ABSTRACT
The level of anthocyanins in plants vary widely among cultivars, developmental stages and environmental stimuli. Previous studies have reported that the expression of various MYBs regulate anthocyanin pigmentation during growth and development. Here we examine the activity of 3 novel R2R3-MYB transcription factor (TF) genes, PamMybA.1, PamMybA.3 and PamMybA.5 from Prunus americana. The anthocyanin accumulation patterns mediated by CaMV double35S promoter (db35Sp) controlled expression of the TFs in transgenic tobacco were compared with citrus-MoroMybA, Arabidopsis-AtMybA1 and grapevine-VvMybA1 transgenics during their entire growth cycles. The db35Sp-PamMybA.1 and db35Sp-PamMybA.5 constructs induced high levels of anthocyanin accumulation in both transformed tobacco calli and the regenerated plants. The red/purple color pigmentation induced in the PamMybA.1 and PamMybA.5 lines was not uniformly distributed, but appeared as patches in the leaves, whereas the flowers showed intense uniform pigmentation similar to the VvMybA1 expressing lines. MoroMybA and AtMybA1 showed more uniform pink coloration in both vegetative and reproductive tissues. Plant morphology, anthocyanin content, seed viability, and transgene inheritance were examined for the PamMybA.5 transgenic plants and compared with the controls. We conclude that these TFs alone are sufficient for activating anthocyanin production in plants and may be used as visible reporter genes for plant transformation. Evaluating these TFs in a heterologous crop species such as citrus further validated that these genes can be useful for the metabolic engineering of anthocyanin production and cultivar enhancement.
KEYWORDS: Anthocyanin, CaMV double35S promoter, citrus, R2-R3 MYB transcription factor, transgenic plants, tobacco
INTRODUCTION
The pigments that color most flowers, fruits, and seeds are flavonoids. Flavonoids belong to a group of plant products with variable phenolic structures and play an important role in protection against biotic and abiotic stresses (Stitt et al., 2010; Winkel-Shirley 2001). These secondary metabolites, widely distributed in plants, are mainly classified into 6 major subgroups: chalcones, flavones, flavonols, flavandiols, anthocyanins, and proanthocyanidins or condensed tannins (Winkel-Shirley 2006). Anthocyanins form a large subclass of flavonoids conferring different colors typically red, purple, or blue in fruits and flowers. (Winkel-Shirley 2001; Tanaka et al., 2008). The structural genes involved in the anthocyanin biosynthetic pathway of plants include chalcone synthase, chalcone isomerase, flavanone 3-hydroxylase, flavonoid 3,5-hydroxylase, dihydroflavonol 4-reductase, anthocyanidin synthase, leucoanthocyanidin dioxygenase and UDP-glucose: flavonoid 3-O-glucosyltransferase (Borevitz et al., 2000). These genes are well characterized in model plants and fruits including grape, apple and litchi. In addition, numerous studies have demonstrated that anthocyanin accumulation is largely regulated by MYB transcriptional factors, which manipulate the expression of the structural genes in the anthocyanin biosynthetic pathway (Boss et al., 1996; Honda et al., 2002; Niu et al., 2010; Petroni et al., 2011; Springob et al., 2003; Wei et al., 2011).
Anthocyanin biosynthesis is typically regulated at the transcriptional level. Three types of transcription factors: R2R3-MYB, basic helix–loop–helix (bHLH) and WD40 proteins, interact together to form a MYB-bHLH-WD40 (MBW) complex. This complex regulates the structural genes of the anthocyanin biosynthesis, modification and transport in many plant species (Allan et al., 2008; Ramsay and Glover 2005; Butelli et al., 2008). MYB proteins are characterized by the highly conserved MYB DNA-binding domain (MYB domain). They have diverse roles in secondary metabolism, development, signal transduction and stress response processes (Dubos et al., 2010) and are categorized into 4 classes based on the number of MYB domains (Dubos et al., 2010). The R2R3 class is regarded as the largest with 137 R2R3-MYB members reported in Arabidopsis (Feller et al., 2011).
Specific MYB transcription factors are a major determinant of anthocyanin activation and accumulation in plants (Zhou et al., 2015; Boase et al., 2015; Allan et al., 2008; Hichri et al., 2011). Numerous studies have demonstrated that R2R3-MYB TFs in various plant species are primarily responsible for the regulation of anthocyanin biosynthetic pathways (Winkel-Shirley 2001). When overexpressed in heterologous systems, these transcription factors resulted in an enhanced accumulation of anthocyanins (Borevitz et al., 2000; Gonzali et al., 2009; Meng et al., 2014). Subgroup 6 clade MYBs such as PAP1 and PAP2, in Arabidopsis and their orthologs in other plant species are known to be key regulators of anthocyanin biosynthesis (Borevitz et al., 2000). Overexpression of these transcription factors induces anthocyanin production throughout the transgenic plants (Borevitz et al., 2000; Dubos et al., 2010; Qiu et al., 2014). Orthologues of PAP1 and PAP2 in other plant species have also been shown to be key regulators of anthocyanin biosynthesis. For example the Myb-related genes such as VlMybA1–1, VlMybA1–2, and VlMybA2 regulate anthocyanin biosynthesis in Kyoho, a black-skinned cultivar of Vitis labruscana (Azuma et al., 2007; Kobayashi et al., 2004; Kobayashi et al., 2005). In grape, white cultivars are thought to have arisen from different red or black cultivars by independent mutations. It has also been shown that a retrotransposon-induced mutation in VvMybA1 is associated with the loss of pigmentation in white cultivars of V. vinifera (This et al., 2007). In over 200 accessions of V. vinifera, the insertion of the retrotransposon Gret1 in the promoter region of VvMybA1 was in strong association with the white-fruited phenotype (This et al., 2007). Recently, Sicilian blood orange was shown to have arisen by insertion of a Copia-like retrotransposon adjacent to a gene encoding CsRuby, a MYB transcriptional activator of anthocyanin production. The retrotransposon controls Ruby expression (and thus anthocyanin accumulation) through induction by cold stress conditions (Butelli et al., 2012). However, the induction process is generally incomplete and leads to fruit which is mottled purple in appearance (Butelli et al., 2012).
Efficient induction of anthocyanin biosynthesis in some cases such as by the MdMyb10 is dependent on the co-expression of a specific bHLH protein. In petunia, PhAn1, a bHLH TF directly activates the expression of the biosynthetic gene DFR and is regulated by MYB factor PhAn2 (Spelt et al., 2000). This difference might result from the characteristics of the exogenous MYBs and the interactions with endogenous tobacco bHLH factors. Therefore, it was concluded that the exogenous MYB induces the expression of tobacco bHLH transcription factor NtAn1b and anthocyanin biosynthetic genes for anthocyanin accumulation (Huang et al., 2013b; Lai et al., 2014). In another example the upregulation of NtAn1b in response to Litchi LcMyb1 overexpression was shown to be essential for anthocyanin accumulation in the leaf and pedicel (Lai et al., 2014). These results suggest that some MYBs require co-expression of the bHLH at the same time to be functional while other MYBs alone are sufficient to induce anthocyanin accumulation.
Thus, understanding the regulatory roles of R2R3-MYB TFs in anthocyanin biosynthesis is essential for performing metabolic engineering of the anthocyanin biosynthetic pathway in food crops such as citrus. In this respect, R2R3-MYB TFs appear to be crucial for the metabolic engineering of the anthocyanin biosynthetic pathway to produce an altered expression profile and, hence, differential pigmentation of leaves, flowers, roots or fruit. In this study, 3 novel plum (Prunus americana) TFs, PamMybA.1, PamMybA.3 and PamMybA.5, were isolated and a synthetic version of CsRuby named MoroMybA containing a sequence modification was generated. Arabidopsis AtMybA1 which is the genomic version of well characterized PAP1 (AtMYB75) and a genomic version of the grape MybA1, VvMybA1 (This et al., 2007) were used as controls and for comparison with the novel plum MybAs in this study. The capacity of these transcription factors to activate anthocyanin accumulation was evaluated in vegetative as well as reproductive tissues. The utility of these TF genes as potential biotechnological tools for engineering anthocyanin accumulation and to produce healthy fruits and vegetables or for the production of ornamental plants is also discussed. The novel PamMybAs characterized in this work may prove to be useful tools for future engineering since they solely are sufficient for inducing the anthocyanin biosynthetic pathway, without requiring exogenous accessory factors.
RESULTS
Isolation of Mybs and Sequence Analysis
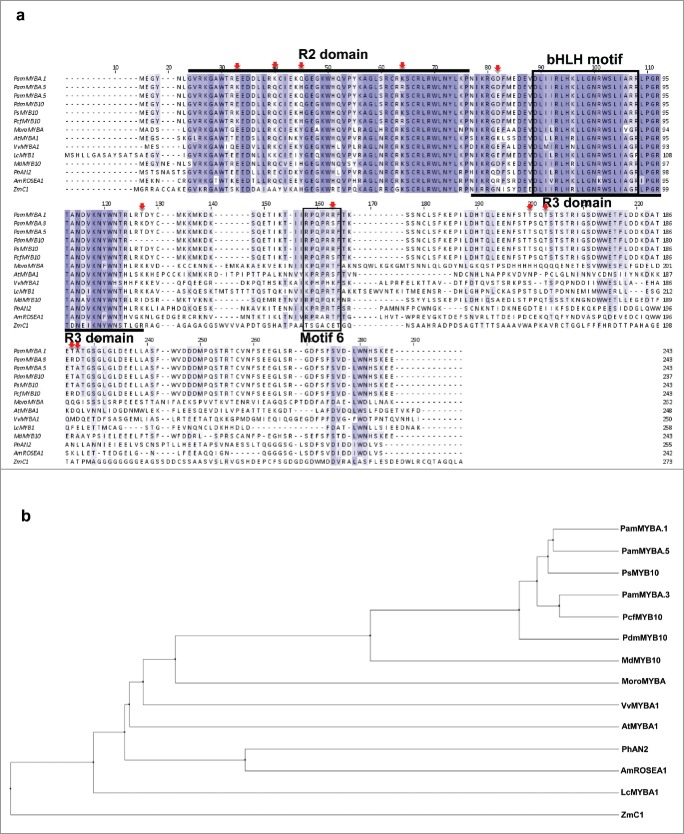
Using primers that aligned to the domestic plum MybA and the Arabidopsis MybA, 3 different PamMybA genes were PCR amplified from plum (Prunus americana) genomic DNA. The PamMybA.1, PamMybA.3, PamMybA.5 genes including their native introns were cloned into a pUC vector and sequenced. The sequences of PamMybA.1 (2018bp), PamMybA.3 (2061bp), PamMybA.5 (2017bp), all encode 243 amino acid proteins, just slightly larger than the domestic plum PdmMyb10 gene, which encodes a 237 amino acid protein (Lin-Wang K et al., 2010). The PamMYBA.1, PamMYBA.3 and PamMYBA.5 proteins each contain highly conserved R2 and R3 MYB domains in the N-terminal regions (Fig. 1a) and are potentially paralogues to one another. In the R2 domain, all the plum MYB proteins contain residues that are predicted to allow interaction with bHLH transcription factors. Binding of R2R3-MYB proteins with bHLH and WD repeat proteins generates a functional transcriptional activation complex that drives the production of anthocyanins through the coordinated transcription of the anthocyanin biosynthetic genes (Zimmermann et al., 2004; Stracke et al., 2001; Takos et al., 2006). In the variable region (marked as motif 6 in Fig. 1a), the RPQPR[R/S]F sequence is conserved in all the plum MYBs analyzed (Fig. 1a). This small motif is part of a larger motif that was previously reported as KPRPR[S/F]F (motif 6) in Stracke et al. (2001).
FIGURE 1.
Alignment and phylogenetic analysis of the PamMYBA and other R2R3-MYB proteins. (a) Protein sequence alignment of the amino acid sequences of PamMYBA.1, PamMYBA.3, PamMYBA.5 (DNA sequence in supplemental text S1), domestic plum (Prunus domestics) PdmMYB10 (EU153580.1), Japanese plum (Prunus salicina) PsMYB10 (EU155161), Cherry-plum (Prunus cerasifera) PcfMYB10 (EU153583), citrus MoroMYBA (DNA sequence in supplemental text. S1), Arabidopsis AtMYBA1 (AF325123.1), Merlot grape VvMYBA1 (AB111101), Litchi LcMYB1 (AGC14139.1), Apple MdMYB10 (DQ267896), Petunia PhAN2 (AAF66727), Snapdragon AmROSEA1 (ABB83826), Maize ZmC1 (AAK81903) are shown. Identical amino acids are shaded in dark blue and similar in light blue. The R2 and R3 MYB domains are underlined. The bHLH-binding motif is boxed in black within the R3 domain. Motif 6 is highlighted in a black box. (b) Phylogenetic analysis of PamMYBs and the selected R2R3-MYB proteins from other plant species that are known to regulate the flavonoid pathway are shown. The tree was constructed using Average Distance from the BLOSUM62 Clustal alignment.
PamMYBA.5 shares 98% identity with PamMYBA.1 and 94% identity with PamMYBA.3 (Fig. 1b). The PamMYBA.1 and PamMYBA.5 proteins are nearly identical with 2 semi-conservative amino acid differences (a Q or K in the R2 domain and an E or D in the R3 domain; Fig. 1a). However, PamMYBA.3 has 9 differences with PamMYBA.1 and PamMYBA.5 (marked by red arrows in Fig. 1a). PamMYBA.5 shares 97% identity with domestic plum PdmMYB10 (Fig. 1b) and is 98% identical to Japanese plum PsMYB10 and 96% identical to cherry plum PcfMYB10 (Ling-Wang et al., 2010). A single 6 amino acid insertion (LR[K/T]DYC) immediately following the conserved R2R3 region in the PamMYB.5, PsMYB10 and PcfMYB10, is what primarily differentiates these sequences from the domestic PdmMYB10 sequence. The phylogenetic analysis of PamMYB.5 further indicates that it is closely related to the Japanese plum PsMYB10 followed by cherry plum PcfMYB10 and then domestic plum PdmMYB10 (Fig. 1b). PamMYB.1 is 98% identical to domestic plum PdmMYB10, 98% identical to Japanese plum PsMYB10 and 96% identical with cherry plum PcfMYB10. PamMYB.3 on the other hand is 94% identical to domestic plum PdmMYB10 and 97% identical to Japanese plum PsMYB10. Protein sequence alignment further revealed that cherry plum PcfMYB10 and PamMYBA.3 are 100% identical with their nucleotide sequences being 96% identical (Ling-Wang et al., 2010). PamMYB.3 is most likely derived/descended from the PcfMYB10 as it shares such high sequence identity.
Alignment of the anthocyanin promoting MYB sequences from various plant species show that this group of MYBs contain highly conserved R2R3, bHLH and motif 6 domains on their N-terminal halves, but are much less well conserved within their C-terminal regions (Fig. 1a). Overall the Prunus MYBs make a relatively tight clade with the apple MYB10 and citrus MoroMybA being the next most closely related (Fig. 1b). MoroMybA is a synthetic version of the citrus CsRuby (Butelli et al., 2012) coding sequence (Materials and Methods, Supplemental text. S1), which encodes a 262 amino acid protein.
Transgenic Tobacco Plants Constitutively Overexpressing the Different Myb Genes
The ability of MYBs to activate anthocyanin biosynthesis was examined by expression under the control of the constitutive CaMV db35S promoter (Fig. 2). Tobacco leaf discs were transformed using Agrobacterium and the resulting undifferentiated callus and regenerated tissues were visually examined for anthocyanin accumulation. Images of the transgenic callus and regenerated shoots are shown in (Fig. 3). All transformation constructs except for the one containing the PamMybA.3 gene, clearly produced visibly pigmented tissues. AtMybA1, MoroMybA and VvMybA1 were chosen as controls due to their wide difference in anthocyanin accumulation patterns. As expected, the intensity and patterns of anthocyanin accumulation varied between the constructs with the VvMybA1 transgenic lines showing the darkest coloration and the MoroMybA transgenic tissues exhibiting the lightest pink coloration. Kanamycin resistant regenerated shoots were transferred to root induction medium and rooted plantlets were transferred to soil and grown in the greenhouse. The presence of the MybA transgene was molecularly confirmed using genomic PCR analysis (Fig. 4a). For all constructs except PamMybA.3, the kanamycin resistant shoots displayed visible colored tissues.
FIGURE 2.
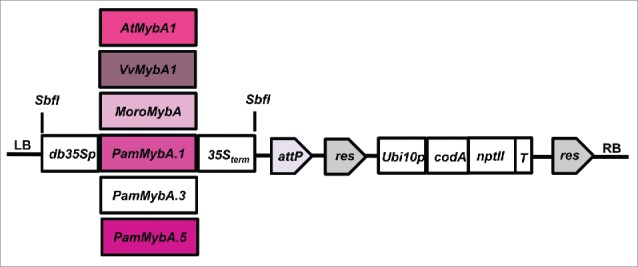
Schematic representation of the Agrobacterium binary vector T-DNA carrying the MybA genes. The binary vector pCTAG7 is a pCAMBIA 0390 derivative harboring the codA-nptII fusion gene under the control of the Arabidopsis Ubiquitin10 promoter (Ubi10p) and nopaline synthase terminator (T). MybA genes are expressed by the double enhanced CaMV 35S promoter (db35Sp) and 35S terminator. The MybA genes used here are from Arabidopsis (AtMybA1), Merlot grape (VvMybA1), Citrus (MoroMybA) and plum (PamMybA.1, PamMybA.3, PamMybA.5). The attP and res are recombinase recognition sites for the Bxb1 and CinH recombinase enzymes respectively. LB and RB designate the Agrobacterium left border and right border respectively.
FIGURE 3.
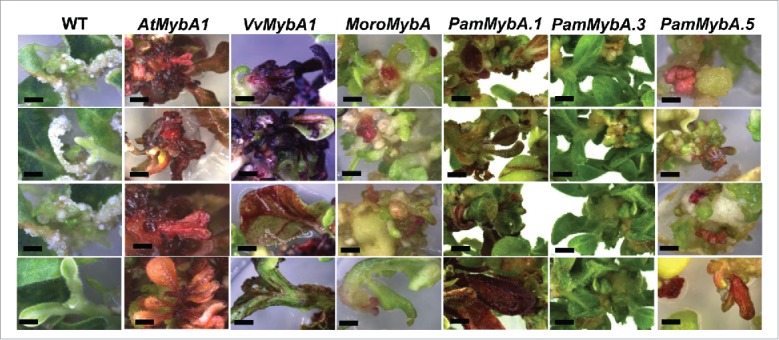
Regenerating shoots from callus in Agrobacterium mediated tobacco leaf disc transformation. The wild type control (WT) is in the left column which shows no anthocyanin accumulation in the shoots followed by AtMybA1, VvMybA1, MoroMybA, PamMybA.1, PamMybA.3 and PamMybA.5 transgenic tobacco tissues. Images were taken 4 weeks after co-cultivation.
FIGURE 4.

Phenotypic and molecular analyses of the MybA transgenic tobacco plants. (a) PCR amplification showing the transgene's presence in independent lines. The no template (−), wild type (WT) and positive (+) plasmid DNA controls are also shown. The expected amplicon sizes are shown in supplemental Fig. S1. (b) Wild type and (c) representative AtMybA1 (d) VvMybA1 (e) MoroMybA (f) PamMybA.1 (g) PamMybA.3 and (h) PamMybA.5 mature T0 transgenic plants are shown.
Distinct Anthocyanin Accumulation Patterns Observed in the Transgenic Tobacco
Fifteen independent lines (T0 generation) harboring db35Sp-MybA constructs were obtained and further characterized for their anthocyanin accumulation pattern. We observed differential pigmentation in the various MybA construct lines (Fig. 4b-h). The AtMybA1 and MoroMybA plants displayed a uniform and homogenously consistent light purplish coloration throughout the entire plant. The VvMybA1 plants exhibited very high levels of anthocyanin accumulation with variegated purple patterns in leaves, dark red flowers and mostly green stems, consistent with previously published results (Li et al., 2011). The PamMybA.1 & PamMybA.5 transgenic lines showed mosaic purple coloration in their leaves, stems and flowers, similar to the VvMybA1 expression pattern. As shown in Fig. 4f and h, the PamMybA.1 & PamMybA.5 expressing lines resulted in dark pink colored flowers and strong anthocyanin accumulation in leaves but not in the stem. As the PamMYBA.1 protein shared 99% identity with PamMYBA.5 and exhibited an indistinguishable pattern of pigmentation, only the PamMybA.5 transgenic lines were characterized further in this study.
Fifteen independent transgenic PamMybA.3 lines were also carefully examined for anthocyanin accumulation, but all failed to show any signs of pigmentation different than what is observed in wild type plants (Fig. 4b, g). This lack of anthocyanin accumulation in the PamMybA.3 transgenic lines is presumably due to the amino acid differences when compared with PamMybA.5 (Fig. 1a) and suggests that PamMYBA.3 requires additional cofactors such as a bHLH for activation of anthocyanin accumulation. This is consistent with the results observed for the transient expression of PcfMYB10 in tobacco where it was shown to induce the anthocyanin pathway only when co-expressed with bHLHs (Lin-Wang et al., 2010). Since PamMYBA.3 and PcfMYB10 are identical proteins and do not activate anthocyanin accumulation in tobacco without an exogenous coactivator, we did not study the PamMybA.3 lines further.
The growth of some of the VvMybA1 lines was slower than wild type plants and seed set was measurably lower as well, suggesting that high levels of anthocyanin accumulation may have deleterious effects on plant growth and development. In contrast the vegetative growth and seed set of the MoroMybA as well as the PamMybA transgenic lines were not noticeably different than wild type suggesting that they are suitable candidates for biotech applications. These results further indicate that there is no requirement for the presence of an exogenous bHLH partner for the plum MYBA.1 and MYBA.5 transcription factors to achieve full functionality in this heterologous system, unlike other MYBs from cherry plum, litchi and apple (Lai et al., 2016; Espley et al., 2007; Li et al., 2007; Takos et al., 2006).
MybA Anthocyanin Accumulation is Heritable in the Next Generation
To further examine the differences in anthocyanin accumulation among the transgenic lines, and to confirm the phenotype was heritable in the next generation in a heterologous system, we obtained T1 generation seed from each T0 transgenic lines and observed the phenotypic features of the T1 plants during 3 months of plant growth. T1 plants were selected for antibiotic resistance and grown to maturity in a growth chamber. Appearance of the anthocyanin pigment in transgenic leaves varied among the lines depending on the Myb gene used. As shown in Fig. 5a, at 14 d post germination, MoroMybA and AtMybA1 T1 transgenic seedlings exhibited anthocyanin pigmentation of their cotyledons and leaves (Fig. 5a). The MoroMybA overexpressing lines accumulated more anthocyanins in its cotyledons than in the first true leaves. AtMybA1 seedlings were similar to the MoroMybA but were unique in that they also accumulated anthocyanins in their roots (marked with black arrows Fig. 5a). Relatively low levels of anthocyanins accumulated in cotyledons of the PamMybA.5 and VvMybA1 transgenic seedlings with anthocyanin pigmentation generally becoming visible in the midrib of developing leaves. Wild type seedlings exhibited no detectable anthocyanin pigmentation. After 25 d of growth, PamMybA.5 and VvMybA1 transgenic plants had regions of intense anthocyanin accumulation spreading out from the midrib region of the leaf (Fig. 5b).
FIGURE 5.
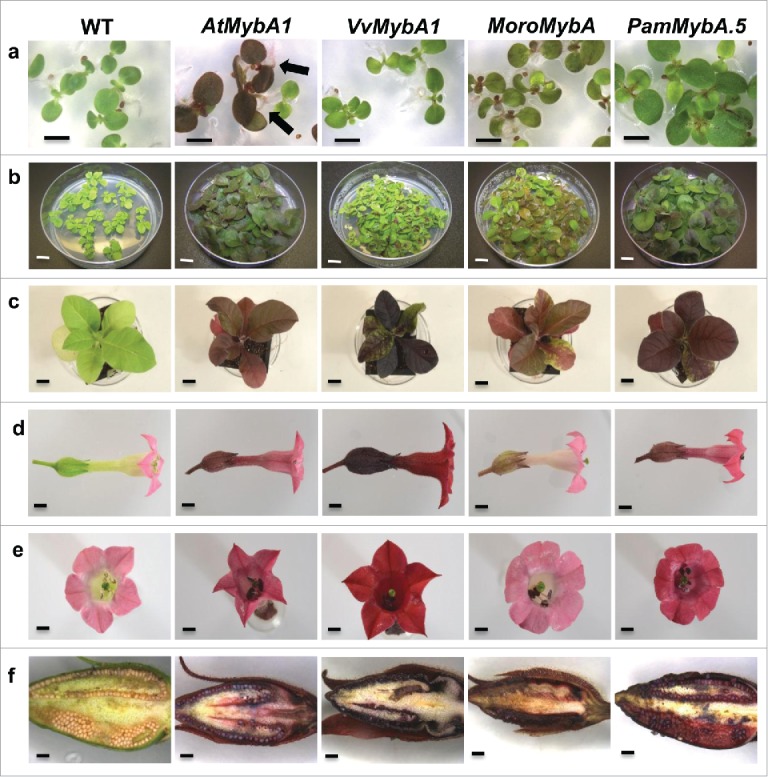
Comparison of vegetative and reproductive growth in T1 transgenic tobacco plants expressing the MybAs. (a) Segregation of 14 day old transgenic tobacco seedlings on kanamycin selection. MoroMybA and AtMybA1 seedlings exhibit visible anthocyanin accumulation in their leaves and the AtMybA1 seedlings accumulate anthocyanins in their leaves and roots (black arrows) (b) 28 day old seedlings on kanamycin selection plates (c) 50 day old transgenic plants in soil (d) and (e) flowers from wild type and transgenic plants and (f) sections of developing tobacco seed pods.
There were also obvious differences in anthocyanin accumulation levels and patterns among the 50 day old transgenic T1 plants grown in soil (Fig. 5c; Fig. S2). Accumulation was restricted to certain sectors of the leaves for the MoroMybA, PamMybAs and VvMybA1 lines in a random mosaic pattern throughout. Typically the younger leaves of MoroMybA plants exhibited a more uniform pattern of anthocyanin pigment throughout the surface area of the leaf tissue only becoming mosaic as the plant matured, as was observed in the T0 generation. The VvMybA1 transgenics exhibited slow growth, short internodes, and delayed flowering. Occasionally, growth of the VvMybA1 plants that exhibited the strongest anthocyanin accumulation were extremely stunted and did not set seed. In contrast, the PamMybA.5 lines exhibited a more uniform accumulation pattern in the leaves similar to what was observed for the MoroMybA lines, however the stem was green, again consistent with the coloration observed in the T0 plants (Fig. 5c).
The flowers of the MoroMybA plants had light pink sepals and pedicel compared with the green tissues in wild type tobacco plants, but the petals were only a slightly darker pink color. The flowers of the PamMybA.5 and AtMybA1 plants were a light red, clearly different from wild type plants and the VvMybA1 lines exhibited vibrant dark red petals, sepals and pedicel (Fig. 5d, e). Upon maturation, the seed pods of each of the analyzed MybA overexpressing lines were sectioned to examine anthocyanin accumulation. Wild type plants did not show any purple coloration, but all of the MybA overexpressing lines had anthocyanin accumulation within the seed pod epidermal layer as well as the seeds, indicating that the MybA transgenes activated anthocyanin accumulation in these tissues as well (Fig. 5f).
To further examine the level of anthocyanin production in the various MybA transgenic tobacco plants, the total anthocyanin content in leaf tissue from representative 50 day old plants was tested. Leaf tissue samples had their total anthocyanins extracted and analyzed using spectrometric measurement. The anthocyanin content in the VvMybA1 sample was the highest as expected based on the visibly dark coloration phenotype. The PamMybA.5 and AtMybA1 samples had slightly lower but similar levels of anthocyanins and the MoroMybA sample contained the lowest amount of total anthocyanins. The non-transgenic wild type leaf sample contained a negligible quantity of total anthocyanins (Fig. 6a). These results further validate that PamMybA.5 and MoroMybA.5 constructs induce the production of anthocyanins in the heterologous system of tobacco.
FIGURE 6.
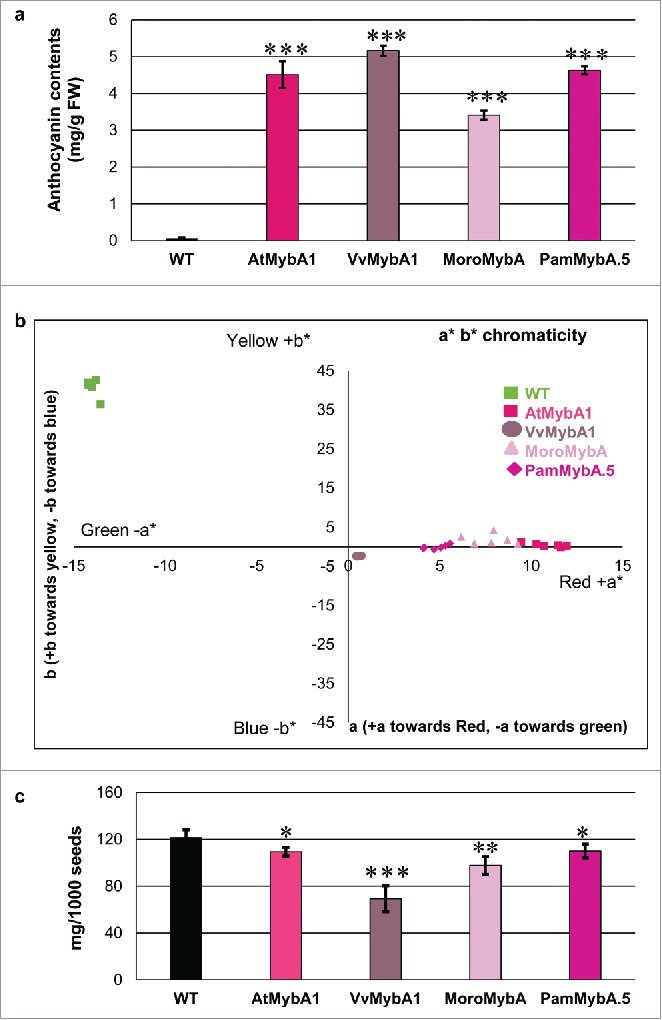
Phenotype analysis of the MybA transgenic tobacco lines (a) Anthocyanin quantification of leaf extracts from 50 day old transgenic tobacco plants. The total anthocyanin contents were spectrophotometrically determined by measuring the absorption of the extracts at wavelengths of 530nm and 657nm. The anthocyanin content of the transgenic samples were significantly higher than wild type (P<0.005) (b) Colorimetric analysis of transgenic tobacco leaves using leaf color measurement as a parameter to compare the intensity of anthocyanin accumulation. (c) Average seed yield represented as weight of 1000 seeds from wild type and transgenic tobacco lines. The error bars indicate the Standard Error of the 12 biological replicates and the Student's t test values are indicated * = P<0.5, ** = P<0.05 and *** = P<0.005.
Leaf color spectrophotometric measurement was further used to compare the anthocyanin content from leaf disks from the youngest leaves of the 50 day old plants. Leaves were sampled for color measurement with a Konica-Minolta CM-700d/600d spectrophotometer according to the CIE L* a* b* system (Hunter 1948). Both PamMybA.5 and MoroMybA clustered in the +a +b quadrant indicating accumulation of red color pigment. As a control, the bright red color of a strawberry extract was also measured (not shown on graph). The strawberry samples had an average +a value of 46.75 and +b value of 19.89. The VvMybA1 samples clustered +a-b values close to the [0,0] x,y value consistent with the almost black coloration on the leaves as shown in Fig. 6b. The Arabidopsis MybA samples displayed a slightly higher +a average value of 10.71 and +b average value of 0.42 than the PamMybA or MoroMybA samples indicating a more pinkish hue. PamMybA.5 displayed +a average value of 4.81 and +b average value of −0.02, whereas MoroMybA gave +a average value of 7.83 and +b average value of 1.98. As expected, the wild type leaf samples produced an -a+b (−13 and +40 respectively) value indicating a greenish yellow coloration (Fig. 6b). These results independently quantify the visibly distinct anthocyanin accumulation colors observed in the transgenic plants for the different MybA constructs.
Finally, we also examined thousand seed weight of the transgenic lines. The AtMybA1 and PamMybA.5 lines had lower seed weight than wild type (Student's t-test P<0.5), whereas the MoroMybA tobacco plants produced a significantly lower test weight (Fig. 6c) (Student's t-test P<0.05). The AtMybA1 and PamMybA.5 seed weight was 90% relative to wild type whereas MoroMybA was 80% relative to wild type. Seed weight was even more dramatically affected in VvMybA1 transgenic lines with only 57% seed weight relative to WT (Fig. 6c) suggesting a negative correlation between anthocyanin accumulation and reproductive fecundity.
Transgenic Citrus Plants Carrying Different Myb Genes Driven by the db35S Promoter Show Varying Patterns of Anthocyanin Accumulation
The MybA genes from plum, citrus and grape were transformed into citrus and used to validate and compare their potential for activating anthocyanin accumulation. VvMybA1 under the db35S promoter in in vitro transgenic citrus plants showed anthocyanin accumulation in the leaves, stem and roots (Fig. 7). These plants exhibited intense pigmentation, curled leaves, reduced regeneration capacity from tissue culture and reduced rates of growth.
FIGURE 7.
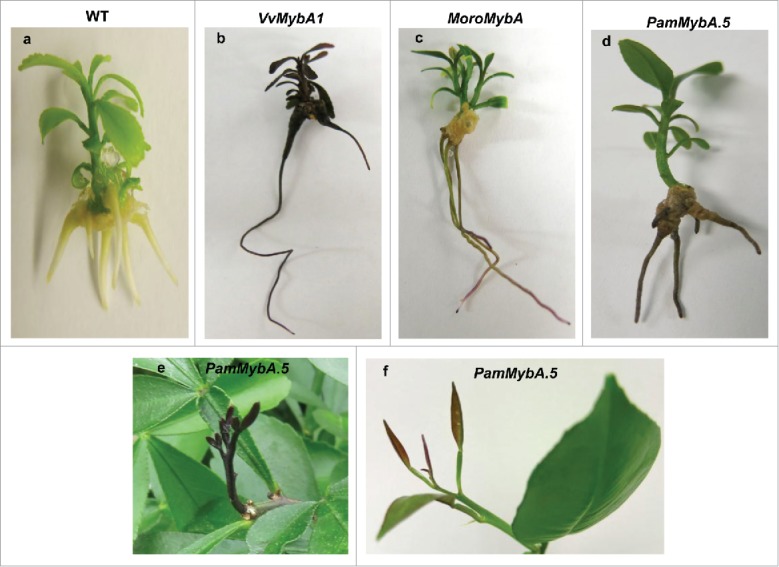
Comparison of the visible phenotypes in the MybA transgenic plantlets and acclimated citrus lines (a) 75 day old in vitro Carrizo regenerated plantlet (control) (b) 75 day old in vitro VvMybA1 regenerated plantlet with visible anthocyanin accumulation in leaves, stem, and roots (c) 75 day old in vitro PamMybA.5 regenerated plantlets expressing faint anthocyanin expression in leaves and roots (d) 75 day old in vitro MoroMybA regenerated plantlet with faint anthocyanin coloration in roots (e) Shoot apex in a PamMybA.5 Carrizo transgenic line showing strong anthocyanin accumulation in a new developing shoot (f) Anthocyanin accumulation in young leaves of the shoot apex in a greenhouse grown PamMybA.5 Hamlin plant.
A similar phenotype has been previously reported in transgenic Hamlin sweet orange and Mexican lime expressing VvMybA1, which resulted in variable expression and was not suitable as a screenable marker (Stover et al., 2013; Dutt et al., 2016). Our work with the transgenic tobacco plants overexpressing the VvMybA1 confirms a detrimental effect on more than half of the regenerated transgenic lines with reduced seed weight (Fig. 6). It is possible that there is an anthocyanin production threshold in plants beyond which high levels of anthocyanin inhibits growth and development. However, citrus transformed with PamMybA.5 and MoroMybA exhibited a high rate of transformation and plant regeneration. Transformation of Hamlin with the VvMybA1 was inefficient at a rate of only 2.5%, while the PamMybA.5 and MoroMybA constructs showed rates of transformation at 22% and 31%, respectively (Table 1). The in vitro PamMybA.5 transgenic plantlets typically exhibited light purple coloration in adventitious roots and faint anthocyanin accumulation in young regenerated leaves (Fig. 7) that disappears as the tissues mature. Greenhouse grown PamMybA.5 plants produced purple apices (Fig. 7, 8) similar to previously published findings for CsRuby transgenic citrus (Dutt et al., 2016). These results are similar to but distinct from that seen in tobacco where the PamMybA.5 transgenic plants produced a mosaic of anthocyanin accumulation in the leaves while MoroMybA plants had a weaker but more uniform coloration. These results clearly indicate the need to individually investigate the function of the MybA transcription factors in the crop species of interest to determine their desirability for engineering anthocyanin accumulation phenotypes.
TABLE 1.
Effect of MybA gene selectable marker on Agrobacterium-mediated transformation in ‘Hamlin’ sweet orange.
| Construct | Number of explants | Number of regenerated plants | Number of plants with purple young leaves | Number of PCR positive plants | Transformation efficiency (%) |
|---|---|---|---|---|---|
| 35Sp-MoroMybA | 80 | 25 | 18 | 25 | 31.2 |
| 35Sp-PamMybA.5 | 80 | 18 | 9 | 18 | 22.5 |
| 35Sp-VvMybA1 | 80 | 2 | 2 | 2 | 2.5 |
FIGURE 8.
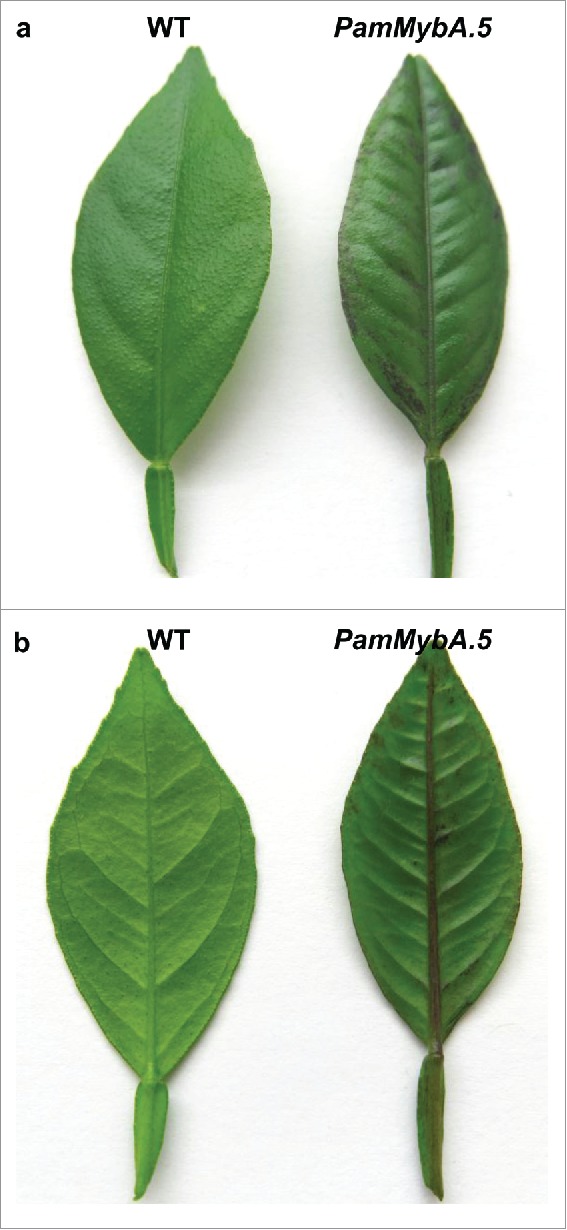
(a) Adaxial and (b) Abaxial surfaces in young leaves in a Hamlin sweet orange non transgenic plant (left), and a PamMybA.5 expressing transgenic plant (right).
DISCUSSION
Anthocyanins are important for various fundamental functions in plants. They've been shown to deter pathogens and predators, provide protection against various stresses and help in recruiting insect pollinators (Hernandez et al., 2009; Kutchan 2001; Karageorgou and Manetas 2006). Nutritionally, anthocyanins provide a major source of antioxidants in the diet to combat diseases. Due to these beneficial properties, identifying, isolating and characterizing new MybA genes can aid metabolic engineering efforts to produce anthocyanins within various plants. Plum is a commercial fruit grown and consumed worldwide, thus characterizing PamMybA TFs provides useful crop-derived molecular tools for biotechnological applications by providing novel genes which confer differential patterns of anthocyanin accumulation and coloration.
Our results show that the overexpression of PamMybA.1 and PamMybA.5 produce obvious anthocyanin pigmentation in the leaves and other tissues of transgenic tobacco and citrus. The leaves typically had a mosaic pattern of accumulation, whereas the sepals, petals, seed pods and seeds showed a more uniform pigmentation (Fig. 4, 5). Even though the same cauliflower mosaic virus db35S promoter was consistently used to express each of the MybA genes, different anthocyanin accumulation patterns were observed (Figs. 3–5). The phenotypic differences in pigment accumulation and the distribution within the transgenic plants may be due to differences in the availability and binding capacity of endogenous regulatory proteins, such as bHLH and WD40, when complexed with the MYB proteins to form a functional complex (Butelli et al., 2008). Our results show that the PamMYBA.1 and PamMYBA.5 TFs exhibit distinct functional interactions with the native tobacco anthocyanin gene expression complex, activating unique patterns of biosynthesis and accumulation compared with previous characterized MybA genes from other species. Interestingly, even proteins with very similar sequences result in noticeably different patterns of anthocyanin accumulation in the tissues, suggesting that subtle changes in the protein structure can have dramatic functional effects. It has been demonstrated that the MdMYB10R6 gene from apple is capable of self-regulating in heterologous host species and enhancing organ/tissue specific anthocyanin pigmentation. But, the activity of MdMYB10 is dependent on endogenous protein partners. Stable transgenic petunias containing MdMYB10R6 lacked foliar pigmentation but had colored flowers. The absence of foliar pigmentation in vegetative tissues was likely due to the failure of the MdMYB10R6 gene to form a functional complex within those cell types. This implies that additional protein partners are required for MdMYB10 to form the correct activation complex and target the genes in this heterologous system (Boase et al., 2015). Alternatively, differences in intron sequences may confer differential tissue specificity or mRNA stability of the MybA transgenes and thus change the anthocyanin accumulation pattern. The MoroMybA transgene has 97bp and 795bp introns, while PamMybA.5 has 305bp and 980bp introns respectively (Fig. S1). The grape MybA has 87bp and 119bp introns while the Arabidopsis MybA has 540bp and 89bp introns (Fig. S1). The size of the second introns is significantly smaller in grape and Arabidopsis compared with MoroMybA and PamMybA.5. It is possible that these intronic differences could alter transgene expression or the efficiency of splicing and thus lead to differences in the anthocyanin accumulation as observed in our work.
In the present study, the grape gene induced a stronger phenotype than either the citrus or the PamMybA.5 gene, as evident by the obviously different pigmentation within the leaves and flowers (Fig. 5). AtMybA1 and PamMybA.5 genes exhibited similar flower color phenotype. Interestingly both the grape and plum MybAs also exhibited the strongest mosaic expression pattern within the leaves. The Arabidopsis and citrus MybAs on the other hand displayed a weaker but more uniform accumulation pattern within the leaves especially during the later developmental stages. In this context, comparing the MybAs in a heterologous system clearly demonstrated the specific phenotypic pattern that can be expected from the various MybAs for biotechnology utilization. Another important aspect for consideration is the deleterious effect of excess anthocyanin accumulation in plants. It has been shown that overexpression of maize anthocyanin regulatory gene Lc has a detrimental effect on plant growth with reduced transformation efficiency (Bradley et al., 1998). Similar growth inhibition was also observed in both transgenic tomato and Arabidopsis lines transformed with the Lc gene (Goldsbrough et al., 1996; Lloyd et al., 1994). This clearly suggests that there is some threshold level at which the transcription factor or anthocyanin accumulation becomes detrimental to the plants. One possible explanation for such phenotypes can be crosstalk between cell wall formation and anthocyanin accumulation. Bhargava et al showed that MYB75/PAP1 has an additional role in regulating secondary cell wall formation in the Arabidopsis stem. Reduced expression or loss of MYB75 function results in the channeling of carbon toward the lignin pathway with increased accumulation in the secondary cell walls (Bhargava et al., 2010). MYB75 was shown to interact with transcription factor such as KNAT7, which plays a role in secondary cell wall formation in Arabidopsis. On the other hand overexpression of MYB75 leads to enhanced carbon flow into the flavonoid pathway with activation of anthocyanin biosynthesis (Li 2009; Bhargava et al., 2010). Lignin provides structural support for land plants and plays important role in plant integrity, growth and development. Reduced amount of lignin causes loss in plant structural integrity and hence explains the detrimental phenotype observed in some of our VvMybA1 overexpressing lines (Fig 6). This suggests that there exists a fine balance of carbon flow between anthocyanin and lignin accumulation necessary for normal plant growth and development.
Recently, Huang et al. (2013a) showed that overexpression of R2R3-MYB EsMybA1 from Epimedium sagittatum herb alone induces strong anthocyanin accumulation in transgenic tobacco and Arabidopsis via upregulation of the main flavonoid biosynthetic pathway genes. This result indicates that the expression of a single R2R3-MYB regulatory gene alone is sufficient for the activation of anthocyanin biosynthesis without the need for the transgenic co-expression of a bHLH protein. This will likely be a significant advantage of engineering anthocyanins in economically important plants. MYB TFs such as IbMYB1a, VlMYBA2, and EsMYBA1 do not require transgenic expression of a bHLH partner for anthocyanin pigmentation in Arabidopsis and tobacco (Huang et al., 2013a; Mano et al., 2007; Azuma et al., 2012). Our results are consistent with these findings where transgenic overexpression of the plum, citrus, Arabidopsis and grape MybA TF's alone were sufficient for anthocyanin accumulation. However in some cases, only petals and seeds accumulate anthocyanins as seen in apple MdMyb1 and Chinese bay berry MrMyb1 transgenic tobacco (Huang et al., 2013b; Ban et al., 2007).
More characterization of novel MYBs will help us better understand the mechanisms behind such interactions and how to control them in a tissue-specific manner. Our results clearly show that the novel plum PamMYA.1 and PamMYBA.5 we identified can function without requiring any other exogenous cofactors such as bHLH in the heterologous systems of tobacco and citrus (Fig. 5, 7, 8). This opens up the possibility for the development of modified fruits and vegetables using these plum genes. For example, it has recently been shown that transgenic ‘Mexican’ lime plants overexpressing the VvMybA1 or the CsRuby transcription factor, resulted in the production of anthocyanins in a citrus cultivar that does not produce anthocyanin naturally (Dutt et al., 2016). However, these lines were produced using the 35S promoter and relied on positional effects to produce anthocyanin accumulation specifically in the fruit. Alternative approach to generating a desirable phenotype would be through the use of fruit-specific promoters such as the tomato E8 or PG promoters (Kneissl et al., 1996; Montgomery et al., 1993). Other organ-specific promoters could also be potentially used to produce ornamental plants with novel and pleasing pigmentation.
An important potential biotech application for the MybA genes are their use as a visual marker in transgenic plants. Several of the MybA genes have already been identified as possible candidates for use this way, and can serve as a simple screenable phenotype indicating plant transformation (Kortstee et al., 2011; Geekiyanage et al., 2007; Goldsbrough et al., 1996; Kim et al., 2010; Li et al., 2011; Ludwig et al., 1990). Lim et al, showed that co-transformation with the R2R3 MYB mPAP1 gene from Arabidopsis and the basic helix loop helix B-Peru gene from maize has potential utility as an alternative visible selectable marker system for transgenic tobacco (Lim et al., 2012). Unlike other commonly used reporter genes, an anthocyanin activating transcription factor gene does not require an exogenous substrate and/or ex vivo assays (Li et al., 2011). For example the most commonly used reporter gene in plants (uidA) requires the addition of the β-glucuronidase enzymatic substrate, while an anthocyanin marker is directly capable of providing a detectable and quantifiable indicator of expression. This allows the detection of promoter activity in a non-destructive and real-time manner. Similarly, detection of GFP expression, another commonly used reporter, relies on excitation with UV or blue light and the detection of fluorescence with specialized detection equipment, but observation of anthocyanin accumulation only requires a simple visible inspection. Thus, using anthocyanin accumulation as a reporter would enable time-lapse imaging of plant samples for studying transgene expression activity (Zhou et al., 2005) and can be potentially used as an alternative method for the selection of transgenic plants instead of antibiotic or herbicide resistance.
CONCLUSIONS
Our present study shows that the PamMybA.1 or the PamMybA.5 genes are functional in tobacco and citrus to enhance anthocyanin biosynthesis, providing a desirable and consistent phenotype without requiring any additional cofactors. PamMybA.1 and PamMybA.5 genes are also promising candidates for various biotech applications such as generating blood orange-like cultivars with fruit that uniformly accumulates anthocyanins, but no longer requires specific environmental conditions to generate their appearance. This study demonstrates that these MybAs can also be used as simple visible markers for plant transformation in these 2 species. Based on our results, expression of the PamMYBA.1 or PamMYBA.5 in tobacco or citrus could be used to identify transgenic plants via the production of visible pigment accumulation in tissue culture. In our experiments, all the primary tobacco transformants and their progeny containing the PamMybA.5 gene, displayed anthocyanin pigmentation in tobacco (Fig. 4, 5, 6, S2). This gene has further been demonstrated to produce anthocyanin accumulation in citrus as well (Fig. 7).
The utilization of the MybA genes not only enables simple, versatile selection of transformed tissues through visualization (Fig. 3), but also can be used to increase the nutritional or aesthetic value of a transgenic plant by increasing its anthocyanin levels. As the understanding of the potential health benefits conferred by anthocyanins has increased over the past decade, interest in consuming anthocyanin-rich plant tissues has also increased dramatically.
MATERIALS AND METHODS
Isolation, Phylogenetic Analysis and Cloning of the Myb Genes
MoroMybA is a synthetic version of CsRuby with an inconvenient restriction site removed. The sequence was generated by GenScript (GenScript USA Inc.), and is available in GenBank accession JN4002334 (also Supplemental text. S1). The MoroMybA sequence was PCR (Polymerase Chain Reaction) amplified, cloned at the SbfI restriction site into the pCTAG7 binary vector (GenBank KT992772) under control of the double enhanced CaMV 35S promoter. The pCTAG7 binary vector is a novel plant genetic transformation platform that contains recombinase recognition sites that enables recombinase mediated cassette exchange (RMCE) for targeted integration of gene(s) of interest and removal of the antibiotic resistance marker (Wang et al., 2011).
Primers that aligned to the domestic plum MybA, PdmMyb10 (EU153580.1) and the Arabidopsis PAP1 gene (AF325123.1) were designed and used to amplify the MybA genes from feral plum genomic DNA isolated from a tree grown in El Cerrito, CA. The primers used are Plum MybA F62 5′- ATGGAGGGATATAACTTGGGTGTGAGAAAAGGAG- 3′ and Plum Myb R61 5′-CTATTCTTCTTTTGAATGATTCCAAAGGTCCACGC- 3′. The DNA fragments were sequenced (GenBank accessions KT992773–6) and cloned at the SbfI restriction site into the pCTAG7 binary vector (Supplemental text S1 for the 3 Plum MybA sequences).
Using gene specific primers, a 1.3kb genomic fragment was amplified from Arabidopsis Col-0 genomic DNA (AtMybA1 F62 5′- ATGGAGGGTTCGTCCAAAGGGC- 3′, AtMybA1 R61 5′-CTAATCAAATTTCACAGTCTCTCCATCGAAAAGACTCC- 3′) and a 0.96kb fragment of the merlot grape VvMybA1 gene (VvMybA1 F60 5′- ATGGAGAGCTTAGGAGTTAGAAAGGGTG- 3′, VvMybA1 R60 5′- ACTAGTCTAGTTCAGATCAAGTGATTTACTTGTGTG- 3′) from the pDEAT plasmid (a gift of Dennis Gray). AtMybA1 is the genomic sequence of Arabidopsis MybA1 (cDNA version is referred to as PAP1). The primers for amplifying the VvMybA1 were designed based on the available sequence (AB111101; Li et al., 2011; Borevitz et al., 2000; This et al., 2007). The resulting PCR fragments were fused to the double enhanced CaMV 35S promoter in the binary vector pCTAG7 at the SbfI restriction site and sequence confirmed. All pCTAG7-db35Sp-MybA-35St binary vectors were transformed into Agrobacterium tumefaciens strain GV3101 and selected on LB plates supplemented with kanamycin and gentamycin at 100 mg/L.
For phylogenetic analysis of the MYB protein sequences, a multiple sequence alignment was performed using Jalview 2.8.2 (http://www.jalview.org/; Waterhouse et al., 2009). The amino acid sequences of Myb genes were retrieved from GenBank database, and their accession numbers are embedded in Fig. 1. ClustalWS alignment program was used for multiple alignment (Thompson et al., 1994). The phylogenetic tree was calculated using BLOSUM62 Average Distance from the ClustalWS alignment. The percent sequence identity was calculated using a pairwise alignment. The default parameters for ClustalWS and BLOSUM62 were used.
Plant Transformation and Transgenic Plant Regeneration
Tobacco (Nicotiana tabacum L. cv. Petit Havana SR1) was used for leaf disk transformation (Horsch and Klee 1986). Agrobacterium cells were grown to an optical density of 1.0 at 600 nm (OD600), and a final suspension at OD600 of 0.5 was used for plant infection. Young, healthy green leaves were cut into pieces approximately 10 mm in length, and the leaf segments were incubated in an Agrobacterium suspension for 30 minutes. The leaf segments were then blotted dry on sterile filter paper for 5 min and placed onto MS co-cultivation medium (Sigma, St. Louis, MO) in sterile Petri dishes and kept in the growth chamber at 25°C for 3 d in the dark. The infected leaf explants were then transferred to regeneration/selection medium (24°C with 16 h of light and 8 h of dark at 20°C). Primary tobacco transformants were selected on MS medium, with 100 mg/mL kanamycin. After 2–3 weeks, the infected leaf explants were transferred onto fresh regeneration/selection media. Separate shoots from explants were excised carefully and transferred into plant culture dishes containing rooting medium. Rooted plants were grown in Sunshine potting mix (Sun Gro Horticulture Ltd. Agawam, MA) in the greenhouse with 16 h of light at 150 photosynthetic photon flux density (μmol photons m−2 s−1) at 23°C and 8 h of dark at 20°C with 70% humidity. Twenty kanamycin resistant lines were obtained from the T0 generation for each construct. T1 seeds were collected and selected on an MS plate supplemented with kanamycin 100 mg / mL. After 25 days, T1 plants were transferred from plates to soil, characterized and dried seed pods were collected for measuring seed weight per 1000 seeds from individual plants.
The transgenic ‘Carrizo’ citrange and ‘Hamlin’ sweet orange lines were generated using epicotyl explants protocol as described previously by Oliveira et al. (2009).
Polymerase Chain Reaction (PCR) Analysis
Genomic DNA was extracted by grinding a 1 cm2 piece of tobacco leaf in 400 μL of buffer (200 mm Tris–HCl pH 7.8, 250 mm NaCl, 25 mm EDTA, 0.5% SDS). After centrifugation and isopropanol precipitation, the pellet was washed with 70% ethanol and resuspended in 50 μL of water with 1mM RNaseA. PCR amplification was performed using 2 μL of genomic DNA in reactions with a total volume of 25 μL. Presence of the transgene was confirmed by PCR using transgene specific primers specifically binding to the cauliflower mosaic virus (CaMV) double35S promoter (hereafter defined as db35S) promoter and 35S terminator region in Fig. 2 and Fig. S1 (35S F62 5′-GGATTGATGTGATATCTCCACTGACGTAAGG- 3′ and 35S Term R60 5′-CCTTATCTGGGAACTACTCACACATTATTCTGGAG- 3′).
Imaging of Transgenic Plants
The photographs of the plants were recorded using a Nikon D7000 digital camera with an AF Micro Nikkor 60 mm 1:2.8 D lens or AF-S Nikkor 18–70 mm DX lens (Nikon Inc., Melville, NY) under tungsten lamps (Philips, 120 V, 300 W). The camera was set manually for all parameters including ISO sensitivity, focus, f-stop and time. A photography gray card was used as a reference to get the correct exposure. Individual flowers were set lengthwise on a round frosted glass to view stem color differences. The top side of flowers were prepared to keep them upright by setting individual flowers stem-down into the barrel of a 10 mL syringe with the inside bore cut out. Flower preparations were set upon an 8×10” piece of glossy photo paper (Epson Premium Glossy Photo Paper) as a clean, white background, to view and compare the flower and stem color.
The callus images in petri plates were observed and photographed in a Leica MZ16-F (Leica Microsystems, Inc., Buffalo Grove, IL) stereo zoom light microscope equipped with a QImaging Retiga 2000 R fast cooled, digital color camera. For seed pods, the outer bracts were removed from one side before photographing.
Extraction of Anthocyanin and Measurement by Spectrophotometry
Leaf tissue samples from each of the independent tobacco lines were collected, weighed and ground in liquid nitrogen. Total anthocyanin was extracted from finely ground plant material using methanolic HCl and measured spectrophotometrically at wavelengths 530 nm and 657 nm as described by Chu et al. (2013). All samples were measured in duplicates with 6 biological replicates for each independent line. The error bars indicate the standard error (SE) of the average anthocyanin content. The anthocyanin content is expressed as absorbance (Abs) in mg/g FW which is calculated as described in Michael and Chory 1998 and Chu et al. (2013).
Quantification of Leaf Color Using Colorimeter
The color of tobacco leaf discs collected from the center of the leaves was characterized using a color reflectance measurement instrument. A Konica-Minolta CM-700d/600d spectrophotometer was used, equipped with CM-S100w SpectraMagic NX software, D65 illuminant. The spectrophotometer recorded an integrated color, the product of the reflectance of the different colors reflected in the same measurement and a direct function of the color of leaf disc. The numeric information on color is in standard color spaces (CIE L*a*b*) (Hunter 1948). Use of the CIELAB system enables estimation of 3 classical color parameters: L* a*, and b*, where L* represents lightness, a* represents the position between red and green on the redness (+a) to greenness (-a) axis, and b* represents the position between yellow and blue on the yellowness (+) to blueness (−) axis. Six biological reps were used for each independent representative line. For each plant, 3 young leaves were selected and leaf discs from the center of the leaves were used for color measurements.
Supplementary Material
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
The authors declare that they have no conflict of interest.
ACKNOWLEDGMENTS
We would like to thank De Wood for her contribution in imaging the plants and Dennis Gray for the pDEAT plasmid vector.
FUNDING
This research was financially supported by California Citrus Research Board 5200–141. This research was also supported by USDA Agricultural Research Service CRIS projects 5325–21000–018, 5325–21000–020, 6618–21000–014–00. Mention of trade names or commercial products is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. USDA is an equal opportunity provider and employer.
AUTHOR CONTRIBUTION STATEMENT
The authors have made the following declarations regarding their contributions: JT conceived the project; KD designed and performed the experiments with tobacco: MLO and ES designed and performed the experiments with citrus. KD, JT and RT analyzed the data and wrote the manuscript.
REFERENCES
- Allan AC, Hellens RP, Laing WA. MYB transcription factors that color our fruit. Trends Plant Sci 2008; 13:99-102; PMID:18280199; http://dx.doi.org/ 10.1016/j.tplants.2007.11.012 [DOI] [PubMed] [Google Scholar]
- Azuma A, Yakushiji H, Koshita Y, Kobayashi S. Flavonoid biosynthesis-related genes in grape skin are differentially regulated by temperature and light conditions. Planta 2012; 236:1067-80; PMID:22569920; http://dx.doi.org/ 10.1007/s00425-012-1650-x [DOI] [PubMed] [Google Scholar]
- Azuma A, Kobayashi S, Yakushui H, Yamada M, Mitani N, Sato A. VvmybA1 genotype determines grape skin color. Vitis 2007; 46(3):154-5. [Google Scholar]
- Ban Y, Honda C, Hatsuyama Y, Igarashi M, Bessho H, Moriguchi T. Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol 2007; 48:958-70; PMID:17526919; http://dx.doi.org/ 10.1093/pcp/pcm066 [DOI] [PubMed] [Google Scholar]
- Bhargava A, Mansfield SD, Hall HC, Douglas CJ, Ellis BE. MYB75 functions in regulation of secondary cell wall formation in the Arabidopsis inflorescence stem. Plant Physiol 2010; 154:1428-38; PMID:20807862; http://dx.doi.org/ 10.1104/pp.110.162735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley JM, Davies KM, Deroles SC, Bloor SJ, Lewis DH. The maize Lc regulatory gene up-regulates the flavonoid biosynthetic pathway of Petunia. Plant J 1998; 13(3):381-92; http://dx.doi.org/ 10.1046/j.1365-313X.1998.00031.x [DOI] [Google Scholar]
- Boase MR, Brendolise C, Wang L, Ngo H, Espley RV, Hellens RP, Schwinn KE, Davies KM, Albert NW. Failure to launch: The self-regulating Md -MYB10R6 gene from apple is active in flowers but not leaves of Petunia. Plant Cell Rep 2015; 34:1817-23; PMID:26113165; http://dx.doi.org/ 10.1007/s00299-015-1827-4 [DOI] [PubMed] [Google Scholar]
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 2000; 12:2383-94; PMID:11148285; http://dx.doi.org/ 10.1105/tpc.12.12.2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss PK, Davies C, Robinson SP. Analysis of the expression of anthocyanin pathway genes in developing Vitis vinifera L. cv Shiraz grape berries and the implications for pathway regulation. Plant Physiol 1996; 111:1059-66; PMID:12226348; http://dx.doi.org/ 10.1104/pp.111.4.1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butelli E, Licciardello C, Zhang Y, Liu J, Mackay S, Bailey P, Reforgiato-Recupero G, Martin C. Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell 2012; 24:1242-55; PMID:22427337; http://dx.doi.org/ 10.1105/tpc.111.095232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butelli E, Titta L, Giorgio M, Mock HP, Matros A, Peterek S, Schijlen EGWM, Hall RD, Bovy AG, Luo J, et al.. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nature Biotechnol 2008; 26:1301-8; http://dx.doi.org/ 10.1038/nbt.1506 [DOI] [PubMed] [Google Scholar]
- Chu H, Jeong JC, Kim WJ, Chung DM, Jeon HK, Ahn YO, Kim SH, Lee HS, Kwak SS, Kim CY. Expression of the sweet potato R2R3-type IbMYB1a gene induces anthocyanin accumulation in Arabidopsis. Physiol Plant 2013; 148:189-99; PMID:23039825; http://dx.doi.org/ 10.1111/j.1399-3054.2012.01706.x [DOI] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C. MYB transcription factors in Arabidopsis. Trends Plant Sci 2010; 15:573-81; PMID:20674465; http://dx.doi.org/ 10.1016/j.tplants.2010.06.005 [DOI] [PubMed] [Google Scholar]
- Dutt M, Stanton D, Grosser JW. Ornacitrus: Development of genetically modified anthocyanin-expressing citrus with both ornamental and fresh fruit potential. J Amer Soc Hort Sci 2016; 141(1):54-61. [Google Scholar]
- Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC. Red coloration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J 2007; 49(3):414-27; PMID:17181777; http://dx.doi.org/ 10.1111/j.1365-313X.2006.02964.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller A, Machemer K, Braun EL, Grotewold E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J 2011; 66:94-116; PMID:21443626; http://dx.doi.org/ 10.1111/j.1365-313X.2010.04459.x [DOI] [PubMed] [Google Scholar]
- Geekiyanage S, Takase T, Ogura Y, Kiyosue T. Anthocyanin production by over-expression of grape transcription factor gene VlmybA2 in transgenic tobacco and Arabidopsis. Plant Biotechnol Rep 2007; 1:11-8; http://dx.doi.org/ 10.1007/s11816-006-0001-4 [DOI] [Google Scholar]
- Goldsbrough AP, Tong Y, Yoder JI. LC as a non-destructive visual reporter and transposition excision marker gene for tomato. Plant J 1996; 9:927-33; http://dx.doi.org/ 10.1046/j.1365-313X.1996.9060927.x [DOI] [Google Scholar]
- Gonzali S, Mazzucato A, Perata P. Purple as a tomato: Towards high anthocyanin tomatoes. Trends Plant Sci 2009; 14:237-41; PMID:19359211; http://dx.doi.org/ 10.1016/j.tplants.2009.02.001 [DOI] [PubMed] [Google Scholar]
- Hernandez I, Alegre L, Breusegem FV, Munne-Bosch S. How relevant are flavonoids as antioxidants in plants. Trends Plant Sci 2009; 14:125-32; PMID:19230744; http://dx.doi.org/ 10.1016/j.tplants.2008.12.003 [DOI] [PubMed] [Google Scholar]
- Hichri I, Barrieu F, Bogs J, Kappel C, Delrot S. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J Exp Bot 2011; 62:2465-83; PMID:21278228; http://dx.doi.org/ 10.1093/jxb/erq442 [DOI] [PubMed] [Google Scholar]
- Honda C, Kotoda N, Wada M, Kondo S, Kobayashi S, Soejima J, Zhang Z, Tsuda T, Moriguchi T. Anthocyanin biosynthetic genes are coordinately expressed during red coloration in apple skin. Plant Physiol Biochem 2002; 40:955-62; http://dx.doi.org/ 10.1016/S0981-9428(02)01454-7 [DOI] [Google Scholar]
- Horsch RB, Klee HJ. Rapid assay of foreign gene expression in leaf discs transformed by Agrobacterium tumefaciens: role of T-DNA borders in the transfer process. Proc Natl Acad Sci U S A 1986; 83:4428-32; PMID:16593716; http://dx.doi.org/ 10.1073/pnas.83.12.4428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Sun W, Lv H, Luo M, Zeng S, Pattanaik S, Yuan L, Wang Y. A R2R3-MYB transcription factor from Epimedium sagittatum regulates the glavonoid biosynthetic pathway. PLoS ONE 2013a; 8(8):e70778; PMID:23936468; http://dx.doi.org/ 10.1371/journal.pone.0070778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YJ, Song S, Allan AC, Liu X, Yin XR, Xu CJ, Chen KS. Differential activation of anthocyanin biosynthesis in Arabidopsis and tobacco over-expressing an R2R3 MYB from Chinese bayberry. Plant Cell Tiss Org Cult 2013b; 113:491-9; http://dx.doi.org/ 10.1007/s11240-013-0291-5 [DOI] [Google Scholar]
- Hunter RS. Photoelectric Color-Difference Meter. JOSA 1948; 38(7):661 (Proceedings of the Winter Meeting of the Optical Society of America). [Google Scholar]
- Karageorguo P, Manetas Y. The importance of being red when young: anthocyanins and the protection of young leaves of Quercus coccifera from insect herbivory and excess light. Tree Physiol 2006; 26(5):613-21; PMID:16452075; http://dx.doi.org/ 10.1093/treephys/26.5.613 [DOI] [PubMed] [Google Scholar]
- Kutchan TM. Ecological arsenal and developmental dispatcher. The paradigm of secondary metabolism. Plant Physiol 2001; 125:58-60; PMID:11154296; http://dx.doi.org/ 10.1104/pp.125.1.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CY, Ahn YO, Kim SH, Kim YH, Lee HS, Catanach AS, Jacobs JM, Conner AJ, Kwak SS. The sweet potato IbMYB1 gene as a potential visible marker for sweet potato intragenic vector system. Physiol Plantar 2010; 139:229-40; http://dx.doi.org/ 10.1111/j.1399-3054.2010.01353.x [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Yamamoto NG, Hirochika H. Retrotransposon-induced mutations in grape skin color. Science 2004; 304:982; PMID:15143274; http://dx.doi.org/ 10.1126/science.1095011 [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Yamamoto NG, Hirochika H. Association of VvmybA1 gene expression with anthocyanin production in grape (Vitis vinifera) skin - color mutants. J Jpn Soc Hort Sci 2005; 74:196-203; http://dx.doi.org/ 10.2503/jjshs.74.196 [DOI] [Google Scholar]
- Kortstee AJ, Khan SA, Helderman C, Trindade LM, Wu Y, Visser RG, Brendolise C, Allan AC, Schouten HJ, Jacobsen E. Anthocyanin production as a potential visual selection marker during plant transformation. Transgenic Res 2011; 20:1253-64; PMID:21340526; http://dx.doi.org/ 10.1007/s11248-011-9490-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai B, Li X-J, Hu B, Qin YH, Huang XM, Wang HC, Hu GB. LcMYB1 is a key determinant of differential anthocyanin accumulation among genotypes, tissues, developmental phases and ABA and light stimuli in Litchi chinensis. PLoS ONE 2014; 9(1):e86293; PMID:24466010; http://dx.doi.org/ 10.1371/journal.pone.0086293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai B, Du LN, Liu R, Hu B, Su WB, Qin YH, Zhao JT, Wang HC, Hu GB. Two LcbHLH transcription factors interacting with LcMYB1 in regulating late structural genes of anthocyanin biosynthesis in Nicotiana and Litchi chinensis during anthocyanin accumulation. Front Plant Sci 2016; 7:166; PMID:26925082; http://dx.doi.org/ 10.3389/fpls.2016.00166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E. Identification and characterization of regulatory genes associated with secondary wall formation in Populus and Arabidopsis thaliana. PhD thesis University of British Columbia, Vancouver, Canada: 2009. [Google Scholar]
- Lim SH, Sohn SH, Kim DH, Kim JK, Lee JY, Kim YM, Ha SH. Use of an anthocyanin production phenotype as a visible selection marker system in transgenic tobacco plant. Plant Biotech Rep 2012; 6(203):203-11; http://dx.doi.org/ 10.1007/s11816-012-0215-6 [DOI] [Google Scholar]
- Lin-Wang K, Bolitho K, Grafton K, Kortstee A, Karunairetnam S, McGhie TK, Espley RV, Hellens RP, Allan AC. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol 2010; 10:50-66; PMID:20302676; http://dx.doi.org/ 10.1186/1471-2229-10-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Flachowsky H, Fischer TC, Hanke MV, Forkmann DG, Treutter , Schwab W, Hoffmann T, Szankowski I. Maize Lc transcription factor enhances biosynthesis of anthocyanins, distinct proanthocyanidins and phenylpropanoids in apple (Malus domestica Borkh.). Planta 2007; 226(5):1243-54; PMID:17618453; http://dx.doi.org/ 10.1007/s00425-007-0573-4 [DOI] [PubMed] [Google Scholar]
- Li ZT, Dhekney SA, Gray DJ. Use of the VvMybA1 gene for non-destructive quantification of promoter activity via color histogram analysis in grapevine (Vitis vinifera) and tobacco. Transgenic Res 2011; 20:1087-97; PMID:21229312; http://dx.doi.org/ 10.1007/s11248-010-9482-6 [DOI] [PubMed] [Google Scholar]
- Lloyd AM, Schena M, Walbot V, Davis RW. Epidermal cell fate determination in Arabidopsis: patterns defined by a steroid-inducible regulator. Science 1994; 266: 436–439. [DOI] [PubMed] [Google Scholar]
- Ludwig SR, Bowen B, Beach L, Wessler SR. A regulatory gene as a novel visible marker for maize transformation. Science 1990; 247:449-450; PMID:17788612; http://dx.doi.org/ 10.1126/science.247.4941.449 [DOI] [PubMed] [Google Scholar]
- Mano H, Ogasawara F, Sato K, Higo H, Minobe Y. Isolation of a regulatory gene of anthocyanin biosynthesis in tuberous roots of purple-fleshed sweet potato. Plant Physiol 2007; 143:1252-68; PMID:17208956; http://dx.doi.org/ 10.1104/pp.106.094425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng XB. Yin HL, Feng S, Zhang XQ, Liang QW Meng. Overexpression of R2R3-MYB gene leads to accumulation of anthocyanin and enhanced resistance to chilling and oxidative stress. Biol Plant 2014; 58(1):121-30; http://dx.doi.org/ 10.1007/s10535-013-0376-3 [DOI] [Google Scholar]
- Neff MM, Chory J. Genetic interactions between Phytochrome A, Phytochrome B, and Cryptochrome 1 during Arabidopsis development. Plant Physiol 1998; 118(1):27-35; PMID:9733523; http://dx.doi.org/ 10.1104/pp.118.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneissl ML, Deikman J. The tomato E8 gene influences ethylene biosynthesis in fruit but not in flowers. Plant Physiol 1996; 112:537-47; PMID:12226407; http://dx.doi.org/ 10.1104/pp.112.2.537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery J, Pollard V, Deikman J, Fischer RL. Positive and negative regulatory regions control the spatial distribution of polygalacturonase transcription in tomato fruit pericarp. Plant Cell 1993; 5(9):1049-62; PMID:8400876; http://dx.doi.org/ 10.1105/tpc.5.9.1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu SS, Xu CJ, Zhang WS, Zhang B, Li X, Lin-Wang K, Ferguson IB, Allan AC, Chen KS. Coordinated regulation of anthocyanin biosynthesis in Chinese bayberry Myrica rubra fruit by a R2R3 MYB transcription factor. Planta 2010; 231:887-99; PMID:20183921; http://dx.doi.org/ 10.1007/s00425-009-1095-z [DOI] [PubMed] [Google Scholar]
- Oliveira MLPD, Febres VJ, Costa MGC, Moore GA, Otoni WC. High-Efficiency Agrobacterium-Mediated Transformation of Citrus via Sonication and Vacuum Infiltration. Plant Cell Rep 2009; 28:387-95; PMID:19048258; http://dx.doi.org/ 10.1007/s00299-008-0646-2 [DOI] [PubMed] [Google Scholar]
- Petroni K, Tonelli C. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci 2011; 181:219-29; PMID:21763532; http://dx.doi.org/ 10.1016/j.plantsci.2011.05.009 [DOI] [PubMed] [Google Scholar]
- Qiu J, Sun S, Luo S, Zhang J, Xiao X, Zhang L, Wang F, Liu S. Arabidopsis AtPAP1 transcription factor induces anthocyanin production in transgenic Taraxacum brevicorniculatum. Plant Cell Rep 2014; 33(4):669-80; PMID:24556963; http://dx.doi.org/ 10.1007/s00299-014-1585-8 [DOI] [PubMed] [Google Scholar]
- Ramsay NA, Glover BJ. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci 2005; 10:63-70; PMID:15708343; http://dx.doi.org/ 10.1016/j.tplants.2004.12.011 [DOI] [PubMed] [Google Scholar]
- Spelt C, Quattrocchio F, Mol J, Koes R. ANTHOCYANIN1 of petunia controls pigment synthesis, vacuolar pH, and seed coat development by genetically distinct mechanisms. Plant Cell 2000; 14:2121-35; http://dx.doi.org/ 10.1105/tpc.003772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springob K, Nakajima J, Yamazaki M, Saito K. Recent advances in the biosynthesis and accumulation of anthocyanins. Nat Prod Rep 2003; 20:288-303; PMID:12828368; http://dx.doi.org/ 10.1039/b109542k [DOI] [PubMed] [Google Scholar]
- Stitt M, Sulpice R, Keurentjes J. Metabolic networks: how to identify key components in the regulation of metabolism and growth. Plant Physiol 2010; 152:428-44; PMID:20018593; http://dx.doi.org/ 10.1104/pp.109.150821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover E, Avila Y, Li ZT, Gray D. Transgenic expression in citrus of Vitis MybA1 from a bi directional promoter resulted in variable anthocyanin expression and was not suitable as a screenable marker without antibiotic selection. Proc Fla State Hort Soc 2013; 126:84-88. [Google Scholar]
- Stracke R, Werber M, Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 2001; 4:447-56; PMID:11597504; http://dx.doi.org/ 10.1016/S1369-5266(00)00199-0 [DOI] [PubMed] [Google Scholar]
- Takos AM, Jaffe FW, Jacob SR, Bogs J, Robinson SP, Walker AR. Light induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol 2006; 142:1216-32; PMID:17012405; http://dx.doi.org/ 10.1104/pp.106.088104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Sasaki N, Ohmiya A. Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J 2008; 54:733-49; PMID:18476875; http://dx.doi.org/ 10.1111/j.1365-313X.2008.03447.x [DOI] [PubMed] [Google Scholar]
- This P, Lacomb T, Cadle-Davidson M, Owens C. Wine grape (Vitis vinifera L.) color associates with allelic variation in the domestication gene VvmybA1. Theor Appl Genet 2007; 114:723-30; PMID:17221259; http://dx.doi.org/ 10.1007/s00122-006-0472-2 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 1994; 22:4673-80; PMID:7984417; http://dx.doi.org/ 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yau YY, Perkins-Balding D, Thomson JG. Recombinase technology: applications and possibilities. Plant Cell Rep 2011; 30(3):267-85; PMID:20972794; http://dx.doi.org/ 10.1007/s00299-010-0938-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009; 25:1189-91; PMID:19151095; http://dx.doi.org/ 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei YZ, Hu FC, Hu GB, Li XJ, Huang XM, Wand HC. Differential expression of anthocyanin biosynthetic genes in relation to anthocyanin accumulation in the pericarp of Litchi chinensis Sonn. PLoS ONE 2011; 6(4):319455; http://dx.doi.org/ 10.1371/journal.pone.0019455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. Flavonoid biosynthesis: a colorful model for genetics, biochemistry, cell biology and biotechnology. Plant Physiol 2001; 126:485-93; PMID:11402179; http://dx.doi.org/ 10.1104/pp.126.2.485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. The biosynthesis of flavonoids, in The Science of Flavonoids, ed Grotewold E, editor. (New York, NY: Springer; ) 2006; 71-95. [Google Scholar]
- Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J 2004; 40:22-34; PMID:15361138; http://dx.doi.org/ 10.1111/j.1365-313X.2004.02183.x [DOI] [PubMed] [Google Scholar]
- Zhou X, Carranco R, Vitha S, Hall TC. The dark side of green fluorescent protein. New Phytol 2005; 168:313-22; PMID:16219071; http://dx.doi.org/ 10.1111/j.1469-8137.2005.01489.x [DOI] [PubMed] [Google Scholar]
- Zhou H, Lin-Wang K, Wang H, Gu C, Dare AP, Espley RV, He H, Allan AC, Han Y. Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J 2015; 82(1):105-21; PMID:25688923; http://dx.doi.org/ 10.1111/tpj.12792 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.