Abstract
Sesame (Sesamum indicum L.) has high oil content, a small diploid genome and a short growth period, making it an attractive species for genetic studies on oilseed crops. With the advancement of next-generation sequencing technology, genomics and functional genomics research of sesame has developed quickly in the last few years, and large amounts of data have been generated. However, these results are distributed in many different publications, and there is a lack of integration. To promote functional genomics research of sesame, we collected genetic information combined with comprehensive phenotypic information and integrated them in the web-based database named SesameFG. The current version of SesameFG contains phenotypic information on agronomic traits of 705 sesame accessions, de novo assembled genomes of three sesame varieties, massive numbers of identified SNPs, gene expression profiles of five tissues, gene families, candidate genes for the important agronomic traits and genomic-SSR markers. All phenotypic and genotypic information in SesameFG is available for online queries and can be downloaded freely. SesameFG provides useful search functions and data mining tools, including Genome Browser and local BLAST services. SesameFG is freely accessible at http://ncgr.ac.cn/SesameFG/. SesameFG provides valuable resources and tools for functional genomics research and the molecular breeding of sesame.
Introduction
Sesame (Sesamum indicum L.) is a major oilseed crop and is widely grown around the world, with concentrations in Asia and Africa1. With excellent characteristics, such as high oil content (~55%), strong drought resistance, a short growing season (~90 d), a large propagation coefficient (3,000–10,000 seeds per plant), and a small diploid genome (~350 Mb), sesame is regarded as an attractive model for genetic research of oilseed crops2. In addition, more than 35,000 accessions of sesame have been collected worldwide3, and massive genome variations have been identified in the sesame population4, 5. The rich levels of phenotypic and genotypic diversity of sesame are also valuable for functional genomics research.
With the development of next-generation sequencing (NGS) technology, genetic research on sesame has developed quickly in the last few years. Genome sequencing of both the variety and landrace sesame have been published5, 6, large amounts of polymorphism molecular markers have been developed7–10, several genetic linkage maps have been constructed, and many quantitative trait loci (QTL) have been identified11–15. Additionally, some gene families were investigated16, 17, gene expression profiles in different tissues were detected by transcriptome sequencing6, 8, 18, 19, and more than 5.4 million single nucleotide polymorphisms (SNPs) were identified from large-scale genome re-sequencing2. Candidate genes that are related to oil production and quality were explored in genome-wide association studies (GWAS), providing precise clues for uncovering the genetic mechanism of important sesame agronomic traits. These achievements in sesame genetic research provide a valuable basis for functional genomics research. However, these results are distributed over different publications and lack integration. It is difficult for researchers to utilize the previous results and data for future sesame research.
Three databases that are related to sesame genomics have been constructed in the last years, including Sinbase (http://ocri-genomics.org/Sinbase/)20, the Sesame Genome Project (http://www.sesamegenome.org/)21, and SesameHapMap (http://ncgr.ac.cn/SesameHapMap/)2, 5. The genome sequences of several sesame varieties and SNPs in the population are provided in these databases. The genomics-related databases laid the foundation for the construction of a functional genomics database. By combining the genomics data with newly released functional genomics data and integrated phenotype information, a comprehensive sesame functional genomics database can be constructed. Comprehensive and integrated databases for functional genomics research have been constructed in several other crops, such as rice, tomato, and foxtail millet22–24. These databases have been widely used in functional genomics research and have greatly promoted the basic research and molecular breeding of these crops25–27. Therefore, an integrated database will likely play an important role in sesame functional genomics research.
Germplasm collections contain superior alleles that can be uncovered by functional genomics research and used in crop breeding28. Generally, crop improvement relies on the utilization of superior alleles contained in the germplasm29. Collection of various germplasms and identification of admirable germplasms is the basis for crop functional genomics research and molecular breeding. To identify valuable germplasm with superior alleles, the precise phenotypic information of the germplasm is required. Many germplasm resources have been collected in several sesame germplasm banks30–32, and the important agronomic traits of these germplasms have been observed. However, little phenotypic information of these germplasms is available online. As far as we know, there is no database that contains detailed phenotype information of various sesame germplasms. Consequently, a database which includes comprehensive and detailed phenotype information of sesame germplasms is desired for researchers.
We have established the SesameFG (Sesame Functional Genomics Database, http://ncgr.ac.cn/SesameFG/) to provide comprehensive genetic information, phenotypic information and bioinformatics analysis for sesame functional genomics research. The published data, which were useful for gene identification and gene functional research in sesame, were collected and analyzed in our database, including materials information, genome sequences, genome variations, gene families, gene expression, candidate genes and simple sequence repeat (SSR) markers. The detailed phenotype of sesame core collections that precisely investigated by our group were also submitted and integrated into the database. In addition, the gene functional analysis tools, Genome Browser and BLAST, are available in the database. The goal of the database is to build a user-friendly and widely used repository that covers comprehensive functional genomics-related resources and that will be updated with newly released data regarding sesame functional genomics research in the future. In this study, we introduce the current version of SesameFG.
Methods
Data collection
SesameFG was constructed using large-scale genetic and phenotypic sesame resources that came from public databases, literature and sesame functional genomics consortium inputs (Fig. 1). The genetic and phenotypic information collected and integrated in SesameFG mainly includes germplasm information, phenotypes, plant photos, genome sequences and variations, population SNPs, gene families, gene expression profiles, candidate gene of agronomic traits, SSR loci and polymorphic SSR markers (Table 1).
Figure 1.
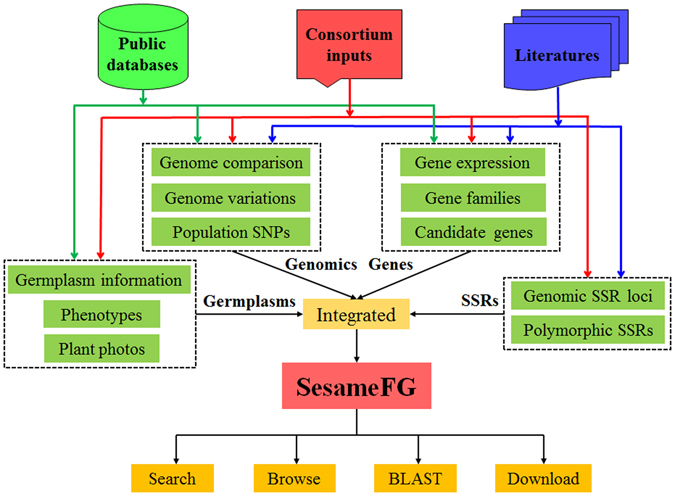
Flowchart of the construction of SesameFG. SesameFG is a comprehensive database for browsing and retrieving resources about sesame functional genomics and related data. SesameFG collected information mainly from three sources: i) genetic resources from public databases; ii) detailed curation of functional genomics research from the literature; iii) public contributions from the sesame functional research consortium and unpublished data from our group. All of the information stored in the database can be browsed in a user-friendly manner and downloaded through any web browser.
Table 1.
Summary of the data sources.
| Category | Description | Detail | Source | References |
|---|---|---|---|---|
| Genome | Zhongzhi13 | 274 Mb | Sinbase | 6, 27 |
| Mishuozhima | 254 Mb | SesameHapMap | 5 | |
| Baizhima | 267 Mb | SesameHapMap | 5 | |
| Genome variation | SNPs | 1,332,025 SNPs | Publication | 5 |
| Indels | 506,245 Indels | Publication | 5 | |
| Transposons | 525,537 transposons | Publication | 5 | |
| Population SNPs | 5,407,981 SNPs | SesameHapMap | 2 | |
| Germplasm information | Core collections | 705 accessions | Publication | 2 |
| Phenotype | Agronomic traits | 56 traits | Zhang’s group | |
| Gene expression | Difference in tissues | 27,148 genes in 5 tissues | Zhang’s group and NCBI (SRA122023) | 6 |
| Root under waterlogging stress | 27,148 genes in 5 stress points | NCBI (SRX1406650) | 18 | |
| Stem tip and leaf of determinate growth plant | 27,148 genes in 2 developmental stages | Zhang’s group | ||
| Developing seeds | 27,148 genes in 5 developmental stages | Zhang’s group | ||
| Gene family | MADS-box, AP2 and Hsf | 219 genes | Publications | 16, 17, 37 |
| Transcription factor and common gene families | 3,867 genes of 26 gene families | Zhang’s group | ||
| Candidate gene | Oil quality and production related genes | 47 genes | Publication | 2 |
| SSR | Genomic SSRs | 104,836 SSRs | Zhang’s group and publication | 9 |
| Polymorphism SSRs | 218 SSRs | Publications | 7, 8, 9 |
Germplasm information and phenotypes
Sesame core collection information was collected in SesameFG, using accession name, geographic origin, ecotype, sequencing coverage and group information. The sesame core collection consists of 705 accessions from 29 countries around the world33, representing the most genetic diversity of all of the germplasms conserved in the Chinese Sesame Genebank of the Oil Crops Research Institute. All of these materials had their genomes re-sequenced with an average of ~2.6-fold genome coverage, and the results were used in GWAS research2. Phenotype values of fifty-six agronomic traits of the 705 sesame accessions in four environments were collected into SesameFG. The fifty-six traits included yield-related, disease resistance, quality, growth cycle related, and morphological traits. The four phenotyping sites included Luohe in Henan province (114.02E, 33.56N), Wuhan in Hubei province (114.30E, 30.60N), Nanning in Guangxi province (108.33E, 22.84N) and Sanya in Hainan province (109.31E, 18.14N). Moreover, photos of the flowering stage of each accession were also included.
Genome sequences and variations
The genome sequence of “Zhongzhi13”, a widely grown sesame variety, was downloaded from Sinbase (http://ocri-genomics.org/Sinbase/) and used as a reference genome6. The other two assembled genome sequences of sesame landraces, “Mishuozhima” and “Baizhima”, were obtained from SesameHapMap (http://ncgr.ac.cn/SesameHapMap/). In addition, 5,407,981 sesame population SNPs were also downloaded from SesameHapMap. These population SNPs were identified from the re-sequencing of 705 sesame core collections2.
Gene families
Gene families in the sesame genome were identified using several analysis tools, such as Pfam, HMM, BLAST and SMART34–36. In addition, all published sesame gene families, including MADS-box, AP2 and Hsf genes, were collected into SesameFG16, 17, 37.
Gene expression
The gene expression profiles of different tissues in several sesame accessions were collected6. These profiles included expression information for the capsule, leaf, seeds and stem of the sesame variety “Zhongzhi13”, for the root of the waterlogging-tolerant variety “ZZM2541”, for the stem tip of the typical determinate growth sesame accession “ZZM3305” and for the developing seeds of the high oil content variety “Zhongfengzhi1”. All gene expression levels were indicated by the Reads Per Kilobases per Million reads (RPKM) values, which were calculated from the transcriptome sequencing data.
Candidate genes of agronomic traits
Candidate genes that are related to important agronomic traits were collected and provided in SesameFG, such as yield, lipid metabolism, beginning flowering date and disease resistance. These genes were discovered in genetic analysis and molecular experiments2.
SSR markers
We identified SSR loci in sesame genomes using the microsatellite identification software (MISA)38 and designed SSR markers for each locus using Primer339. One to six nucleotide motifs were considered, and the minimum repeat unit was defined as ten reiterations for mononucleotides, six reiterations for dinucleotides, and five reiterations for other repeat units. In total, 104,836 SSR markers were developed from the sesame genome, and all were submitted into SesameFG. In addition, the polymorphic SSRs that were detected from previous experimental research were also collected7–9.
Data preprocessing
The assembled contigs of “Mishuozhima” and “Baizhima” were aligned on the “Zhongzhi13” genome by MUMmer40 to identify the homologous genome regions. The genome variants in “Mishuozhima” and “Baizhima” were detected using the diffseq program in the EMBOSS package41. The SNPs in each gene were identified, and the SNP effect was annotated using the reference genome. The transposons in “Mishuozhima” and “Baizhima” were identified using RepeatMasker (http://repeatmasker.org) and annotated by aligning with the “Zhongzhi13” genome.
To use whole-genome SNPs conveniently, each SNP of the population SNPs was labeled with a unique identifier (ID, e.g., Sis0000000011). The SNPs present in each gene were identified using a self-customized Perl script. The SNP number and their allele frequency in landraces, varieties, south and north groups were summarized. Moreover, the SNPs between each two accessions of the core collection were identified. The linkage group location, position, and numbers and frequency in different groups of SNPs were also identified. Moreover, the SNPs between each two accessions, which were valuable for identification of the variations in the genomes of the different sesame germplasms, were also analyzed and provided in the database.
The Arabidopsis genome was downloaded from the TAIR database42. All genes in the sesame genome were analyzed against Arabidopsis genes using BLAST and were annotated using homologous Arabidopsis genes34. For the gene expression profiles, the gene expression of each accession was integrated based on the gene ID in the sesame genome. Furthermore, the SSR loci in the sesame genome around each gene was identified and provided in the database.
The gene structure, tRNA genes, microRNA genes, transposons and other genome components downloaded from Sinbase and the population SNPs downloaded from SesameHapMap were integrated into the Genome Browser43. Detailed information of the genome components and population SNPs, such as the ID, position, length and sequences, were linked into the Browser.
Database implementation
The SesameFG database was developed using Perl/CGI, Python and JavaScript on a platform with the MySQL 5.0 database management system. The web interfaces were constructed using PHP (Version 5.6), a popular scripting language for dynamic webpages. JavaScript and jQuery were used to enhance the website interface and to improve the user experience. A navigation tool bar containing several links is also contained in each webpage. The database is powered by an Apache server running Ubuntu Linux 15.04.
Results
The database feature of SesameFG
To promote sesame functional genomics research, SesameFG has collected both the comprehensive genetic and phenotypic information of sesame and endeavors to provide all necessary resources and tools for sesame functional genomics research (Fig. 2). The phenotypic information is included in the Material section, while the genotypic resources are contained in the Genome related, Gene and SSR sections. The BLAST and Genome Browser functions are included in the Tool section. The Download section provides the genome sequences and genomic variations. The introduction and user guidelines of the database can be found in the Home and Help sections.
Figure 2.
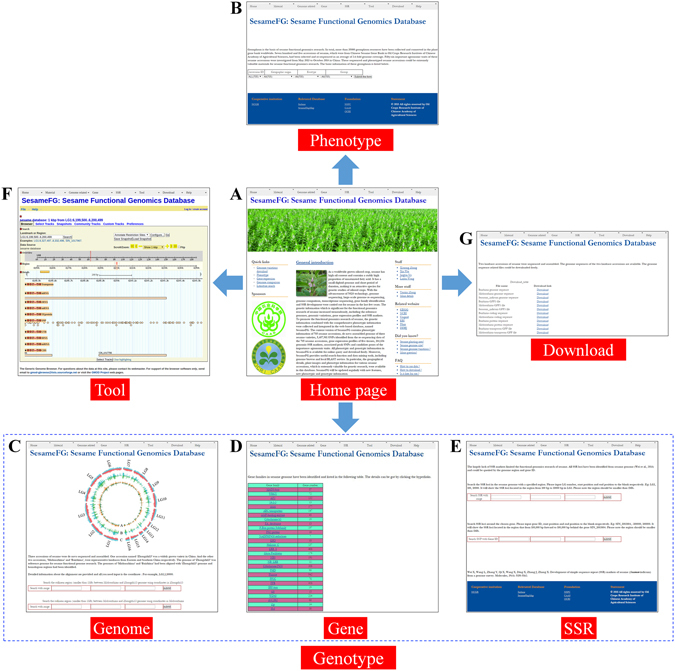
Main contents and interfaces of SesameFG. (A) The home page of SesameFG. (B) Basic information of the 705 sesame germplasm. (C) Collinear regions and genome variations in sesame genomes. (D) Gene families identified from the sesame genome. (E) SSR loci identified from the sesame genome. (F) The Genome Browser for locating gene structures, SNPs, and RNAs. (G) Genome variations of 705 sesame accessions for downloading.
The basic information of the sesame core collections contains 705 sesame accessions that are available on the Germplasm page, including the accession name, geographic origin, ecotype, sequencing coverage and group (Fig. 2B). For each information cluster, when clicked, the database provides a drop-down menu with a list of selectable options. It is easy to determine the information for any group of the sesame core collections on this page, which makes it convenient to get the materials for functional genomics research in further studies. The phenotypes of 56 agronomic traits for these sesame accessions are provided on the Phenotype page. The minimum, maximum and average values of all traits in the four environments were calculated and are provided on the Query by trait page. A fuzzy search function was developed and can be used in the inquiry of phenotypes. Phenotypic value of each trait in each accession can be queried freely on the Query by accession page. The plant photos of the accessions can also be viewed on the Phenotype photo page.
SesameFG provides three genomes and massive genome variations in the Genome related section. Based on the alignment of the three genomes, collinear regions between the sesame landrace and variety genomes can be queried by limiting the genomic coordinates of the “Zhongzhi13” genome on the Genome comparison page. The SNPs, Indels and transposons that were identified from the genome comparison of the three sesame genomes are available on the SNP query, Indel query and Transposon query pages, respectively. All of these genome variations can be easily searched by limiting the genomic coordinates of the sesame genome (Fig. 2C). The linkage group location, position, number in the different groups and frequency in different groups of population SNPs are also available. The SNPs in a specific region, around a chosen SNP and around a chosen gene can be queried on the Population SNP query page. Moreover, the SNPs between each of the two accessions are provided on the Query SNP between two accessions page.
The sesame gene families, gene expression profiles and candidate genes of important agronomic traits are available in the Gene section. Detailed information regarding the gene families is provided on the Gene family page (Fig. 2D). Thus far, there are 29 gene families with 4,085 members that can be found in the database, including transcription regulators, kinase protein-encoding genes, cytochrome P450 proteins, and lipid metabolism enzymes. Detailed information regarding the gene families can be used in functional analyses of the gene clusters. For example, the identified MADS-box genes are involved in the photo-period regulation of sesame flowering17. Expression profiles of sesame genes in the root, stem, leaf, capsule and seeds, which are valuable for gene identification, are all available on the Gene expression page. These gene expression results are crucial for gene family analysis and gene function validation. For instance, gene expression profiling of the Hsf gene family had been analyzed based on the data provided in this database37. Detailed information of the candidate genes with important agronomic traits can be obtained on the Candidate genes page, including the Gene IDs and locations of the genes in sesame genome, the related traits, the peak SNP associated with the related traits, the major allele of the peak SNP, the major allele frequency of the SNP and annotation of the candidate genes. Since only a few genes have been validated by population mapping and molecular experiments in the sesame genome, these candidate genes can be valuable resources and greatly promote sesame functional genomics research.
The SSR loci in the sesame genome, SSR markers design function and polymorphic SSR markers all can be found in the SSR section. All 104,836 SSR loci and primer sequences are available on the SSR Query page (Fig. 2E). The SSR markers can be queried and designed over a random range or for a chosen gene. The polymorphic SSRs that were detected from previous experimental research are all provided on the Polymorphic SSRs page.
SesameFG provides tools to facilitate bench work and further analysis in the Tool section, containing Genome Browser and BLAST. We implemented 10 tracks in the Genome Browser, including gene structures, tRNA genes, microRNA genes, transposons and population SNPs. Users can browse detailed information of each feature of each track on the Genome Browser page (Fig. 2F). Detailed information of the genome components and population SNPs can be obtained by clicking the corresponding hyperlink of the feature. Take the population SNPs as an example, the SNP ID, position, reference SNP and alternative SNP, major allele and major allele frequencies in landrace and variety, and the number of SNPs in landrace and variety are all shown in the database. A standard NCBI BLAST software package was embedded in SesameFG, providing a similar sequence search function for the users. Not only is the reference sesame genome available, but the genome sequences of the two landrace accessions are provided on the BLAST page. The queried nucleic acid or amino acid sequences can be uploaded in a file or pasted in the search box directly. Several BLAST programs are available for different sequence types. The BLAST function aids users in extracting homologous genome components and annotations of query sequences by quick match.
SesameFG also offers users the capability to download and use the sesame genome sequence and variation data in the Download section. The genome sequences include commonly used and requested data sets such as Genome FASTA, Generic Feature Format 3 (GFF3) containing the annotated gene models, CDS FASTA, protein FASTA and transposon GFF of the sesame landrace accessions. These genome sequences can be downloaded from the Genome sequences page (Fig. 2G). On the Genome variations page, the population SNPs that were generated from the re-sequencing of 705 sesame accessions are provided for downloading.
Application of SesameFG
Seed coat color is one of the most important characters of sesame seeds. There are two major colors of sesame seed: black and white. It has been reported that the PPO gene, which encodes polyphenol oxidase and produces black pigments is the key regulatory gene of sesame seed coat color5. Here we show that the SesameFG can be easily used in the cloning of PPO (Fig. 3).
Figure 3.
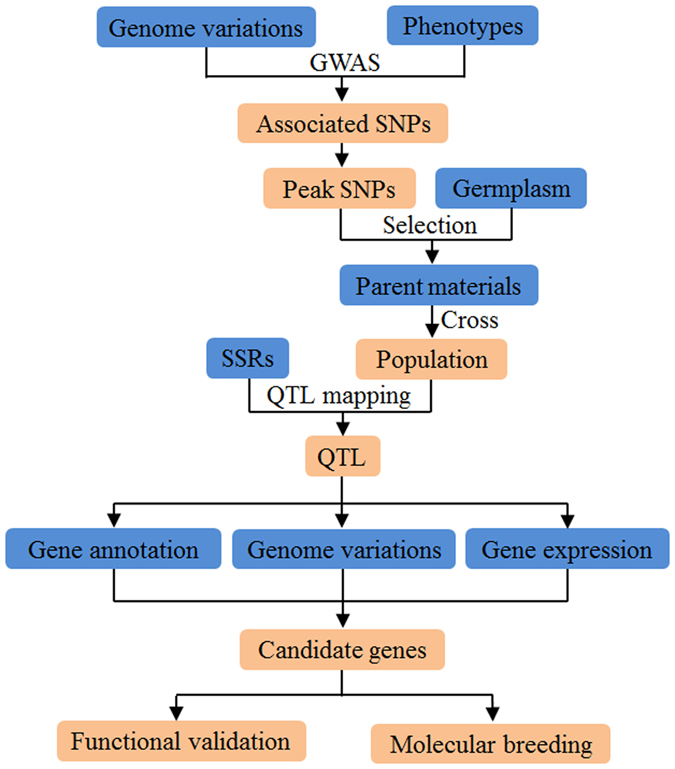
Flowchart of the identification of seed coat color genes from sesame with SesameFG. SesameFG provides user-friendly phenotypic and genotypic information and analysis tools for gene discovery and molecular breeding of important agronomic traits. The blue color represents the information that is available in SesameFG. The orange color indicates the analysis and experimental results.
The sesame coat colors of 705 sesame accessions can be obtained from Phenotype query, and SNPs of the sesame accessions can be downloaded from Genome variations. GWAS of sesame coat color can be performed using phenotypes and SNPs. Then, the associated SNPs with high degrees of confidence will be gained. Detailed information of the associated SNPs can be queried in the Population SNP query. It was revealed that the associated peak SNP at the 11,607,534 bp in linkage group 4, which had a maximal P value (P = 9.33 × 10−130), had a C/A mutation; the C allele was related to the black seed coat2. Accessions with the black (C) and white (A) alleles can be selected, and F2 populations can be developed by crossing the accessions. SSR markers around the peak SNP (from 100 kb forward to 100 kb behind) can be queried from the SSR query. QTL fine mapping of sesame seed coat color can be performed based on SSR markers, and a major QTL containing the peak SNP can be focused into a small region. Then, genes in this QTL can be annotated on the Literature annotation page, and the expression profiles of sesame seed genes can be queried on the Gene expression page. Since the variety “Zhongzhi13” is a white seed accession and the landrace “Mishuozhima” is a black seed accession, variations of the genes can be found using BLAST. Gene annotation, expression and variation analyses showed that only the PPO gene was related to the sesame seed coat color. The SSR markers that closely linked to seed coat color can be useful molecular markers for the molecular-assisted breeding of sesame varieties with black seed. Therefore, SesameFG can be effectively used in candidate gene identification and validation of important agronomic traits for sesame. To help new users who want to quickly become acquainted with SesameFG, a step-by-step tutorial of the gene cloning is provided in the Help section, with seed coat color is used as an example.
Discussion
To our knowledge, SesameFG is the only website that provides comprehensive resources related to sesame functional genomics research supported by user-friendly interfaces and an easy-to-used system for the mapping and cloning of important genes. SesameFG was constructed using both phenotypic and genotypic information and based on the first version of the sesame genome6. Although three sesame genomics-related databases have been constructed, only the “Zhongzhi13” genome sequence and population SNPs are provided. The genomic-related data available in the databases were lack of integration and functional genomics data, such as QTLs, gene expression profiles, SSRs, and important genes, were not included. The data in SesameFG were collected from these databases and other public databases, literature and research results of the sesame functional genomic research consortium. To improve the usability of the massive amount of data, the sesame genomics and functional genomics-related data were analyzed and integrated in SesameFG. We hypothesized that these data could be conveniently and effectively utilized in discovering the genes of important agronomic traits and developing effective markers for the molecular breeding of sesame.
Compared with cereal crops, such as rice, wheat and maize, functional genomics research of oilseed crops is quite limited. However, the consumption and market demand of vegetable oil has increased greatly in the last decades44. With its high oil content (58% in seeds) and small diploid genome (350 Mb), sesame is regarded as an attractive model species for oilseed crops functional genomics research. Previous studies revealed that the major loci underlying oil content in sesame are not always the enzymes in the oil biosynthetic pathway. The genes regulating the non-oil components (mainly protein and dietary fiber) in oilseeds may have important indirect effects on the oil content2. This result provides a new strategy in the improvement of the oil content in sesame and other oilseed crops. Therefore, large-scale genetic resources and a database that can be convenient used will provide strong support to the functional genomics research of all oilseed crops.
Although more than 35,000 accessions of sesame have been conserved in the major crop germplasm genebanks of the world and their important agronomic traits have been investigated30–32, little phenotypic information of the sesame germplasm is available online. Therefore, it is quite difficult for sesame researchers to find a superior germplasm used for functional genomics research from the existing gene banks. A database containing detailed phenotype information of various germplasms is greatly needed for the sesame researchers. To data, this is the first time that the phenotypes of sesame germplasms have been freely available on a website. The detailed phenotypic information of these materials will be useful for sesame researchers in selecting materials. In addition, all of the germplasms in this database have been genome sequenced and their SNPs are also provided, making the germplasm easily selectable for use in GWAS, evolution, QTL mapping, gene cloning and molecular breeding studies. Of note, this database contains the most comprehensive phenotypic and genomic information for oilseed crop functional genomics research published to date and is expected to have a lasting impact on the genomics research and genetic improvement of oilseed crops.
In SesameFG, three high-quality assembled sesame genomes are available. A genome from a single variety does not adequately represent the diversity contained within a species; several accurate genomic sequences are critical for functional genomics research45. The three sequenced accessions, “Zhongzhi13”, “Mishuozhima” and “Baizhima”, can be used as model parents of artificial populations. With the three reference genomes and identified genomic variations, the map-based cloning of genes related to complex traits will be greatly accelerated5. Heterosis, which refers to higher yield in F1 hybrids compared to the parents, could reach 30–60% in sesame46, 47. However, the genetic mechanism of sesame heterosis remains unknown. The availability of three genomes from three diverse sesame accessions provides the opportunity to explore the biological basis of heterosis in sesame. Genomic variations between each two accessions in the sesame core collections have been identified and are available in the SesameFG. This information can also be used in map-based gene cloning when the sesame core collections are selected to be the parents of some populations. These genomes and genomic variations will be very useful for sesame genetic improvement to help meet human’s increasing consumption of vegetable oil.
In addition to the genomes and genome variations that can provide clues for gene cloning, the genetic resources available in the Gene section also provide valuable information for identifications of gene function. Combined with the gene families and gene expression profiles, genes involved in the regulation of some agronomic traits, such as flowering, waterlogging resistance, and drought tolerance, can be discovered16–18, 37. The candidate sesame oil-related genes in the database (e.g., SiNST1, a gene involved in lignin and cellulose biosynthesis) likely also play important roles in other oilseed species, offering the opportunity to look for genes with common functions in these other species.
Although SSR markers are widely used in high-throughput genotyping and map construction for high-abundance, high-polymorphism and stable co-dominance, -SSR makers for sesame functional genomics research are lacking. Based on the first sequenced and assembled sesame genome, all genomic SSR loci were surveyed and identified in the SesameFG. The polymorphic SSR markers have also been collected and are available. Among these SSR markers, thirty-two SSR markers were selected as the core SSR markers and have been successfully used in the genealogical analysis of sesame elites9. The SSR design tool, which is included in the database, will substantially accelerate the QTL and gene fine mapping of sesame. These SSR markers can provide useful resources for genetic linkage map construction, genetic diversity detection, evolution analysis and marker-assisted selection breeding of sesame.
Seed coat color is an important agronomic trait in sesame. Through the utilization of the phenotypic and genotypic resources in the database, the candidate gene PPO, which encodes polyphenol oxidase and produces black pigments through a browning reaction, was identified as the key regulation gene of seed coat color in sesame. Several indels were identified in the coding regions of PPO, resulting in loss of function of the PPO enzyme, which caused the inability of black pigments to accumulate in the seed coat5. In fact, besides the PPO gene, several other causative genes related to important agronomic traits have been identified with the help of SesameFG. For example, SiACS was identified as the causative gene of capsule number per axil2. A GWAS revealed that the peak SNP (P = 1.02 × 10−128) can explain up to 60% of the phenotypic variation and is located in the SiACS. The SNP resulted in a phenylalanine mutated to a serine at the 284th amino acid of the SiACS protein. SSR markers have been designed with SesameFG and used in the QTL mapping of the F2 population, which was generated from crossing the “Zhongzhi13” (three-capsule allele) and “Baizhima” (one-capsule allele) varieties. The QTL region was successfully focused into 79 kb. BLAST results showed that the SNP in SiACS was the only coding variant around the QTL region between “Zhongzhi13” and “Baizhima”. For ongoing efforts in the gene cloning projects of more complex agronomic traits for functional genomics research in sesame, this database may provide valuable information and may prove to be an effective assistant system.
Molecular breeding is considered to be the best option for crop breeders to improve breeding efficiency48. Three steps require the assistance of massive molecular markers: gene or QTL identifications, formulation of the ideal genotype, and efficient molecular breeding49–51. The SNPs that were identified in the genome comparison were collected and used as effective markers in the molecular breeding of rice, soybean and peanut48, 52, 53. However, because of the lack of an efficient genotyping system, the molecular breeding of sesame has been difficult in recent years. The SSR markers and SNPs that are closely linked or associated with the agronomic traits provided in SesameFG can be easily obtained and used in molecular breeding of sesame. A high-density SNP genotyping array for sesame molecular breeding can also be designed from the genomic SNPs in SesameFG.
We will continue to make efforts to improve and update the database with newly released sesame phenotypic and genotypic information. The reference genome used in SesameFG will be updated according to new versions of the sesame reference genome sequence. The size of the database will continue to expand with the addition of more sesame genome sequences, genome variations, gene expression profiles, gene families and candidate genes. We are also planning to add metabolomics data into SesameFG, making it a more comprehensive database for sesame functional genomics studies. We will also strive to make the database more user-friendly and more efficient following reflection and feedback on the first version of SesameFG. We hope that the accumulation and spread of sesame phenotypic, metabolomic and genetic information in SesameFG will greatly facilitate sesame functional genomics research and accelerate sesame genetic improvement in the future.
Conclusion
We developed SesameFG, an integrated repository of comprehensive genotype-phenotype information, to promote sesame functional genomics research. The currently available dataset in SesameFG consists of information on materials and phenotype values, genome sequences, genome variations, gene families, gene expression, candidate genes and SSR markers. The data and functions of SesameFG include valuable material and genetic resources for sesame linkage and association mapping, gene cloning, gene functional validation and evolutionary research. SesameFG will facilitate further functional genomics research, molecular breeding and genetic improvements of sesame, and may also provide useful information in the genetic research of other closely-related oilseed species, such as rapeseed.
Acknowledgements
We thank Mr. Tao Huang from the National Center for Gene Research of the Chinese Academy of Sciences for his technical support in database establishment and Mr. Komivi Dossa from the Oil Crops Research Institute of the Chinese Academy of Agricultural Sciences for providing the AP2 genes. This work was funded by the National Natural Science Foundation of China (31401412 and 31671282), the Agricultural Science and Technology Innovation Project of the Chinese Academy of Agricultural Sciences (CAAS-ASTIP-2013-OCRI), and the Fundamental Research Funds for Central Non-profit Scientific Institution (1610172014003).
Author Contributions
X.Z. and X.W. contributed to the design of this research. X.W., H.G., J.Y., P.L., L.W., and Y.Z. participated in the data collection, data analysis and database establishment. X.W. drafted the manuscript. All authors read and approved the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Xin Wei and Hao Gong contributed equally to this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Weiss, E. A. Oilseed crops (Blackwell Science, 2000).
- 2.Wei X, et al. Genetic discovery for oil production and quality in sesame. Nat Commun. 2015;6:8609. doi: 10.1038/ncomms9609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hodgkin, T. et al. Developing sesame core collections in China and India. In Core Collections for Today & Tomorrow (eds Johnson, R. C. & Hodgkin, T.) (International Plant Genetic Resources Institute, Roma, 1999).
- 4.Wang L, et al. Deep resequencing reveals allelic variation in Sesamum indicum. BMC Plant Biol. 2014;14:225. doi: 10.1186/s12870-014-0225-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei X, et al. Identification of sesame genomic variations from genome comparison of landrace and variety. Front Plant Sci. 2016;7:1169. doi: 10.3389/fpls.2016.01169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang L, et al. Genome sequencing of the high oil crop sesame provides insight into oil biosynthesis. Genome Biol. 2014;15:R39. doi: 10.1186/gb-2014-15-2-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L, Zhang Y, Qi X, Gao Y, Zhang X. Development and characterization of 59 polymorphic cDNA-SSR markers for the edible oil crop Sesamum indicum (Pedaliaceae) Am J Bot. 2012;99:e394–e398. doi: 10.3732/ajb.1200081. [DOI] [PubMed] [Google Scholar]
- 8.Wei W, et al. Characterization of the sesame (Sesamum indicum L.) global transcriptome using Illumina paired-end sequencing and development of EST-SSR markers. BMC Genomics. 2011;12:451. doi: 10.1186/1471-2164-12-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei X, et al. Development of simple sequence repeat (SSR) markers of sesame (Sesamum indicum) from a genome survey. Molecules. 2014;19:5150–62. doi: 10.3390/molecules19045150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang H, Wei L, Miao H, Zhang T, Wang C. Development and validation of genic-SSR markers in sesame by RNA-seq. BMC Genomics. 2012;13:316. doi: 10.1186/1471-2164-13-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L, et al. Updated sesame genome assembly and fine mapping of plant height and seed coat color QTLs using a new high-density genetic map. BMC Genomics. 2016;17:31. doi: 10.1186/s12864-015-2316-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei W, et al. Association analysis for quality traits in a diverse panel of Chinese sesame (Sesamum indicum L.) germplasm. J Integr Plant Biol. 2013;55:745–58. doi: 10.1111/jipb.12049. [DOI] [PubMed] [Google Scholar]
- 13.Wu K, et al. High-density genetic map construction and QTLs analysis of grain yield-related traits in sesame (Sesamum indicum L.) based on RAD-Seq techonology. BMC Plant Biology. 2014;14:274. doi: 10.1186/s12870-014-0274-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang H, et al. Genetic analysis and QTL mapping of seed coat color in sesame (Sesamum indicum L.) PLoS One. 2013;8:e63898. doi: 10.1371/journal.pone.0063898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, et al. Construction of a high-density genetic map for sesame based on large scale marker development by specific length amplified fragment (SLAF) sequencing. BMC Plant Biol. 2013;13:141. doi: 10.1186/1471-2229-13-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dossa K, et al. Insight into the AP2/ERF transcription factor superfamily in sesame and expression profiling of DREB subfamily under drought stress. BMC Plant Biol. 2016;16:171. doi: 10.1186/s12870-016-0859-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei X, et al. Genome-wide identification and analysis of the MADS-box gene family in sesame. Gene. 2015;569:66–76. doi: 10.1016/j.gene.2015.05.018. [DOI] [PubMed] [Google Scholar]
- 18.Wang L, et al. Tolerant and susceptible sesame genotypes reveal waterlogging stress response patterns. PLoS One. 2016;11:e0149912. doi: 10.1371/journal.pone.0149912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L, et al. Global gene expression responses to waterlogging in roots of sesame (Sesamum indicum L.) Acta Physiol Plant. 2012;34:2241–2249. doi: 10.1007/s11738-012-1024-9. [DOI] [Google Scholar]
- 20.Wang L, Yu J, Li D, Zhang X. Sinbase: an integrated database to study genomics, genetics and comparative genomics in Sesamum indicum. Plant Cell Physiol. 2015;56:e2. doi: 10.1093/pcp/pcu175. [DOI] [PubMed] [Google Scholar]
- 21.Zhang H, et al. Genome sequencing of the important oilseed crop Sesamum indicum L. Genome Biol. 2013;14:401. doi: 10.1186/gb-2013-14-1-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fei Z, et al. Tomato Functional Genomics Database: a comprehensive resource and analysis package for tomato functional genomics. Nucleic Acids Res. 2011;39:D1156–D1163. doi: 10.1093/nar/gkq991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu T, et al. RICD: a rice indica cDNA database resource for rice functional genomics. BMC Plant Biol. 2008;8:118. doi: 10.1186/1471-2229-8-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.You Q, et al. SIFGD: Setaria italica Functional Genomics Database. Mol Plant. 2015;8:967–970. doi: 10.1016/j.molp.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 25.Kusano M, Fukushima A. Current challenges and future potential of tomato breeding using omics approaches. Breed Sci. 2013;63:31–41. doi: 10.1270/jsbbs.63.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JM, et al. Combined transcriptome, genetic diversity and metabolite profiling in tomato fruit reveals that the ethylene response factor SlERF6 plays an important role in ripening and carotenoid accumulation. Plant J. 2012;70:191–204. doi: 10.1111/j.1365-313X.2011.04863.x. [DOI] [PubMed] [Google Scholar]
- 27.Wang L, et al. A dynamic gene expression atlas covering the entire life cycle of rice. Plant J. 2010;61:752–766. doi: 10.1111/j.1365-313X.2009.04100.x. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Q. Strategies for developing Green Super Rice. Proc Natl Acad Sci USA. 2007;104:16402–16409. doi: 10.1073/pnas.0708013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang X, et al. Genomic analysis of hybrid rice varieties reveals numerous superior alleles that contribute to heterosis. Nat Commun. 2015;6:6258. doi: 10.1038/ncomms7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bisht IS, Mahajan RK, Loknathan TR, Agrawal RC. Diversity in Indian sesame collection and stratification of germplasm accessions in different diversity groups. Genet Resour Crop Ev. 1998;45:325–335. doi: 10.1023/A:1008652420477. [DOI] [Google Scholar]
- 31.Morris JB. Characterization of sesame (Sesamum indicum L.) germplasm regenerated in Georgia, USA. Genet Resour Crop Ev. 2009;56:925–936. doi: 10.1007/s10722-009-9411-9. [DOI] [Google Scholar]
- 32.Zhang Y, et al. Genetic diversity assessment of sesame core collection in China by phenotype and molecular markers and extraction of a mini-core collection. BMC Genet. 2012;13:102. doi: 10.1186/1471-2156-13-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X, et al. Establishment of sesame germplasm core collection in China. Genet Resour Crop Ev. 2000;47:273–279. doi: 10.1023/A:1008767307675. [DOI] [Google Scholar]
- 34.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finn RD, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dossa K, Diouf D, Cissé N. Genome-wide investigation of Hsf genes in sesame reveals their segmental duplication expansion and their active role in drought stress response. Front Plant Sci. 2016;7:1522. doi: 10.3389/fpls.2016.01522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thiel T, Michalek W, Varshney RK, Graner A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.) Theor Appl Genet. 2003;106:411–422. doi: 10.1007/s00122-002-1031-0. [DOI] [PubMed] [Google Scholar]
- 39.Kortt AA, Caldwell JB, Lilley GG, Higgins TJ. Amino acid and cDNA sequences of a methionine-rich 2S protein from sunflower seed (Helianthus annuus L.) Eur J Biochem. 1991;195:329–334. doi: 10.1111/j.1432-1033.1991.tb15710.x. [DOI] [PubMed] [Google Scholar]
- 40.Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–7. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 42.Huala E, et al. The Arabidopsis Information Resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res. 2001;29:102–105. doi: 10.1093/nar/29.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Donlin, M. J. Using the Generic Genome Browser (GBrowse). Curr Protoc Bioinformatics Chapter 9, 9.9.1–9.9.24 (2009). [DOI] [PubMed]
- 44.Faostat. Statistical Databases. Food and Agriculture Organization of the United Nations (2015).
- 45.Zhang J, et al. Extensive sequence divergence between the reference genomes of two elite indica rice varieties Zhenshan 97 and Minghui 63. Proc Natl Acad Sci USA. 2016;113:E5163–E5171. doi: 10.1073/pnas.1611012113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang W, et al. Studies on the heterosis of the dynamic growth of hybrid sesame (Sesamum indicum L.) Acta Agronomica Sinica. 1997;23:440–445. [Google Scholar]
- 47.Murty DS. Heterosis, combining ability and reciprocal effects for agronomic and chemical characters in sesame. Theor Appl Genet. 1975;45:294–299. doi: 10.1007/BF00276682. [DOI] [PubMed] [Google Scholar]
- 48.Chen H, et al. A high-density SNP genotyping array for rice biology and molecular breeding. Mol Plant. 2014;7:541–553. doi: 10.1093/mp/sst135. [DOI] [PubMed] [Google Scholar]
- 49.Flavell R. From genomics to crop breeding. Nat Biotechnol. 2010;28:144–145. doi: 10.1038/nbt0210-144. [DOI] [PubMed] [Google Scholar]
- 50.Morrell PL, Buckler ES, Ross-Ibarra J. Crop genomics: advances and applications. Nat Rev Genet. 2011;13:85–96. doi: 10.1038/nrg3097. [DOI] [PubMed] [Google Scholar]
- 51.Varshney RK, Terauchi R, McCouch SR. Harvesting the promising fruits of genomics: applying genome sequencing technologies to crop breeding. PLoS Biol. 2014;12:e1001883. doi: 10.1371/journal.pbio.1001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Janila P, et al. Molecular breeding for introgression of fatty acid desaturase mutant alleles (ahFAD2A and ahFAD2B) enhances oil quality in high and low oil containing peanut genotypes. Plant Sci. 2016;242:203–213. doi: 10.1016/j.plantsci.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 53.Joshi T, et al. Soybean knowledge base (SoyKB): a web resource for integration of soybean translational genomics and molecular breeding. Nucleic Acids Res. 2014;42:D1245–D1252. doi: 10.1093/nar/gkt905. [DOI] [PMC free article] [PubMed] [Google Scholar]


