast proteins; (g, h, i) three replicated experiments of mta chloroplast proteins. The blue, red and green gel images were labeled with fluorescent Cy2, Cy5 and Cy3 dyes, respectively.
(PDF)
(PDF)
Blue color represents number of SNPs, and red color represents number of genes with SNP.
(PDF)
Q30 (base content of clean reads with a quality no less than 30).
(XLSX)
(XLSX)
(XLSX)
a Spot no are given in S6 Fig. b Fold change: average volume ratio (mta/ WT). c Mascot score: more than 20 is significant (P≤0.05) from MS data. d SC: sequence coverage. e Molecular weights (MW, kDa) and isoelectric point (pI) of spots from the gel.
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All sequencing reads from the four libraries have been submitted to the Sequence Read Archive (SRA), National Center for Biotechnology Information (NCBI) with an accession of SRP106763 (BioProject ID of PRJNA386075; Four run ID of SRR5520556, SRR5527113, SRR5527114 and SRR5527116 represent mta_R1, mta_R2, WT_R1 and WT_R2 sample reads, respectively).