ABSTRACT
Transposon-insertion sequencing methods are finding their way into the molecular toolbox of many fields of microbiology. These methods can identify the genomic locations and density of transposon insertions in saturated transposon mutant libraries and can be used to make inferences on gene function. For example, where no insertions or very few insertions are identified within a gene in a mutant library grown under permissive conditions, the gene may be essential. Furthermore, where mutations are enriched or lost in a gene after passaging the library through a selective process, the gene is likely to be involved in phenotypes linked to the process. Typically, a fitness based selection such as a stress condition is used in these experiments and the processed sequencing data are used to identify genes required for fitness under the selection. Our research team recently expanded the utility of the transposon directed insertion sequencing (TraDIS) method by applying a physical separation of a transposon mutant library mediated by fluorescence activated cell sorting, rather than a fitness-based selection. This approach, which we have named “TraDISort” is significant because it allows the study of phenotypes that are not linked to cell survival. The TraDISort approach has a broad range of future applications, in drug development, metabolic engineering and in studies of basic bacterial cell physiology.
KEYWORDS: biosensor, cell sorting, FACS, fluorescence, reporter construct, transposon sequencing
Introduction
The advent of high-throughput sequencing has sparked a new era in microbiology where genome scale experiments have become routine in most laboratories. The impact of these sequencing methods can be clearly seen in advances made to the fields of comparative genomics and metagenomics, and has driven the development of technologies that can elucidate gene function on a genome wide scale, such as transcriptomics, ChIP-seq (chromatin immunoprecipitation followed by sequencing), and most recently, transposon insertion sequencing on a genome wide scale.1-5 High-throughput sequencing provided a basis for “transposon insertion sequencing” methods to easily profile high-density libraries of individual random transposon mutants, allowing the insertion sites across the mutant population to be mapped and the relative abundance of each mutant to be determined. Using these methods the insertion site profile of a mutant library that has been subjected to a selection can be compared with the profile of the same library grown without the selection, to infer which genes and genetic elements are acted on either positively or negatively by the selection and thus the possible functions of these genes.
Several sample preparation protocols have been developed for mapping transposon insertion sites, and distinct studies using each method were first published in 2009. These include transposon directed insertion-site sequencing (TraDIS),6 transposon sequencing (Tn-seq),7 insertion sequencing (INseq)8 and high-throughput insertion tracking by deep sequencing (HITS).9 The specific differences of each method, as well as advantages and disadvantages are described in several excellent recent reviews.4,5
The majority of transposon-insertion sequencing studies that have been conducted to date have applied fitness based methods to differentially select between mutants in the population, e.g. growth during exposure to a stress condition. These can be conceptualised as massive scale competition experiments between all the mutant strains in the library; a loss of mutations in a particular gene across the library suggests that the gene is important for growth, survival or competitive fitness under the stress, an increase in mutations in a gene suggests that the gene is detrimental to growth, survival or fitness under the stress, and no change in the frequency of mutants for a particular gene suggests that the gene is not important for the fitness of the strain under the stress (Fig. 1).
Figure 1.
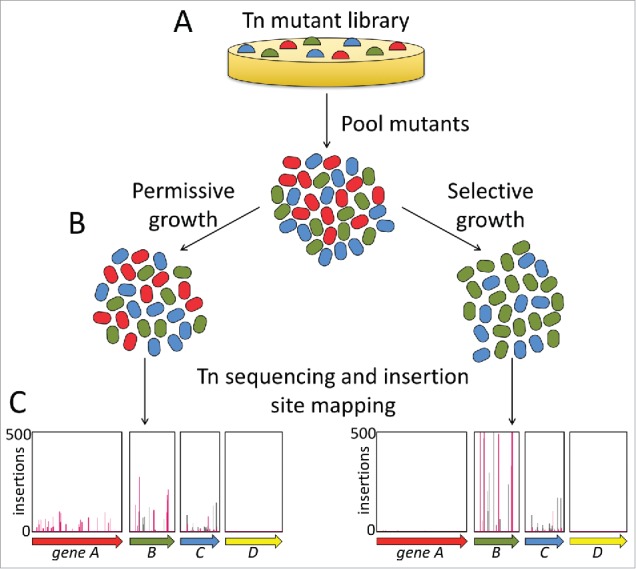
Overview of a typical transposon insertion sequencing fitness-based experiment. (A) A mutant library in the strain of interest is constructed and plated on media permissive to growth. (B) The mutants are pooled and then cultured in permissive and selective conditions, e.g., laboratory media without or with an antimicrobial, respectively. (C) DNA is isolated from the cultured mutant populations and the transposon insertion sites and frequencies are determined using transposon sequencing. A reduction in the frequency of mutants carrying insertions in a particular gene grown under selective conditions compared with permissive conditions indicates that the gene is required for bacterial fitness under the selective condition (e.g., gene A). An increase in the frequency of mutants carrying insertions in a gene grown under selective conditions relative to permissive conditions suggests that there is an advantage to inactivating the gene under the selection used (e.g., gene B). An equal frequency of mutants with insertions in a gene in populations grown under both selective and permissive conditions suggests that the gene does not influence bacterial fitness under the selective condition (e.g., gene C). For some genes (typically around 10% of annotated genes in bacterial genome) there will be no insertions, or very few insertions in the initial mutant pool and after permissive growth (e.g., gene D). These genes are likely to be essential for bacterial survival even under permissive laboratory growth conditions and typically encode housekeeping functions, such as DNA replication. Overall, the information gathered in these experiments allows hypotheses on gene function to be formulated.
Due to the huge potential to identify novel gene functions, fitness based transposon sequencing experiments have found a place in the molecular toolbox of many fields of microbiology including microbial ecology, industrial microbiology and medical microbiology. For example, these experiments have been used to identify genes that are involved in small molecule resistance or tolerance, including antibiotics in the opportunistic human pathogens Klebsiella pneumoniae10 and Staphylococcus aureus,11 and industrial chemicals in Escherichia coli.12 Transposon sequencing experiments have been used to identify genes required for bacterial colonisation of animals, both in the context of symbionts, such as Snodgrassella alvi colonisation of honey bee guts,13 and in pathogenic interactions, such as Salmonella enterica serovar Typhimurium colonisation of food-producing animals14 and Acinetobacter baumannii colonisation of insect larvae as a virulence model.15 Furthermore, transposon sequencing experiments have been used to identify genes required for bacterial transitions between growth states, such as persister cell formation in uropathogenic E. coli16 and spore formation in Clostridium difficile17 and Bacillus subtilis.18 A very recent study used transposon sequencing to examine bacterial fitness under hundreds of unique selective conditions, including carbon and nitrogen utilization and chemical stress conditions, in 25 bacterial strains.19 Through this massive study, the authors report the identification of potential phenotypes for close to 8,500 proteins of unknown function, demonstrating the huge potential of these methods for assigning function to novel genes.19
Physical enrichment for mutants of interest using “TraDISort”
In contrast to the fitness based mutant selection approaches used in other studies, we recently became interested in whether transposon sequencing methods could be used to directly probe for genes that define physical traits of bacterial cells that may not heavily influence their relative fitness under an easily imposed selective pressure; could we physically separate mutants of interest and then use transposon-insertion sequencing? Fluorescence activated cell sorting (FACS) is a well-established method that can be used to very rapidly screen millions of living cells (or other small particles) for their size, granularity and fluorescence, and sort these cells according to user-defined physical characteristics. Therefore, FACS provided an ideal method to impose a physical gating for mutant cell enrichment.
In our initial study, we set out to identify mutants of the human pathogen Acinetobacter baumannii that differentially accumulated the fluorescent dye ethidium bromide.20 The UV fluorescence of ethidium bromide increases significantly when it is intercalated into nucleic acids, a property that has been exploited for several decades in the highly sensitive detection of nucleic acids following gel electrophoresis. This property of ethidium bromide also means that it is differentially fluorescent inside and outside of cells—more fluorescent inside due to the high nucleic acid content. Therefore, the fluorescence associated with individual cells can be used as a measure of the amount of ethidium inside the cell. This method has been used extensively to study the function of multidrug efflux pumps that recognize ethidium as a substrate.21
A large population (> 100,000) of unique transposon insertion mutants of A. baumannii was incubated with a low concentration of ethidium bromide for a sufficient time to allow their intracellular ethidium bromide concentrations to reach equilibrium, i.e., when the rate of accumulation was equal to the rate of efflux. The cells were then subjected to FACS using a BD Influx flow sorter to enrich for sub-populations of mutants that contained the highest and the lowest concentrations of ethidium bromide, the top and bottom 2% of cells based on ethidium bromide fluorescence, respectively. We then used TraDIS sequencing protocols and analyses tools22 to profile the transposon insertion sites across these differentially fluorescent populations and compare them to the insertion site profile of the total mutant library pool that had been grown in parallel to FACS. The data showed that insertions in genes encoding various multidrug efflux systems, particularly adeABC and amvA, were positively and negatively selected in the high and low fluorescent pools, respectively (Fig. 2). This fits with the notion that inactivation of these pumps by transposon insertion would reduce the overall rate of ethidium bromide efflux and result in a higher equilibrium concentration of ethidium in these mutant cells. Therefore, these cells would be far less likely to show low fluorescence and be selected in the low fluorescent pool, but far more likely to show high fluorescence and be selected in the high fluorescent pool. In line with the differential selection for mutations in genes encoding efflux pumps, cells carrying mutations in efflux pump regulator genes were also highly differentially selected. Insertions in efflux pump activator genes showed similar patterns of selection to the efflux pump genes themselves, whereas, insertions in efflux pump repressor genes were much less abundant in the high fluorescent pool, since their inactivation leads to overexpression of efflux pumps, a higher rate of ethidium bromide efflux and a lower equilibrium concentration of ethidium.20
Figure 2.
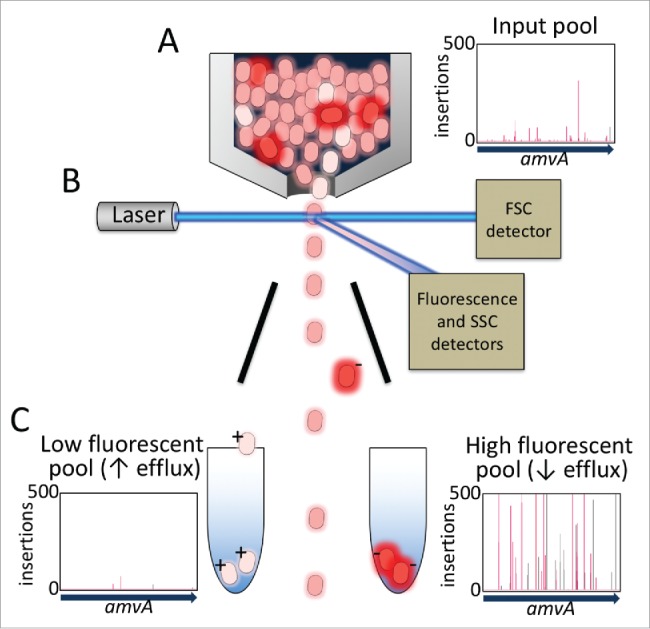
Overview of the TraDISort method for the physical enrichment of A. baumannii transposon mutants that have differentially accumulated ethidium bromide. (A) A mutant library pool is incubated with a low concentration of ethidium bromide and loaded onto a FACS instrument. The plot shows the density and frequency of insertions in the amvA gene, which encodes a major multidrug efflux pump, within the starting mutant pool. (B) Cells flow past the laser detection system and are screened for their ethidium content, size and granularity based on their light scattering and fluorescence properties. After screening the droplets containing only single cells break off from the flow stream and the droplets are differentially charged based on the fluorescence of the cell inside. (C) Cell droplets are sorted based on charge by deflection plates into high and low fluorescent pools, such that the most highly fluorescent cells (top 2%) are collected in one tube, and the most weakly fluorescent cells (bottom 2%) are collected in a second tube for extraction and TraDIS analysis. The plots show the locations and frequencies of insertions in amvA in the high and low fluorescent mutant pools.
The total amount of ethidium bromide in a cell will reflect not only its equilibrium concentration (i.e., the sum of accumulation and efflux), but also the total cell volume, where larger cells will have more ethidium bromide, and possibly nucleic acids, and therefore typically higher fluorescence than smaller cells. Additionally, many bacterial strains, including A. baumannii BAL062 used in our initial study, can form aggregates in planktonic culture and the total fluorescence of these particles would be the sum of the aggregated cells. Consequently, we applied a gating procedure during our FACS cell enrichments to exclude any cell aggregates or large cells with division defects before fluorescence based sorting. As a result of this gating we observed a significant reduction in mutants carrying insertions in several cell division genes, and conversely a significant increase in mutants carrying insertions in genes that may promote aggregation, such as the csu type I pilus genes.20 Thus, another clear application of TraDISort is directly identifying cells involved in regulating and maintaining cellular size and shape, which could also be done with gating alone and without fluoresence.
Collectively, the data generated in these experiments show that flow sorting is a viable approach to enrich for mutants showing altered cellular phenotypes of interest before transposon insertion sequencing. Due to the combination of TraDIS sequencing and flow sorting, we have called this approach “TraDISort”.20 The TraDISort approach expands the utility of transposon insertion sequencing because it provides an opportunity to study phenotypes that are not directly linked to cell survival or regeneration, i.e., where there may be little significant difference in the fitness of mutants within the population, or where the mutants of interest may be less fit than the average.
Future directions using TraDISort
Following from our study examining ethidium accumulation into A. baumannii cells, TraDISort has significant potential as a new tool for investigating bacterial multidrug efflux pumps. In addition to ethidium bromide, there are many small molecule fluorophores that can be used to monitor the activity of multidrug efflux pumps.23 These compounds differ in their chemistry and their sites of accumulation within cells, so can be used to examine the function of different sub-sets of multidrug efflux pumps. For example, similar to ethidium, Hoechst 33342, and 4′-6-diamidino-2-phenylindole (DAPI) are differentially fluorescent when intercalated into nucleic acids and total cell fluorescence could be used as a proxy for the intracellular concentrations of these dyes.24 However, whereas ethidium, and Hoeschst 33342 are monovalent, DAPI is a bivalent compound so may be recognized by a different set of efflux pumps.25 Other dyes accumulate in the periplasm of Gram-negative bacteria rather than penetrating the cytosol and could be used in TraDISort to target the functions of efflux pumps that specifically capture their substrates from the periplasm.23,26 Finally, the genes involved in controlling the accumulation of fluorescent antibiotics, such as tetracyclines and fluoroquinolones, should be identifiable using the TraDISort approach and may also identify specific toxin transporter systems in addition to efflux. However, protocols are currently being fine-tuned to account for the low fluorescence of these compounds and the lack of differential fluorescence inside and outside bacterial cells. Once efflux systems that recognize these diverse substrates have been identified, TraDISort could further be used to identify the targets for efflux pump inhibitor compounds by treating mutant populations with these compounds in combination with fluorescent efflux pump substrates. Comparison of the mutants selected with and without the inhibitor should identify the pumps being inhibited and potentially the extent of inhibition.
A range of commercially available fluorescent dyes can be used as indicators for the intracellular concentrations of various free ions, such as sodium, iron, zinc and protons.27,28 Typically, these dyes undergo a fluorescence change, such as a shift in their excitation or emission spectra, or a shift in their fluorescence intensity when bound to their cognate indicator ion. TraDISort could be used in combination with these dyes to probe for the systems involved in homeostasis of these important ions.
Rather than monitoring fluorescence based on a small molecule fluorophore, the TraDISort approach may also be used in combination with genetic fluorescent reporter constructs. Fluorescent reporter systems incorporating a gene encoding a fluorescent protein have been used for many decades as tools to examine levels of gene and protein expression in various biological systems. Mutant libraries built around strains carrying promoter fusion fluorescent reporter constructs could be used to identify novel regulators that recognize sequence elements in the cloned promoter region. In these experiments mutants showing increased or decreased expression of the fluorescent reporter could be enriched and the insertion sites mapped. The enriched mutants may harbour mutations in the regulators of the gene of interest.
TraDISort has significant potential to be used as a tool in metabolic engineering and synthetic biology. The goal of a typical metabolic engineering project is to develop bacterial strains that can be used as factories to produce a small molecule or molecules of interest, which may be native to the producing strain, or the product of introduced metabolic pathways. A major challenge is to channel metabolic energy toward production of the small molecule(s). The development of a commercially viable production strain typically involves many rounds of rational design, synthetic strain construction and product yield testing. Random mutants, including transposon mutants have been used to assist in the design of high yielding strains. However, these studies have traditionally relied on the isolation and characterization of single isolated mutants, and are thus prone to complications with high numbers of false positives. With recent advances in the development of fluorescent biosensors, saturation transposon mutant libraries could be readily applied in metabolic engineering in a highly-streamlined approach using TraDISort. Fluorescent biosensors responsive to the product of interest would allow mutants that produce higher levels of the product to be enriched by flow sorting approaches in TraDISort and avoid potential for false positives. Such mutants would typically have a lower overall fitness, due to the burden of channelling more metabolic energy into compound production rather than growth, but may be identified using the TraDISort approach because mutant enrichment is separated from cell fitness.
Conclusions
Transposon insertion sequencing technologies have brought about a new age in genome-wide investigations of gene function. The relative ratios of mutants in a large transposon mutant library can be examined before and after exposure to a selective condition to infer which genes are required for fitness under the selection. Rather than applying a fitness based selection, mutants can also be enriched based on physical characteristics by FACS, as used in the TraDISort approach. By separating mutant enrichment from fitness, TraDISort allows transposon sequencing to be used in a broader range of experiments. For example, some of the most significant impacts of this new technology may be within metabolic engineering and synthetic biology, where fluorescent biosensors could be used with TraDISort to identify mutants that produce superior yields of compounds of interest.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
Parts of this work were supported by Project Grants from the National Health and Medical Research Council of Australia to ITP and KAH (GNT1060895 and GNT1120298). KAH thanks the EU for the award of a Marie Skłodowska-Curie Research Fellowship (706499). Sequencing was supported by the Wellcome Trust (WT098051). A.K.C. was supported by the Medical Research Council (G1100100).
ORCID
Karl A. Hassan: http://orcid.org/0000–0003–2031–9679
References
- [1].Marguerat S, Bahler J. RNA-seq: From technology to biology. Cell Mol Life Sci 2010; 67:569-79; PMID:19859660; https://doi.org/ 10.1007/s00018-009-0180-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wooley JC, Godzik A, Friedberg I. A primer on metagenomics. PLoS Comput Biol 2010; 6:e1000667; PMID:20195499; https://doi.org/ 10.1371/journal.pcbi.1000667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Park PJ. ChIP-seq: Advantages and challenges of a maturing technology. Nat Rev Genet 2009; 10:669-80; PMID:19736561; https://doi.org/ 10.1038/nrg2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Barquist L, Boinett CJ, Cain AK. Approaches to querying bacterial genomes with transposon-insertion sequencing. RNA Biol 2013; 10:1161-9; PMID:23635712; https://doi.org/ 10.4161/rna.24765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].van Opijnen T, Camilli A. Transposon insertion sequencing: A new tool for systems-level analysis of microorganisms. Nat Rev Microbiol 2013; 11:435-42; PMID:23712350; https://doi.org/ 10.1038/nrmicro3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Langridge GC, Phan MD, Turner DJ, Perkins TT, Parts L, Haase J, Charles I, Maskell DJ, Peters SE, Dougan G, et al.. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res 2009; 19:2308-16; PMID:19826075; https://doi.org/ 10.1101/gr.097097.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].van Opijnen T, Bodi KL, Camilli A. Tn-seq: High-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods 2009; 6:767-72; PMID:19767758; https://doi.org/ 10.1038/nmeth.1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, Lozupone CA, Knight R, Gordon JI. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe 2009; 6:279-89; PMID:19748469; https://doi.org/ 10.1016/j.chom.2009.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gawronski JD, Wong SM, Giannoukos G, Ward DV, Akerley BJ. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc Natl Acad Sci U S A 2009; 106:16422-7; PMID:19805314; https://doi.org/ 10.1073/pnas.0906627106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Jana B, Cain AK, Doerrler WT, Boinett CJ, Fookes MC, Parkhill J, Guardabassi L. The secondary resistome of multidrug-resistant Klebsiella pneumoniae. Sci Rep 2017; 7:42483; PMID:28198411; https://doi.org/ 10.1038/srep42483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Rajagopal M, Martin MJ, Santiago M, Lee W, Kos VN, Meredith T, Gilmore MS, Walker S. Multidrug intrinsic resistance factors in Staphylococcus aureus identified by profiling fitness within high-diversity transposon libraries. mBio 2016; 7:e00950-16; PMID:27531908; https://doi.org/ 10.1128/mBio.00950-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Rau MH, Calero P, Lennen RM, Long KS, Nielsen AT. Genome-wide Escherichia coli stress response and improved tolerance towards industrially relevant chemicals. Microb Cell Fact 2016; 15:176; PMID:27737709; https://doi.org/ 10.1186/s12934-016-0577-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Powell JE, Leonard SP, Kwong WK, Engel P, Moran NA. Genome-wide screen identifies host colonization determinants in a bacterial gut symbiont. Proc Natl Acad Sci U S A 2016; 113:13887-92; PMID:27849596; https://doi.org/ 10.1073/pnas.1610856113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chaudhuri RR, Morgan E, Peters SE, Pleasance SJ, Hudson DL, Davies HM, Wang J, van Diemen PM, Buckley AM, Bowen AJ, et al.. Comprehensive assignment of roles for Salmonella typhimurium genes in intestinal colonization of food-producing animals. PLoS Genet 2013; 9:e1003456; PMID:23637626; https://doi.org/ 10.1371/journal.pgen.1003456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gebhardt MJ, Gallagher LA, Jacobson RK, Usacheva EA, Peterson LR, Zurawski DV, Shuman HA. Joint transcriptional control of virulence and resistance to antibiotic and environmental stress in Acinetobacter baumannii. mBio 2015; 6:e01660-15; PMID:26556274; https://doi.org/ 10.1128/mBio.01660-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Molina-Quiroz RC, Lazinski DW, Camilli A, Levy SB. Transposon-sequencing analysis unveils novel genes involved in the generation of persister cells in uropathogenic Escherichia coli. Antimicrob Agents Chemother 2016; 60:6907-10; PMID:27550350; https://doi.org/ 10.1128/AAC.01617-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Dembek M, Barquist L, Boinett CJ, Cain AK, Mayho M, Lawley TD, Fairweather NF, Fagan RP. High-throughput analysis of gene essentiality and sporulation in Clostridium difficile. mBio 2015; 6:e02383; PMID:25714712; https://doi.org/ 10.1128/mBio.02383-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Meeske AJ, Rodrigues CD, Brady J, Lim HC, Bernhardt TG, Rudner DZ. High-throughput genetic screens identify a large and diverse collection of new sporulation genes in Bacillus subtilis. PLoS Biol 2016; 14:e1002341; PMID:26735940; https://doi.org/ 10.1371/journal.pbio.1002341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Price MN, Wetmore KM, Waters RJ, Callaghan M, Ray J, Kuehl JV, Melnyk RA, Lamson JS, Suh Y, Esquivel Z, et al.. Deep annotation of protein function across diverse bacteria from mutant phenotypes. bioRxiv 2016; 072470; https://doi.org/ 10.1101/072470 [DOI] [Google Scholar]
- [20].Hassan KA, Cain AK, Huang T, Liu Q, Elbourne LD, Boinett CJ, Brzoska AJ, Li L, Ostrowski M, Nhu NT, et al.. Fluorescence-based flow sorting in parallel with transposon insertion site sequencing identifies multidrug efflux systems in Acinetobacter baumannii. mBio 2016; 7:e01200-16; PMID:27601573; https://doi.org/ 10.1128/mBio.01200-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hassan KA, Brzoska AJ, Wilson NL, Eijkelkamp BA, Brown MH, Paulsen IT. Roles of DHA2 family transporters in drug resistance and iron homeostasis in Acinetobacter spp. J Mol Microbiol Biotechnol 2011; 20:116-24; PMID:21430390; https://doi.org/ 10.1159/000325367 [DOI] [PubMed] [Google Scholar]
- [22].Barquist L, Mayho M, Cummins C, Cain AK, Boinett C, Page AJ, Langridge GC, Quail MA, Keane JA, Parkhill J. The TraDIS toolkit: Sequencing and analysis for dense transposon mutant libraries. Bioinformatics 2016; 32:1109-11; PMID:26794317; https://doi.org/ 10.1093/bioinformatics/btw022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Blair JM, Piddock LJ. How to measure export via bacterial multidrug resistance efflux pumps. mBio 2016; 7: e00840-16; PMID:27381291; https://doi.org/ 10.1128/mBio.00840-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hassan KA, Skurray RA, Brown MH. Transmembrane helix 12 of the Staphylococcus aureus multidrug transporter QacA lines the bivalent drug binding pocket. J Bacteriol 2007; 189:9131-4; PMID:17951386; https://doi.org/ 10.1128/JB.01492-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Paulsen IT, Brown MH, Littlejohn TG, Mitchell BA, Skurray RA. Multidrug resistance proteins QacA and QacB from Staphylococcus aureus: Membrane topology and identification of residues involved in substrate specificity. Proc Natl Acad Sci U S A 1996; 93:3630-5; PMID:8622987; https://doi.org/ 10.1073/pnas.93.8.3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Tal N, Schuldiner S. A coordinated network of transporters with overlapping specificities provides a robust survival strategy. Proc Natl Acad Sci U S A 2009; 106:9051-6; PMID:19451626; https://doi.org/ 10.1073/pnas.0902400106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Han J, Burgess K. Fluorescent indicators for intracellular pH. Chem Rev 2010; 110:2709-28; PMID:19831417; https://doi.org/ 10.1021/cr900249z [DOI] [PubMed] [Google Scholar]
- [28].Dean KM, Qin Y, Palmer AE. Visualizing metal ions in cells: An overview of analytical techniques, approaches, and probes. Biochim Biophys Acta 2012; 1823:1406-15; PMID:22521452; https://doi.org/ 10.1016/j.bbamcr.2012.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]


