Abstract
MdmX contains an intramolecular binding motif that mimics the binding of the p53 tumor suppressor. This intramolecular binding motif is connected to the p53 binding domain of MdmX by a conserved flexible linker that is 85 residues long. The sequence of this flexible linker has an identity of 51% based on multiple protein sequence alignments of 52 MdmX homologs. We used polymer statistics to estimate a global KD value for p53 binding to MdmX in the presence of the flexible linker and the intramolecular binding motif by assuming the flexible linker behaves as a wormlike chain. The global KD estimated from the wormlike chain modeling was nearly identical to the value measured using isothermal titration calorimetry. According to our calculations and measurements, the intramolecular binding motif reduces the apparent affinity of p53 for MdmX by a factor of 400. This study promotes a more quantitative understanding of the role that flexible linkers play in intramolecular binding and provides valuable information to further studies of cellular inhibition of the p53/MdmX interaction.
Main Text
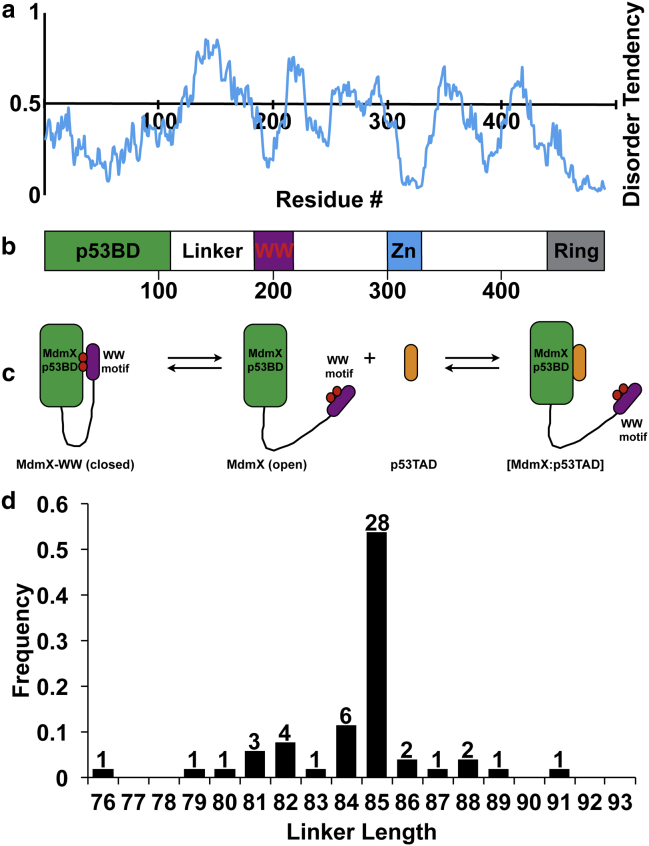
Mdm2 and MdmX (also known as Mdm4) are arguably the most important regulators of p53, being responsible for generating the dynamic characteristics of the p53 signaling pathway (1). Mdm2 is a classic p53 target gene and it was recently shown that p53 also induces MdmX transcription (2, 3). Both Mdm2 and MdmX form negative feedback loops that regulate p53 activity using different mechanisms and mouse models show that Mdm2 and MdmX are necessary for controlling p53 activity (4, 5, 6, 7, 8, 9). Mdm2 and MdmX contain N-terminal domains that bind tightly to residues 17–29 of the p53 transcriptional activation domain (p53TAD). MdmX also contains a short linear binding motif between residues 195–206 that resembles residues 17–29 of p53TAD. This motif interacts with the p53 binding domain (p53BD) of MdmX and interferes with p53 binding (10, 11). We recently showed that a short peptide containing MdmX residues 195–206 binds to the p53BD of MdmX with a KD = 8.3 ± 0.1 μM (11). Based on this previous work, we hypothesize the intramolecular binding motif at residues 195–206, referred to as the WW motif because it contains two tryptophan residues, is connected to the p53-binding domain of MdmX by a long flexible linker that controls the intramolecular binding affinity. This hypothesis is supported by the IUPred disorder prediction of human MdmX shown in Fig. 1 a (12). IUPred correctly predicts the p53 binding domain of MdmX is ordered (13, 14). After the p53 binding domain is a long segment from residues 112–181 predicted to be disordered, and a short segment from residues 182–211, which includes the WW motif, with disorder tendency values that dip below 0.5. Previous studies have shown that short protein segments with a disorder tendency <0.5 will often correspond to linear binding motifs (15, 16, 17). In the case of MdmX, the binding of this linear motif is intramolecular. It is also worth noting that ANCHOR correctly predicts the WW motif is a protein binding site (18).
Figure 1.
MdmX has a conserved flexible linker. (a) Plot showing IUPred disorder prediction of human MdmX. (b) Domain schematic of human MdmX showing position of p53 binding domain (p53BD), the linker, and the WW motif. The residue numbers in the disorder prediction are aligned with the domain schematic. (c) Model showing competition between the WW motif and p53TAD for the p53BD. (d) Histogram showing the distribution of lengths for the flexible linkers of 52 MdmX homologs based on a multiple protein sequence alignment that included 10 primates, 24 other mammals, 4 birds, 1 amphibian, 1 reptile, and 12 fish. The number of species in each bin is shown above the bars.
The disorder prediction shown in Fig. 1 a provides a useful global assessment of disorder propensity but it lacks the resolution to determine the physical boundaries of the MdmX linker at the single amino acid level. To determine the C-terminal boundary of the linker, we analyzed a recent study published by Fersht and colleagues, where NMR spectroscopy was used to characterize the interaction between the p53BD of MdmX (residues 1–111) and a 15N-labeled peptide containing the WW motif (residues 181–209) (10). In the 15N-HSQC spectrum of this peptide bound to the p53BD of MdmX, resonances for residues 195–204 disappear but the resonance for residue 194 is still detectable. It is not as strong as the resonance in the free peptide, but it is clearly visible and does not shift its position. Based on these observations, we conclude the C-terminal boundary of the linker is at residue 194. To determine the N-terminal boundary of the linker, we examined the structure of human MdmX (residues 23–111) bound to a p53 peptide (residues 15–29; PDB: 3DAB) (14). Residue 109 of MdmX is the last amino acid observed in this structure. This means that residues 110 and 111 are flexible and do not produce coherent scattering in the x-ray diffraction pattern. Based on this observation, starting the linker at residue 110 seems reasonable. Including residues 110 and 194 makes the linker 85 residues long. It is no great surprise that the last two residues in a crystal structure are not resolved, but as we show below, changing this length by ±4 residues has a minimal effect on the predicted strength of intramolecular binding.
Using the linker boundaries described above, we performed multiple protein sequence alignments on 52 MdmX homologs (Fig. S1). Based on the sequence identity matrix calculated from the alignments, the overall sequence identity is 51%. A histogram showing the length distribution of the linkers from the 52 homologs is presented in Fig. 1 d. A total of 48 of the 52 (92%) homologs have a length of 85 ± 4 residues. The shortest linker length is 76 residues (frog) and the longest is 91 residues (opossum). Based on this analysis, we conclude both the length and sequence of the linker is highly conserved in vertebrates.
Flexible linkers represent a functionally important class of intrinsically disordered regions. They can modulate the distance between multiple inter- or intramolecular protein-binding sites and this, in turn, can modulate binding affinity. In some cases, this behavior can be predicted by the wormlike chain (WLC) model initially proposed by Zhou and co-workers (19, 20, 21, 22, 23, 24). The WLC model describes semiflexible polymers like IDPs using two variables—a contour length (Lc) and a persistence length (Lp). For a fixed chain length, higher values of Lc and Lp lead to more extended conformations (20, 22). Lc is the length of a fully extended chain with a certain number of monomers. Lp is a measure of polymer stiffness and corresponds to the distance necessary for the direction of the chain to become uncorrelated. For our modeling, Lc = Nres × 3.8 Å, where Nres is the number of residues in the linker, and 3.8 Å is the distance per residue and Lp = 3 Å. These values provide a good approximation for the behavior of polypeptides (20). We used the WLC model to estimate the probability of end-to-end distances for the MdmX linker region and the effective concentration (Ceff) for the WW motif at the p53BD of MdmX (Supporting Material, Eqs. S1–S3). Ceff represents the concentration of one linker end at a fixed distance from the site of tethering. For a fixed distance, Ceff gradually increases with linker length up to a maximal value, and then decreases slowly for longer linkers (20, 22).
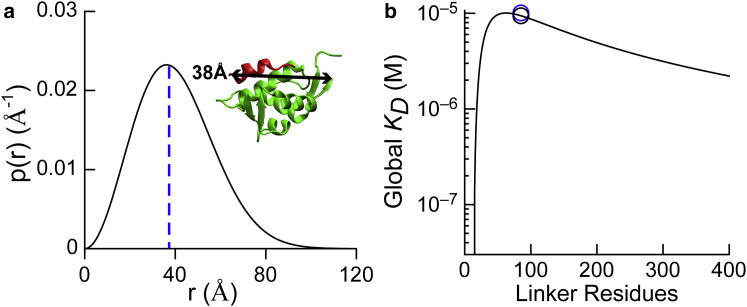
The probability distribution of end-to-end distances, p(r), for an 85-residue linker had a maximum at 36 Å (Fig. 2). If we assume a binding orientation for the WW motif that is similar to the p53TAD peptide shown in Fig. 2 a, 36 Å is very close to the observed separation of 38 Å between the end of the p53BD (residue 109) and the putative beginning of the WW motif (14). Because 92% of the homologs used to make the histogram in Fig. 1 d have linker lengths from 81 to 89 residues, we calculated p(r) for 81- and 89-residue linkers. The values are 35.5 and 37 Å, respectively, which are also very close to the experimentally measured distance of 38 Å.
Figure 2.
Modeling MdmX intramolecular interactions using polymer statistics. (a) Shown here is the probability distribution of end-to-end distances for the MdmX linker region (residues 110–194). The distribution peaks at 36 Å are given. The blue dashed line marks 38 Å, the measured distance between the end of the MdmX domain and the beginning of the WW motif considering the same binding orientation as p53TAD, measured from PDB: 3DAB. The ribbon model shows a structure from PDB: 3DAB. (b) Shown here is the predicted global KD as a function of linker length for a direct binding orientation model. Black circle: global KD predicted by the WLC model for an 85-residue linker; blue circle: experimentally measured global KD (see Fig. 3).
To estimate the effective concentration (Ceff) of the WW motif at the p53BD of MdmX, we used the most probable linker length of 85 residues and a separation distance between the end of the p53 binding domain and the beginning of the WW motif of 38 Å (Supporting Material; Eq. S3). Using these values, we calculated a Ceff value of 2.12 mM. This Ceff value, along with the previously determined bimolecular association constant for a short peptide corresponding to the WW motif binding to the p53BD of MdmX (11), was used to calculate the intramolecular association constant, KA′, and the fraction of the MdmX molecules that are in the closed conformation shown in Fig. 1 c (Supporting Material; Eqs. S4 and S5). According to these equilibrium calculations, the fraction of MdmX molecules in the closed conformation is 0.996, which corresponds to a KA′ value of 255. Using KA′, we calculated the global dissociation constant for the binding of p53TAD to a fragment of MdmX containing the p53BD, the linker, and the WW motif (residues 23–210) (Supporting Material; Eqs. S6 and S7). For this calculation, we considered the two equilibrium reactions illustrated in Fig. 1 c. The first equilibrium reaction is between the closed and open conformations where the WW motif is respectively bound and not bound to the p53BD. In the second equilibrium reaction, MdmX23–210 is in the open conformation and p53TAD is bound to the p53BD. The predicted global KD reached a maximum value for a linker length of 62 residues (Fig. 2 b). This is the length at which p53 binding is maximally compromised by the autoinhibitory effects of the WW motif binding to the p53BD. Using a linker length of 85 residues, we calculate a global KD = 9.4 ± 1.5 μM for p53TAD binding to MdmX23–210. The global KD values we estimated for 81 and 89 residue linkers are 9.6 and 9.3 μM, respectively. Our modeling suggests that the intramolecular binding affinity achieved by an 85 residue linker is close to the maximum and that the range of linker lengths observed for the homologs will all have a similar autoinhibitory effect on p53 binding.
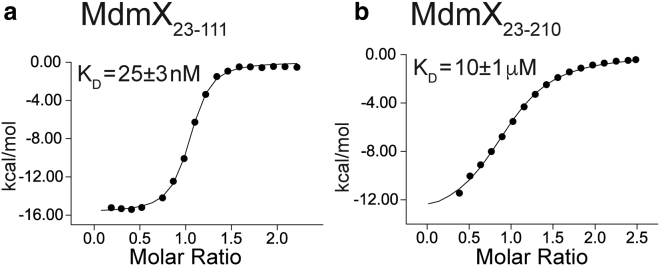
Fig. 3 shows the results from isothermal titration calorimetry (ITC) experiments for two fragments of MdmX binding to a fragment of the p53 transactivation domain that contains residues 1–73 (p53TAD). Fig. 3 a shows the fitted ITC data for p53TAD binding to MdmX23–111. We observe a KD = 25 ± 3 nM. p53TAD binds more tightly to MdmX23–111 than the short peptide corresponding to p53 residues 17–28 used for previous structural studies (25, 26, 27). This tighter binding is due to additional interactions between MdmX23–111 and p53 residues 29–32 (10). Fig. 3 b shows the KD for p53TAD binding to MdmX23–210 dramatically increases to a value of 10 ± 1 μM due to the addition of the WW motif. Fig. S2 shows ITC data for longer MdmX fragments that lack the WW motif. As expected, the WW motif interferes with the binding of p53TAD to MdmX, reducing the apparent binding affinity by 400-fold. The experimentally obtained KD value is in very close agreement with the KD of 9.4 ± 1.5 μM predicted by the Ceff values obtained using the WLC model. The agreement between the experimentally measured KD and the value predicted using the WLC model suggests that the MdmX linker behaves as a flexible polymer that enhances the intramolecular binding of the WW motif. This intramolecular binding competes with the binding of p53, reducing its apparent affinity for MdmX by two orders of magnitude. The fact the 92% of the homologs we analyzed have a linker length that is within four amino acids of the human homolog suggests the linker length and the intramolecular affinity enhancement are conserved. These results have important implications for our understanding of p53 regulation and help to explain previous results from the Chen group showing that CK1α copurifies with MdmX by binding to the acidic domain and stimulating MdmX-p53 binding (28). It is not clear exactly where CK1α binds to MdmX, but we hypothesize it is positioned to interfere with intramolecular binding of the WW motif, allowing p53TAD to access the p53BD of MdmX (11). Our work also suggests that the MdmX linker has evolved to promote optimal positioning of the WW motif for binding to the p53BD. Finally, we think our work adds support for the use of WLC modeling of disordered regions that act mainly as flexible tethers (21, 22, 23, 24). To improve the accuracy of this modeling, additional variables, such as sequence patterning (see CIDER analysis in Fig. S3) and tethering interactions, must ultimately be considered (29, 30).
Figure 3.
Plots showing p53TAD binding to fragments of MdmX with and without the flexible linker and WW motif. Black circles show enthalpy per mole of injectant, measured using isothermal titration calorimetry, plotted as a function [p53TAD]/[MdmX]. Black lines show the fit to the data using a single site binding model. (a) Shown here is p53TAD binding to MdmX23–111. (b) Shown here is p53TAD binding to MdmX23–210.
Author Contributions
W.B. performed experiments, analyzed data, and wrote the manuscript. L.C. generated expression plasmids. J.C. designed experiments and edited the manuscript. L.B.C. performed polymer statistics calculations and edited the manuscript. G.W.D. managed the project, designed experiments, wrote the manuscript, and edited the manuscript.
Acknowledgments
This work is supported in part by National Institutes of Health (NIH) grants Nos. GM115556 and CA141244 to G.W.D., NIH grants Nos. CA109636, CA141244, and CA186917 and Florida Department of Health grant No. 4BB14 to J.C., and Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT) grant No. PICT-2013-1895 to L.B.C. The H. Lee Moffitt Cancer Center & Research Institute is a National Cancer Institute-designated Comprehensive Cancer Center through grant No. P30-CA076292.
Editor: Rohit Pappu.
Footnotes
Supporting Materials and Methods and three figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30433-2.
Contributor Information
Lucía B. Chemes, Email: lchemes@leloir.org.ar.
Gary W. Daughdrill, Email: gdaughdrill@usf.edu.
Supporting Material
References
- 1.Wade M., Li Y.C., Wahl G.M. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat. Rev. Cancer. 2013;13:83–96. doi: 10.1038/nrc3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li B., Cheng Q., Chen J. p53 inactivation by MDM2 and MDMX negative feedback loops in testicular germ cell tumors. Cell Cycle. 2010;9:1411–1420. doi: 10.4161/cc.9.7.11255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phillips A., Teunisse A., Jochemsen A.G. HDMX-L is expressed from a functional p53-responsive promoter in the first intron of the HDMX gene and participates in an autoregulatory feedback loop to control p53 activity. J. Biol. Chem. 2010;285:29111–29127. doi: 10.1074/jbc.M110.129726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Finch R.A., Donoviel D.B., Zhang N. Mdmx is a negative regulator of p53 activity in vivo. Cancer Res. 2002;62:3221–3225. [PubMed] [Google Scholar]
- 5.Grier J.D., Xiong S., Lozano G. Tissue-specific differences of p53 inhibition by Mdm2 and Mdm4. Mol. Cell. Biol. 2006;26:192–198. doi: 10.1128/MCB.26.1.192-198.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones S.N., Roe A.E., Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 7.Migliorini D., Lazzerini Denchi E., Marine J.C. Mdm4 (Mdmx) regulates p53-induced growth arrest and neuronal cell death during early embryonic mouse development. Mol. Cell. Biol. 2002;22:5527–5538. doi: 10.1128/MCB.22.15.5527-5538.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Montes de Oca Luna R., Wagner D.S., Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 9.Parant J.M., Reinke V., Lozano G. Organization, expression, and localization of the murine mdmx gene and pseudogene. Gene. 2001;270:277–283. doi: 10.1016/s0378-1119(01)00432-2. [DOI] [PubMed] [Google Scholar]
- 10.Bista M., Petrovich M., Fersht A.R. MDMX contains an autoinhibitory sequence element. Proc. Natl. Acad. Sci. USA. 2013;110:17814–17819. doi: 10.1073/pnas.1317398110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen L., Borcherds W., Chen J. Autoinhibition of MDMX by intramolecular p53 mimicry. Proc. Natl. Acad. Sci. USA. 2015;112:4624–4629. doi: 10.1073/pnas.1420833112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dosztányi Z., Csizmók V., Simon I. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J. Mol. Biol. 2005;347:827–839. doi: 10.1016/j.jmb.2005.01.071. [DOI] [PubMed] [Google Scholar]
- 13.Sanchez M.C., Renshaw J.G., Vogtherr M. MDM4 binds ligands via a mechanism in which disordered regions become structured. FEBS Lett. 2010;584:3035–3041. doi: 10.1016/j.febslet.2010.05.058. [DOI] [PubMed] [Google Scholar]
- 14.Popowicz G.M., Czarna A., Holak T.A. Structure of the human Mdmx protein bound to the p53 tumor suppressor transactivation domain. Cell Cycle. 2008;7:2441–2443. doi: 10.4161/cc.6365. [DOI] [PubMed] [Google Scholar]
- 15.Oldfield C.J., Cheng Y., Dunker A.K. Coupled folding and binding with α-helix-forming molecular recognition elements. Biochemistry. 2005;44:12454–12470. doi: 10.1021/bi050736e. [DOI] [PubMed] [Google Scholar]
- 16.Davey N.E., Van Roey K., Gibson T.J. Attributes of short linear motifs. Mol. Biosyst. 2012;8:268–281. doi: 10.1039/c1mb05231d. [DOI] [PubMed] [Google Scholar]
- 17.Mészáros B., Dosztányi Z., Simon I. Disordered binding regions and linear motifs—bridging the gap between two models of molecular recognition. PLoS One. 2012;7:e46829. doi: 10.1371/journal.pone.0046829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mészáros B., Simon I., Dosztányi Z. Prediction of protein binding regions in disordered proteins. PLoS Comput. Biol. 2009;5:e1000376. doi: 10.1371/journal.pcbi.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou H.X. Loops in proteins can be modeled as worm-like chains. J. Phys. Chem. B. 2001;105:6763–6766. [Google Scholar]
- 20.Zhou H.X. Polymer models of protein stability, folding, and interactions. Biochemistry. 2004;43:2141–2154. doi: 10.1021/bi036269n. [DOI] [PubMed] [Google Scholar]
- 21.Zhou H.X. Quantitative relation between intermolecular and intramolecular binding of pro-rich peptides to SH3 domains. Biophys. J. 2006;91:3170–3181. doi: 10.1529/biophysj.106.090258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bertagna A., Toptygin D., Barrick D. The effects of conformational heterogeneity on the binding of the Notch intracellular domain to effector proteins: a case of biologically tuned disorder. Biochem. Soc. Trans. 2008;36:157–166. doi: 10.1042/BST0360157. [DOI] [PubMed] [Google Scholar]
- 23.Krishnamurthy V.M., Semetey V., Whitesides G.M. Dependence of effective molarity on linker length for an intramolecular protein-ligand system. J. Am. Chem. Soc. 2007;129:1312–1320. doi: 10.1021/ja066780e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Valen D., Haataja M., Phillips R. Biochemistry on a leash: the roles of tether length and geometry in signal integration proteins. Biophys. J. 2009;96:1275–1292. doi: 10.1016/j.bpj.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pazgier M., Liu M., Lu W. Structural basis for high-affinity peptide inhibition of p53 interactions with MDM2 and MDMX. Proc. Natl. Acad. Sci. USA. 2009;106:4665–4670. doi: 10.1073/pnas.0900947106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phan J., Li Z., Chen J. Structure-based design of high affinity peptides inhibiting the interaction of p53 with MDM2 and MDMX. J. Biol. Chem. 2010;285:2174–2183. doi: 10.1074/jbc.M109.073056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Popowicz G.M., Dömling A., Holak T.A. The structure-based design of Mdm2/Mdmx-p53 inhibitors gets serious. Angew. Chem. Int. Ed. Engl. 2011;50:2680–2688. doi: 10.1002/anie.201003863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen L., Li C., Chen J. Regulation of p53-MDMX interaction by casein kinase 1α. Mol. Cell. Biol. 2005;25:6509–6520. doi: 10.1128/MCB.25.15.6509-6520.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Das R.K., Pappu R.V. Conformations of intrinsically disordered proteins are influenced by linear sequence distributions of oppositely charged residues. Proc. Natl. Acad. Sci. USA. 2013;110:13392–13397. doi: 10.1073/pnas.1304749110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sherry K.P., Johnson S.E., Barrick D. Effects of linker length and transient secondary structure elements in the intrinsically disordered Notch RAM region on Notch signaling. J. Mol. Biol. 2015;427:3587–3597. doi: 10.1016/j.jmb.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.