Abstract
The potato psyllid, Bactericera cockerelli (Sulc) (Hemiptera: Triozidae), is a pest of potato and other solanaceous crops in North and Central America and New Zealand. Previous genotyping studies have demonstrated the presence of three different haplotypes of B. cockerelli in the United States corresponding to three geographical regions: Central, Western, and Northwestern. These studies utilized psyllids collected in the western and central United States between 1998 and 2011. In an effort to further genotype potato psyllids collected in the 2012 growing season, a fourth B. cockerelli haplotype was discovered corresponding to the Southwestern United States geographical region. High-resolution melting analyses identified this new haplotype using an amplicon generated from a portion of the B. cockerelli mitochondrial cytochrome coxidase subunit I gene. Sequencing of this gene, as well as use of a restriction enzyme assay, confirmed the identification of the novel B. cockerelli haplotype in the United States.
Keywords: potato disease, zebra chip, liberibacter, psyllid haplotype
The potato psyllid, Bactericera cockerelli (Sulc) (Hemiptera: Triozidae), is an economically important pest of solanaceous crops such as potatoes, where it has been linked to psyllid yellows disease and zebra chip (ZC) disease. Studies of the psyllid yellows disease of potatoes suggest it is caused when the psyllid nymph releases a toxin upon feeding that triggers yellowing or purpling of the leaves and in severe cases can decrease tuber yield and size ( Wallis 1955 , Rondon et al. 2012 ). Of greater economic importance as of late, the potato psyllid has been linked to ZC, which has now been identified across the western and central United States, Mexico, Central America, and New Zealand ( Munyaneza 2012 , Munyaneza and Henne 2012 ). This disease is attributed to infection with the bacterium “ Candidatus Liberibacter solanacearum” (Lso) that is vectored by the potato psyllid and causes purpling, curling, and chlorosis of leaves, shortened internodes, swollen nodes, aerial tuber formation, and rapid plant decline ( Munyaneza et al. 2007 , Crosslin et al. 2010 , Munyaneza 2012 ). Tubers on potato plants affected by ZC exhibit dark patterns of necrosis and streaking of the medullary ray tissues, which are exacerbated upon processing into fries or chips, thereby rendering them unacceptable, resulting in significant economic loss ( Munyaneza et al. 2007 ; Crosslin et al. 2010 ; Munyaneza 2010 , 2012 ).
Population mapping studies have identified three distinct potato psyllid haplotypes within the United States that are associated with three different geographical regions: Central, Western, and Northwestern ( Liu et al. 2006 ; Swisher et al. 2012 , 2013 ). Psyllids of the Central haplotype have been identified across a large geographical range, spanning from eastern Mexico up to Texas, Kansas, Colorado, Nebraska, and Wyoming, all the way to North Dakota ( Swisher et al. 2012 , 2013 ). Likewise, psyllids of the Western haplotype were identified across a large geographical range from California and New Mexico up to Washington, Oregon, and Idaho ( Swisher et al. 2012 , 2013 ). To date, the Northwestern haplotype has only been identified in the Northwestern states of Washington, Oregon, and Idaho ( Swisher et al. 2012 , 2013 ; data not shown).
For psyllid population mapping, original studies using intersimple sequence repeat markers, internal transcribed spacer 2, and mitochondrial cytochromecoxidase I gene comparison identified the Western and Central psyllid populations ( Liu et al. 2006 ). Subsequently, studies have utilized the novel, high-resolution melting (HRM) analysis technique for comparative genotyping of a portion of the mitochondrial cytochrome c oxidase subunit I-like gene (CO1; Chapman et al. 2010 , 2012 ; Swisher et al. 2012 , 2013 ). In a 500-bp portion of the CO1 gene, only one single-nucleotide polymorphism (SNP) exists between the Western and Central haplotypes, whereas 16 and 17 SNPs exist between the Northwestern haplotype and the Central and Western psyllid haplotypes, respectively ( Swisher et al. 2012 ).
Previous studies have looked at psyllid genotypes from the year 2011 and before. This study began in an effort to genotype potato psyllids collected in the 2012 potato growing season in the western and central United States by HRM analysis, and surprisingly, identified a novel potato psyllid haplotype within the United States. To confirm this initial finding, additional individual psyllids were analyzed by HRM analysis and confirmed by DNA sequencing evidence. A restriction enzyme assay was developed as a tool to further discriminate this novel haplotype from the three previously defined haplotypes, which can be used to support DNA melting analyses.
Materials and Methods
Origin of Psyllids
All potato psyllids were collected as part of an on-going psyllid population study from research plots or commercial fields near Farmington, NM; Alamosa and Wray, CO; Scottsbluff, NE, and Garden City, KS, between May and August of the 2012 potato growing season. Psyllids were collected from yellow sticky traps placed throughout each field (see Goolsby et al. 2012 for collection, processing, and sorting details). Because these samples were part of an on-going population study to determine Lso incidence, no voucher samples are available. Control psyllids for the Western, Central, and Northwestern haplotypes originated from the initial 2011 psyllid dataset ( Swisher et al. 2012 ).
Nucleic Acid Extraction
All samples were processed in the same manner as previously described for HRM analysis ( Swisher et al. 2012 ). The cetyl trimethyl ammonium bromide (CTAB) extraction as described by Crosslin et al. (2006) was used to extract the total nucleic acids within 1–2 wk following initial collection and processing.
Primers, Polymerase Chain Reaction, and DNA Sequencing
Primers specific to the B. cockerelli mitochondrial cytochromecoxidase subunit I gene were used as previously described for both HRM and DNA sequencing analysis ( Swisher et al. 2012 , 2013 ). Species specificity has been previously determined for these primers ( Chapman et al. 2012 , Swisher et al. 2013 ). Briefly, primers CO1 F3 and CO1 meltR generate a 94-bp amplicon and were used in conjunction with primers CO1 meltF and CO1 meltR, which generate a 67-bp amplicon, for HRM analysis ( Chapman et al. 2010 , Swisher et al. 2013 ).
Primers CO1 F3 and CO1 R3 generate a 500-bp CO1 amplicon and were utilized for DNA sequencing and the SfcI restriction enzyme assay. Conventional polymerase chain reaction (PCR) was used as described by Crosslin et al. (2011) to generate the 500-bp amplicon, following which the PCR products were purified using the Wizard PCR Clean Up Kit (Promega, Madison, WI). Purified PCR products were sequenced directly using the CO1 F3 and CO1 R3 primers. For amino acid translation, the SIB ExPASy Bioinformatics Resources Portal was used with the genetic code setting for invertebrate mitochondria ( Artimo et al. 2012 ). The Clustal Omega (EMBL-EBI, Cambridgeshire, UK) sequence alignment tool was subsequently used to align the amino acid sequences.
HRM Analysis
All HRM analyses were performed as previously described by Swisher et al. (2013) . Briefly, the LightCycler 480 (Roche Applied Science, Indianapolis, IN) was used for real-time quantitative PCR (qPCR) and HRM analysis. For the primers CO1 F3/CO1 meltR and CO1 meltF/CO1 meltR, a touchdown qPCR program was utilized as done previously ( Swisher et al. 2013 ). Analysis of all melting results was done using the LightCycler 480 Gene Scanning Software (Roche Applied Science) as previously described ( Swisher et al. 2012 , 2013 ).
SfcI Digestion
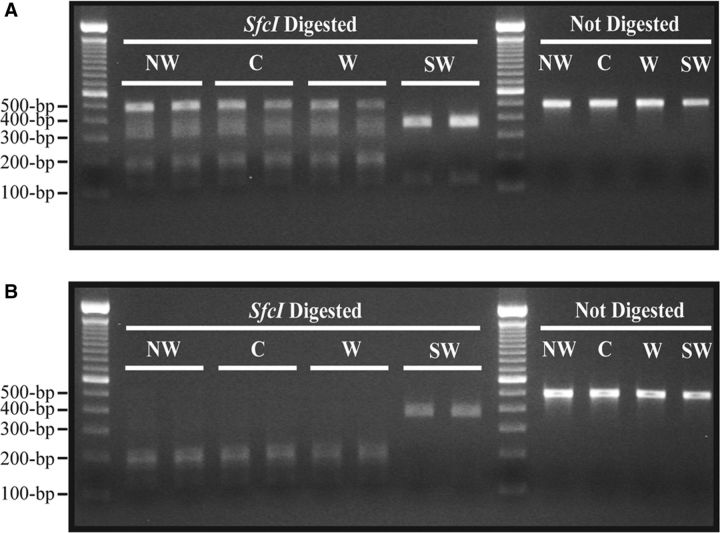
Sequencing results for the 500-bp CO1 amplicon identified a unique SfcI site in the Northwestern, Western, and Central haplotypes, enabling the Southwestern haplotype to be differentiated by a restriction enzyme assay. Purified 500-bp PCR products generated with the CO1 F3 and CO1 R3 primers were used for this assay. For partial digestion ( Fig. 4 A), a total volume of 30 µl was used, consisting of 15 µl of purified 500-bp CO1 PCR product, 3 µl of 10× NEBuffer 4 (New England BioLabs Inc., Ipswich, MA), 0.5 µl of 100× BSA (New England BioLabs Inc.), 0.5 µl of SfcI enzyme (New England BioLabs Inc.), and 11 µl of Ultra Pure H 2 O (Invitrogen, Carlsbad, CA). For complete digestion of the 500-bp CO1 amplicon ( Fig. 4 B), the amount of purified PCR product added to the 30 µl total volume was reduced to 5 µl. The ultrapure H 2 O (Invitrogen) volume was then raised to 21 µl. All samples were then incubated at 37°C for 1 h, followed by a 20-min SfcI heat inactivation step at 65°C. Digestion products were separated by electrophoresis on a 2.2% agarose gel and stained with ethidium bromide for visualization.
Fig. 4.
Digestion of CO1 with the restriction enzyme, SfcI . Partial (A) and complete (B) digestion of the 500-bp CO1 amplicon by the restriction enzyme, SfcI , was used to distinguish the Southwestern (SW) haplotype from the Central (C), Western (W), and Northwestern (NW) haplotypes based on digestion pattern. SfcI digestion at CTAC/TAG generates three fragments of 201-, 177-, and 122-bp for the Central, Western, and Northwestern haplotypes. In contrast, SfcI digestion generates only two fragments of 378- and 122-bp for the Southwestern haplotype; 500-bp undigested samples for each haplotype are loaded on the right side of the 2.2% agarose gel. Marker is a 100-bp ladder (Invitrogen).
Results
HRM Analysis
The melting behavior of the B. cockerelli mitochondrial cytochromecoxidase subunit I (CO1) gene was analyzed from individual potato psyllid samples collected during the 2012 potato growing season from the southwestern and central United States. Previous HRM analyses have utilized several CO1 amplicons of different sizes to differentiate between the three known psyllid haplotypes ( Chapman et al. 2012 ; Swisher et al. 2012 , 2013 ). Initial HRM analyses of psyllids collected in the southwestern United States during the 2012 growing season utilized a 353-bp CO1 amplicon. This amplicon generates double-peak melting profiles that are shifted to different temperatures for each of the three known psyllid haplotypes ( Swisher et al. 2013 ). However, the melting behavior of a subset of the southwestern samples gave ambiguous results, thereby preventing these samples from being clearly labeled as the Central, Western, or Northwestern haplotype (data not shown). As a result, both the 94- and 67-bp CO1 amplicons that were initially used on archived psyllid samples ( Swisher et al. 2013 ) were used to identify the haplotypes of the southwestern psyllid samples. The LightCycler Gene Scanning software (Roche Applied Science) was used to analyze the raw fluorescent data produced from HRM analysis of the 94- and 67-bp CO1 amplicons ( Fig. 1 ).
Fig. 1.
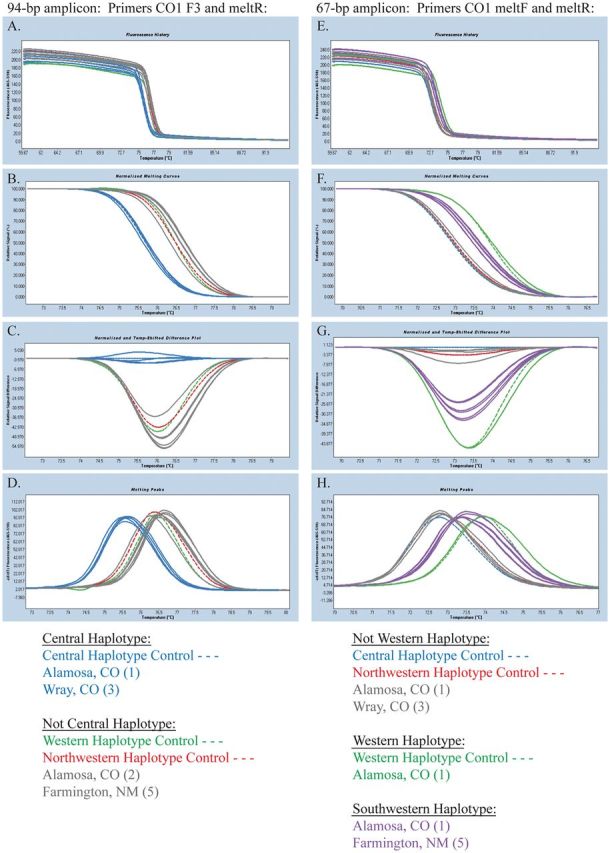
Genotypic analysis of 94-bp and 67-bp CO1 amplicons by HRM. HRM analysis of psyllids collected in New Mexico and Colorado during the 2012 potato growing season was conducted using the 94-bp CO1 amplicon (A–D) and the 67-bp CO1 amplicon (E–H). Individual psyllids of the known Central, Western, and Northwestern haplotypes were used as controls from the original 2011 psyllid dataset ( Swisher et al. 2012 , dashed blue, green, and red lines, respectively). (A and E) Original fluorescence for the specific amplicons obtained from the melting analysis. (B and F) Melt curves generated upon normalization of the raw fluorescent data. (C and G) Representation of the normalized data as differences in signal intensity upon increasing temperature as set to the Central haplotype control. (D and H) Melting peaks generated from the raw fluorescent data. Gray lines represent samples that can only be differentiated as “not Central haplotype” by HRM analysis of the 94-bp CO1 amplicon (A–D) and as “not Western haplotype” by HRM analysis of the 67-bp CO1 amplicon (E–H). The Central haplotype is represented by blue lines, the Western by green lines, the Northwestern by red lines, and the Southwestern by purple lines.
Previously, the 94-bp CO1 amplicon was shown to isolate the central haplotype from the Northwestern and Western haplotypes ( Swisher et. al 2013 ). Using samples from the 2011 dataset as controls ( Swisher et al. 2012 ), analysis of the normalized DNA melting curves again isolated the Central haplotype control ( Fig. 1 A–C, dashed blue line). Representation of the raw fluorescent data as melting peaks clearly showed the Central haplotype peak shifted to a lower melting temperature when compared with the Western and Northwestern controls ( Fig. 1 D). Data from a representative sampling of psyllids collected in the southwestern and central United States during 2012 also identified three samples from Wray, CO, and one sample from Alamosa, CO, to be the Central haplotype ( Fig. 1 A–D). Here, two Alamosa, CO, samples and five Farmington, NM, samples were identified as a haplotype other than Central.
HRM analysis using the 67-bp CO1 amplicon previously demonstrated the isolation of the Western haplotype from the Central and Northwestern haplotypes ( Swisher et al. 2013 ). Coupling this result with the isolation of the Central haplotype using the 94-bp CO1 amplicon, samples of the Northwestern haplotype were deduced ( Swisher et al. 2013 ). For analysis of samples collected in 2012 from the southwestern and central United States, samples from the 2011 dataset were used as controls for the Central, Western, and Northwestern haplotypes ( Swisher et al. 2012 ). Interestingly, analysis of the normalized DNA melting curves not only isolated the Western haplotype control (dashed green line) from the Central and Northwestern haplotypes but it also generated a third group with a different melting curve ( Fig. 1 E–G). From the representative sampling of 2012 psyllids, one Alamosa, CO, and five Farmington, NM, samples were among this third group of melting curves, termed the Southwestern haplotype (purple lines). Representation of the raw fluorescent data as melting peaks clearly showed three distinct, although largely overlapping peaks ( Fig. 1 H). Of the representative samples collected in 2012, one Alamosa, CO, psyllid was identified as the Western haplotype ( Fig. 1 E–H).
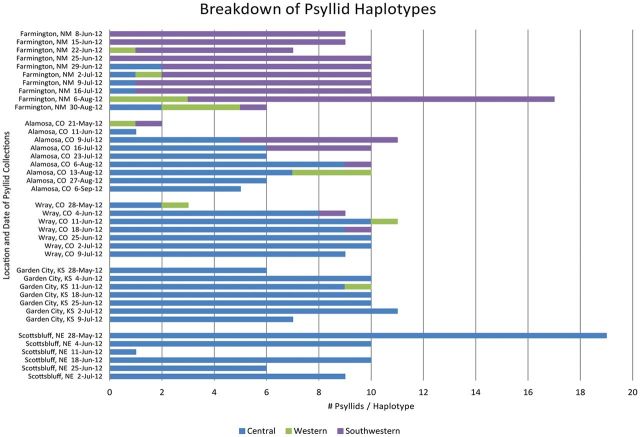
Figure 2 summarizes the HRM data obtained from analysis of 340 individual potato psyllids collected from Farmington, NM; Alamosa and Wray, CO; Garden City, KS; and Scottsbluff, NE, during the 2012 potato growing season. Of the 340 samples tested, 97 were identified as the novel Southwestern haplotype by HRM analysis. Interestingly, these Southwestern haplotype samples were not identified outside of New Mexico and Colorado.
Fig. 2.
Chart representation of the HRM analysis genotyping results. In total, 340 individual psyllids collected in Farmington, NM; Alamosa and Wray, CO; Garden City, KS, and Scottsbluff, NE, were analyzed by HRM analysis to determine their haplotype. The chart provides visual representation of the number of psyllids identified for each haplotype from the specific locations and dates of collection. The Central haplotype is represented in blue, the Western haplotype in green, and the new, Southwestern haplotype in purple. No psyllids of the Northwestern haplotype were identified from these collection sites.
Sequencing Analysis
To confirm the identification of a fourth B. cockerelli haplotype in the United States, and to determine how distinct this Southwestern haplotype might be, a 500-bp portion of the CO1 gene of several individual psyllid samples collected in the southwestern and central United States were sequenced and compared with previously identified sequences of the Central, Western, and Northwestern haplotypes ( Swisher et al. 2012 ). DNA sequencing analysis supported the HRM results, as it identified a fourth, clearly distinct psyllid haplotype: the Southwestern haplotype (GenBank accession number KC305359; Fig. 3 ). Within the 500-bp portion of CO1 , there are 6 SNPs between the Southwestern and the Central haplotypes and 7 SNPs between the Southwestern and Western haplotypes. The Southwestern and the Northwestern haplotypes show the greatest difference in DNA sequence, containing 15 SNPs. Of all the SNPs present within this 500-bp region of CO1 , the Southwestern haplotype contained 2 SNPs that were not seen in any of the other three haplotypes ( Fig. 3 ).
Fig. 3.
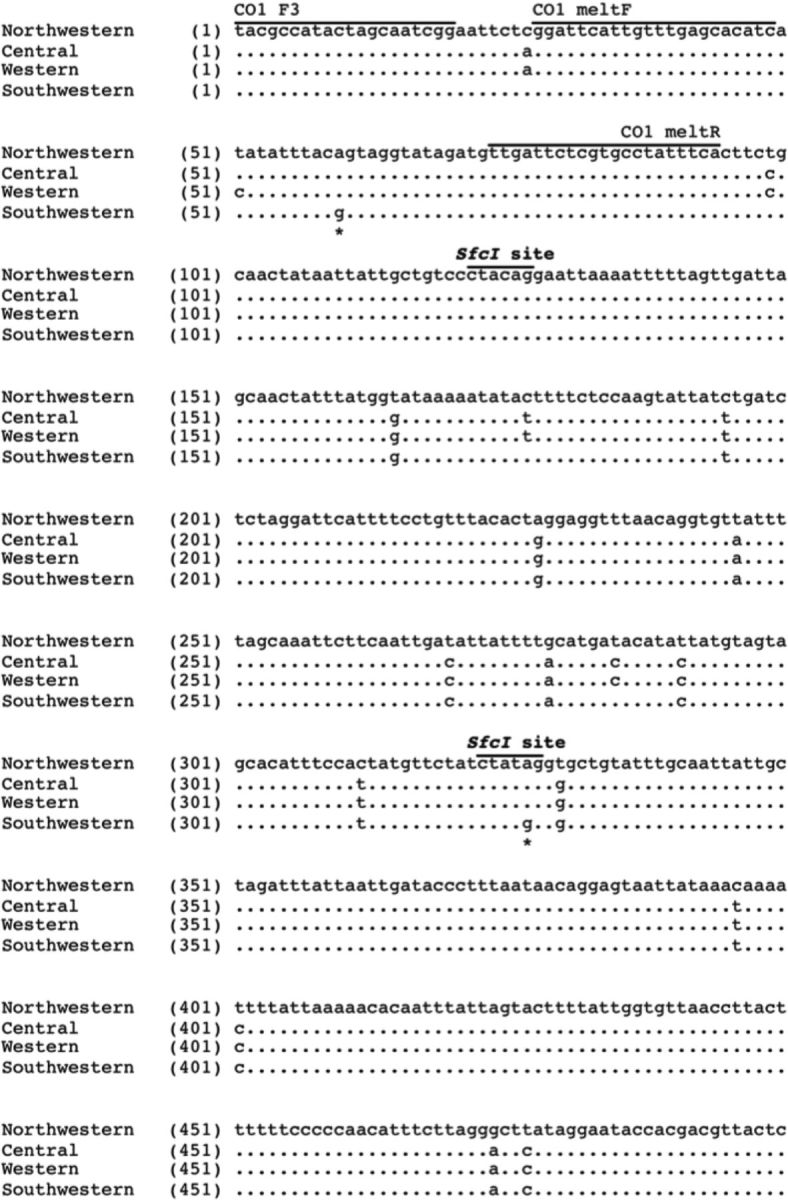
DNA sequencing analysis of a 500-bp portion of the CO1 gene identifies a fourth potato psyllid haplotype. A 500-bp portion of the CO1 gene of the novel psyllid haplotype was sequenced for comparison to the DNA sequence of the three known haplotypes: Central, Western, and Northwestern. Sequencing analysis identified 6 SNPs between the Central and fourth, Southwestern haplotype, 7 SNPs between the Western and Southwestern haplotypes, and 15 SNPs between the Northwestern and Southwestern haplotypes. The Southwestern haplotype contained 2 new SNPs not present in the other three haplotypes (*). SfcI digestion sites are noted above the specific CTAC/TAG sequences.
In total, CO1 partial sequences from 12 psyllids were determined. Eight psyllids from Farmington, NM, were sequenced and identified as the Southwestern haplotype. Two of these eight psyllids did not contain the SNP found at nucleotide 60 but contained all other SNPs. The absence of this SNP did not lead to any change in the amino acid sequence. Two psyllids from Wray, CO, were sequenced and identified as the Southwestern haplotype. One psyllid from Alamosa, CO, was sequenced and identified as the Southwestern haplotype, although it contained an additional SNP at nucleotide 480, which was not present in any psyllid haplotype identified to date. This SNP did not cause an amino acid change. Finally, one psyllid from Garden City, KS, was sequenced and identified as the Central haplotype.
Following the DNA sequencing analysis, amino acid sequences were predicted using the translation code for invertebrate mitochondrial DNA. The amino acid codes were identical between the Southwestern, Western, and Central haplotypes. Interestingly, the presence of 15 SNPs between the Northwestern and Southwestern haplotypes amounted to a single amino acid difference in the Northwestern haplotype.
SfcI Digestion
HRM analyses could not confidently distinguish between the Western haplotype and the Southwestern haplotype for ∼4% of the samples. As a result, and to eliminate the need to sequence the samples giving ambiguous results by HRM analysis, a restriction enzyme assay was used to distinguish the Southwestern haplotype from the other three haplotypes. The enzyme SfcI has the restriction site 5′ … C*T A/G C/T A G … 3′. Within the 500-bp CO1 amplicon of a Southwestern haplotype psyllid, this endonuclease should cleave only one time, generating two fragments that are 378- and 122 bp in size. For psyllids of the Western, Central, and Northwestern haplotypes, this endonuclease should cleave two times, generating three fragments that are 201-, 177-, and 122 bp in size. Figure 4 shows the expected result of a partial SfcI digestion ( Fig. 4 A) and a complete SfcI digestion ( Fig. 4 B), where the two Southwestern haplotype samples show a different digestion pattern than the other three haplotypes. Control samples taken from the 2011 dataset were used for the Northwestern, Central, and Western haplotypes ( Swisher et al. 2012 ) and show digested fragments of 200 bp or smaller. The Southwestern haplotype samples, verified by sequencing prior to their digestion, show two expected bands with one just under 400 bp and the other just over 100 bp. The SfcI restriction enzyme assay is an effective tool to provide further support of the HRM analysis results.
Geographical Distribution
Of all 340 individual potato psyllids tested, the new, fourth potato psyllid haplotype was only present in New Mexico and Colorado, thus giving it the name, Southwestern haplotype ( Fig. 5 ). Approximately 85% of the psyllids tested from collections made near Farmington, NM, were of this Southwestern haplotype population ( Figs. 2 and 5 ). Likewise, ∼20% of the psyllids tested from nearby Alamosa, CO, were Southwestern, whereas <4% of the psyllids tested from collections made closer to the central United States in Wray, CO, were Southwestern. Psyllids tested from collections made near Scottsbluff, NE, and Garden City, KS, overwhelmingly represented the Central haplotype ( Figs. 2 and 5 ).
Fig. 5.
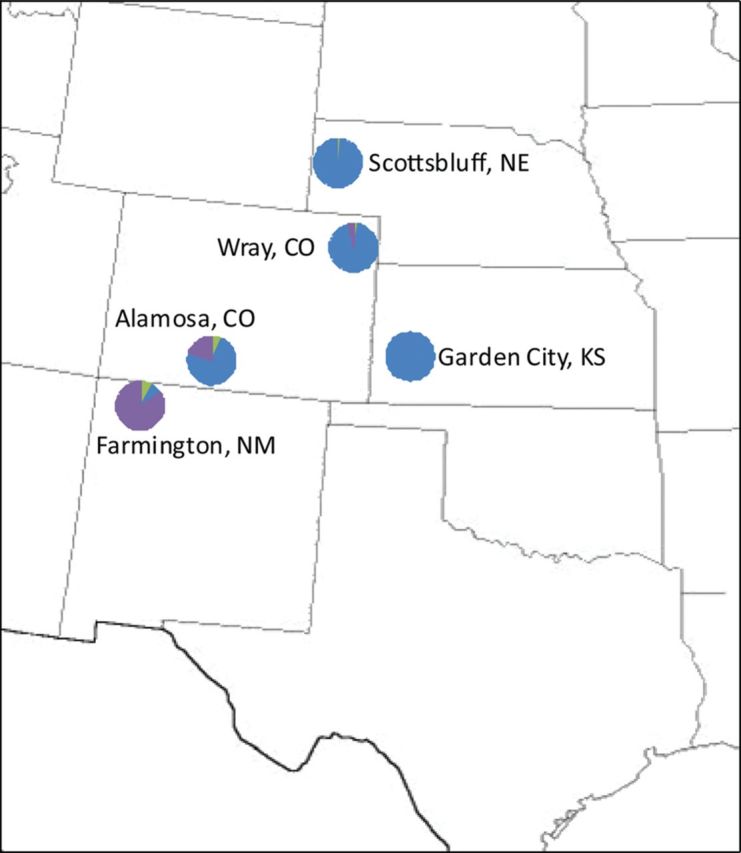
Geographical distribution of the four psyllid haplotypes. Haplotyping results of potato psyllids collected in New Mexico, Colorado, Nebraska, and Kansas are depicted on the U.S. map by pie charts. These pie charts depict the percentage of each haplotype identified at each location, independent of psyllid number. The Central haplotype is depicted in blue, the Western in green, and the new, Southwestern haplotype in purple.
Discussion
In previous studies, HRM analysis of the B. cockerelliCO1 gene was used as a tool to genotype individual potato psyllid samples ( Swisher et al. 2012 , 2013 ). Here, this technique effectively identified a fourth potato psyllid haplotype within the United States. This new haplotype was designated as the Southwestern haplotype because it was predominantly identified in the southwestern region of the United States, particularly in New Mexico, but also in Southern Colorado ( Fig. 5 ). A small percentage of the Southwestern haplotype psyllids were also identified from Northeastern Colorado, but no Southwestern psyllids were identified in the samples tested from Nebraska and Kansas.
Previous research looked at archived psyllid samples from the northwestern and central United States to gain insight into the psyllid population dynamics over time ( Swisher et al. 2013 ). Potentially, application of this same strategy to the southwestern region of the United States could provide an even greater idea of psyllid population dynamics and shine light on the origin of this novel Southwestern haplotype. Unfortunately, at present, the oldest archived psyllid samples available that originate from New Mexico only date back to 2010. However, HRM analysis was used to haplotype these two psyllids and identified them as the Southwestern haplotype (data not shown). Although this sample number is far too small to draw any substantial conclusions, it is clear that the Southwestern haplotype has been present in the southwestern region of the United States prior to the 2012 potato growing season.
It is of particular interest that although the Southwestern haplotype seems to have a significant difference in nucleic acid sequence compared with the Western and Central haplotypes, this difference does not exist at the amino acid level ( Fig 3 ; data not shown). Between the Southwestern haplotype and the Central and Western haplotypes, there were six and seven SNPs, respectively. However, none of these SNPs altered the amino acid code, raising the question as to the evolutionary significance of these SNPs within the Southwestern psyllid haplotype.
In addition, of the samples that were identified as the Southwestern haplotype by HRM analysis, none were found to be Lso positive by standard PCR testing (data not shown). Therefore, it remains to be seen if the Southwestern haplotype psyllids are a vector of Lso. However, it would be surprising if the Southwestern psyllids did not become infected by the bacterium or become a carrier capable of transmitting Lso to solanaceous crops.
The identification of the Southwestern psyllid haplotype increases the knowledge of the potato psyllid population dynamics within the United States and raises the possibility that new psyllid haplotypes may emerge or be detected at different times, and in different geographical regions. It also, along with the Northwestern haplotype, suggests that populations of psyllids might exist, which do not migrate large distances, such is believed to occur with the Western and Central haplotype psyllids (Wallis 1995, Liu et al. 2006 , Swisher et al. 2012 ). Unfortunately, in a time where greenhouse plants are rapidly transported by human means across the country, and even the globe, these pockets of psyllid populations may not exist for long.
In previous biological studies, it has been demonstrated that the Western psyllid population has a lower growth index and survivorship, as well as a slower rate of development compared with the Central population ( Liu and Trumble 2007 ). However, the biological ramifications of the genotypic differences in the Northwestern psyllid population, as well as the novel, Southwestern haplotype identified in this study, have yet to be determined. Biological differences between the four known B. cockerelli haplotypes could complicate the psyllid eradication methods currently in use and lead to the spread of potato psyllid-induced diseases such as ZC. In light of this, understanding the biological differences between the haplotypes should be a priority in this field of study.
Acknowledgments
We thank Launa Hamlin for her technical assistance in the laboratory (United States Department of Agriculture (USDA), Agricultural Research Service, Prosser, WA). This research was supported by the USDA-SCRI Project 2009-51181-20176 and the USDA-RAMP Project 2009-51101-05892. The use of trade, firm, or corporation names in this publication is for the information and convenience of the reader. Such use does not constitute an official endorsement or approval by the United States Department of Agriculture or the Agricultural Research Service of any product or service to the exclusion of others that may be suitable. USDA is an equal opportunity provider and employer.
References Cited
- Artimo P., Jonnalagedda M., Arnold K., Baratin D., Csardi G., de Castro E., Duvaud S., Flegel V., Fortier A., Gasteiger E., et al. . 2012. . ExPASy: SIB bioinformatics resource portal . Nucleic Acids Res. 40 : 597 – 603 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman R. I., Macias-Velasco J. F., Arp A. P., Bextine B. . 2012. . Using quantitative real time PCR melt curve analysis of partial CO1 sequence for rapid biotype differentiation of Bactericera cockerelli (Hemiptera: Triozidae) . Southwest. Entomol. 37 : 475 – 484 . [Google Scholar]
- Chapman R. I., Strube L., Bextine B. . 2010. . Population genetics of the potato psyllid: impacts on zebra chip epidemiology, pp. 64–68 . In Proceedings, 10th Annual Zebra Chip Reporting Session, 7–10 November 2010, Texas AgriLife, College Station, TX . [Google Scholar]
- Crosslin J. M., Munyaneza E., Brown J. K., Liefting L. W. . 2010. . Potato zebra chip disease: a phytopathological tale . Plant Health Prog. doi:10.1094/PHP-2010-0317-01-RV . [Google Scholar]
- Crosslin J. M., Vandemark G. J., Munyaneza J. E. . 2006. . Development of a real-time, quantitative PCR for detection of the Columbia Basin potato purple top phytoplasma in plants and beet leafhoppers . Plant Dis. 90 : 663 – 667 . [DOI] [PubMed] [Google Scholar]
- Crosslin J. M., Lin H., Munyaneza J. E. . 2011. . Detection of ‘ Candidatus Liberibacter solanacearum’ in the potato psyllid, Bactericera cockerelli (Sulc), by conventional and real-time PCR . Southwest. Entomol. 36 : 125 – 135 . [Google Scholar]
- Goolsby J. A., Adamczyk J., Crosslin J. M., Troxclair N., Anciso J., Bester G., Bradshaw J., Bynum E., Carpio L., Henne D., et al. . 2012. . Seasonal population dynamics of the potato psyllid (Hemiptera: Triozidae) and its associated pathogen “ Candidatus Liberibacter solanacearum” in potatoes in the southern Great Plains of North America . J. Econ. Entomol. 105 : 1268 – 1276 . [DOI] [PubMed] [Google Scholar]
- Liu D., Trumble J. T. . 2007. Comparative fitness of invasive and native populations of the potato psyllid ( Bactericera cockerelli ) . Entomol. Exp. Appl. 123 : 35 – 42 . [Google Scholar]
- Liu D., Trumble J. T., Stouthamer R. . 2006. . Genetic differentiation between eastern populations and recent introductions of potato psyllid ( Bactericera cockerelli ) into western North America . Entomol. Exp. Appl. 118 : 177 – 183 . [Google Scholar]
- Munyaneza J. E. 2010. . Psyllids as vectors of emerging bacterial diseases of annual crops . Southwest. Entomol. 35 : 471 – 477 . [Google Scholar]
- Munyaneza J. E. 2012. . Zebra chip disease of potato: biology, epidemiology, and management . Am. J. Potato Res. 89 : 329 – 350 . [Google Scholar]
- Munyaneza J. E., Henne D. C. . 2012. . Leafhopper and psyllid pests of potato, pp. 65–102 . InGiordanengo P., Vincent C., Alyokhin A. (eds.), Insect pests of potato: global perspectives on biology and management . Academic Press; , San Diego, CA: . [Google Scholar]
- Munyaneza J. E., Crosslin J. M., Upton J. E. . 2007. . Association of Bactericera cockerelli (Homoptera: Psyllidae) with “Zebra Chip,” a new potato disease in Southwestern United States and Mexico . J. Econ. Entomol. 100 : 656 – 663 . [DOI] [PubMed] [Google Scholar]
- Rondon S., Schreiber A., Jensen A., Hamm P., Munyaneza J., Nolte P., Olsen N., Wenninger E., Henne D., Wohleb C., et al. . 2012. . Potato psyllid vector of Zebra Chip Disease in the Pacific Northwest: biology, ecology, and management . PNW Ext. Pub. 633 : 1 – 8 . [Google Scholar]
- Swisher K. D., Munyaneza J. E., Crosslin J. M. . 2012. . High resolution melting analysis of the cytochrome oxidase I gene identifies three haplotypes of the potato psyllid in the United States . Environ. Entomol. 41 : 1019 – 1028 . [Google Scholar]
- Swisher K. D., Munyaneza J. E., Crosslin J. M. . 2013. . Temporal analysis of potato psyllid haplotypes in the United States . Environ. Entomol. 42 : 381 – 393 . [DOI] [PubMed] [Google Scholar]
- Wallis R. L. 1955. . Ecological studies on the potato psyllid as a pest of potatoes . United States Department of Agriculture Technical Bulletin (1107) , Washington, DC: . [Google Scholar]