Abstract
Introduction
There is a need to develop imaging methods sensitive to axonal injury in multiple sclerosis (MS), given the prominent impact of axonal pathology on disability and outcome. Advanced multi-compartmental diffusion models offer novel indices sensitive to white matter microstructure. One such model, neurite orientation dispersion and density imaging (NODDI), is sensitive to neurite morphology, providing indices of apparent volume fractions of axons (vin), isotropic water (viso) and the dispersion of fibers about a central axis (orientation dispersion index, ODI). NODDI has yet to be studied for its sensitivity to spinal cord pathology. Here, we investigate the feasibility and utility of NODDI in the cervical spinal cord of MS patients.
Methods
NODDI was applied in the cervical spinal cord in a cohort of 8 controls and 6 MS patients. Statistical analyses were performed to test the sensitivity of NODDI-derived indices to pathology in MS (both lesion and normal appearing white matter NAWM). Diffusion kurtosis imaging (DKI) and diffusion tensor imaging (DTI) analysis were also performed to compare with NODDI.
Results
A decrease in NODDI-derived vin was observed at the site of the lesion (p < 0.01), whereas a global increase in ODI was seen throughout white matter (p < 0.001). DKI-derived mean kurtosis (MK) and radial kurtosis (RK) and DTI-derived fractional anisotropy (FA) and radial diffusivity (RD) were all significantly different in MS patients (p < 0.02), however NODDI provided higher contrast between NAWM and lesion in all MS patients.
Conclusion
NODDI provides unique contrast that is not available with DKI or DTI, enabling improved characterization of the spinal cord in MS.
Keywords: NODDI, Multiple sclerosis, Diffusion, Spinal cord, Axon, Volume fraction
Highlights
-
•
NODDI was applied to the human spinal cord in vivo in MS patients at 3T.
-
•
Unlike DTI and DKI, NODDI can provide metrics more specific to neurite morphology.
-
•
NODDI-derived vin and ODI are sensitive to known pathology in MS.
-
•
NODDI shows higher contrast between lesions and white matter than DTI and DKI.
1. Introduction
Multiple sclerosis (MS) is a chronic disease of the central nervous system (CNS). Demyelination, inflammation, gliosis and axonal loss are all cardinal pathological aspects of the disease (Dendrou et al., 2015), however, it is the presence of axonal injury that results in accumulation of irreversible neurological impairment (Brück, 2005). Damage in the spinal cord is known to be prominently eloquent in MS as functional deficits can be directly associated to damage of specific spinal cord tracts (Rossignol et al., 2006). However, current radiological tools have shown little sensitivity for spinal cord tissue injury and even lower specificity in accurately distinguishing potential pathogenic mechanisms at a microscopic level (Bot and Barkhof, 2009). There is a need for advanced noninvasive magnetic resonance imaging (MRI) techniques that are sensitive to specific aspects of MS pathology in the spinal cord to understand its development and progression in vivo.
Diffusion imaging has become a rapidly growing area of study, offering significant insight into the microstructural abnormalities in MS. With diffusion, the MRI signal is sensitive to the random motion (displacement) of water molecules, which is restricted and/or hindered by fibrous structures or barriers (e.g. cell and axon membranes and myelin sheaths). Therefore, diffusion MRI offers an opportunity to indirectly probe microstructural integrity, however, the majority of diffusion MRI in the literature relies on the utilization of a model that summarizes all water compartments inside the tissue of interest. Clinically, the most conventionally used model is the diffusion tensor (diffusion tensor imaging, DTI), which models heterogeneous water displacement with a single three-dimensional tensor (Basser et al., 1994). While DTI has demonstrated sensitivity to changes in tissue microstructure, such as demyelination and axonal loss (Budde et al., 2008, Oh et al., 2012, Song et al., 2005), a major drawback is that DTI does not account for structural heterogeneity and is affected by multiple confounding tissue properties, resulting in reduced specificity of the derived indices to pathologic variations of clinical importance. Additionally, DTI assumes that the underlying probability distribution function of diffusion is Gaussian, which is not true due to the complexity of white matter microstructure, especially when higher b-values are used (Assaf and Cohen, 2000, Kärger, 1985). Diffusion kurtosis imaging (DKI) has been introduced to quantify the degree of non-Gaussianity of the probability density function with the kurtosis (Jensen et al., 2005), but it still suffers from limitations of probing specific tissue compartments.
Other groups have developed multi-compartmental diffusion models in order to more accurately characterize the diffusion signal in the presence of multiple tissue compartments or milieu (Alexander, 2008, Assaf and Basser, 2005, Assaf et al., 2008, Behrens et al., 2003, Chiang et al., 2014, Stanisz et al., 1997, Xu et al., 2014, Zhang et al., 2012). Unlike DTI or DKI, these methods can provide indices related to specific compartments of white matter microstructure. Among these methods, neurite orientation dispersion and density imaging (NODDI) has been recently proposed as a clinically feasible (i.e. can be performed on regular clinical MRI systems without significant modification and with relatively short scan times) protocol to provide more tissue-specific indices, such as the apparent intra-neurite volume fraction and orientation dispersion index. With NODDI, the observed diffusion signal is decomposed into three compartments: (i) an intracellular pool, which is modeled as restricted sticks, or hard cylinders with zero radius, (ii) the extracellular pool, or anisotropically hindered diffusion, and (iii), free water, such as areas containing cerebrospinal fluid (CSF), modeled as isotropic diffusion. NODDI has been demonstrated in brain tumors (Wen et al., 2015), neurofibromatosis (Billiet et al., 2014), focal cortical dysplasia (Winston et al., 2014) and the brain in multiple sclerosis (Schneider et al., 2014).
To date, most applications using NODDI focus on the brain, but recently, the feasibility of performing NODDI in the healthy human spinal cord in vivo was reported (Grussu et al., 2015b). There are, however, no studies evaluating NODDI in the spinal cord with pathology such as MS. Therefore, our main objectives were (1) to study the feasibility of NODDI in the spinal cord of patients with MS and (2) to study the usefulness of NODDI to provide distinct information from DKI and conventional DTI in a patient population. We hypothesize that NODDI may provide the unique ability to offer richer and more model-specific information about the pathological changes known to occur in the spinal cord of MS patients, potentially improving upon DKI and conventional DTI.
2. Methods
2.1. MRI experiments
Eight healthy controls (mean age ± standard deviation = 29.0 ± 5.0 years, 5 M/3F) participated in this study, four of whom had a rescan within a month to assess reproducibility. Six relapsing-remitting MS (RRMS) patients (6F, 39.3 ± 6.1 years) were recruited for this study. Patients' disability was rated using the Expanded Disability Status Scale (EDSS) score (Kurtzke, 1983) in the Vanderbilt University Multiple Sclerosis Clinic (patient EDSS range = 0–6). Table 1 lists specific clinical demographics of the patients. Local institutional review board approval and written informed consent were obtained prior to imaging.
Table 1.
Clinical and demographic characteristics of patients.
| Patient | Age (years) | Sex | MS type | MS duration (years) | EDSS | Lesion (in diffusion volume)a |
|---|---|---|---|---|---|---|
| MS 1 | 46 | F | RRMS | 17 | 2 | Y (LLC) |
| MS 2 | 36 | F | RRMS | 8 | 2.5 | Y (LLC, DC) |
| MS 3 | 45 | F | RRMS | 4 | 3.5 | Y (RLC, LLC, DC) |
| MS 4 | 34 | F | RRMS | 10 | 6 | Y (diffuse) |
| MS 5 | 32 | F | RRMS | 1 | 0 | Y (LLC) |
| MS 6 | 43 | F | RRMS | 2 | 1 | Y (RLC, LLC) |
Y = yes, N = no.
RLC = right lateral column, LLC = left lateral column, DC = dorsal column.
All experiments were performed on a 3.0T whole body MR scanner (Philips Achieva, Best, Netherlands). A quadrature body coil was used for excitation and a 16-channel SENSE neurovascular coil was used for reception. The maximum gradient strength of the system was 80 mT/m at a slew rate of 100 mT/m/s.
For each subject, a high-resolution (0.65 × 0.65 × 5 mm3) multi-slice, multi-echo gradient echo (mFFE) anatomical image (Held et al., 2003) was acquired (TR/TE/∆TE = 753/7.1/8.8 ms, α = 28°, number of slices = 14, 6:12 min) for co-registration and to serve as a reference image for segmentation.
The diffusion sequence consisted of a cardiac-triggered, spin echo sequence with single-shot echo planar imaging (EPI) readout with the following parameters: TR/TE = 3 beats (~ 3000 ms)/65 ms, resolution = 1.25 × 1.25 mm2, slice thickness = 10 mm, slices = 1, FOV = 68 × 52 mm, SENSE (AP) = 1.5 and NEX = 3. Reduced field-of-view was applied using an outer volume suppression technique (Wilm et al., 2007) and fat suppression was achieved using SPIR. A multi-shell acquisition, similar to the previously published NODDI protocol in the brain (Zhang et al., 2012) and the one implemented in the spinal cord (Grussu et al., 2015b) was used with uniform sampling: (i) b = 711 s/mm2 with 32 directions and (ii) b = 2855 s/mm2 with 64 directions, with constant gradient times of ∆ (separation between gradients) = 31.8 ms and δ (gradient duration) = 21.0 ms. A non-diffusion weighted scan (b = 0 s/mm2 or b0) was acquired with each shell. Total scan time was 18:11 min. All images were centered at the C3/C4 level, except for one MS patient that was centered at the C4/C5 level where more lesions were detected. Images were acquired in the axial plane for both the anatomical and diffusion images.
2.2. Image analysis and processing
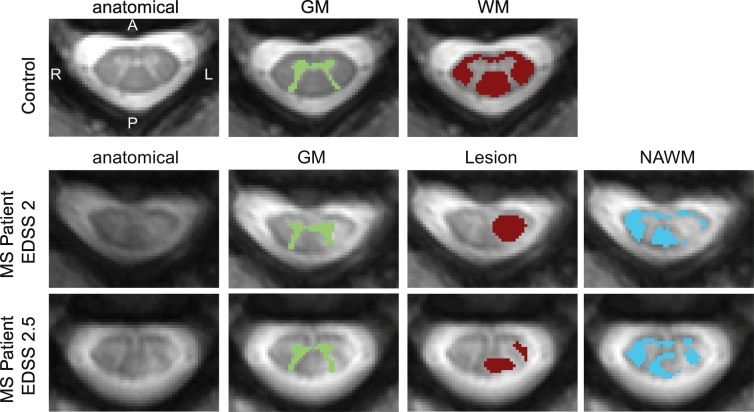
All diffusion-weighted volumes were co-registered to the anatomical (mFFE). First, the b0 images from each shell was diffeomorphically registered to the anatomical using ANTS (Avants et al., 2011). All other diffusion-weighted volumes were then registered to its b0 image using an affine transformation to correct for eddy current distortions (Haselgrove and Moore, 1996). Finally, regions of interest (ROIs) of white matter (WM) and gray matter (GM) were automatically segmented from the co-registered anatomical image using a slice-based groupwise multi-atlas procedure designed specifically for the spinal cord (Asman et al., 2014). The ROIs were eroded by a disk-shaped structuring element with a radius of four voxels in the white matter and one voxel in the gray matter to avoid inaccurate voxels at the boundaries, potentially arising from misregistration or partial volume effects. For patients, the same groupwise multi-atlas segmentation procedure was performed for white and gray matter. Additionally, we refined the segmentation by manually delineating lesions on the anatomical image, and then normal appearing white matter (NAWM) was any of the segmented white matter not containing manually drawn lesion voxels. It is possible that some gray matter voxels either contain gray matter lesions themselves, or are partial volumed with white matter lesions. For the latter, this is somewhat unavoidable due to the reliance on automatic gray matter segmentation, which cannot differentiate lesions from gray matter (similar contrast) when they are in close proximity to one another; for the former, we expect that gray matter is damaged in some participants and thus the gray matter values we report are a combination of both normal appearing gray matter and gray matter with lesions. Examples of ROI identification for a representative control (WM, GM) and two MS patients (lesion, NAWM) are shown in Fig. 1. Note that all images are displayed on the radiological coordinate system.
Fig. 1.
Examples of ROI identification. Representative control (top row) includes the anatomical (left) from which the GM (second column) and WM (third column) were automatically segmented. For MS patients (middle and bottom row), the GM (second column) and WM are automatically segmented the same way as controls. WM, however, is separated into manually delineated lesions (third column) and any WM voxels containing no lesion was considered NAWM (fourth column). Note for all images the radiological coordinate system is used.
NODDI fitting was performed using the NODDI MATLAB Toolbox (http://nitrc.org/projects/noddi_tolbox). Diffusion coefficients for the intra-axonal and isotropic compartments were fixed with values of d|| = 1.7 μm2/ms and diso = 3.0 μm2/ms respectively as in (Zhang et al., 2012) and (Grussu et al., 2015b). From this fitting, the derived NODDI indices included: the apparent intra-axonal volume fraction vin, representing the fraction of dendrites and axons; the isotropic volume fraction viso, representing the fraction of free water such as CSF; and the orientation dispersion ODI, a measure of how nonparallel axons disperse about a central orientation by assuming a cylindrically symmetric Watson distribution (Zhang et al., 2012).
For comparison, using all of the same data as NODDI, DKI fitting was performed with a weighted linear least squares estimator (Veraart et al., 2013) using the freely available NYU DKI toolbox (Veraart et al., 2011). The resulting DKI maps included the mean kurtosis (MK), axial kurtosis (AK), and radial kurtosis (RK). For clarity, the kurtosis estimates the non-Gaussian nature of a distribution, where MK indicates the average diffusion kurtosis over all directions; AK indicates the diffusion kurtosis along the primary axis of the kurtosis tensor and is typically low in healthy white matter tissue since the diffusion along axons is relatively unrestricted; RK indicates the diffusion kurtosis along the axis perpendicular to the primary axis of the spinal cord and is typically high in healthy tissue due to the more heterogeneous pattern in the presence of myelin sheaths (Steven et al., 2013). Conventional DTI fitting was also performed, using only the data from the b = 711 s/mm2 shell. The tensor was calculated using a nonlinear fit from Camino (Cook et al., 2006) and the fractional anisotropy (FA), mean diffusivity (MD), axial diffusivity (AD) and radial diffusivity (RD) maps were estimated from the diffusion tensor.
2.3. Repeatability
The reproducibility of the NODDI-, DKI-, and DTI-derived indices were assessed using Bland-Altman (Bland and Altman, 1986) for white matter. In the Bland Altman analysis, each subject's mean index within the automatically segmented ROI (white matter) entered reproducibility analysis as a single data point. The 95% confidence interval (CI) for the mean difference was calculated and if the 95% CI overlaps zero, the indication is that there is no significant difference between scan 1 and scan 2 at α = 0.05. The normalized Bland Altman (DBA) was also used as an estimate of reliability to be used for comparison across derived indices, calculated as:
| (1) |
where D12 is the mean difference between the two sessions and M is the mean diffusion-derived index of interest. A nonparametric Wilcoxon signed rank was performed, in addition to the Bland Altman analysis, to determine whether a significant difference exists between the mean of each diffusion-derived index for each of the two scans at α = 0.05.
Histograms of the NODDI-fitted parameters were created over all of the healthy control white matter voxels in the scan and rescan to visualize whether any deviations from one another were noticeable. For vin, a bin width of 2% over a range of 25 to 95% was used; for viso a bin width of 2% with a range of 0 to 80% was used; for ODI, a bin width of 0.008 with a range of 0 to 0.15 was used.
2.4. Group comparison
A cross-sectional analysis was conducted to determine white matter differences in NODDI-derived indices between the healthy and MS cohorts. A nonparametric Wilcoxon rank sum test was performed on the mean vin, viso, and ODI values in healthy white matter (within healthy controls), NAWM, and lesions for patients at a significant threshold of α = 0.05. The same comparison was performed for DKI and DTI. To account for multiple comparisons (n = 3 for each metric), a Bonferroni correction was used, resulting in an α = 0.017. For NODDI, statistical differences between healthy gray matter (within healthy controls) and normal appearing gray matter (MS patients) were also assessed using a nonparametric Wilcoxon rank sum test; no Bonferroni correction was necessary for the one comparison, and thus, a significant threshold of α = 0.05 was used.
2.5. Analysis of maps
Image quality in the NODDI-, DKI- and DTI-derived maps was assessed by the image contrast between lesions and NAWM in the MS patients. Contrast was defined as:
| (2) |
where μx represents the mean over the specified region of interest.
3. Results
3.1. Feasibility of NODDI in controls
3.1.1. Mean maps over controls
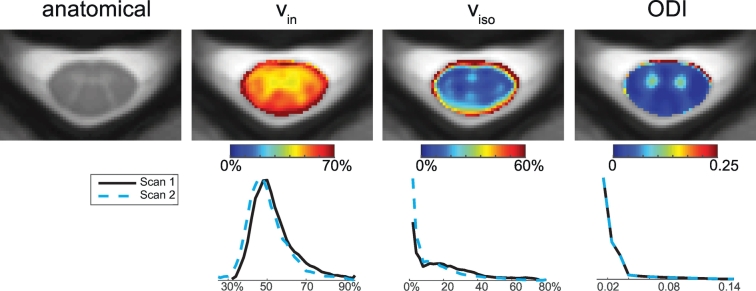
Fig. 2 shows the NODDI-derived maps co-registered and averaged over all healthy controls, along with the averaged anatomical image (mFFE). The vin maps show high contrast between gray and white matter, with lower vin values in the gray matter compared to white matter, as expected. The averaged image's (from Fig. 2) contrast between white matter and gray matter is 0.23; the image contrast from the maps of individual controls is smaller (mean over all controls ± standard deviation = 0.13 ± 0.07). The viso map highlights the central sulcus and central canal, along with some gray and white matter contrast (averaged image's contrast: 0.53, mean contrast over all controls = 0.42 ± 0.36). Lastly, the ODI maps provide the most significant contrast between gray and white matter (averaged image's contrast: 1.19, mean contrast over all controls = 1.08 ± 0.19), which is expected since the orientations of gray matter dendrites are much more non-uniformly distributed compared to close-to-uniformly oriented axons in the white matter tracts. It is important to point out that the high value voxels at the rim of the spinal cord in both the vin and ODI maps are presumably due to registration errors, and are exaggerated when taking the average over all healthy control subjects. For viso, high viso levels at the boundary of the spinal cord and cerebrospinal fluid (CSF) can be attributed to higher partial volume effects, in addition to the registration errors.
Fig. 2.
Maps and histograms of fitted parameters using NODDI in controls. Top row: Mean maps over all of the controls were calculated for the anatomical (left), vin, viso, and ODI (right). Bottom row: Histograms over all white matter voxels for scan 1 and scan 2 for vin, viso, and ODI (right).
3.1.2. Reproducibility in controls
To highlight any outliers and demonstrate the overlap between scan and rescan, the bottom row of Fig. 2 includes the histograms over all white matter voxels for all of the controls in scan 1 and scan 2. For vin, both histograms overlap one another significantly. For viso, although both of the histograms largely overlap, there is a higher frequency of voxels with low viso (< 10%) in scan 2 than scan 1. For ODI, the histograms overlay onto each other directly, indicating high reproducibility. Reproducibility histograms for individual controls (data not shown) demonstrated the same trends as the group histograms (Fig. 2), and were consistent across subjects, yielding mean percent differences between scan and rescan over all controls of 8.22%, 45.5% and 5.09% for vin, viso, and ODI respectively. Table 2 lists the reproducibility metrics for each NODDI-derived index. The reproducibility for the DKI and DTI indices for the same scans is also listed. It is important to note that the 95% confidence intervals for the mean difference of all of the derived indices (NODDI, DKI and DTI) overlap 0 and the Wilcoxon sign rank p-value is > 0.05, indicating that the metrics are not significantly different from one another in the two different time points. Both vin and ODI yield DBA under 10%, however viso yields a high DBA of 35.5%, indicating that the variability between the two scans is large for viso. All DKI- and DTI-derived metrics resulted in a DBA under 10%, with DKI metrics yielding slightly lower DBA than the comparing DTI metrics.
Table 2.
Reproducibility Metrics for NODDI, DKI and DTI.
| Scan 1 | Scan 2 | Bland-Altman |
WSR | |||
|---|---|---|---|---|---|---|
| Mean ± std | Mean ± std | Difference | 95% CI | DBA (%) | p-Value | |
| NODDI | ||||||
| vin | 54.3 ± 3.53% | 50.4 ± 8.19% | 3.88 | [− 4.52, 12.3] | 7.47 | 0.38 |
| viso | 21.8 ± 4.84% | 15.2 ± 8.30% | 6.59 | [− 3.07, 16.2] | 35.5 | 0.13 |
| ODI | 0.021 ± 0.007 | 0.021 ± 0.003 | − 0.0005 | [− 0.013, 0.012] | 2.27 | 0.86 |
| DKI | ||||||
| MK | 0.81 ± 0.04 | 0.78 ± 0.10 | 0.03 | [− 0.13, 0.19] | 3.89 | 1 |
| AK | 0.53 ± 0.04 | 0.56 ± 0.02 | − 0.03 | [− 0.10, 0.04] | 5.39 | 0.25 |
| RK | 1.56 ± 0.16 | 1.49 ± 0.20 | 0.07 | [− 0.25, 0.40] | 4.77 | 0.63 |
| DTI | ||||||
| FA | 0.70 ± 0.06 | 0.71 ± 0.03 | − 0.01 | [− 0.10, 0.08] | 1.34 | 0.88 |
| MDa | 1.15 ± 0.04 | 1.08 ± 0.09 | 0.07 | [− 0.01, 0.16] | 6.49 | 0.13 |
| ADa | 2.28 ± 0.21 | 2.15 ± 0.14 | 0.13 | [− 0.01, 0.27] | 5.81 | 0.13 |
| RDa | 0.58 ± 0.06 | 0.54 ± 0.07 | 0.04 | [− 0.11, 0.19] | 7.81 | 0.38 |
Units of μm2/ms.
3.2. Application in MS cohort
3.2.1. NODDI in MS cohort
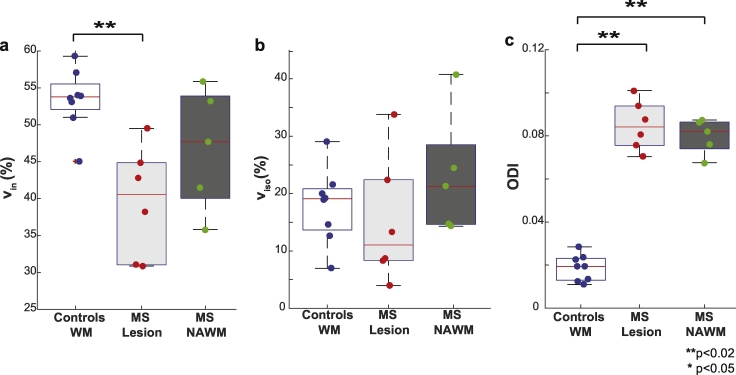
In Fig. 3, boxplots summarize the observed trends for vin, viso, and ODI over healthy white matter for controls (white) and, in the MS patients, lesions (identified from the anatomical) and NAWM in light and dark gray boxes respectively. The median (red line) and interquartile range (whiskers) over each cohort is displayed, along with the mean value for each individual as it enters the boxplot. Note, only five points are plotted for the NAWM in the MS cohort, as one MS patient had a diffuse lesion over the entire cord at the C3/C4 level. For vin (Fig. 3a), lesions show a significant decrease (p = 0.001) compared to healthy control white matter. There is no detectable difference between NAWM and control white matter (p = 0.171) or between NAWM and lesions (p = 0.247), but a trend is observed where NAWM values fall between lesions and control white matter tissue. No significant differences were observed in viso across cohorts in either tissue type (Fig. 3b). Importantly, Fig. 3c shows a global increase in ODI in lesions (p < 0.001) and NAWM (p = 0.002) compared to healthy volunteer white matter. When comparing gray matter of healthy controls to MS patients (data not shown), a significant decrease in vin was observed (p = 0.04) along with a significant increase in ODI (p = 0.003), but no change was observed in viso (p = 0.22).
Fig. 3.
Comparison of vin, viso, and ODI. Boxplots highlighting the median, 25th and 75th percentiles over controls (WM) and MS patients (lesions and NAWM) for (a) vin, (b) viso and (c) ODI. Mean values from each subject plotted and asterisks indicate significant differences between the groups, where double asterisks indicate significance after Bonferroni correction.
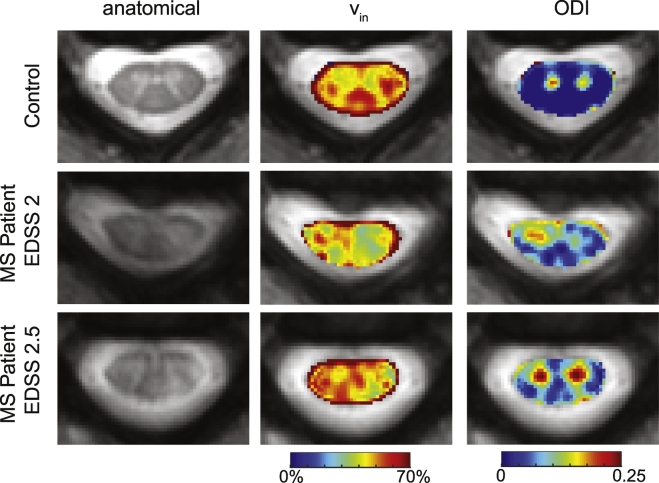
Fig. 4 shows examples of the anatomical and NODDI-derived maps in a healthy volunteer and two MS patients (patient 1: EDSS = 2, duration of disease = 17 years, patient 2: EDSS = 2.5, duration of disease = 8 years). For succinctness, only the NODDI-derived maps that indicated sensitivity to MS pathology are shown. For patient 1, a lesion in the left lateral column (as shown in Fig. 1) can be observed in the anatomical and corresponds to areas of decreased intra-axonal fractions in the vin maps. Additionally, this patient shows decreased vin in areas where the anatomical looks otherwise normal, such as the right dorsal column. In patient 2, smaller lesions are seen in the left dorsal and left lateral column on the anatomical (as shown in Fig. 1), and similarly, a decreased vin is observed. The ODI maps in both MS participants demonstrate changes not localized at the site of the lesion, but rather, diffuse increases throughout the cord's white matter and in MS patient 2, a large increase in the gray matter signal. This indicates that ODI may have the ability to probe the subtle microstructural changes in NAWM before they can be detected by conventional MRI such as the mFFE.
Fig. 4.
Example images from NODDI. Representative control is shown in the first row, followed by examples from two different MS patients (same as Fig. 1). From left to right, the anatomical, vin, viso, and ODI are shown.
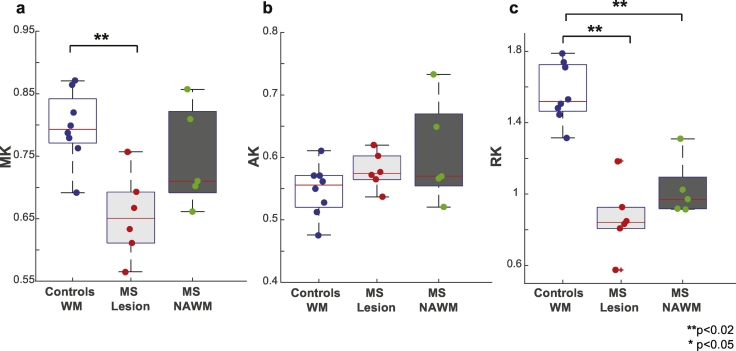
3.2.2. Comparison to DKI
Group comparisons for the DKI-derived metrics are shown in Fig. 5. A decrease in MK in the lesions of MS patients is detected (p = 0.003). No significant differences were observed between healthy white matter and MS white matter for AK. For RK, a significant decrease was observed between healthy white matter and lesions in MS patients (p < 0.001). Additionally, a decrease was observed between healthy white matter and NAWM in MS patients (p = 0.016), albeit to a lesser degree than what was observed in lesions.
Fig. 5.
Comparison of MK, AK and RK. Boxplots highlighting the median, 25th and 75th percentiles over controls (WM) and MS patients (lesions and NAWM) for (a) MK, (b) AK and (c) RK. Mean values from each subject plotted and asterisks indicate significant differences between the groups. Kurtosis metrics are unitless.
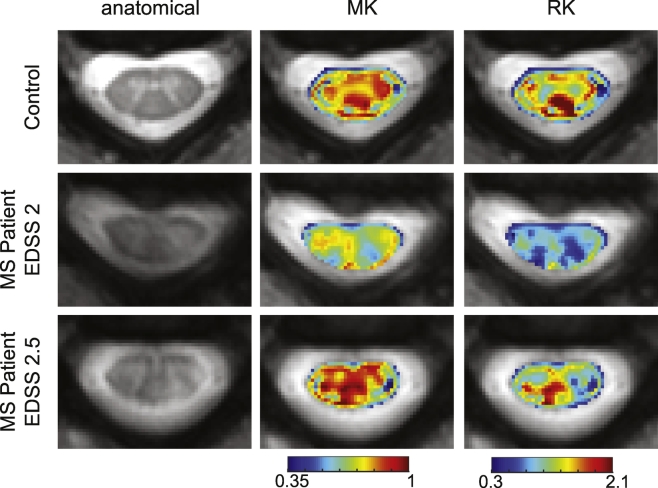
Fig. 6 shows the derived DKI images from the same control and MS patients as in Fig. 3. In the healthy control, it is difficult to delineate the white matter from gray matter in both the MK (mean image contrast = 0.02) and RK (mean image contrast = 0.11) maps, as there are areas of inconsistent estimation in the white matter, particularly in the dorsal and lateral columns. There is, however, a noticeable decrease in MK and RK at the site of the lesions. For MS patient 1 (middle row), a larger decrease in MK and RK is observed in other areas outside the lesion.
Fig. 6.
Example images from DKI. Representative control is shown in the first row, followed by examples from two MS patients. From left to right, the anatomical, MK and RK are shown. The same control and patients from Fig. 4 are included.
3.2.3. Comparison to DTI
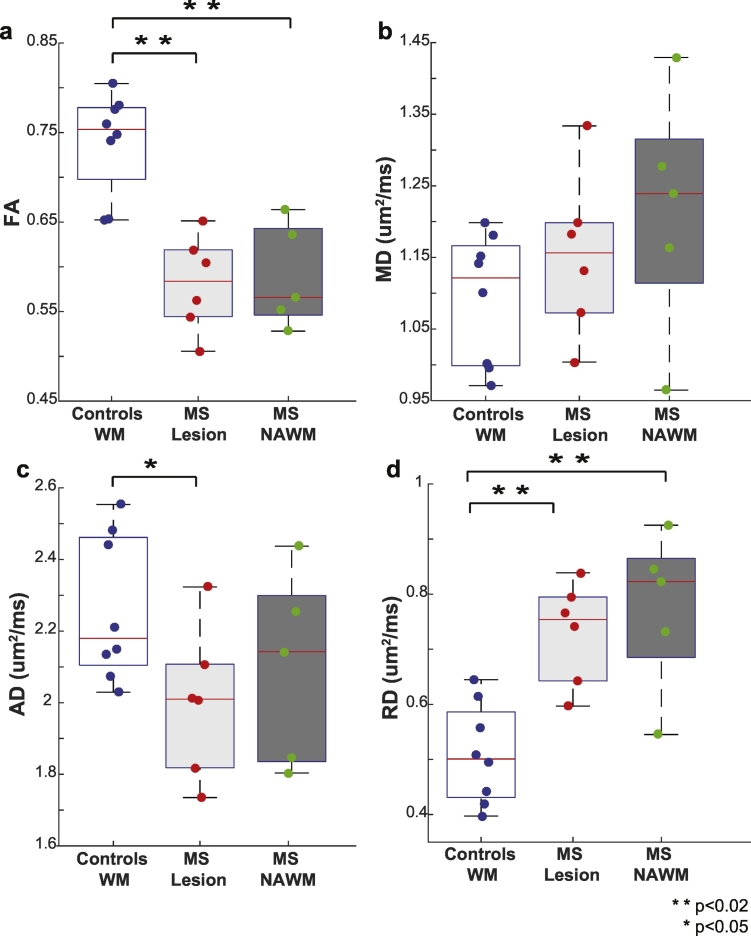
Fig. 7 compares the median and interquartile range for DTI-derived indices over all volunteers. FA (Fig. 7a) is reduced in both lesions (p < 0.001) and NAWM. No significant differences are detected for MD (Fig. 7b) or AD (Fig. 7c) after Bonferroni correction, however there is a trend towards an increased MD and decreased AD (p = 0.03). Lastly, differences in RD were also seen (Fig. 7d) in both lesions (p = 0.005) and NAWM (p = 0.01).
Fig. 7.
Comparison of FA, MD, AD and RD. Boxplots highlighting the median, 25th and 75th percentiles over controls (WM) and MS patients (lesions and NAWM) for (a) FA, (b) MD, (c) AD, and (d) RD. Mean values from each subject plotted and asterisks indicate significant differences between the groups.
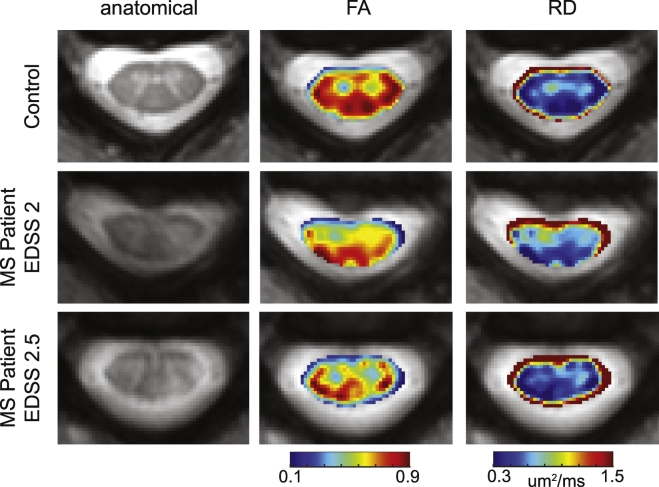
Fig. 8 shows the FA and RD maps for the same control and MS patients as shown in Fig. 4. While changes are observed in the FA and RD maps of the MS patients respective to the control, it is difficult to delineate the lesions from NAWM.
Fig. 8.
Example images from DTI. Representative control is shown in the first row, followed by examples from two MS patients. From left to right, the anatomical, FA and RD are shown. The same control and patients from Fig. 4 are included.
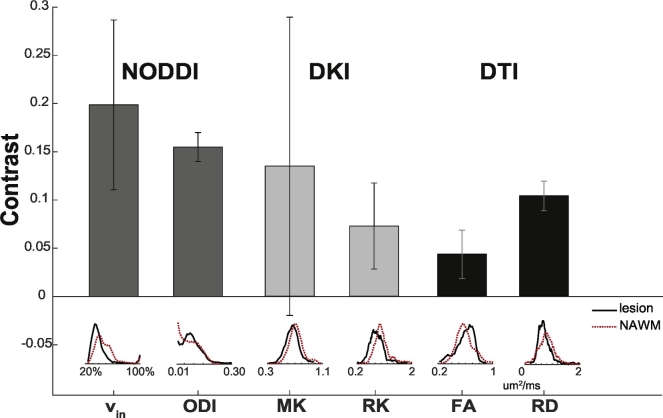
Fig. 9 compares the mean contrast, along with the standard deviation, between lesion and NAWM over all of the patients for NODDI-derived vin and ODI, DKI-derived MK and RK, and DTI-derived FA and RD. It is apparent that NODDI provides the highest contrast between NAWM and lesions than both DKI and DTI. Below the bar plot, a histogram of all the voxels delineating lesions (black) and NAWM (dashed red) over all patients is shown. In all cases, the contrast between lesion and NAWM from the NODDI indices, either vin (0.20. ± 0.08) or ODI (0.15 ± 0.02), was greater than any of the DKI (MK: 0.13 ± 0.15, RK: 0.07 ± 0.0.05) or DTI indices (FA: 0.04 ± 0.02, RD: 0.10 ± 0.02). Furthermore, the histograms demonstrate that the distributions of the lesion and NAWM voxels mostly overlap one another for the DKI and DTI indices, whereas the histograms of the NODDI indices indicate greater deviation from one another. In particular, the vin histogram for the NAWM highlights a bump to the right of the lesion histogram, indicating a large frequency of increased vin values in the NAWM. With ODI, a global increase throughout the cord was observed, so there is overlap in the histograms for NAWM and lesion voxels, but a more distinct peak is seen in the lesion histogram.
Fig. 9.
Contrast in diffusion maps. Mean contrast between lesions and NAWM over all patients is shown, with error bars indicating the standard deviation over all of the patients.
4. Discussion
This work demonstrates (1) the feasibility of deriving high-quality NODDI indices in the spinal cord of patients with MS and (2) the sensitivity improvement of NODDI over DKI and DTI when focusing on spinal cord lesions in MS. Specifically, we show that vin and ODI provide high intra-cord contrast in patients with MS, which allows for unique characterization of damage that the spinal cord undergoes along the evolution of MS.
Recently, the first application and feasibility of NODDI in the cervical spinal cord was reported with healthy volunteers only (Grussu et al., 2015b). In this current study, we observed that our NODDI-derived indices fall within the range of those observed by Grussu et al., who report a vin of 57 ± 9% (our study: 54 ± 4%), viso of 12 ± 11% (18 ± 7%), and ODI of 0.027 ± 0.0.003 (0.018 ± 0.006). Additionally, our reproducibility measurements are in good agreement to those previously reported, where vin and ODI show reliable measurements but the reproducibility of viso is poor. As a result, viso did not provide a reliable contrast when applied to the MS cohort, and would be more difficult to follow in longitudinal studies. However, we observed a statistically significant decrease in vin in lesions, a finding that is consistent with the expected pathology in chronic MS lesions with known axonal injury and loss. This also corresponds well with findings from an ex vivo spinal cord of MS study (Grussu et al., 2015a), where decreased vin was observed in lesions in comparison to the NAWM and correlated well with histological metrics. Interestingly, in some patients in our study, a decreased vin was also observed in surrounding NAWM, which may indicate the potential of the index to detect microstructural changes of white matter without obvious inflammation, which is not detectable using conventional MRI. An increased ODI throughout the white matter was observed, suggesting pervasive changes in the spinal cord in the presence of MS. These findings also align well with the ex vivo study, where ODI was higher in NAWM than in lesions (Grussu et al., 2015a), and may indicate that ODI has the potential to be used as a prognostic indicator even before new lesions appear. Finally, significant differences in gray matter of healthy controls and normal appearing gray matter in MS patients were observed for vin and ODI, which may be indicative of pathological changes known to occur in gray matter lesions, such as axonal transection, in addition to neuronal, glial and synaptic loss (Geurts and Barkhof, 2008). One limitation of this study, however, is that fixed diffusivities of d|| = 1.7 μm2/ms and diso = 3.0 μm2/ms for the intra-axonal and isotropic compartments respectively, as implemented in the NODDI toolbox (Zhang et al., 2012) and in the previous study of NODDI in the healthy spinal cord (Grussu et al., 2015b). While the DTI-derived indices from this study indicated that these estimates were reasonable and similar to literature values (Oh et al., 2012) for the spinal cord in both healthy and MS cohorts, future studies should investigate the effect of fixing the diffusivities to known values for a specific cohort.
Furthermore, NODDI-derived vin and ODI provide unique contrast between lesions and NAWM that is not detectable in DKI- and DTI-derived indices (and even anatomical mFFE imaging). Only the b = 711 s/mm2 shell was used in the DTI tensor calculation since at higher b-values, the signal is more sensitive to slow diffusing time components, and it is well known that the diffusion signal no longer follows a Gaussian approximation (Farrell et al., 2008). While DTI-derived FA and RD both showed sensitivity to MS pathology, the generated contrast between NAWM and lesion is much lower in comparison to the NODDI-derived indices. This is consistent with previous findings that point to improved quality of fit with NODDI versus DTI (Grussu et al., 2015b) and we hypothesize this is due to the non-specificity of the tensor model. For example, it has been demonstrated that a decrease in FA could be due to a decrease in vin or increase in ODI (Grussu et al., 2015b, Zhang et al., 2012). Parameters such as vin and ODI may allow assessment of specific pathological changes with minimized confounding influence (i.e. from CSF), which may be advantageous in longitudinal tracking of the disease more accurately.
It is important, however, to note that the DTI model did not use all of the data in the NODDI acquisition, and consequently, was not a scan time equivalent comparison. Previous work has demonstrated that the underlying measurement error in the DTI-derived indices in the spinal cord is negligible when increasing the number of averages from one to two for a 32-direction scheme (By et al., 2016). Therefore, we do not expect different conclusions in our DTI and NODDI comparison, if we instead were to use a scan time equivalent comparison for DTI. However, we also chose to compare NODDI with the DKI model in order to have a comparison using all of the same data. This comparison has never been made for in vivo spinal cord imaging, but answers a relevant question of whether multi-compartmental and biophysically based models are indeed more sensitive than conventional single-compartment models, or whether the source of sensitivity arises from acquiring multiple b-shells. Interestingly, a significant change was observed in MK at the site of the lesion, whereas no change was detected with MD. This may indicate DKI's ability to overcome some of the limitations of DTI. Nonetheless, NODDI is particularly advantageous over both of these diffusion signal models in that more specific information regarding microstructural components may be obtained.
There has recently been an increased interest of applying advanced diffusion models in the human cervical spinal cord in vivo (Duval et al., 2015, Farrell et al., 2008, Grussu et al., 2015b, Murphy et al., 2015) and to our knowledge, no multi-compartmental diffusion models have been used to measure specific microstructural information of spinal cord of patients with MS. A main technical concern that has hampered this application is the difficulty in registering diffusion-weighted volumes well. With the high b-values (b > 1000 s/mm2) needed in many of these protocols, the signal is often too low for registration. Previous studies have interleaved non diffusion-weighted volumes throughout the acquisition, and relied on applying the transformation of the nearest interleaved volume to the diffusion-weighted volumes (Cohen-Adad et al., 2008, Grussu et al., 2015b). This method, however, assumes that negligible motion has occurred in between non diffusion-weighted volumes, which is impractical in the presence of patient movement such as swallowing. With sufficient signal-to-noise ratio (SNR), we were able to achieve reliable affine registration of the individual diffusion-weighted volumes, which also enabled eddy current correction (Mohammadi et al., 2010). Furthermore, we note that no healthy subjects or patients had to be excluded from the study because of motion artifacts.
Only one slice of the cervical spinal cord was acquired in the current study, because the main goal was to investigate the feasibility and sensitivity of NODDI in MS patients, and therefore, rather than optimizing for acquisition time, we chose to implement a sequence that minimized any dependencies of SNR and focused only on one level of the cord. In future studies, simultaneous multi-slice excitation imaging will help improve slice coverage without the expense of additional acquisition time (Setsompop et al., 2012). Optimal direction schemes may also be investigated for the spinal cord specifically to allow for additional decreases in acquisition time.
Future studies involving a larger cohort and investigating the correlation of the NODDI-derived parameters with clinical disability are warranted. While no effect on age was observed on the NODDI-derived metrics in this study, Taso et al. have previously reported an association with age and decreased DTI-derived metrics (Taso et al., 2016); however, their results indicate that an effect on age is only significant when comparing groups younger than 50 to groups older than 50 years old, which was not the case for this current study. However, future studies involving age-matched controls are worthwhile. Lastly, a longitudinal study utilizing multi-parametric MRI may help gain confidence of the NODDI-derived indices, in addition to further disentangling the pathological processes occurring in MS (Cohen-Adad et al., 2011).
In conclusion, we demonstrated the feasibility and initial results of NODDI in the cervical spinal cord of MS patients. NODDI maps provide distinguishable contrast that is not seen in DKI or DTI maps, and may reflect underlying microstructural changes known to occur in MS.
Acknowledgements
This study was funded, in part, by the National Multiple Sclerosis Society NMSS RG-1501-02840 (SAS). Additional funding sources include NIH/NINDS R21 NS087465-01 (SAS), NIH/NEI R01 EY023240 (SAS) and DoD W81XWH-13-0073 (SAS). We would like to thank Ms. Kristen George-Durrett, Ms. Clair Jones, Ms. Leslie McIntosh, and Mr. Chris Thompson, who have provided invaluable assistance with scheduling and subject assistance.
References
- Alexander D.C. A general framework for experiment design in diffusion MRI and its application in measuring direct tissue-microstructure features. Magn. Reson. Med. 2008;60:439–448. doi: 10.1002/mrm.21646. [DOI] [PubMed] [Google Scholar]
- Asman A.J., Bryan F.W., Smith S.A., Reich D.S., Landman B.A. Groupwise multi-atlas segmentation of the spinal cord's internal structure. Med. Image Anal. 2014;18:460–471. doi: 10.1016/j.media.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaf Y., Basser P.J. Composite hindered and restricted model of diffusion (CHARMED) MR imaging of the human brain. NeuroImage. 2005;27:48–58. doi: 10.1016/j.neuroimage.2005.03.042. [DOI] [PubMed] [Google Scholar]
- Assaf Y., Cohen Y. Assignment of the water slow-diffusing component in the central nervous system using q-space diffusion MRS: implications for fiber tract imaging. Magn. Reson. Med. 2000;43:191–199. doi: 10.1002/(sici)1522-2594(200002)43:2<191::aid-mrm5>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Assaf Y., Blumenfeld-Katzir T., Yovel Y., Basser P.J. AxCaliber: a method for measuring axon diameter distribution from diffusion MRI. Magn. Reson. Med. 2008;59:1347–1354. doi: 10.1002/mrm.21577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants B.B., Tustison N.J., Song G., Cook P.A., Klein A., Gee J.C. A reproducible evaluation of ANTs similarity metric performance in brain image registration. NeuroImage. 2011;54:2033–2044. doi: 10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basser P.J., Mattiello J., LeBihan D. MR diffusion tensor spectroscopy and imaging. Biophys. J. 1994;66:259–267. doi: 10.1016/S0006-3495(94)80775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens T.E., Woolrich M.W., Jenkinson M., Johansen-Berg H., Nunes R.G., Clare S., Matthews P.M., Brady J.M., Smith S.M. Characterization and propagation of uncertainty in diffusion-weighted MR imaging. Magn. Reson. Med. 2003;50:1077–1088. doi: 10.1002/mrm.10609. [DOI] [PubMed] [Google Scholar]
- Billiet T., Mädler B., D'Arco F., Peeters R., Deprez S., Plasschaert E., Leemans A., Zhang H., Van den Bergh B., Vandenbulcke M. Characterizing the microstructural basis of “unidentified bright objects” in neurofibromatosis type 1: a combined in vivo multicomponent T2 relaxation and multi-shell diffusion MRI analysis. NeuroImage: Clinical. 2014;4:649–658. doi: 10.1016/j.nicl.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bland J.M., Altman D.G. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1:307–310. [PubMed] [Google Scholar]
- Bot J.C., Barkhof F. Spinal-cord MRI in multiple sclerosis: conventional and nonconventional MR techniques. Neuroimaging Clin. N. Am. 2009;19:81–99. doi: 10.1016/j.nic.2008.09.005. [DOI] [PubMed] [Google Scholar]
- Brück W. The pathology of multiple sclerosis is the result of focal inflammatory demyelination with axonal damage. J. Neurol. 2005;252:v3–v9. doi: 10.1007/s00415-005-5002-7. [DOI] [PubMed] [Google Scholar]
- Budde M.D., Kim J.H., Liang H.F., Russell J.H., Cross A.H., Song S.K. Axonal injury detected by in vivo diffusion tensor imaging correlates with neurological disability in a mouse model of multiple sclerosis. NMR Biomed. 2008;21:589–597. doi: 10.1002/nbm.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- By S., Smith A.K., Dethrage L.M., Lyttle B.D., Landman B.A., Creasy J.L., Pawate S., Smith S.A. Quantifying the impact of underlying measurement error on cervical spinal cord diffusion tensor imaging at 3T. J. Magn. Reson. Imaging. 2016;44:1608–1618. doi: 10.1002/jmri.25308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C.W., Wang Y., Sun P., Lin T.H., Trinkaus K., Cross A.H., Song S.K. Quantifying white matter tract diffusion parameters in the presence of increased extra-fiber cellularity and vasogenic edema. NeuroImage. 2014;101:310–319. doi: 10.1016/j.neuroimage.2014.06.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Adad J., Descoteaux M., Rossignol S., Hoge R.D., Deriche R., Benali H. Detection of multiple pathways in the spinal cord using q-ball imaging. NeuroImage. 2008;42:739–749. doi: 10.1016/j.neuroimage.2008.04.243. [DOI] [PubMed] [Google Scholar]
- Cohen-Adad J., El Mendili M.M., Lehéricy S., Pradat P.F., Blancho S., Rossignol S., Benali H. Demyelination and degeneration in the injured human spinal cord detected with diffusion and magnetization transfer MRI. NeuroImage. 2011;55:1024–1033. doi: 10.1016/j.neuroimage.2010.11.089. [DOI] [PubMed] [Google Scholar]
- Cook P.A., Bai Y., Nedjati-Gilani S., Seunarine K.K., Hall M.G., Parker G.J., Alexander D.C. 2006. Camino: Open-source Diffusion-MRI Reconstruction and Processing, Joint Annual Meeting ISMRM-ESMRMB; p. 2759. (Proc. Int. Soc. Magn. Reson. Med.). (Seattle, WA) [Google Scholar]
- Dendrou C.A., Fugger L., Friese M.A. Immunopathology of multiple sclerosis. Nat. Rev. Immunol. 2015;15:545–558. doi: 10.1038/nri3871. [DOI] [PubMed] [Google Scholar]
- Duval T., McNab J.A., Setsompop K., Witzel T., Schneider T., Huang S.Y., Keil B., Klawiter E.C., Wald L.L., Cohen-Adad J. In vivo mapping of human spinal cord microstructure at 300 mT/m. NeuroImage. 2015;118:494–507. doi: 10.1016/j.neuroimage.2015.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell J.A., Smith S.A., Gordon-Lipkin E.M., Reich D.S., Calabresi P.A., van Zijl P.C. High b-value q-space diffusion-weighted MRI of the human cervical spinal cord in vivo: feasibility and application to multiple sclerosis. Magn. Reson. Med. 2008;59:1079–1089. doi: 10.1002/mrm.21563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts J.J.G., Barkhof F. Grey matter pathology in multiple sclerosis. Lancet Neurol. 2008;7:841–851. doi: 10.1016/S1474-4422(08)70191-1. [DOI] [PubMed] [Google Scholar]
- Grussu F., Schneider T., Yates R., Tachrount M., Newcombe J., Zhang H., Alexander D., DeLuca G., Wheeler-Kingshott C. Joint Annual Meeting ISMRM-ESMRMB. 2015. Histological metrics confirm microstructural characteristics of NODDI indices in multiple sclerosis spinal cord; p. 909. (Proc. Intl. Soc. Mag. Reson. Med.). (Toronto, Canada) [Google Scholar]
- Grussu F., Schneider T., Zhang H., Alexander D.C., Wheeler-Kingshott C.A. Neurite orientation dispersion and density imaging of the healthy cervical spinal cord in vivo. NeuroImage. 2015;111:590–601. doi: 10.1016/j.neuroimage.2015.01.045. [DOI] [PubMed] [Google Scholar]
- Haselgrove J.C., Moore J.R. Correction for distortion of echo-planar images used to calculate the apparent diffusion coefficient. Magn. Reson. Med. 1996;36:960–964. doi: 10.1002/mrm.1910360620. [DOI] [PubMed] [Google Scholar]
- Held P., Dorenbeck U., Seitz J., Fründ R., Albrich H. MRI of the abnormal cervical spinal cord using 2D spoiled gradient echo multiecho sequence (MEDIC) with magnetization transfer saturation pulse. A T2* weighted feasibility study. J. Neuroradiol. 2003;30:83–90. [PubMed] [Google Scholar]
- Jensen J.H., Helpern J.A., Ramani A., Lu H., Kaczynski K. Diffusional kurtosis imaging: the quantification of non-Gaussian water diffusion by means of magnetic resonance imaging. Magn. Reson. Med. 2005;53:1432–1440. doi: 10.1002/mrm.20508. [DOI] [PubMed] [Google Scholar]
- Kärger J. NMR self-diffusion studies in heterogeneous systems. Adv. Colloid Interf. Sci. 1985;23:129–148. [Google Scholar]
- Kurtzke J.F. Rating neurologic impairment in multiple sclerosis: an expanded disability status scale (EDSS) Neurology. 1983;33:1444–1452. doi: 10.1212/wnl.33.11.1444. [DOI] [PubMed] [Google Scholar]
- Mohammadi S., Moller H.E., Kugel H., Muller D.K., Deppe M. Correcting eddy current and motion effects by affine whole-brain registrations: evaluation of three-dimensional distortions and comparison with slicewise correction. Magn. Reson. Med. 2010;64:1047–1056. doi: 10.1002/mrm.22501. [DOI] [PubMed] [Google Scholar]
- Murphy R.K., Sun P., Xu J., Wang Y., Sullivan S., Gamble P., Wagner J., Wright N.N., Dorward I.G., Riew D., Santiago P., Kelly M.P., Trinkaus K., Ray W.Z., Song S.K. Magnetic resonance imaging biomarker of axon loss reflects cervical spondylotic myelopathy severity. Spine (Phila Pa 1976) 2015 doi: 10.1097/BRS.0000000000001337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J., Zackowski K., Chen M., Newsome S., Saidha S., Smith S.A., Diener-West M., Prince J., Jones C.K., Van Zijl P.C. Multiparametric MRI correlates of sensorimotor function in the spinal cord in multiple sclerosis. Mult. Scler. J. 2012:427–435. doi: 10.1177/1352458512456614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol S., Dubuc R., Gossard J.P. Dynamic sensorimotor interactions in locomotion. Physiol. Rev. 2006;86:89–154. doi: 10.1152/physrev.00028.2005. [DOI] [PubMed] [Google Scholar]
- Schneider T., Brownlee W., Zhang H., Ciccarelli O., Miller D., Wheeler-Kingshott C. 2014. Application of Multi-shell NODDI in Multiple Sclerosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setsompop K., Gagoski B.A., Polimeni J.R., Witzel T., Wedeen V.J., Wald L.L. Blipped-controlled aliasing in parallel imaging for simultaneous multislice echo planar imaging with reduced g-factor penalty. Magn. Reson. Med. 2012;67:1210–1224. doi: 10.1002/mrm.23097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S.K., Yoshino J., Le T.Q., Lin S.J., Sun S.W., Cross A.H., Armstrong R.C. Demyelination increases radial diffusivity in corpus callosum of mouse brain. NeuroImage. 2005;26:132–140. doi: 10.1016/j.neuroimage.2005.01.028. [DOI] [PubMed] [Google Scholar]
- Stanisz G.J., Szafer A., Wright G.A., Henkelman R.M. An analytical model of restricted diffusion in bovine optic nerve. Magn. Reson. Med. 1997;37:103–111. doi: 10.1002/mrm.1910370115. [DOI] [PubMed] [Google Scholar]
- Steven A.J., Zhuo J., Melhem E.R. Diffusion kurtosis imaging: an emerging technique for evaluating the microstructural environment of the brain. AJR Am. J. Roentgenol. 2014;202:W23–W33. doi: 10.2214/AJR.13.11365. [DOI] [PubMed] [Google Scholar]
- Taso M., Girard O.M., Duhamel G., Le Troter A., Feiweier T., Guye M., Ranjeva J.-P., Callot V. Tract-specific and age-related variations of the spinal cord microstructure: a multi-parametric MRI study using diffusion tensor imaging (DTI) and inhomogeneous magnetization transfer (ihMT) NMR Biomed. 2016;29:817–832. doi: 10.1002/nbm.3530. [DOI] [PubMed] [Google Scholar]
- Veraart J., Poot D.H.J., Van Hecke W., Blockx I., Van der Linden A., Verhoye M., Sijbers J. More accurate estimation of diffusion tensor parameters using diffusion kurtosis imaging. Magn. Reson. Med. 2011;65:138–145. doi: 10.1002/mrm.22603. [DOI] [PubMed] [Google Scholar]
- Veraart J., Sijbers J., Sunaert S., Leemans A., Jeurissen B. Weighted linear least squares estimation of diffusion MRI parameters: strengths, limitations, and pitfalls. NeuroImage. 2013;81:335–346. doi: 10.1016/j.neuroimage.2013.05.028. [DOI] [PubMed] [Google Scholar]
- Wen Q., Kelley D.A.C., Banerjee S., Lupo J.M., Chang S.M., Xu D., Hess C.P., Nelson S.J. Clinically feasible NODDI characterization of glioma using multiband EPI at 7 T. NeuroImage: Clinical. 2015;9:291–299. doi: 10.1016/j.nicl.2015.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilm B.J., Svensson J., Henning A., Pruessmann K.P., Boesiger P., Kollias S.S. Reduced field-of-view MRI using outer volume suppression for spinal cord diffusion imaging. Magn. Reson. Med. 2007;57:625–630. doi: 10.1002/mrm.21167. [DOI] [PubMed] [Google Scholar]
- Winston G.P., Micallef C., Symms M.R., Alexander D.C., Duncan J.S., Zhang H. Advanced diffusion imaging sequences could aid assessing patients with focal cortical dysplasia and epilepsy. Epilepsy Res. 2014;108:336–339. doi: 10.1016/j.eplepsyres.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Li H., Harkins K.D., Jiang X., Xie J., Kang H., Does M.D., Gore J.C. Mapping mean axon diameter and axonal volume fraction by MRI using temporal diffusion spectroscopy. NeuroImage. 2014;103:10–19. doi: 10.1016/j.neuroimage.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Schneider T., Wheeler-Kingshott C.A., Alexander D.C. NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. NeuroImage. 2012;61:1000–1016. doi: 10.1016/j.neuroimage.2012.03.072. [DOI] [PubMed] [Google Scholar]