Abstract
5-Fluorouracil (5FU)-based adjuvant therapy is the first-line therapy for treating stage II and III colon cancer after surgery. However, its therapeutic efficacy is limited because of chemoresistance, especially in deficient mismatch repair (dMMR) colon cancer. Here, we first used laser capture microdissection to obtain purified cells from four dMMR and four proficient mismatch repair (pMMR) colon cancer tissues. Second, microRNA (miRNA) microarray chips were used to identify miRNAs that are differentially expressed between these two classes of tumors. Third, we analyzed their differential expression by qRT-PCR in a panel of 5-FU-resistant colon cancer cell lines. We identified that miR-1290 was one of the most upregulated miRNAs in both dMMR colon cancer tissues and 5-FU-resistant cells. We also found that miR-1290 was positively correlated with dMMR status and predicted poor prognosis in stage II and III colon cancer patients who received 5-FU-based chemotherapy. Furthermore, we demonstrated that inhibition of the expression of miR-1290 enhanced sensitivity to 5-FU treatment in vitro and in tumor xenografts in vivo by direct targeting hMSH2. Our study indicates that miR-1290 may become a promising biomarker of dMMR colon cancer and predicts the prognosis of stage II and III patients who receive 5-FU-based adjuvant therapy.
Keywords: miR-1290, dMMR, 5-fluorouracil, chemoresistance, colon cancer
Introduction
Colon cancer is one of the most prevalent cancers, with a high recurrence and cancer-related mortality rate worldwide.1 5-Fluorouracil (5-FU)-based adjuvant therapy is the first-line therapy for treating colon cancer and improves overall and disease-free survival of patients with advanced colon cancer.2 However, the response rates of patients with advanced colon cancer are only 10%–50% because of chemoresistance.2 In addition to the tumor stage, the mismatch repair (MMR) status is also an important factor, which affects the chemosensitivity of colon cancer to 5-FU.3
The MMR system is involved in the recognition and repair of DNA base mismatches.4 The human MMR protein family members responsible for the recognition of mispaired DNA include human mutS homolog 2 (hMSH2), human mutL homolog 1 (hMLH1), hMSH3, hMSH6, hMLH3, and hPMS2.5 Among them, hMSH2 and hMLH1 are core proteins in the MMR system. Deficient MMR (dMMR) colon cancer is caused by the inactivation of the DNA MMR genes mentioned above.3 dMMR is present in ∼15% of sporadic colon cancer cases.6 Some retrospective studies indicated that patients with dMMR colon cancer weakly or did not respond to 5-FU-based adjuvant therapy compared with patients with proficient MMR (pMMR).3 Although efforts have been made, the molecular mechanisms are still unclear. Therefore, the mechanism of 5-FU-based chemoresistance in patients with dMMR colon cancer still needs to be explored.
MicroRNAs (miRNAs) are a class of small (20–22 nt in length), non-protein-coding RNAs that act as negative regulators of gene expression by partially binding to complementary sites of the 3′ UTRs of target mRNAs.7 miRNAs regulate more than 30% of mRNAs and are involved in many fundamental processes, including development, differentiation, cell proliferation, and apoptosis.8 Thus, miRNAs participate in a variety of human diseases, including cancer.9 Moreover, various miRNAs play critical roles in regulating resistance to chemotherapeutic drugs in human tumors.10, 11
Although prior studies reported miRNA signatures of colon cancer, few studies have analyzed miRNA profiles in dMMR colon cancer. In this study, miR-1290 was elevated in both dMMR cell types and 5-FU-resistant colon cancer cell lines. To determine its role in chemoresistance of colon cancer, we performed the current study to evaluate the association between miR-1290 deregulation with 5-FU resistance of colon cancer both in vitro and in vivo and investigate its relationship with the prognosis of patients with stage II and III colon cancer who received 5-FU-based adjuvant therapy.
Results
miRNA Profiles in dMMR and pMMR Colon Cancer Cells
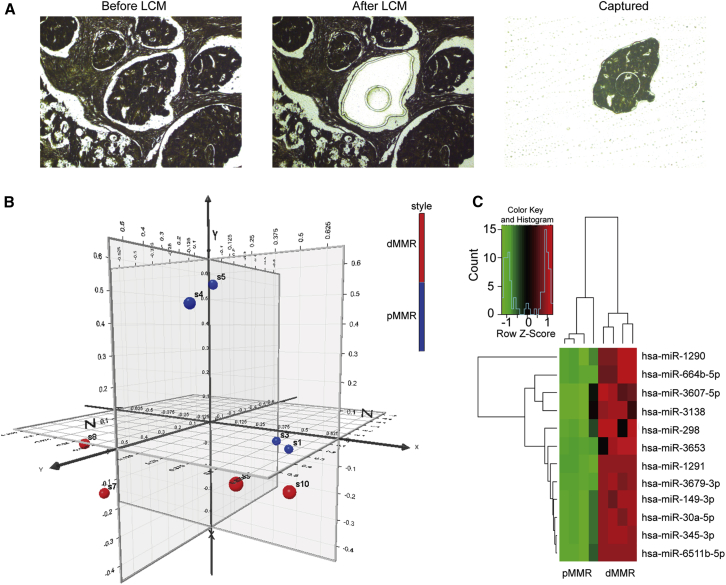
miRNA microarray technology can provide enormous information about cellular miRNA expression and is widely used to identify miRNAs that are key to prognosis and therapy for colon cancer. However, the heterogeneity of surgical tumor tissue samples, which involves different cell types, may confound the measurement of miRNA expression and, in turn, significantly impacts the subsequent statistical analysis. Thus, laser capture microdissection (LCM) was used to capture purified cell populations from heterogeneous tumor tissues, and precise information regarding the miRNA expression profile of defined cell types could be obtained. We used LCM to obtain purified dMMR and pMMR cells from tumor tissues of four dMMR and four pMMR cases (Figure 1A). Clinicopathological data regarding these eight patients are shown in Table 1. The miRNA expression microarray analysis was conducted on purified dMMR and pMMR cells. Principal-component analysis (PCA) showed that dMMR and pMMR cells clustered distinctly (Figure 1B). 37 human mature miRNAs were differentially expressed between dMMR and pMMR cells, including 35 upregulated miRNAs and 2 downregulated miRNAs. Using p < 0.05 as cutoff value, we identified 12 upregulated miRNAs (including miR-1290, miR-664b-5p, miR-3607-5p, miR-3138, miR-298, miR-3653, miR-1291, miR-3679-3p, miR-149-3p, miR-30a-5p, miR-345-3p, and miR-6511b-5p) and no downregulated miRNAs (Table S2). Unsupervised hierarchical clustering of miRNA expression profiles distinctly separated dMMR and pMMR cells (Figure 1C).
Figure 1.
Microdissection and miRNA Microarray Analysis of dMMR and pMMR Colon Cancer Cells
(A) dMMR and pMMR colon cancer cells were captured from fixed tissue-sections by laser capture microdissection. (B) Principal-component analysis shows the dMMR and pMMR cells were distinctly clustered. (C) Unsupervised hierarchical clustering analysis of the 12 miRNAs differentially expressed between the dMMR and pMMR colon cancer cells. Higher intensities of red indicate higher expression levels, while lower intensities of green indicate lower expression levels.
Table 1.
Association between miR-1290 Expression and Clinicopathologic Characteristics in 291 Stage II and III Colon Cancer Patients
| Variable | Total (n = 291) | miR-1290 Expression |
p Value | |
|---|---|---|---|---|
| Low (n = 155) | High (n = 136) | |||
| Age | ||||
| < 65 y | 129 | 67 | 62 | 0.686 |
| ≥65 y | 162 | 88 | 74 | |
| Gender | ||||
| Male | 112 | 65 | 47 | 0.761 |
| Female | 179 | 90 | 89 | |
| Location | ||||
| Right | 86 | 43 | 43 | 0.012a |
| Transverse | 17 | 15 | 2 | |
| Left | 188 | 97 | 91 | |
| T Stage | ||||
| T1+T2 | 17 | 9 | 8 | 0.978 |
| T3+T4 | 274 | 146 | 128 | |
| N Stage | ||||
| N0 | 155 | 100 | 55 | < 0.001a |
| N1+N2 | 136 | 55 | 81 | |
| AJCC Stage | ||||
| II | 155 | 100 | 55 | < 0.001a |
| III | 136 | 55 | 81 | |
| Differentiation | ||||
| Well | 144 | 86 | 58 | 0.029a |
| Poor | 147 | 69 | 78 | |
| Vascular Invasion | ||||
| No | 277 | 150 | 127 | 0.177 |
| Yes | 14 | 5 | 9 | |
| MMR Status | ||||
| dMMR | 48 | 10 | 38 | < 0.001a |
| pMMR | 243 | 145 | 98 | |
p values are based on chi-square test or or Fisher’s exact test if necessary.
p < 0.05 was considered significant.
miR-1290 Is Upregulated in 5-FU-Resistant Colon Cancer Cell Lines and Reduces the Sensitivity to 5-FU In Vitro
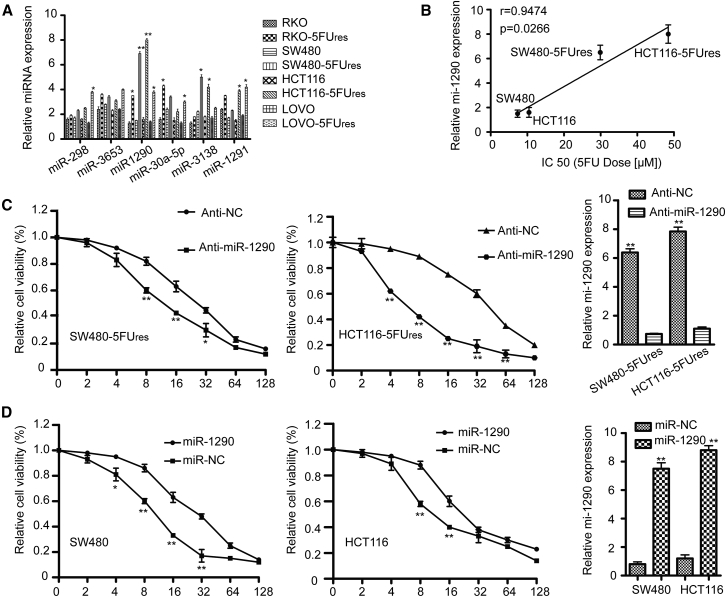
The biology underlying dMMR status has been linked to chemoresistance to 5-FU.3 As we identified several miRNAs upregulated in dMMR colon cancer cells, the role of these miRNAs in the context of 5-FU chemoresistance in colon cancer aroused our interest. Therefore, we established a series of 5-FU-resistant colon cancer cell lines (RKO-5Fures, SW480-5Fures, HCT116-5Fures, and LoVo-5Fures) and then selected six miRNAs, which have been previously studied in cancer (miR-1290, miR-1291, miR-298, miR-30a-5p, miR-3653, and miR-3138), to analyze their differential expression by qRT-PCR. miR-1290 was overexpressed in all 5-FU-resistant variants when using a panel of colon cancer cell lines (RKO, SW480, HCT116, and LoVo) (Figure 2A). The highest expression of miR-1290 was detected in SW480-5Fures and HCT116-5Fures cell lines. Therefore, SW480, SW480-5Fures, HCT116, and HCT116-5Fures cells were selected for the subsequent functional studies.
Figure 2.
miRNA Expression in Colon Cancer Cell Lines and the Influence of miR-1290 on the Sensitivity to 5-FU In Vitro
(A) The relative expression of six selected miRNAs (miR-298, miR-3653, miR-1290, miR-30a-5p, miR-3138, and miR-1291) was validated in four pairs of 5-FU-sensitive and resistant colon cancer cells by qRT-PCR. U6 small nuclear RNA was used as an internal control. All experiments were repeated three times. *p < 0.05, **p < 0.01. (B) The relationship between miR-1290 expression and IC50 of SW480, SW480-5Fures, HCT116 and HCT116-5Fures cells (7.34 ± 0.36 μM, 29.89 ± 1.89 μM, 10.45 ± 0.61 μM, and 48.45 ± 2.88 μM, respectively) was assessed by Spearman rank correlation analysis. (C) Rhe 5-FU resistant colon cancer cells (SW480-5Fures and HCT116-5Fures) transfected with miR-1290 inhibitor (anti- miR-1290) or negative control (anti-NC) were treated with increasing concentrations of 5-FU. After 48 hr, cell viability assays were performed using a CCK-8 kit. The group not treated with 5-FU was presented as 100% viable cells and was used as an internal control for comparison. (D) The cell viability of miR-1290 mimics or negative control transfected SW480 and HCT116 cells were treated as in (C). Histograms on the right show the level of miR-1290 expression. Experiments were repeated three times (*p < 0.05, **p < 0.01).
In order to investigate the biological functions of miR-1290 in 5-FU resistance of colon cancer cells, IC50 values (the concentration of 5-FU that reduced cell viability by 50%) were calculated after 5-FU treatment using CCK8 assays (SW480, 7.34 ± 0.36 μM; SW480-5Fures, 29.89 ± 1.89 μM; HCT116, 10.45 ± 0.61 μM; HCT116-5Fures, 48.45 ± 2.88 μM). miR-1290 expression was positively correlated with the IC50 values of 5-FU in the cell lines (p = 0.0266, r = 0.9474) (Figure 2B). To confirm the association between 5-FU resistance and miR-1290 expression, SW480-5Fures and HCT116-5Fures were transfected with miR-1290 inhibitor (anti-miR-1290) or negative control (anti-NC). Cells were then treated with increasing concentrations of 5-FU (0, 2, 4, 8, 16, 32, 64, and 128 μM) for 48 hr. The viability assay clearly revealed that cell viability was significantly reduced in cells transfected with miR-1290 inhibitor (anti-1290) as compared with negative control cells (Figure 2C). In contrast, the cell viability of SW480 and HCT116 cells transfected with miR-1290 mimics was significantly higher than that of control cells (Figure 2D). These results suggest that miR-1290 expression levels are negatively correlated with the sensitivity of colon cancer cells to 5-FU.
miR-1290 Is Positively Correlated with dMMR Status and Predicts Poor Prognosis in Patients with Stage II and III Colon Cancer Who Received 5-FU-Based Chemotherapy
To determine whether miR-1290 expression correlated with dMMR status, the expression of miR-1290 was examined by qRT-PCR and dMMR status was determined by immunohistochemistry (IHC) in 291 patients with stage II and III colon cancer who received 5-FU-based chemotherapy after radical colectomy. As shown in Table 1, miR-1290 was positively correlated with dMMR status (p < 0.001). In addition, upregulated miR-1290 expression was closely correlated with tumor location (p = 0.029), N stage (p < 0.001), American Joint Committee on Cancer (AJCC) stage (p < 0.001), and tumor differentiation (p = 0.029) in patients with colon cancer.
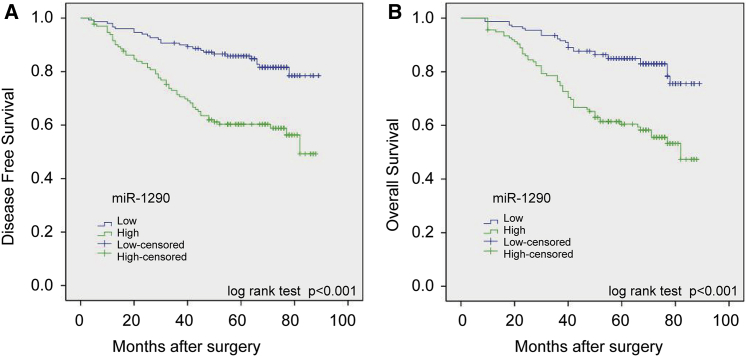
Kaplan-Meier and univariate Cox proportional hazard regression analyses showed that miR-1290 expression was significantly associated with survival of patients with stage II and III colon cancer who received 5-FU-based chemotherapy after radical colectomy. Kaplan-Meier analysis showed that high miR-1290 expression correlated with lower overall survival (OS) (p < 0.001) and disease-free survival (DFS) (p < 0.001) (Figures 3A and 3B). Univariate analysis indicated that decreased OS and DFS were associated with N stage, AJCC stage, tumor differentiation, vascular invasion, miR-1290 expression, and MMR status (Tables S4 and S5). Additionally, patients with pMMR tumors had a better OS (hazard ratio [HR] = 0.44; 95% confidence interval [CI], 0.27–0.71; p = 0.001) (Table S4) and DFS (HR = 0.45; 95% CI, 0.27–0.74; p = 0.002) (Table S5) compared with patients with dMMR tumors, which is consistent with previous studies.3, 12 Moreover, multivariate Cox proportional hazard regression analyses revealed that miR-1290 expression is an independent prognostic factor for OS (HR = 1.48; 95% CI, 0.85–2.90; p = 0.008) (Table S4) and DFS (HR = 1.59; 95% CI, 1.32–2.95; p = 0.016) (Table S5). In addition, MMR status was also a significant independent prognostic factor for OS (HR = 0.51; 95% CI, 0.37–0.93; p = 0.021) (Table S4) and DFS (HR = 0.43; 95% CI, 0.21–0.87; p = 0.018) (Table S5).
Figure 3.
miR-1290 Expression Correlates with OS and DFS in Patients with Stage II and III Colon Cancer Who Received 5-FU-Based Chemotherapy
(A and B) The patients were separated into two groups based on low or high miR-1290 expression levels. Kaplan-Meier survival curves and log-rank tests were used to compare (A) DFS and (B) OS between the two groups.
hMSH2 Is the Direct Target of miR-1290 and Inversely Correlates with miR-1290 in Both Colon Cancer Cell Lines and Tumor Tissues
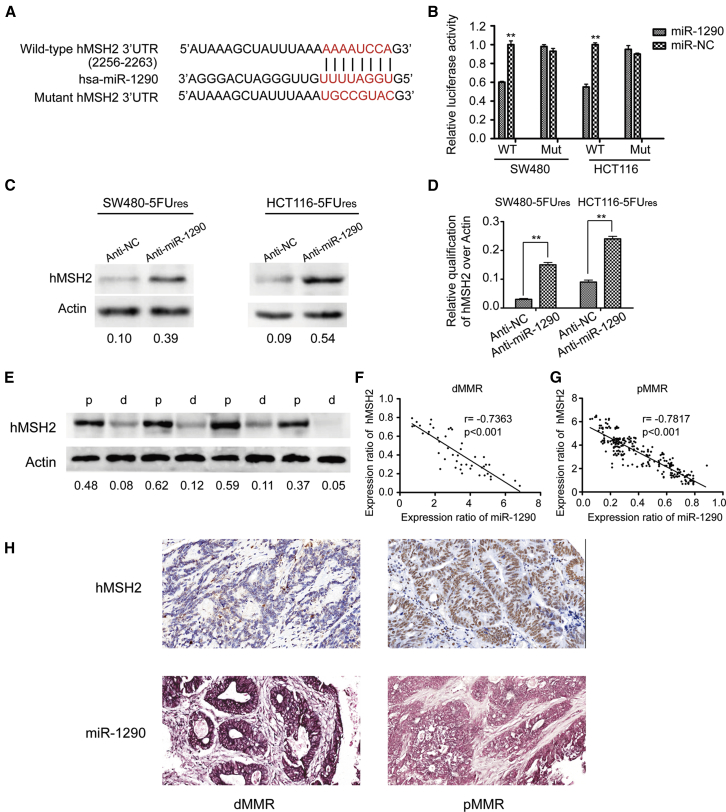
hMSH2 and hMLH1 are core proteins in the MMR system.13, 14 Their reduced or silenced expression is associated with dMMR status.14 As miR-1290 is upregulated in dMMR cells, we hypothesized that miR-1290 induced dMMR status by directly targeting hMSH2 and/or hMLH1. Three miRNA-target-predicting programs (Target-Scan, miRWalk, and miRanda) were used to determine whether hMSH2 and/or hMLH1 were candidate targets of miR-1290. All three programs predicted hMSH2, but not hMLH1, as the candidate target of miR-1290.
To confirm that hMSH2 is targeted by miR-1290, a luciferase reporter containing the complimentary seed sequence of miR-1290 at the 3′ UTR of hMSH2 mRNA was constructed (Figure 4A). Luciferase reporter assay indicated that the activity of wild-type hMSH2 3′-UTR reporter was significantly reduced by co-transfection with miR-1290 mimics in SW480 and HCT116 cells. In contrast, the activity of the hMSH2 control reporter containing a mutated sequence of the same fragment was not affected by co-transfection with miR-1290 mimics (Figure 4B). These results indicated that the miR-1290 seed region binds to complementary sites in the 3′ UTR of hMSH2 mRNA, suggesting that hMSH2 is the direct target of miR-1290. Western blot assays were also performed to detect the expression levels of hMSH2 protein in SW480-5Fures and HCT116-5Fures cells transfected with lentiviral vectors containing miR-1290 inhibitor. The expression levels of hMSH2 protein were remarkably upregulated in SW480-5Fures and HCT116-5Fures cells stably knocked down for miR-1290 (Figures 4C and 4D). Overall, miR-1290 negatively regulated hMSH2 expression in vitro.
Figure 4.
miR-1290 Suppresses hMSH2 Expression by Directly Targeting Its 3′ UTR
(A) RNA sequence alignment showing that the 3′ UTR of hMSH2 mRNA contains a complementary site for the seed region of miR-1290. hMSH2 mutant sequence is used as a negative control. (B) Dual luciferase reporter assay was performed using SW480 and HCT116 cells transfected with wild-type or mutated hMSH2 reporters plus miR-1290 mimics. All values are presented as mean ± SD (**p < 0.01). Experiments were repeated three times. (C) Inhibition of miR-1290 expression upregulated the expression of hMSH2 protein in both SW480-5Fures and HCT116-5Fures cell lines. (D) Grayscale values were evaluated. Histograms show the relative expression of hMSH2 over actin (n = 3, **p < 0.01). (E) Western blot analysis of hMSH2 protein expression in four dMMR and four pMMR representative colon tumor tissues. (F and G) The relationship between miR-1290 and hMSH2 mRNA expression in dMMR and pMMR colon cancer tissues was assessed by Spearman rank correlation analysis. (H) Representative images of miR-1290 and hMSH2 expression in dMMR and pMMR colon cancer tissues using ISH and IHC staining, respectively.
In addition, we observed that the expression of miR-1290 was upregulated in dMMR colon cancer tissues and downregulated in pMMR samples. In contrast to miR-1290, western blot analyses revealed that hMSH2 protein expression was downregulated in dMMR colon cancer tissues compared with pMMR tissues (Figure 4E). Moreover, the Spearman correlation analysis clearly showed that miR-1290 levels were inversely correlated with hMSH2 mRNA expression in dMMR and pMMR colon cancer tissues (Figures 4F and 4G). Furthermore, we analyzed miR-1290 and hMSH2 expression in these samples using in situ hybridization (ISH) and IHC staining, respectively. Results revealed that dMMR colon cancer tissues presented high miR-1290 and low hMSH2 expression, while pMMR tissues showed low miR-1290 and high hMSH2 expression (Figure 4H).
Decreased Expression of miR-1290 Increased the Sensitivity of Colon Cancer Cells to 5-FU by Promoting Apoptosis via hMSH2 Targeting
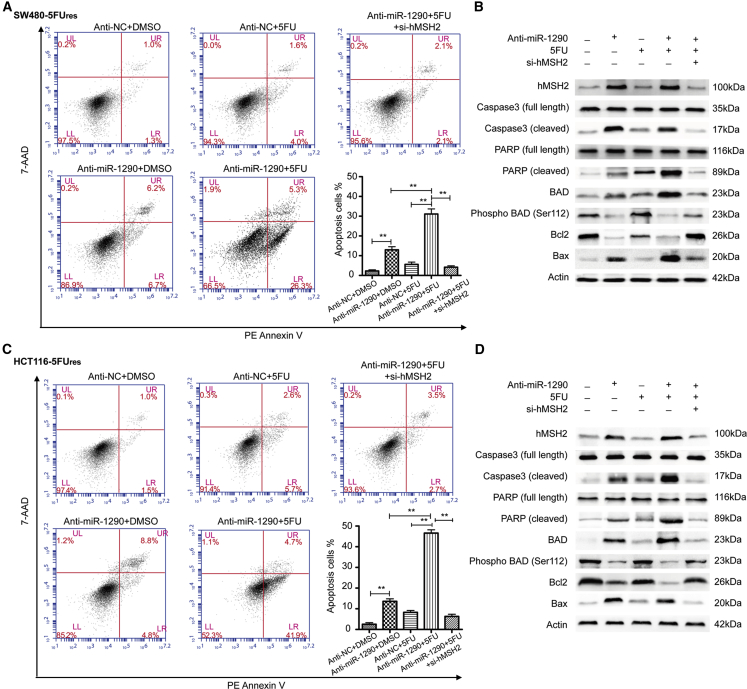
The mechanism by which hMSH2 promotes the sensitivity of colon cancer cells to 5-FU has been described by Tajima et al.15 One recent study reported that miR-21 induces resistance to 5-FU by directly targeting hMSH2.10 As miR-1290 inhibits the sensitivity of colon cancer cells to 5-FU and directly targets hMSH2, we hypothesized that miR-1290 decreases the sensitivity to 5-FU through downregulation of hMSH2. To confirm this hypothesis, we performed flow cytometry assays using PE Annexin V and 7-amino-actinomycin (7-AAD) double staining to examine the effects of miR-1290 and 5-FU on apoptosis of SW480-5Fures and HCT116-5Fures cells. All cells were treated with 10 μM 5-FU or the same volume of DMSO as a control and incubated for 48 hr. Results showed that the early apoptotic rates (PE Annexin V+/7-AAD- events) of miR-1290 inhibitor (anti-miR-1290) transfected SW480-5Fures cells (6.7% and 26.3% in DMSO and 5-FU-treated cells, respectively) were significantly higher than the corresponding negative control (anti-NC) cells (1.3% and 4.0%, respectively; Figure 5A). Among these different treatment groups, SW480-5Fures cells, which were both transfected with miR-1290 inhibitor and treated with 5-FU, had the highest proportion of apoptotic cells (26.3%). Moreover, hMSH2 knockdown by si-hMSH2 transfection restored the apoptotic rates of miR-1290 inhibitor transfected SW480-5Fures cells treated with 5-FU (2.1%; p < 0.001). These results indicated that decreased miR-1290 expression increased the sensitivity to 5-FU by promoting apoptosis via hMSH2 targeting in 5-FU-resistant colon cancer cells.
Figure 5.
Decreased Expression of miR-1290 Increased the Sensitivity to 5-FU in SW480-5Fures and HCT116-5Fures Cells by Targeting hMSH2, Resulting in Altered Expression of Apoptosis-Related Proteins
(A and C) Representative flow cytometry analysis by using PE Annexin V and 7-Amino-Actinomycin (7-AAD) double staining in SW480-5Fures and HCT116-5Fures cells, which were transfected with miR-1290 inhibitor (anti-miR-1290), negative control (anti-NC), or miR-1290 plus si-hMSH2 and treated with 10 μM 5-FU or the same volume of DMSO as a control and incubated for 48 hr. Histograms represent the proportion of apoptotic cells presented as the mean ± SD from three independent experiments (**p < 0.01). (B and D) Representative western blots using the indicated antibodies in SW480-5Fures and HCT116-5Fures cells transfected and treated as described above.
To further explore the molecular mechanisms underlying the anti-apoptotic role of miR-1290, we performed western blot assays to examine several biochemical markers of apoptosis. Total protein was extracted from cells stably transfected with miR-1290 inhibitor or negative control after treatment with 10 μM 5-FU or the same volume of DMSO for 48 hr. Cleaved poly(ADP-ribose) polymerase (PARP) and caspase-3 were then detected in the miR-1290 inhibitor (anti-miR-1290) transfected SW480-5Fures cell lines. 5-FU treatment induced more cleavage of PARP and caspase-3 expression than DMSO treatment. Conversely, in the negative control (anti-NC) cell lines, which were resistant to apoptosis, most PARP and caspase-3 proteins were not cleaved (Figure 5B). These results indicated that decreased expression of miR-1290 may promote sensitivity to 5-FU-induced apoptosis. The B cell lymphoma protein Bcl-2 is an anti-apoptotic protein, while Bax is a proapoptotic protein.16 Western blot assays confirmed that the protein level of Bcl-2 was markedly decreased, while Bax protein expression was significantly upregulated in the miR-1290 inhibitor (anti-miR-1290) transfected SW480-5Fures cell lines, especially when simultaneously treated with 5-FU. In addition, as a proapoptotic member of the BCL-2 family, BAD plays a key role by interacting with Bcl-2 or Bcl-xL.17 However, it dissociates from the complexes with BCL-2 and BCL-xL when it is phosphorylated, which prevents the release of cytochrome c from mitochondria and suppresses apoptosis.18 As shown in Figure 5B, a significant decrease in phosphorylated BAD (p-BAD) was observed in miR-1290 inhibitor (anti-miR-1290) transfected SW480-5Fures cells treated with 5-FU. In the negative control (anti-NC) cell lines, a significant increase in p-BAD was observed. Moreover, hMSH2 knockdown by si-hMSH2 transfection reversed the expression level of apoptosis-related proteins in miR-1290 inhibitor transfected SW480-5Fures cells treated with 5-FU. Collectively, these results suggested that decreased expression of miR-1290 increased the sensitivity to 5-FU by promoting apoptosis via hMSH2 targeting in 5-FU-resistant colon cancer cells. Similar results were observed in HCT116-5Fures cell lines (Figures 5C and 5D).
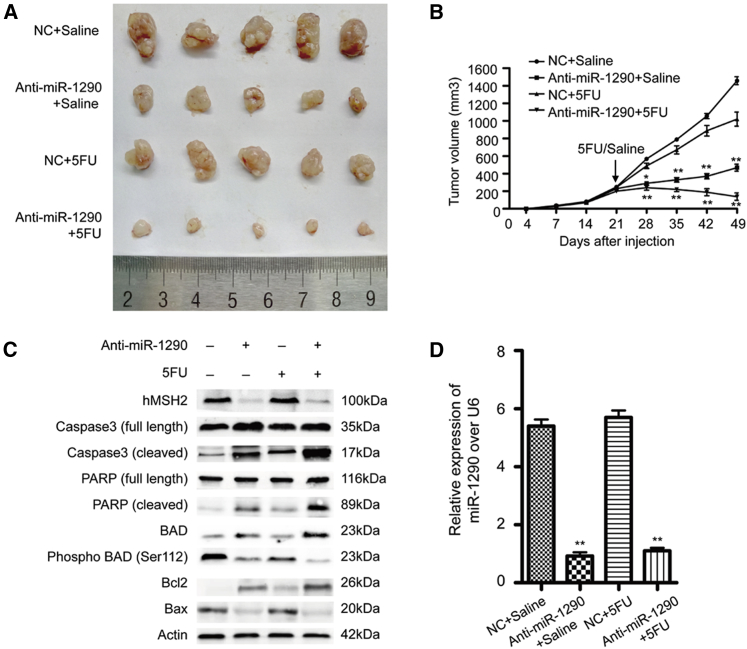
Decreased Expression of miR-1290 Promotes Tumor Sensitivity to 5-FU In Vivo
Our studies suggested that anti-miR-1290 promoted 5-FU-induced apoptosis in different colon cancer cell lines in vitro. To better evaluate the biologic function of anti-miR-1290 in vivo, HCT116-5Fures cells stably expressing miR-1290 inhibitor (anti-miR-1290) or vector control (NC) (2 × 106 cells) were subcutaneously injected into each flank of each nude mice simultaneously. Once xenografts reached a palpable volume (>5 mm in diameter), they were randomly assigned into 5-FU or saline treatment groups. There were no significant differences in initial tumor volumes (within 3 weeks) among the four groups. 5-FU (50 mg·kg−1·d−1) or the same volume of saline was intraperitoneally injected for five consecutive days per week for 4 weeks. The mice were euthanized on day 49, and tumors were immediately harvested (Figure 6A). Total RNA and proteins were extracted from the representative tumor mass. The tumor volume of the tumors expressing the anti-miR-1290 and presenting high expression of hMSH2 was significantly smaller than that of NC tumors (Figures 6A–6C). These results are consistent with previous studies and suggest that MMR-proficient cells respond better to 5-FU therapy. The combination of miR-1290 inhibitor and 5-FU treatment induced the most significant reduction of tumor growth among the four groups (Figures 6A and 6B). The miR-1290 expression levels of the four groups are shown in Figure 6D. In addition, a significant increase in cleaved PARP, cleaved caspase-3, BAD, and Bcl-2 and a decrease in p-BAD and Bax were observed in tumors expressing the anti-miR-1290, especially in the 5-FU-treated group (Figure 6C). This revealed that decreased expression of miR-1290 increased 5-FU-induced apoptosis in vivo. Altogether, our results suggested that miR-1290 promotes resistance to 5-FU by downregulating hMSH2, a core MMR protein.
Figure 6.
Decreased Expression of miR-1290 Promotes Tumor Sensitivity to 5-FU In Vivo
HCT116-5Fures cell lines stably expressing miR-1290 inhibitor (anti-miR-1290) or vector control (NC) were subcutaneously injected into nude mice. After 3 weeks, 5-FU (50 mg·kg−1·d−1) or the same volume of saline were intraperitoneally injected for five consecutive days per week for 4 weeks. (A) Nude mice were euthanized on day 49, and tumors were immediately peeled off. (B) Tumor size was measured on days 4, 7, 14, 21, 28, 35, 42, and 49 with Vernier calipers (mean ± SD; n = 5). *p < 0.05, **p < 0.01. (C) Western blot analysis of representative excised tumor mass using the indicated antibodies. (D) qRT-PCR analysis of miR-1290 expression from representative excised tumor mass (**p < 0.01).
Discussion
In this study, we demonstrated that miR-1290 is upregulated in both dMMR colon cancer cells and tumor tissues and predicts poor prognosis in patients with stage II and III colon cancer who received 5-FU-based adjuvant therapy. Furthermore, we demonstrated that decreased expression of miR-1290 enhanced the sensitivity to 5-FU treatment in vitro and in tumor xenografts in vivo by directly targeting hMSH2.
Chemoresistance is the main cause of poor prognosis in patients with colon cancer, especially in patients with dMMR colon cancer who respond weakly or not at all to 5-FU-based adjuvant therapy.19 Although recent studies indicate that ectopic miRNAs expression is closely correlated to chemoresistance, the expression of chemoresistance-related miRNAs in dMMR colon cancer is largely unknown.10, 11, 20 First, LCM was applied to obtain purified cell populations from eight heterogeneous colon cancer tissues, including four dMMR and four pMMR samples. Second, miRNA microarray was performed to identify miRNAs that are differentially expressed between these two classes of tumors. Twelve upregulated miRNAs were identified as the most significant ones using p < 0.05 and 2-fold expression difference level as cutoff values. miR-1290 has been indicated as a novel biomarker for early detection, recurrence, and prognosis of colon cancer and mediates chemoresistance to paclitaxel in hepatocellular carcinoma.21 Both miR-1290 and miR-3138 could promote radio resistance of human cervical cancer cells.22 miR-298 increases the chemoresistance to doxorubicin in human breast cancer by downregulating P-gp expression.23 miR-30a-5p suppresses tumor growth and metastasis of colon cancer by targeting DTL and ITGB3, respectively.24 miR-3653 is upregulated in cervical cancer tissues associated with human papillomavirus (HPV) infection as compared with paired normal tissues.25 miR-1291 promotes apoptosis in esophageal squamous cell carcinoma by targeting mucin1.26 Third, these six miRNAs were detected in a panel of 5-FU-resistant colon cancer cell lines by qRT-PCR to identify chemoresistance-related miRNAs. miR-1290 overexpression was detected in all resistant variants, with the highest expression in SW480-5Fures and HCT116-5Fures cell lines. We also confirmed that miR-1290 promoted chemoresistance to 5-FU in vitro. Additionally, we confirmed that miR-1290 is upregulated in patients with dMMR colon cancer using an independent large cohort of colon cancer tissues and demonstrated that it predicts poor prognosis in patients with stage II and III colon cancer who received 5-FU-based chemotherapy. Thus, we believe that miR-1290 is a valuable biomarker to predict the prognosis of patients with dMMR colon cancer and chemoresistance to 5-FU.
Previous studies demonstrated that aberrant miR-1290 expression is involved in the malignant processes of a few cancers, including colon cancer.27 In non-small-cell lung cancer (NSCLC), high expression of miR-1290 was detected in tissues and serum and was closely correlated with lymph node metastasis, tumor/node/metastasis (TNM) stage, and poor prognosis.21, 28 miR-1290 overexpression promotes proliferation, migration, and invasion in esophageal squamous cell carcinoma (ESCC) by targeting NFIX.29 Furthermore, miR-1290 targets the potential target genes of estrogen receptor α (ERα)-positive breast cancer, FOXA1 and NAT1.28 Belian et al. showed that upregulated miR-1290 is involved in drug resistance in gastric cancer cells.30 Moreover, miR-1290 is significantly upregulated in colon cancer tissues and affects the reprogramming of colon cancer cells by activating the Wnt pathway and increasing the related transcription factors, c-Myc and Nanog.31 However, the association between miR-1290 and 5-FU resistance in colon cancer is still unknown. We demonstrated for the first time that miR-1290 is upregulated in dMMR colon cancer tissues and 5-FU-resistant colon cancer cell lines. Furthermore, we found that miR-1290 was positively correlated with dMMR status in an independent cohort and inversely correlated with DFS and OS in patients who received 5-FU-based adjuvant therapy. Collectively, these data indicate that miR-1290 expression can be used as a potential surrogate marker of dMMR colon cancer and that miR-1290 may contribute to poorer prognosis.
Defects in MMR are associated with colon cancer.19 Mutations in the human DNA MMR gene hMSH2 are associated with hereditary nonpolyposis colon cancer as well as a significant proportion of sporadic colon cancer cases.14, 32 hMSH2 inactivation results in the accumulation of somatic mutations in the genome of tumor cells and resistance to a variety of chemotherapeutic agents, including 5-FU. Recent studies indicated that MMR proteins promote DNA-damage-induced apoptosis as part of the cellular response to 5-FU.33 In this study, we demonstrated that miR-1290 directly binds to the 3′ UTR of hMSH2 and suppresses its expression. Furthermore, our study also showed that miR-1290 levels were inversely correlated with hMSH2 mRNA in colon cancer tumor samples. In addition, we demonstrated that decreased miR-1290 expression enhanced cell apoptosis, with upregulated proapoptotic proteins and downregulated anti-apoptotic proteins. Furthermore, hMSH2 knockdown by si-hMSH2 transfection reversed the function of miR-1290 inhibitor. Taken together, these results suggest that miR-1290 enhanced the resistance to 5-FU by downregulating hMSH2. miR-1290 may be an effective therapeutic target and is likely to be an important indicator of 5-FU therapeutic efficacy in colon cancer.
This is the first study to demonstrate that decreased expression of miR-1290 promotes the sensitivity to 5-FU by targeting hMSH2 in both a cellular and xenograft tumor model. We found that tumor volume was significantly reduced after treatment with both anti-miR-1290 and 5-FU compared with the other three groups (NC + saline, NC + 5-FU, and anti-miR-1290 + saline). Moreover, we showed that the prognosis of patients with lower miR-1290 expression was better using an independent large cohort of patients with colon cancer who received 5-FU-based chemotherapy. Recently, specific molecular subtypes get increasing attention in the drug research area and may represent the future of cancer therapeutics.34 dMMR colon cancer is an ideal molecular subtype for new drug development because of the compelling rationale motivated by molecular, clinical, pathological, prognostic, and predictive studies that are already changing the practice of oncology. Thus, miR-1290, which is upregulated in dMMR colon cancer, is expected to become a potential therapeutic target.
Although our study shows that miR-1290 is a promising biomarker for the diagnosis of dMMR colon cancer and the prediction of poor prognosis in patients with stage II and III colon cancer who received 5-FU-based adjuvant therapy, the data shown in the current study have some limitations. Although we examined miR-1290 expression in an independent cohort with 291 patients, this sample size is not sufficient to support the use of miR-1290 in the clinic. Thus, a larger clinical sample size study is required to validate our results regarding miR-1290 expression in patients with colon cancer. Additionally, in order to improve the reliability and convenience of miR-1290 usage in the clinic, preoperative serum samples from patients with colon cancer are required to examine miR-1290 expression.
In conclusion, miR-1290 is a promising biomarker for the detection of dMMR colon cancer and predicts poor prognosis of patients with stage II and III colon cancer who received 5-FU-based adjuvant therapy. miR-1290 downregulation may be useful in combination with 5-FU for treating chemoresistant colon cancer.
Materials and Methods
Human Colon Cancer Tissues and MMR Status Determination
Tissue samples were obtained from patients who underwent radical resection at the General Surgery Department of Shanghai General Hospital. No chemotherapy or radiation therapy was applied to these patients before radical resection. IHC was carried out by using antibodies against MLH1, MSH2, MSH6, and PMS2 proteins, which provided insight into the functionality of the MMR system. Lack of expression of one or more of these proteins is a diagnostic of dMMR, and the expression of these proteins is defined as pMMR. Four dMMR tumors and four pMMR tumor samples were randomly selected to analyze their miRNA profiles using LCM (Table S1). Furthermore, 291 stage II and stage III colon cancer tumor tissues were obtained from patients at the Anal-Colorectal Surgery Department, General Hospital of Ningxia Medical University. These patients received 5-FU-based chemotherapy after radical resection. The diagnosis was confirmed by two pathologists, and cancer staging was determined based on pathological findings according to the AJCC. All patients provided informed consent, and the study was approved by the institutional review board of General Hospital of Ningxia Medical University. The follow-up of this cohort ended on September 15, 2014, and the median duration of follow-up was 57 (range, 9–89) months. DFS and OS rates were defined as the interval from the initial surgery to clinically or radiologically proven recurrence/metastasis and death, respectively.
LCM and miRNA Microarray Analysis
Purified dMMR and pMMR colon cancer cells were obtained using LCM according to the protocol of our previous study.35 Total RNA was then extracted using a miRNeasy mini kit (QIAGEN). Total RNA was sent to BoHao Bio-tech for miRNA microarray analysis. Differentially expressed miRNAs were identified using p < 0.05 as cutoff value. Finally, we identified 12 significant miRNAs, all of which were upregulated (Table S2).
Total RNA Extraction and Quantitative Real-Time PCR Analysis
Total RNA, including miRNA, from colon cancer samples and cell lines was extracted using the miRNeasy Mini kit according to the manufacturer’s instructions (QIAGEN). First-strand cDNA was synthesized with the RevertAid First Strand cDNA Synthesis Kit (QIAGEN) using 1 μg total RNA as the template. miRNAs were measured with a miScript SYBR Green PCR Kit (QIAGEN) using the ABI7500 Real-Time PCR System (Applied Biosystems). Real-time PCR to assess the expression of hMSH2 and miRNAs was carried out using ViiA 7 system (Thermo Fisher Scientific) according to the manufacturer’s instructions. The sequences of all primers used in this study are shown in Table S3. The PCR amplification conditions were as follows: pre-denaturing at 95°C for 10 min, followed by 40 cycles of denaturing at 95°C for 10 s, annealing at 58°C for 20 s, and extension at 72°C for 10 s. The expression level of each miRNA was measured by using the 2−ΔΔCt (Ct, cycle threshold) method. All experiments were performed in triplicate.
Establishment of 5-FU-Resistant Colon Cancer Cell Lines and Transfection
The human colon cancer cell lines RKO, SW480, HCT116, and LoVo were obtained from the Type Culture Collection of the Chinese Academy of Sciences. All cell lines were cultured as described previously.36 5-FU-resistant colon cancer cell lines (RKO-5Fures, SW480-Fures, HCT116-5Fures, and LoVo-5Fures) were developed through a stepwise incremental treatment with 5-FU. 5-FU (Sigma-Aldrich) was dissolved in DMSO (Sigma-Aldrich) and stored in airtight containers protected from light before experiments. The parental cell lines RKO, SW480, HCT116, and LoVo were exposed to increasing concentrations of 5-FU stepwise, starting at 1 μM and ending at 100 μM. 5-FU (1 μM) was included in the culture medium for RKO-5Fures, SW480-5Fures, HCT116-5Fures, and LoVo-5Fures to maintain drug resistance. The cells were maintained in 5-FU-free medium at least 2 weeks before the experiments. Cell lines with stable downregulation of miR-1290 were established by lentiviral transduction using a pCDH plasmid (System Biosciences) carrying miR-1290 inhibitor. All cell lines were cultured at 37°C in a humidified atmosphere with 5% CO2 and maintained in DMEM supplemented with 10% fetal bovine serum (Gibco). SW480, HCT116, SW480-5Fures, and HCT116-5Fures were then transfected with the plasmids using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol.
Plasmid Construction
The miR-1290 mimic and miR-1290 mimic negative controls used for transient transfection were designed and synthesized by BioLink. Small interfering RNA (siRNA) against hMSH2 (si-hMSH2) was synthesized by BioLink and used to silence hMSH2 expression. The sequences of siRNA targeting hMSH2 were as follows: 5′-TGGCAATCTCTCTCAGTTT-3′ (sense), 5′-TTCTCCGAACGTGTCACGT-3′ (antisense). Cells were transfected with miR-1290 mimics, negative control, and siRNA using Lipofectamine 2000 Reagent (Invitrogen) according to the manufacturer’s protocol.
Cell Viability and Apoptosis Assays
Cell Counting Kit-8 (CCK8) assays (Dojindo) were used to measure cell viability according to the manufacturer’s protocol. To analyze the effect of miR-1290 on 5-FU treatment, cells transfected with miR-1290 mimics, miR-1290 inhibitor, or negative control were treated with 0, 2, 4, 8, 16, 32, 64, and 128 μM 5-FU for 48 hr. IC50 values were calculated using SPSS software.
For the apoptosis assay, SW480-5Fures and HCT116-5Fures cells stably transfected with miR-1290 inhibitor, negative control, or hMSH2 siRNA were cultured in 10-cm dishes and treated with 5-FU (10 μM) for 48 hr before harvesting. The cells were then trypsinized and collected into a centrifuge tube, centrifuged at 800 × g for 3 min, washed twice with cold PBS, and resuspended in 100 μL 1 × binding buffer, and the PE Annexin V and 7-AAD (BD Biosciences) double staining method was used to examine cell viability following the manufacturer’s protocol. The frequency of apoptosis was measured using the BD FACS Calibur Flow Cytometer (BD Biosciences) according to the manufacturer’s instructions. The percentage of apoptotic cells was calculated according to the number of cells positive or negative for PE Annexin V and 7-AAD. Results are presented as the percentage of living cells (PE Annexin V−/7-AAD−), early apoptotic cells (PE Annexin V +/7-AAD−), late apoptotic cells, and dead cells (PE Annexin V +/7-AAD +). Each assay was repeated three times.
Western Blot Analysis
Western blot analysis was performed as previously described.36 The following antibodies were used: anti-MLH1, anti-MSH2, anti-MSH6, anti-PMS2, anti-caspase-3, anti-cleaved caspase-3, anti-PARP, anti-cleaved PARP, anti-BAD, anti-pBAD (S112), anti-Bcl2, anti-Bax (Cell Signaling Technology), and anti-actin (Sigma).
Luciferase Reporter Assay
The dual-Luciferase reporter assays (Promega) were performed in SW480 and HCT116 cells according to the manufacturer’s instructions. The putative binding site of miR-1290 in the 3′ UTR of hMSH2 mRNA (WT) or its mutant sequence (MUT) was cloned downstream of the firefly luciferase gene. SW480 and HCT116 cells were co-transfected with 50 nM miR-1290 mimics or miRNA mimics negative control and 0.5 μg psiCHECK-2-hMSH2–3′-UTR-WT or psiCHECK-2-hMSH2–3′-UTR-MUT using Lipofectamine 2000 (Invitrogen). Firefly and Renilla luciferase activity was measured by using the GloMax fluorescence reader (Promega) after 48 hr transfection. The experiments were repeated three times.
In Situ Hybridization and IHC on Tissue Microarray
The tissue microarray (TMA) containing 291 colon cancer tumor specimens was constructed in cooperation with Xin Chao Company. Tumors were resected from patients in the Anal-Colorectal Surgery Department, General Hospital of Ningxia Medical University between June 2004 and February 2007. The end of the follow-up period was September 15th, 2014, and the median duration of follow-up was 57 (range, 9–89) months. The TMA includes specimens from 112 men and 179 women with a mean age of 65.29 years (range, 29–81 years). The expression pattern of miR-1290 in dMMR and pMMR colon cancer tumor tissues was detected by in situ hybridization. The TMA slides were dewaxed in xylene for 15 min twice. After being dehydrated by immersion in 100% ethanol for 5 min, the slides were air-dried and then incubated with pepsin at 37°C for 15 min. The slides were then fixed in 4% paraformaldehyde, dehydrated in 90% ethanol, and incubated with the digoxigenin-labeled probe (Exiqon) complementary to miR-1290 at 37°C overnight, according to the manufacturer’s instructions. The slides were washed twice with 2× saline sodium citrate (SSC) at room temperature and incubated with mouse anti-digoxigenin monoclonal antibody according to the manufacturer’s protocol. miR-1290 expression in the TMA was assessed by two independent pathologists. The proportion of positively stained tumor cells was graded as follows: 0 (no positive cells), 1 (1%–25% positive cells), 2 (26%–50% positive cells), 3 (51%–75% positive cells), and 4 (76%–100% positive cells). The intensity of the staining was recorded as 0 (no staining), 1 (weak staining), 2 (moderate staining), or 3 (strong staining). The staining index (SI) was defined as the proportion of positively stained tumor cells × staining intensity, and the specimens were divided into two groups based on the final score as low (0–6) or high.7, 8, 9, 10, 11, 35 To further confirm that hMSH2 is the target of miR-1290, we assessed hMSH2 protein expression levels by IHC on the same location of the TMA. IHC was also performed using antibodies against MLH1, MSH2, MSH6, and PMS2 proteins to identify dMMR and pMMR colon cancer tumors. The IHC assays on the TMA were performed as previously described.36
In Vivo Colon Cancer Xenograft Mouse Model
Ten 6-week-old male BALB/c nude mice were purchased from the Shanghai Jiaotong University and maintained in specific-pathogen-free (SPF) conditions. HCT116-5Fures cells (2 × 106) transfected with miR-1290 inhibitor or a negative control were injected subcutaneously into each flank of the mice. After 3 weeks, when all xenografts reached a palpable volume (>5 mm in diameter), mice were randomly divided into 5-FU or saline treatment groups, with five mice per group. 5-FU (50 mg·kg−1·d−1) or the same volume of saline was intraperitoneally injected for five consecutive days per week for 4 weeks. The tumors size was measured on days 4, 7, 14, 21, 28, 35, 42, and 49. Tumor volume was calculated using the formula: V (mm3) = width2 (mm2) × length (mm)/2. All nude mice were euthanized by cervical dislocation on day 49, and their tumors were harvested and photographed. All procedures followed the Shanghai Jiao tong University Affiliated Shanghai General Hospital Animal Care guidelines.
Statistical Analysis
The statistical software program SPSS version 22 was used for statistical analysis. The two-tailed Student’s t test was used to determine the significance between groups in the in vitro and in vivo experiments. The χ2 test or Fisher’s exact test was used to analyze the relationship between miR-1290 and clinicopathological features, including MMR status. Survival rates were calculated by using the Kaplan-Meier method, and differences between the survival curves were examined using log-rank tests. Cox proportional hazards models were applied to estimate the effects of miR-1290 expression on survival in univariate and multivariate analyses. p < 0.05 was considered statistically significant.
Author Contributions
L.Y., T.J., H.S., and J.F. contributed to the design of the experiments; Y.L. and T.J. performed the experiments; H.S., L.Z., W.Z., and Y.L. analyzed and interpreted data and statistical analysis; L.Y. and T.J. wrote the manuscript; and T.H., B.Q., X.Z., and J.F. edited the manuscript.
Conflicts of Interest
The authors declare no potential conflicts of interest.
Acknowledgments
This research was supported by the National Natural Science Foundation (grants 81560394 and 81272744), which is funded by the Chinese governmentand a Scientific Research Project grant funded by Ningxia High School (NGY2015096).
Footnotes
Supplemental Information includes one figure and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.omtn.2017.05.006.
Contributor Information
Bingyu Qin, Email: nicolasby@126.com.
Xiaoqing Zhang, Email: zxqkitten@163.com.
Junwei Fan, Email: drjunweifan@163.com.
Supplemental Information
References
- 1.Miller K.D., Siegel R.L., Lin C.C., Mariotto A.B., Kramer J.L., Rowland J.H., Stein K.D., Alteri R., Jemal A. Cancer treatment and survivorship statistics, 2016. CA Cancer J. Clin. 2016;66:271–289. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 2.Longley D.B., Harkin D.P., Johnston P.G. 5-fluorouracil: mechanisms of action and clinical strategies. Nat. Rev. Cancer. 2003;3:330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 3.Sinicrope F.A., Foster N.R., Thibodeau S.N., Marsoni S., Monges G., Labianca R., Kim G.P., Yothers G., Allegra C., Moore M.J. DNA mismatch repair status and colon cancer recurrence and survival in clinical trials of 5-fluorouracil-based adjuvant therapy. J. Natl. Cancer Inst. 2011;103:863–875. doi: 10.1093/jnci/djr153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kunkel T.A., Erie D.A. DNA mismatch repair. Annu. Rev. Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 5.Wheeler J.M., Bodmer W.F., Mortensen N.J. DNA mismatch repair genes and colorectal cancer. Gut. 2000;47:148–153. doi: 10.1136/gut.47.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peltomäki P. Role of DNA mismatch repair defects in the pathogenesis of human cancer. J. Clin. Oncol. 2003;21:1174–1179. doi: 10.1200/JCO.2003.04.060. [DOI] [PubMed] [Google Scholar]
- 7.Gregory R.I., Shiekhattar R. MicroRNA biogenesis and cancer. Cancer Res. 2005;65:3509–3512. doi: 10.1158/0008-5472.CAN-05-0298. [DOI] [PubMed] [Google Scholar]
- 8.Iorio M.V., Croce C.M. MicroRNAs in cancer: small molecules with a huge impact. J. Clin. Oncol. 2009;27:5848–5856. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garofalo M., Croce C.M. microRNAs: master regulators as potential therapeutics in cancer. Annu. Rev. Pharmacol. Toxicol. 2011;51:25–43. doi: 10.1146/annurev-pharmtox-010510-100517. [DOI] [PubMed] [Google Scholar]
- 10.Valeri N., Gasparini P., Braconi C., Paone A., Lovat F., Fabbri M., Sumani K.M., Alder H., Amadori D., Patel T. MicroRNA-21 induces resistance to 5-fluorouracil by down-regulating human DNA MutS homolog 2 (hMSH2) Proc. Natl. Acad. Sci. USA. 2010;107:21098–21103. doi: 10.1073/pnas.1015541107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borralho P.M., Kren B.T., Castro R.E., da Silva I.B., Steer C.J., Rodrigues C.M. MicroRNA-143 reduces viability and increases sensitivity to 5-fluorouracil in HCT116 human colorectal cancer cells. FEBS J. 2009;276:6689–6700. doi: 10.1111/j.1742-4658.2009.07383.x. [DOI] [PubMed] [Google Scholar]
- 12.Sargent D.J., Marsoni S., Monges G., Thibodeau S.N., Labianca R., Hamilton S.R., French A.J., Kabat B., Foster N.R., Torri V. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J. Clin. Oncol. 2010;28:3219–3226. doi: 10.1200/JCO.2009.27.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fishel R. The selection for mismatch repair defects in hereditary nonpolyposis colorectal cancer: revising the mutator hypothesis. Cancer Res. 2001;61:7369–7374. [PubMed] [Google Scholar]
- 14.Yoon Y.S., Yu C.S., Kim T.W., Kim J.H., Jang S.J., Cho D.H., Roh S.A., Kim J.C. Mismatch repair status in sporadic colorectal cancer: immunohistochemistry and microsatellite instability analyses. J. Gastroenterol. Hepatol. 2011;26:1733–1739. doi: 10.1111/j.1440-1746.2011.06784.x. [DOI] [PubMed] [Google Scholar]
- 15.Tajima A., Hess M.T., Cabrera B.L., Kolodner R.D., Carethers J.M. The mismatch repair complex hMutS α recognizes 5-fluorouracil-modified DNA: implications for chemosensitivity and resistance. Gastroenterology. 2004;127:1678–1684. doi: 10.1053/j.gastro.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 16.Harada H., Becknell B., Wilm M., Mann M., Huang L.J., Taylor S.S., Scott J.D., Korsmeyer S.J. Phosphorylation and inactivation of BAD by mitochondria-anchored protein kinase A. Mol. Cell. 1999;3:413–422. doi: 10.1016/s1097-2765(00)80469-4. [DOI] [PubMed] [Google Scholar]
- 17.Sakamaki J., Daitoku H., Ueno K., Hagiwara A., Yamagata K., Fukamizu A. Arginine methylation of BCL-2 antagonist of cell death (BAD) counteracts its phosphorylation and inactivation by Akt. Proc. Natl. Acad. Sci. USA. 2011;108:6085–6090. doi: 10.1073/pnas.1015328108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cotteret S., Jaffer Z.M., Beeser A., Chernoff J. p21-Activated kinase 5 (Pak5) localizes to mitochondria and inhibits apoptosis by phosphorylating BAD. Mol. Cell. Biol. 2003;23:5526–5539. doi: 10.1128/MCB.23.16.5526-5539.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hewish M., Lord C.J., Martin S.A., Cunningham D., Ashworth A. Mismatch repair deficient colorectal cancer in the era of personalized treatment. Nat. Rev. Clin. Oncol. 2010;7:197–208. doi: 10.1038/nrclinonc.2010.18. [DOI] [PubMed] [Google Scholar]
- 20.Garofalo M., Croce C.M. MicroRNAs as therapeutic targets in chemoresistance. Drug Resist. Updat. 2013;16:47–59. doi: 10.1016/j.drup.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim G., An H.J., Lee M.J., Song J.Y., Jeong J.Y., Lee J.H., Jeong H.C. Hsa-miR-1246 and hsa-miR-1290 are associated with stemness and invasiveness of non-small cell lung cancer. Lung Cancer. 2016;91:15–22. doi: 10.1016/j.lungcan.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 22.Zhang B., Chen J., Ren Z., Chen Y., Li J., Miao X., Song Y., Zhao T., Li Y., Shi Y. A specific miRNA signature promotes radioresistance of human cervical cancer cells. Cancer Cell Int. 2013;13:118. doi: 10.1186/1475-2867-13-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bao L., Hazari S., Mehra S., Kaushal D., Moroz K., Dash S. Increased expression of P-glycoprotein and doxorubicin chemoresistance of metastatic breast cancer is regulated by miR-298. Am. J. Pathol. 2012;180:2490–2503. doi: 10.1016/j.ajpath.2012.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baraniskin A., Birkenkamp-Demtroder K., Maghnouj A., Zöllner H., Munding J., Klein-Scory S., Reinacher-Schick A., Schwarte-Waldhoff I., Schmiegel W., Hahn S.A. MiR-30a-5p suppresses tumor growth in colon carcinoma by targeting DTL. Carcinogenesis. 2012;33:732–739. doi: 10.1093/carcin/bgs020. [DOI] [PubMed] [Google Scholar]
- 25.Gao D., Zhang Y., Zhu M., Liu S., Wang X. miRNA expression profiles of HPV-infected patients with cervical cancer in the Uyghur population in China. PLoS ONE. 2016;11:e0164701. doi: 10.1371/journal.pone.0164701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo H., Guo W., Wang F., You Y., Wang J., Chen X., Wang J., Wang Y., Du Y., Chen X. miR-1291 targets mucin 1 inhibiting cell proliferation and invasion to promote cell apoptosis in esophageal squamous cell carcinoma. Oncol. Rep. 2015;34:2665–2673. doi: 10.3892/or.2015.4206. [DOI] [PubMed] [Google Scholar]
- 27.Imaoka H., Toiyama Y., Fujikawa H., Hiro J., Saigusa S., Tanaka K., Inoue Y., Mohri Y., Mori T., Kato T. Circulating microRNA-1290 as a novel diagnostic and prognostic biomarker in human colorectal cancer. Ann. Oncol. 2016;27:1879–1886. doi: 10.1093/annonc/mdw279. [DOI] [PubMed] [Google Scholar]
- 28.Endo Y., Yamashita H., Takahashi S., Sato S., Yoshimoto N., Asano T., Hato Y., Dong Y., Fujii Y., Toyama T. Immunohistochemical determination of the miR-1290 target arylamine N-acetyltransferase 1 (NAT1) as a prognostic biomarker in breast cancer. BMC Cancer. 2014;14:990. doi: 10.1186/1471-2407-14-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belian E., Kurucz R., Treue D., Lage H. Effect of YB-1 on the regulation of micro RNA expression in drug-sensitive and drug-resistant gastric carcinoma cells. Anticancer Res. 2010;30:629–633. [PubMed] [Google Scholar]
- 30.Peltomäki P. Deficient DNA mismatch repair: a common etiologic factor for colon cancer. Hum. Mol. Genet. 2001;10:735–740. doi: 10.1093/hmg/10.7.735. [DOI] [PubMed] [Google Scholar]
- 31.Wu J., Ji X., Zhu L., Jiang Q., Wen Z., Xu S., Shao W., Cai J., Du Q., Zhu Y., Mao J. Up-regulation of microRNA-1290 impairs cytokinesis and affects the reprogramming of colon cancer cells. Cancer Lett. 2013;329:155–163. doi: 10.1016/j.canlet.2012.10.038. [DOI] [PubMed] [Google Scholar]
- 32.Wei W., Liu F., Liu L., Li Z., Zhang X., Jiang F., Shi Q., Zhou X., Sheng W., Cai S. Distinct mutations in MLH1 and MSH2 genes in hereditary non-polyposis colorectal cancer (HNPCC) families from China. BMB Rep. 2011;44:317–322. doi: 10.5483/BMBRep.2011.44.5.317. [DOI] [PubMed] [Google Scholar]
- 33.Meyers M., Wagner M.W., Mazurek A., Schmutte C., Fishel R., Boothman D.A. DNA mismatch repair-dependent response to fluoropyrimidine-generated damage. J. Biol. Chem. 2005;280:5516–5526. doi: 10.1074/jbc.M412105200. [DOI] [PubMed] [Google Scholar]
- 34.O’Connell M.J., Lavery I., Yothers G., Paik S., Clark-Langone K.M., Lopatin M., Watson D., Baehner F.L., Shak S., Baker J. Relationship between tumor gene expression and recurrence in four independent studies of patients with stage II/III colon cancer treated with surgery alone or surgery plus adjuvant fluorouracil plus leucovorin. J. Clin. Oncol. 2010;28:3937–3944. doi: 10.1200/JCO.2010.28.9538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fan J., Peng Z., Zhou C., Qiu G., Tang H., Sun Y., Wang X., Li Q., Le X., Xie K. Gene-expression profiling in Chinese patients with colon cancer by coupling experimental and bioinformatic genomewide gene-expression analyses: identification and validation of IFITM3 as a biomarker of early colon carcinogenesis. Cancer. 2008;113:266–275. doi: 10.1002/cncr.23551. [DOI] [PubMed] [Google Scholar]
- 36.Ye L., Lin S.T., Mi Y.S., Liu Y., Ma Y., Sun H.M., Peng Z.H., Fan J.W. Overexpression of LARP1 predicts poor prognosis of colorectal cancer and is expected to be a potential therapeutic target. Tumour Biol. 2016;37:14585–14594. doi: 10.1007/s13277-016-5332-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.