Abstract
Williams Syndrome (WS) is a rare genetic disorder with unique behavioral features. Yet the rareness of WS has limited the number and type of studies that can be conducted in which inferences are made about how neuroanatomical abnormalities mediate behaviors. In this study, we extracted a WS-specific neuroanatomical profile from structural magnetic resonance imaging (MRI) measurements and tested its association with behavioral features of WS. Using a WS adult cohort (22 WS, 16 healthy controls), we modeled a sparse representation of a WS-specific neuroanatomical profile. The predictive performances are robust within the training cohort (10-fold cross-validation, AUC = 1.0) and accurately identify all WS individuals in an independent child WS cohort (seven WS, 59 children with diverse developmental status, AUC = 1.0). The WS-specific neuroanatomical profile includes measurements in the orbitofrontal cortex, superior parietal cortex, Sylvian fissures, and basal ganglia, and variability within these areas related to the underlying size of hemizygous deletion in patients with partial deletions. The profile intensity mediated the overall cognitive impairment as well as personality features related to hypersociability. Our results imply that the unique behaviors in WS were mediated through the constellation of abnormalities in cortical-subcortical circuitry consistent in child WS and adult WS. The robustness of the derived WS-specific neuroanatomical profile also demonstrates the potential utility of our approach in both clinical and research applications.
Keywords: Williams Syndrome, Neuroanatomy, Social cognition, Magnetic resonance imaging
Highlights
-
•
A Williams Syndrome Specific neuroanatomical profile is extracted to provide both explanatory power and clinical utilities
-
•
Multidimensional features of MRI are summarized into one single score to depict the morphological differences across groups
-
•
The model robustly identify Williams Syndrome individuals despite age differences
-
•
The selected features indicate the mediation paths from brain to cognition
1. Introduction
Williams Syndrome (WS) is a rare multi-system disorder caused by hemideletion of ~ 26 genes on chromosome 7. Although the cognitive impact of WS is evident in general intelligence and visuospatial capabilities, the cardinal feature of WS cognition is overly social behavior (Pober, 2010). WS individuals express heightened social approach behavior and social emotional behavior very early on, distinguishing them from others with disorders that include intellectual impairment (Doyle et al., 2004). This had led to extensive research using magnetic resonance imaging (MRI), in the hope of identifying the mediating neural processes from genetic deletions to social behavioral impact (Martens et al., 2008). Previous MRI studies had found that what distinguishes WS from other genetic disorders with intellectual impairment — e.g., Down syndrome — is not the reduced total brain volume per se, but the aberrant regionalization of the brain (Jernigan and Bellugi, 1990). The most consistent findings are the gyral patterns in the superior parietal regions and orbital frontal cortex, which were found to be different between WS patients and healthy individuals (Gaser et al., 2006, Kippenhan et al., 2005, Meda et al., 2012, Meyer-Lindenberg et al., 2004).
Yet the specificity of these findings to WS and relevance to its distinct behavioral features were left unanswered. Differences in regional cortical surface area, such as in lingual gyrus, post-central gyrus, and temporal poles, were also reported (Thompson et al., 2005, Wang et al., 2013). Abnormalities in the Sylvian fissures (Eckert et al., 2006) and disproportional volumetric changes of subcortical structures were also reported, but not consistent (Meyer-Lindenberg et al., 2004, Martens et al., 2009, Capitao et al., 2011, Meda et al., 2012). Furthermore, the diagnostic process for WS requires that clinicians identify individuals with WS features and use fluorescent in situ hybridization (FISH) to confirm. This precludes identification of individuals who have different deletions in the WS chromosome region (WSCR), resulting in slightly altered profiles of WS features. A recent analysis focused on cases of individuals with atypical deletions in the WSCR suggested that the varying size of the deletion would result in different behavioral profiles (Hoeft et al., 2014), which conceivably would make it difficult to identify those individuals in clinical settings. The rarity of both typical and atypical WS individuals makes the quantitative comparisons across MRI measures and groups impractical.
Here, we re-examined the WS-specific neuroanatomical profile using a novel analytic approach with the aim of developing a scoring system to quantify WS neuroanatomical variations. First, we extracted the WS-specific neuroanatomical profile from an adult WS cohort, using multiple measures derived from structural MRI of cerebrum, including subcortical volumes, cortical surface area (Dale et al., 1999, Fischl et al., 1999), sulcal depth (Kippenhan et al., 2005), and cortical surface geometry (Fan et al., 2015). To deal with the large number of MRI measures and limited sample size, we used an elastic-net model to achieve balance between the robust prediction and sparseness for easy interpretation. The resulting model provides the basis for calculating WS neuroanatomical scores that represent the similarity of an individual's brain to the WS given his/her multimodal MRI features. The generalizability of the WS-specific neuroanatomical profile was then tested in an independent child WS cohort. After establishing the generalizability of the model, we examined whether the WS neuroanatomical scores could reflect the reduced size of genetic deletions in WSCR and whether the scores were associated with the behavioral features of WS.
2. Materials and methods
2.1. Participants
All participants were recruited as part of a multi-project program, including two cohorts in current analyses, one as child cohort and the other as adult cohort. Except time of recruitment, age differences, additional diagnostic groups, and behavior measures, the protocols for inclusion and imaging acquisition were kept the same, which were described in separate publications (Eckert et al., 2006, Mills et al., 2013). Participants were screened based on the following measures: normal or corrected vision/hearing, English native-language speaker, and no remarkable mental health history. Caregivers completed an interview and extensive demographic and family history questionnaires to assess whether participants met the screening criteria. Caregivers and child participants provided consent and assent, respective, for participation. Individuals with intellectual disabilities required a more simple, verbally delivered description for assent along with guardian informed consent. All procedures were explained in person, within the testing environment, with the caregiver present, to show the participants more concretely what to expect. They could choose at any time to withdraw from participation, even after beginning. Study protocols were approved by the Institutional Review Boards at the Salk Institute and at UCSD.
2.1.1. Adult WS cohort
The adult cohort, on which the WS-specific neuroanatomical profile was trained, consisted of 22 individuals with “typical” WS deletions (approximately 26 genes in the WSCR 7q11.23 region) as well as 16 healthy controls (HC) (Table 1). Part of this cohort has been involved in a series of MRI studies for WS that were published elsewhere (Eckert et al., 2006, Van Essen et al., 2006). The diagnosis of WS was based on clinical presentation (WS Diagnostic Score Sheet) and confirmation of meeting genetic criteria for WS using fluorescent in situ hybridization. HCs were screened for a history of neurological disorders, psychiatric illness, and substance abuse. Intellectual functioning was assessed with the age-appropriate version of the Wechsler tests to include the Wechsler Adult Intelligence Scale 3rd Edition, Wechsler Abbreviated Scale of Intelligence (WASI), and Wechsler Intelligence Scale for Children 3rd Edition WISC-III (Wechsler, 2008). Sociability was assessed with the Salk Institute Sociability Questionnaire (SISQ) (Doyle et al., 2004).
Table 1.
Demographics and global MRI measurements of participants in two cohorts.
| Groups | n | Age – years | Gender – male | Full IQ | SISQ – AS | SISQ – ES | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Adult WS cohort | ||||||||||
| WS | 22 | 31.6 | (10.8) | 59% | 66.6 | (5.0) | 5.4 | (1.4) | 5.8 | (0.8) |
| HC | 16 | 25.9 | (7.0) | 37% | 96.7 | (14.6) | 3.6 | (1.2) | 4.4 | (1.0) |
| Atypical WSa | 5 | 17.7 | (2.5) | 20% | 74 | 4.2 | 5.8 | |||
| Child cohort | ||||||||||
| WS | 7 | 11.95 | (1.75) | 29% | ||||||
| TD | 23 | 9.48 | (1.87) | 52% | ||||||
| FL | 8 | 9.73 | (1.26) | 50% | ||||||
| HFA | 14 | 9.83 | (1.45) | 79% | ||||||
| SLI | 8 | 10.09 | (1.48) | 75% | ||||||
WS: Williams Syndrome. HC: healthy controls. TD: typical developed individuals. FL: Individuals with focal lesions in the MRI scans of brain. HFA: high function autism. SLI: specific language impairment.
Behavioral measures on Atypical WS were available for one male teenager. Hence, standard deviations were not shown.
2.1.2. Child cohort
The generalizability of the WS-specific neuroanatomical profile was tested with a cohort of 60 children (age range 6 to 13 years): seven individuals with WS, 23 typical developing children (TD), and 30 individuals with heterogeneous diagnoses to include high-functioning autism (HFA), specific language impairment (SLI), and focal lesions in the brain (FL). The demographic characteristics of each cohort are shown in Table 1. Children with WS were diagnosed using the same criteria as adults with WS. Subjects in the TD group were recruited from the community, had scores on a standardized test of intellectual functioning (WASI) in the normal range and no history of developmental or language delay. Individuals with HFA, SLI, and FL were recruited from populations at a local pediatric neurology clinic and a clinic for speech and language disorders (Mills et al., 2013). Detailed recruiting procedures and diagnostic criteria can be found in previously published studies (Mills et al., 2013).
2.1.3. Individuals with atypical deletions in WSCR
We further examined if the scores from the trained model for WS-specific neuroanatomical profile can identify whose brain phenotypes lie between WS and HC, such as individuals with reduced deletion size on WSCR. We tested our model on five individuals from one family with small deletions on chromosome 7q11.23, sparing regions coding for FZD9, GTF2I, and GTF2IRD1 (Hoeft et al., 2014).
2.1.4. Imaging acquisition and extracting multimodal MRI features
All participants were scanned on a 1.5 Tesla MRI scanner (GE HDxt, echo time (TE) = 3.0 ms, repetition time (TR) = 8.7 ms, inversion time = 270 ms, flip angle = 8°, field of view = 24 cm, voxel size = 1.25 × 1.25 × 1.2 mm). To reduce and prevent possible motion artifacts, real-time prospective motion tracking and correction (PROMO) was used for all participating subjects (White et al., 2010, Brown et al., 2010). Distortions caused by nonlinearity of the spatial encoding gradient fields were corrected with predefined nonlinear transformations (Jovicich et al., 2006). Non-uniformity of signal intensity was reduced with the nonparametric nonuniform intensity normalization method (Sled et al., 1998). After initial image data inspection and quality control, T1-weighted images underwent automated volumetric segmentation and cortical surface reconstruction using methods implemented in Freesurfer software (Dale et al., 1999, Fischl et al., 1999). This automated processing corrects variations in image intensity due to RF coil sensitivity inhomogeneities, registers to a common reference, then segments volumes into cortical and subcortical structures. For each cohort, one staff research associate performed quality control (QC) of the surfaces and segmentations for all MRI images at the same time, blind to age and group identification. Both the child cohort and the adult cohort went through the same QC processes. The segmentations and reconstructed surfaces were inspected for accuracy, manually edited using control points, and iteratively re-processed, blind to age or group labels, to ensure consistent quality across different cohorts.
Four different morphological measures of T1-weighted images were derived, including the volumes of subcortical structures (Dale et al., 1999), sulcal depths of the cortical surface (Kippenhan et al., 2005), cortical surface area (Fischl et al., 1999), and geometric deformations of the cortical surface (Fan et al., 2015). Sulcal depth is the distance from each point on the cortical surface to the average mid-plane of the cortical surface, measuring gyrification of the brain. Cortical surface area expansion is the area surrounding a given cortical surface point relative to total cortical surface area. The geometric deformation is the 3D Cartesian coordinates of the cortical surface, characterizing the folding patterns of the brain. Subcortical structure volumes were divided by total brain volumes, and sulcal depths and geometric deformations were divided by the cubic root of each total brain volume to produce a uniform index, as well as to control for the global brain volume differences. Those imaging features were selected as a comprehensive representation of the neuroanatomical variations of the human brain possible with structural MRI without unnecessary a priori defined regions of interest.
2.1.5. Model training
To characterize the WS-specific neuroanatomical profile from MRI measures, we fit an elastic-net logistic regression using data from the adult cohort and checked their performance with 10-fold cross validation. The index for model performance was area under curve (AUC) in the ROC analysis. The model included all four types of MRI measures; that is, cortical surface area (642 vertices-per-hemisphere), sulcal depths of cortical surface (642 vertices-per-hemisphere), cortical surface geometry (642 vertices-per-hemisphere, each with 3D Cartesian coordinates), and subcortical volumes (thalamus, caudate, putamen, globus pallidum, hippocampus, amygdala, nucleus accumbens, and ventral caudate). To achieve the goal of balancing between predictive power and parsimonious solution, we used the ridge penalties to reduce the problem of rank deficiencies and additional lasso penalties for removing less relevant features (Hastie et al., 2009). The tuning parameters were optimized during the cross-validation.
2.1.6. Model validation
After deriving the WS-specific neuroanatomical profile from the previous training step, the model was applied to the whole child cohort in predicting WS status out of a heterogeneous group. The model was also applied to individuals with atypical deletion size in the WSCR to examine if the scores were in-between the typical WS and HC. Afterward, the relationships between model-predicted scores and behavioral measures were explored using mediation analysis. Within-group variations were examined using Pearson correlations while Sobel tests were used to test whether the group differences were mediated by the neuroanatomical profile.
3. Results
3.1. Deriving the profile and its generalizability
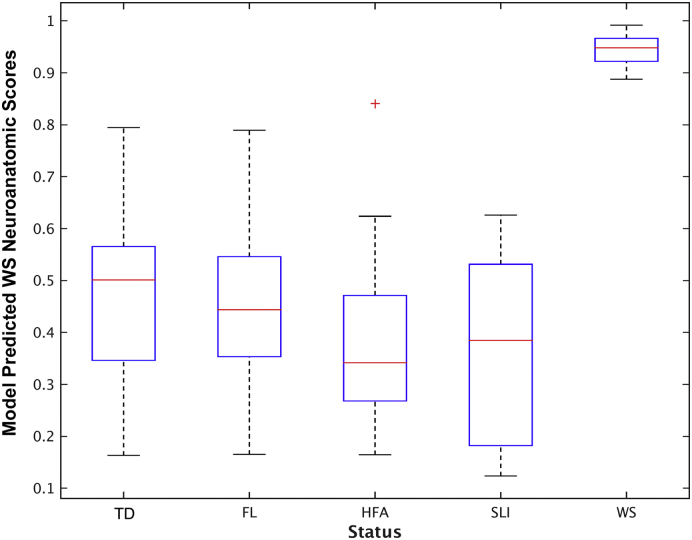
In classifying WS status, the 10-fold cross-validation AUC of the WS-specific neuroanatomical profile achieved 100% in the adult WS cohort (two-tail test for AUC > 0.5, p < 0.05). The model removes 98.4% of the input variables, leaving 412 features from four MRI measures. Among individuals with atypical deletions in WSCR (atypical WS), their predicted scores of the WS-specific neuroanatomical profile lay between typical WS and HC, which is significantly greater than HC (t19 = 9.4, p < 10− 7), and less than patients with typical WS (t25 = − 2.2, p = 0.038). To further test the generalizability of the model, we applied the WS-specific neuroanatomical profile to the whole child cohort. In this independent cohort, the profiling scores have AUC with 1.0 in predicting WS status, achieving 100% sensitivity and 100% specificity with various decision cut-points (Fig. 1).
Fig. 1.
Boxplot of model predicted scores from trained WS-specific neuroanatomical profile across groups in the child cohort. The predicted scores of each group were demonstrated as median and inter-quartile range. The outliers were label as red-cross. Among them, children with WS have higher scores, none overlapping with any other group, that yield AUC of ROC analysis with one.
3.2. Features in the WS-specific neuroanatomical profile
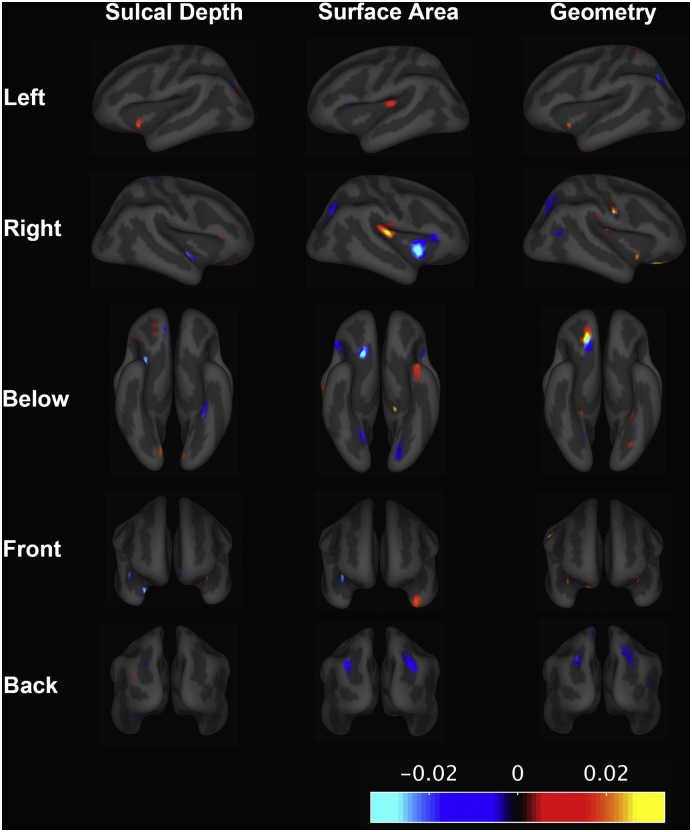
The cortical surface features extracted by the model are shown in Fig. 2. The weights of selected features reflect the relative importance for predicting WS. Selected local features can be observed across different cortical surface regions, yet sparing the dorsal and medial part of the frontal cortex. The orbitofrontal cortex and superior parietal cortex contain predictive features consistently across all three cortical surface measures (Fig. 2). In addition, the cortical surface area contained predictive features in the Sylvian fissure and temporal poles. Two subcortical structures were also selected. Disproportionally decreasing sizes of left putamen (weights = − 0.010) and left nucleus accumbens (weights = − 0.014) were predictive for WS status.
Fig. 2.
Elastic net model learnt features for predicting WS status. The blue or red indicates that the surface measures at that region were selected to be discriminative features. The red represents WS individuals with increased value of measures on that region, whereas the blue represents the decreased value of measures among WS individuals. The magnitude of those colors indicates their relative importance for classifying WS and HC. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. Associations with WS behavioral features
The relationships among WS status, the WS-specific neuroanatomical profile, and behavioral function of WS are illustrated in Table 2. The Sobel tests for mediation indicate that the group differences in general intelligence, SISQ stranger score, and SISQ empathy score are largely explained by the mediating effect of the WS-specific neuroanatomical profile (all p values < 10− 3, Bonferroni corrected). In the within-group analyses, the variations of the WS-specific neuroanatomical profile are significantly associated with SISQ empathy scores, with a “trending” p-value after applying Bonferroni correction for 9 independent tests (corrected p = 0.063).
Table 2.
Mediating effects and within-group correlations between model predicted WS neuroanatomic scores and behavioral measures.
| Mediating effecta | Within HC | Within WS | ||||
|---|---|---|---|---|---|---|
| FIQ | z = − 6.31 | p = 1e − 10 | r = 0.29 | p = 0.34 | r = 0.18 | p = 0.52 |
| SISQ – stranger | z = 3.73 | p = 9e − 5 | r = − 0.01 | p = 0.96 | r = 0.10 | p = 0.74 |
| SISQ – empathy | z = 4.61 | p = 2e − 6 | r = 0.09 | p = 0.74 | r = 0.70 | p = 7e − 3 |
The mediating effect is checked with Sobel test for mediation, treating model predicted WS neuroanatomic scores as the mediator and each behavioral measure as dependent variable.
4. Discussion
In this study, we sought to use a novel approach for characterizing the defining features of a WS-specific neuroanatomy and relating it to behavior. Features within the orbitofrontal cortex, superior parietal cortex, and Sylvian fissures were predictive for WS status across MRI measurements (Fig. 2). Disproportional reductions in the putamen and nucleus accumbens are also important features for predicting WS status. The robust performance of our extracted WS-specific neuroanatomical profile are consistent in both adult and child cohorts (Fig. 1). We also demonstrated that the scores for individuals with atypical deletions on WSCR lay between the WS and HC, and were associated with cardinal behavioral features of WS (Table 2).
MRI studies of WS have focused on localizing the neuroanatomical abnormalities (Eckert et al., 2006, Gaser et al., 2006, Kippenhan et al., 2005, Martens et al., 2008, Meyer-Lindenberg et al., 2006, Van Essen et al., 2006). Although WS individuals have smaller brains in general, early studies have shown that the reductions are not uniformly distributed across brain regions (Jernigan and Bellugi, 1990). Gyrification abnormalities in the orbitofrontal cortex, Sylvian fissures, and superior parietal regions have been reported (Eckert et al., 2006, Gaser et al., 2006, Kippenhan et al., 2005, Van Essen et al., 2006). Some have found that amygdala volumes are disproportionally increased (Martens et al., 2009, Capitao et al., 2011) while others found no significant changes (Meyer-Lindenberg et al., 2004, Meda et al., 2012). The joint relationships across these neuroanatomical features were seldom examined in WS (Martens et al., 2008, Wang et al., 2013). One study had used tensor metrics of cortical surface to predict WS status in adult cohorts (Wang et al., 2013). Different from what they attempted, our study aimed to evaluate all MRI measurements jointly, and the WS-specific neuroanatomical profile achieved 100% AUC in the independent child cohort (Fig. 1).
Our sparse representation of WS profile matched with previously hypothesized causes of the behavioral profile of WS patients (Meyer-Lindenberg et al., 2004, Gaser et al., 2006, Van Essen et al., 2006). The selected features of cortical surface area located at the orbitofrontal, temporal parietal junction, and insula (Fig. 2) are relevant to social functions (Adolphs, 2001, Meyer-Lindenberg et al., 2006, Saxe and Kanwisher, 2003). The superior parietal region has been linked most strongly to the visuospatial processing deficits in WS (Meyer-Lindenberg et al., 2004). Our mediation analyses using the Sobel test showed that the WS-specific neuroanatomical profile explained more variability in the behavioral measures than the WS status itself. This suggests that the WS-specific neuroanatomical profile may capture the underlying neuroanatomical factors that drive the related cognition and social behaviors. Since our behavioral analyses are limited in the WS adult cohort, we envision that longitudinal studies among children can be helpful to further establish the causal relationships between observed neuroanatomical profile and behavioral features of WS. Nevertheless, the robust performance of the WS-specific neuroanatomical profile in our child cohort suggests these features are already expressed during childhood.
Furthermore, case studies have indicated that atypical WS patients with smaller genetic deletions have lower social ratings than typical WS patients (Doyle et al., 2004). The telomere side of WS-related chromosomal regions, which tends to be spared in smaller deletions, contains genes such as GTF2I and GTF2IRD1, which have been associated with social behaviors in mouse models (Tassabehji et al., 2005, Young et al., 2008). Very recently, a study using induced pluripotent stem cells from WS suggested FZD9 may be responsible for aberrant neurodevelopment (Chailangkarn et al., 2016). Our data show that individuals with smaller deletions would have lower WS-specific anatomical scores than typical WS while those scores are positively correlated with hypersociability. These findings suggest that our extracted WS-specific profile of features might relate directly to the underlying genetic cause of hypersociability in WS.
Our study has several limitations. The training samples for the WS-specific neuroanatomical profile are relatively small compared with other machine-learning applications (Hastie et al., 2009). Small sample sizes are common in published studies of WS, considering that the prevalence of WS is rare (Pober, 2010). Direct group comparisons across multiple MRI measures would suffer the burden of multiple hypotheses testing. Our approach for extracting WS-specific features circumvents this limitation of group comparisons. We kept a careful balance between interpretability and predictive power, achieving 100% AUC in both cross-validation of the adult cohort and the independent testing child cohort. Even though the robustness of the predictive performance is ensured, the feature selections are nevertheless constrained by the number of training samples (Hastie et al., 2009). This may explain why some previous reported neuroanatomical abnormalities, such as the amygdala (Meyer-Lindenberg et al., 2004, Martens et al., 2009, Capitao et al., 2011), are not selected as predictive features. The neuroanatomical differences between WS patients and controls are not limited to regions we selected. The differences may be more similar to locally smoothed gradients. Meanwhile, it is also unclear how sensitive the WS-specific neuroanatomical profile is to the scanning protocols. Although our results indicate that our model can identify WS in different age ranges from a very heterogeneous developmental cohort, the MRI images of training and testing samples were obtained and processed with the same protocol. Applying our WS-specific scores in other settings would be a further test of its clinical and research utilities.
Taken together, our novel multidimensional imaging approach captures the widespread differences observed within the neural architecture of individuals with WS. The model can have direct clinical applications, such as measuring the neuroanatomical phenotype of atypical WS with different sizes of deletions on WS chromosomal regions. Furthermore, a major benefit of our analytic strategy is that the extracted features can be readily applied to other imaging datasets. Applications of the extracted features on a large imaging genomic cohort would further inform research on the genetic influences of social behaviors.
Funding
The Williams Syndrome studies were funded by a National Institutes of Health Grant R01 DA038958 and P50 NS022343.
Acknowledgements
Thanks to Ursula Bellugi for leading the recruitment and scientific study of the initial cohort, Doris Trauner for leading the recruitment and scientific study of the validation cohort, and Julie Korenberg for genetic analysis. A.M.D. and E.H. are founders of and hold equity interest in CorTechs Labs, La Jolla, Calif.; A.M.D. also serves on its scientific advisory board. The terms of this arrangement have been reviewed and approved by the University of California, San Diego, in accordance with its conflict-of-interest policies.
Contributor Information
Eric Halgren, Email: ehalgren@ucsd.edu.
Anders M. Dale, Email: amdale@ucsd.edu.
References
- Adolphs R. The neurobiology of social cognition. Curr. Opin. Neurobiol. 2001;11:231–239. doi: 10.1016/s0959-4388(00)00202-6. [DOI] [PubMed] [Google Scholar]
- Brown T.T., Kuperman J.M., Erhart M., White N.S., Roddey J.C., Shankaranarayanan A., Han E.T., Rettman D., Dale A.M. Prospective motion correction of high-resolution magnetic resonance imaging data in children. NeuroImage. 2010;53:135–149. doi: 10.1016/j.neuroimage.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capitao L., Sampaio A., Sampaio C., Vasconcelos C., Fernandez M., Garayzabal E., Shenton M.E., Goncalves O.F. MRI amygdala volume in Williams Syndrome. Res. Dev. Disabil. 2011;32:2767–2772. doi: 10.1016/j.ridd.2011.05.033. [DOI] [PubMed] [Google Scholar]
- Chailangkarn T., Trujillo C.A., Freitas B.C., Hrvoj-Mihic B., Herai R.H., Yu D.X., Brown T.T., Marchetto M.C., Bardy C., McHenry L., Stefanacci L., Järvinen A., Searcy Y.M., DeWitt M., Wong W., Lai P., Ard M.C., Hanson K.L., Romero S., Jacobs B., Dale A.M., Dai L., Korenberg J.R., Gage F.H., Bellugi U., Halgren E., Semendeferi K., Muotri A.R. A human neurodevelopmental model for Williams syndrome. Nature. 2016;536:338–343. doi: 10.1038/nature19067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale A.M., Fischl B., Sereno M.I. Cortical surface-based analysis. I. Segmentation and surface reconstruction. NeuroImage. 1999;9:179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- Doyle T.F., Bellugi U., Korenberg J.R., Graham J. “Everybody in the world is my friend” hypersociability in young children with Williams syndrome. Am. J. Med. Genet. A. 2004;124A:263–273. doi: 10.1002/ajmg.a.20416. [DOI] [PubMed] [Google Scholar]
- Eckert M.A., Galaburda A.M., Karchemskiy A., Liang A., Thompson P., Dutton R.A., Lee A.D., Bellugi U., Korenberg J.R., Mills D. Anomalous sylvian fissure morphology in Williams syndrome. NeuroImage. 2006;33:39–45. doi: 10.1016/j.neuroimage.2006.05.062. [DOI] [PubMed] [Google Scholar]
- Fan C.C., Bartsch H., Schork A.J., Chen C.H., Wang Y., Lo M.T., Brown T.T., Kuperman J.M., Hagler D.J., Jr., Schork N.J., Jernigan T.L., Dale A.M., Pediatric Imaging N., Genetics S. Modeling the 3D geometry of the cortical surface with genetic ancestry. Curr. Biol: CB. 2015;25:1988–1992. doi: 10.1016/j.cub.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B., Sereno M.I., Dale A.M. Cortical surface-based analysis. II: inflation, flattening, and a surface-based coordinate system. NeuroImage. 1999;9:195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- Gaser C., Luders E., Thompson P.M., Lee A.D., Dutton R.A., Geaga J.A., Hayashi K.M., Bellugi U., Galaburda A.M., Korenberg J.R., Mills D.L., Toga A.W., Reiss A.L. Increased local gyrification mapped in Williams syndrome. NeuroImage. 2006;33:46–54. doi: 10.1016/j.neuroimage.2006.06.018. [DOI] [PubMed] [Google Scholar]
- Hastie T., Tibshirani R., Friedman J. Springer; 2009. The Elements of Statistical Learning. [Google Scholar]
- Hoeft F., Dai L., Haas B.W., Sheau K., Mimura M., Mills D., Galaburda A., Bellugi U., Korenberg J.R., Reiss A.L. Mapping genetically controlled neural circuits of social behavior and visuo-motor integration by a preliminary examination of atypical deletions with Williams syndrome. PLoS One. 2014;9 doi: 10.1371/journal.pone.0104088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jernigan T.L., Bellugi U. Anomalous brain morphology on magnetic resonance images in Williams syndrome and Down syndrome. Arch. Neurol. 1990;47:529–533. doi: 10.1001/archneur.1990.00530050049011. [DOI] [PubMed] [Google Scholar]
- Jovicich J., Czanner S., Greve D., Haley E., van der Kouwe A., Gollub R., Kennedy D., Schmitt F., Brown G., Macfall J., Fischl B., Dale A. Reliability in multi-site structural MRI studies: effects of gradient non-linearity correction on phantom and human data. NeuroImage. 2006;30:436–443. doi: 10.1016/j.neuroimage.2005.09.046. [DOI] [PubMed] [Google Scholar]
- Kippenhan J.S., Olsen R.K., Mervis C.B., Morris C.A., Kohn P., Meyer-Lindenberg A., Berman K.F. Genetic contributions to human gyrification: sulcal morphometry in Williams syndrome. J. Neurosci. Off. J. Soc. Neurosci. 2005;25:7840–7846. doi: 10.1523/JNEUROSCI.1722-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens M.A., Wilson S.J., Reutens D.C. Research review: Williams syndrome: a critical review of the cognitive, behavioral, and neuroanatomical phenotype. J. Child Psychol. Psychiatry. 2008;49:576–608. doi: 10.1111/j.1469-7610.2008.01887.x. [DOI] [PubMed] [Google Scholar]
- Martens M.A., Wilson S.J., Dudgeon P., Reutens D.C. Approachability and the amygdala: insights from Williams syndrome. Neuropsychologia. 2009;47(12):2446–2453. doi: 10.1016/j.neuropsychologia.2009.04.017. [DOI] [PubMed] [Google Scholar]
- Meda S.A., Pryweller J.R., Thornton-Wells T.A. Regional brain differences in cortical thickness, surface area and subcortical volume in individuals with Williams syndrome. PLoS One. 2012;7 doi: 10.1371/journal.pone.0031913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Lindenberg A., Kohn P., Mervis C.B., Kippenhan J.S., Olsen R.K., Morris C.A., Berman K.F. Neural basis of genetically determined visuospatial construction deficit in Williams syndrome. Neuron. 2004;43:623–631. doi: 10.1016/j.neuron.2004.08.014. [DOI] [PubMed] [Google Scholar]
- Meyer-Lindenberg A., Mervis C.B., Faith Berman K. Neural mechanisms in Williams syndrome: a unique window to genetic influences on cognition and behaviour. Nat. Rev. Neurosci. 2006;7:380–393. doi: 10.1038/nrn1906. [DOI] [PubMed] [Google Scholar]
- Mills B.D., Lai J., Brown T.T., Erhart M., Halgren E., Reilly J., Appelbaum M., Moses P. Gray matter structure and morphosyntax within a spoken narrative in typically developing children and children with high functioning autism. Dev. Neuropsychol. 2013;38:461–480. doi: 10.1080/87565641.2013.820306. [DOI] [PubMed] [Google Scholar]
- Pober B.R. Williams-Beuren syndrome. N. Engl. J. Med. 2010;362:239–252. doi: 10.1056/NEJMra0903074. [DOI] [PubMed] [Google Scholar]
- Saxe R., Kanwisher N. People thinking about thinking people: the role of the temporo-parietal junction in “theory of mind.”. NeuroImage. 2003;19:1835–1842. doi: 10.1016/s1053-8119(03)00230-1. [DOI] [PubMed] [Google Scholar]
- Sled J.G., Zijdenbos A.P., Evans A.C. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans. Med. Imaging. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- Tassabehji M., Hammond P., Karmiloff-Smith A., Thompson P., Thorgeirsson S.S., Durkin M.E., Popescu N.C., Hutton T., Metcalfe K., Rucka A. GTF2IRD1 in craniofacial development of humans and mice. Science. 2005;310:1184–1187. doi: 10.1126/science.1116142. [DOI] [PubMed] [Google Scholar]
- Thompson P.M., Lee A.D., Dutton R.A., Geaga J.A., Hayashi K.M., Eckert M.A., Bellugi U., Galaburda A.M., Korenberg J.R., Mills D.L., Toga A.W., Reiss A.L. Abnormal cortical complexity and thickness profiles mapped in Williams syndrome. J. Neurosci. 2005;25(16):4146–4158. doi: 10.1523/JNEUROSCI.0165-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen D.C., Dierker D., Snyder A.Z., Raichle M.E., Reiss A.L., Korenberg J. Symmetry of cortical folding abnormalities in Williams syndrome revealed by surface-based analyses. The Journal of Neuroscience: the Official Journal of the Society for Neuroscience. 2006;26:5470–5483. doi: 10.1523/JNEUROSCI.4154-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Yuan L., Shi J., Greve A., Ye J., Toga A.W., Reiss A.L., Thompson P.M. Applying tensor-based morphometry to parametric surfaces can improve MRI-based disease diagnosis. NeuroImage. 2013;74:209–230. doi: 10.1016/j.neuroimage.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler D. NCS Pearson; San Antonio, TX: 2008. Wechsler Adult Intelligence Scale–Fourth Edition (WAIS–IV) [Google Scholar]
- White N., Roddey C., Shankaranarayanan A., Han E., Rettmann D., Santos J., Kuperman J., Dale A. PROMO: real-time prospective motion correction in MRI using image-based tracking. Magn. Reson. Med. 2010;63:91–105. doi: 10.1002/mrm.22176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young E., Lipina T., Tam E., Mandel A., Clapcote S., Bechard A., Chambers J., Mount H., Fletcher P., Roder J. Reduced fear and aggression and altered serotonin metabolism in Gtf2ird1-targeted mice. Genes Brain Behav. 2008;7:224–234. doi: 10.1111/j.1601-183X.2007.00343.x. [DOI] [PMC free article] [PubMed] [Google Scholar]