Abstract
A plethora of evidence suggests that different types of DNA quadruplexes are widely present in the genome of all organisms. The existence of a growing number of proteins that selectively bind and/or process these structures underscores their biological relevance. Moreover, G-quadruplex DNA has been implicated in the alignment of four sister chromatids by forming parallel guanine quadruplexes during meiosis; however, the underlying mechanism is not well defined. Here we show that a G/C-rich motif associated with a meiosis-specific DNA double-strand break (DSB) in Saccharomyces cerevisiae folds into G-quadruplex, and the C-rich sequence complementary to the G-rich sequence forms an i-motif. The presence of G-quadruplex or i-motif structures upstream of the green fluorescent protein-coding sequence markedly reduces the levels of gfp mRNA expression in S. cerevisiae cells, with a concomitant decrease in green fluorescent protein abundance, and blocks primer extension by DNA polymerase, thereby demonstrating the functional significance of these structures. Surprisingly, although S. cerevisiae Hop1, a component of synaptonemal complex axial/lateral elements, exhibits strong affinity to G-quadruplex DNA, it displays a much weaker affinity for the i-motif structure. However, the Hop1 C-terminal but not the N-terminal domain possesses strong i-motif binding activity, implying that the C-terminal domain has a distinct substrate specificity. Additionally, we found that Hop1 promotes intermolecular pairing between G/C-rich DNA segments associated with a meiosis-specific DSB site. Our results support the idea that the G/C-rich motifs associated with meiosis-specific DSBs fold into intramolecular G-quadruplex and i-motif structures, both in vitro and in vivo, thus revealing an important link between non-B form DNA structures and Hop1 in meiotic chromosome synapsis and recombination.
Introduction
Multiple lines of evidence suggest that repetitive DNA motifs in the genome fold into a variety of non-B DNA structures, which, in turn, affect many essential cellular processes including gene expression, DNA replication, and recombination (1, 2, 3). Defects in one or more of these processes can cause mutations and, consequently, lead to genome instability (1, 2). Several different types of non-B DNA structures have been characterized with respect to both their structures and properties. Among them, DNA quadruplex structures have attracted great attention because of their potential physiological significance (1, 2, 3). G-quadruplexes are secondary structures formed in nucleic acids containing tandem arrays of G residues in which the four guanine residues associate via Hoogsteen hydrogen bonding to form a square planar structure (1, 2, 3). Although the existence of G-quadruplexes in vivo is being debated, there is compelling evidence for the occurrence of G4 DNA motifs in nuclear and mitochondrial genomes (4, 5, 6, 7, 8, 9, 10). Furthermore, the presence of a number of proteins/enzymes that selectively bind and/or process these structures is consistent with their possible functional significance (11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21). Such observations have led to the identification of a wide range of natural and synthetic ligands specific for G4 DNA, which, in turn, interfere with cellular processes such as DNA replication, transcription, RNA processing, and translation (22, 23, 24, 25, 26, 27, 28, 29, 30). Together, these studies support the idea that DNA/RNA quadruplex structures regulate processes related to many important biological functions.
Rapid advances in the fields of genetics, bioinformatics, and computational biology have revealed the widespread occurrence of G-quadruplex-forming motifs (hereinafter referred to as “GQs”) in the genomes of different organisms, and their impact on the normal and pathological aspects of nucleic acid metabolism (1, 2, 3, 30, 31). In general, GQs occur more frequently in eukaryotic genomes than in prokaryotes (32, 33). In prokaryotes, GQs are more prevalent in promoter motifs of genes regulating transcription, production of secondary metabolites, and signal transduction (33). Notably, DNA recombination between homeologous pilin genes of Neisseria gonorrheae, resulting in the generation of antigenic variations, is dependent on the GQ structure (34). Bioinformatics analyses have revealed that the human genome contains potential GQ-forming sequences that are implicated in the regulation of gene expression and several human diseases. Estimates for the number of GQ-forming sequences in the human genome range from 35,000 to 55,000 (31, 35, 36). Some examples of their occurrence in regions include immunoglobulin heavy-chain switch motifs (37, 38), telomeric DNA (39, 40), rDNA (41), promoter motifs in wild-type (WT) genes (29, 35, 42), and oncogenes (26, 43, 44, 45). Likewise, a genomewide survey for conserved GQs in Saccharomyces cerevisiae has revealed ∼1400 GQ-forming sequences (6, 46). Interestingly, these motifs are enriched in the promoter regions and rDNA, and also at mitotic and meiosis-specific double-strand break sites (DSBs) (6, 47, 48).
Several studies have reported that the C-rich strand complementary to the G-rich DNA sequence can fold into a distinct quadruplex structure, called the “intercalated motif” (i-motif), the stability of which is strongly dependent on pH (49, 50, 51, 52, 53). Further, i-motif structures were found in C-rich sequences of telomeres (50), centromeres (54), and in the promoters of oncogenes including retinoblastoma (55), BCL2 (56, 57), c-Myc (58, 59), and c-kit (60, 61). Additionally, recent studies have revealed that both GQ DNA and i-motifs are formed simultaneously within the same double-stranded tract in a variety of DNA templates (62, 63). In support of their biological relevance, various studies have found that the i-motif plays a regulatory role in gene expression (64, 65, 66). Indeed, a number of proteins have been shown to bind cytosine-rich strands, possibly i-motif structures. Examples include hnRNP A1 (67, 68), hnRNP K (69, 70), hnRNP LL (56), the Trypanosoma brucei ST-1 protein (71), and the rat nuclear protein qTBP42 (72). On the basis of the reports described above, it is apparent that i-motif structures can serve as attractive drug targets for anti-cancer therapy (56, 73, 74).
Ample evidence suggests that the meiotic recombination in S. cerevisiae is initiated by DNA DSBs, and the genetic exchange between the paternal and maternal genomes in meiosis I is mediated by the synaptonemal complex (SC) (75, 76). The SC is an electron dense, meiosis-specific tripartite proteinaceous structure formed between paired homologous chromosomes (76). Multiple studies suggest that meiotic recombination occurs more frequently in narrow motifs of 1–2 kb called “recombination hotspots”, where the frequency of recombination events is higher than elsewhere in the genome (75, 77). Genomewide mapping and analyses of Spo11-catalyzed DSBs in S. cerevisiae indicate a strong positive correlation between DSBs and high G/C-rich motifs, including when the latter are transposed to DSB-poor genomic locations (6, 48, 78, 79, 80). A similar relationship also exists between high G/C-rich motifs and elevated recombination rates in humans (47, 81). Strikingly, in humans and chimpanzees, the hotspot midpoints occur within poly-pu/py-rich high G/C-rich sequences, indicating that they might be involved in determining hotspot activity (82). Although several characteristics such as locally open chromatin structure and flanking sequences several kilobases away possess the recombination-stimulating potential, emerging evidence supports a significant correlation between GQs and meiosis-specific DSB sites (6, 48, 82). However, despite the genetic correlation, very little is known about the structural features and functional significance of DNA quadruplex motifs and, even less is known about the identity of meiosis-specific proteins that interact with such motifs.
Here, we reveal that the G/C-rich motif (coordinates 1242544–1242567) associated with meiosis-specific DSB formation on chromosome IV in S. cerevisiae (6) (hereafter referred to as “meiosis-specific DSB on chromosome IV”) folds into mutually exclusive GQ DNA and i-motif structures. Furthermore, the presence of these quadruplex structures upstream of the green fluorescent protein (GFP)-coding sequence markedly reduces the levels of gfp mRNA expression in S. cerevisiae cells, with a concomitant decrease in GFP abundance, and blocks primer extension by DNA polymerase, indicating the in vivo functional relevance of these structures. The S. cerevisiae HOP1, a meiosis-specific gene, encodes a major component of the axial/lateral elements of SC, and plays a key role in the synapsis of homologous chromosomes as hop1/hop1 diploids fail to form viable spores (83). Our previous studies revealed that full-length Hop1 is a structure-specific DNA-binding protein and promotes the pairing of synthetic double-stranded DNA helices containing arrays of G-residues (13, 84). Here, we show that full-length Hop1 promotes intermolecular pairing between the G/C-rich DNA associated with meiosis-specific DSB sites. Strikingly, the Hop1 C-terminal domain but not the N-terminal domain exhibit strong i-motif binding activity, implying that the C-terminal domain has a distinct substrate specificity. These results are consistent with the idea that G-quadruplex motifs could mediate the initial pairing between homologous chromosomes that is essential for progression through meiotic prophase I. Furthermore, these findings extend Hop1 function from being just a structural component of SC to having active roles in meiotic chromosome synapsis and recombination.
Materials and Methods
Biochemicals, bacterial strains, enzymes, and DNA oligonucleotides
Fine chemicals were purchased from Wipro GE Healthcare (Bangalore, India), and Sigma-Aldrich (St. Louis, MO). Restriction endonucleases, T4 DNA ligase, and T4 polynucleotide kinase were purchased from New England Biolabs (Ipswich, MA) and Thermo Fisher Scientific (Waltham, MA). All the reagents used were of an analytical grade. Oligonucleotides (ODNs) were synthesized by Sigma-Genosys (Sigma-Aldrich). The [γ-32P]ATP was purchased from the Bhabha Atomic Research Centre (Mumbai, India). Escherichia coli expression strains RIL and BL21∗ and the expression vectors (pET-22b and pET-28a) were purchased from Novagen (Madison, WI). The chromatography media, Ni2+-NTA, SP, and Q Sepharose, were purchased from Qiagen, (Germantown, MD), and Sigma-Aldrich (St. Louis, MO). Fast performance liquid chromatography columns were purchased from Wipro GE Healthcare.
DNA substrates
The ODNs used in this study are listed in Table S1. The ODNs were labeled at the 5′ end using [γ-32P]ATP and T4 polynucleotide kinase (85). Briefly, stoichiometric amounts of purified ODNs were annealed by incubation in 100 μL of 0.3 M sodium citrate buffer (pH 7) containing 3 M NaCl at 95°C, and then by slow cooling to 4°C over a period of 2 h. For an in vitro assay for DNA pairing, the top strand of the duplex DNA was labeled at the 5′ end using [γ-32P]ATP and T4 polynucleotide kinase. The labeled strand was annealed to an equimolar amount of an unlabeled complementary strand. To prepare G4 DNA, the G-rich ODN was labeled at the 5′ end using [γ-32P]ATP and T4 polynucleotide kinase. The labeled ODN was heated at 95°C in 120 mM KCl containing a water-salt solution for 5 min and then cooled to 37°C. Similarly, to prepare i-motif DNA, the C-rich labeled ODN was heated at 95°C in a 50 mM sodium phosphate buffer (pH 5.5) for 5 min and then cooled to 37°C. In each case, the mixtures were subjected to electrophoresis on an 8% (w/v) polyacrylamide gel in a 45 mM Tris-borate buffer (pH 8.3) containing 1 mM EDTA at 10 V/cm for 5 h to 12 h. The bands corresponding to the specific substrates were excised from the gel and eluted into TE buffer (10 mM Tris-HCl (pH 7.5), 1 mM EDTA). The concentrations of DNA substrates were determined by absorbance at 260 nm using the molar extinction coefficient (ε260).
Circular dichroism spectroscopy
Circular dichroism (CD) experiments were conducted on a model No. 810 spectropolarimeter (JASCO, Easton, MD) at a scan speed of 50 nm/min with a response time of 1 s using a quartz cell of a 10-mm optical path length over a wavelength range of 180–300 nm for G4 DNA, and 220–320 nm for the i-motif-forming ODNs. Before the experiment, reaction mixtures containing G-quadruplex-forming sequences were heated in the absence and presence of 120 mM KCl at 95°C for 5 min, and then air cooled to room temperature. An assay was performed in a buffer containing 5 μM of the indicated ODN in 10 mM Tris-HCl (pH 7.5), 0.1 mM EDTA. Similarly, in the case of i-motif-forming DNA, 5 μM C-rich WT or mutant ODNs were heated in a 50 mM sodium phosphate buffer (at pH 4.5, 5.5, 6.5, or 7.5) at 95°C for 5 min, and then air cooled to room temperature. To determine the TM of i-motif DNA, molar ellipticity values were obtained at 286 nm in the temperature range of 20–90°C. The CD spectrum obtained for the buffer alone was subtracted from the spectra of the sample.
NMR spectroscopy
For the NMR experiments, 1 mM ODN d(5′-TTCCCCTCCCCTTCCCCTCCCCTT-3′) was prepared in a 50 mM sodium cacodylate buffer (pH 4.5) containing 10% D2O for locking purposes. The one-dimensional (1D) spectra were recorded at 5°C and 27°C on an 800-MHz AV II spectrometer (Bruker, Billerica, MA) equipped with a cryo-probe. Temperature variation studies were carried out on an AVANCE 500 MHz spectrometer (Bruker) over a temperature range of 5°C to 47°C. The two-dimensional (2D) NMR experiments, TOCSY and NOESY, were carried out on an 800-MHz spectrometer (Bruker). The NOESY spectra were acquired at two mixing times of 200 ms and 250 ms, and at two temperatures, i.e., 5°C and 27°C. Water suppression was achieved by using the excitation sculpting pulse sequence in 1D as well as in 2D experiments. A relaxation time of 1 s was used. A quantity of 4096 points in the t2 dimension and 512 increments in the t1 dimension were used for the 2D TOCSY and NOESY. The DNA sample was subjected to 10 times dilution (with plain water) and the 1D spectrum was recorded again at 27°C. The 2D data were processed using the software TopSpin 2.1 (Bruker). The t1 dimension was zero filled to 2048 data points while processing. Apodization in both dimensions was achieved using a sine-bell with a phase shift of 90°. All the spectra were referenced externally with respect to trimethylsilyl propanoic acid dissolved in D2O. All the data were recorded for various temperatures as indicated on the plots.
Construction of His-tagged Hop1 N-terminal expression plasmid
The nucleotide sequence corresponding to the N-terminal HORMA domain (58–750 bp) was PCR amplified from plasmid pNH54-9 (86). The PCR product was purified from the agarose gel using a gel extraction kit (Qiagen). The PCR product was digested by NdeI and XhoI and directionally cloned into the expression pET-22b vector. The resulting recombinant plasmid was designated “pHORMA”. The identity of the recombinant construct was determined by restriction digestion and DNA sequencing.
DNA binding assays
The reaction mixtures (20 μL) contained 10 mM Tris-HCl (pH 7.5), 0.25 mM ZnCl2 and 0.5 nM of a specified 32P-labeled DNA substrate and increasing concentrations of the specified protein. In the case of DNA binding assays with G4 DNA, the reaction mixtures also contained 50 mM KCl. The reaction mixtures were incubated at 30°C for 30 min. Two microliters of a gel loading dye (0.25% xylene cyanol, 0.25% bromophenol blue in 20% glycerol) were added to stop the reaction. The reaction mixtures were electrophoresed on a 6% native polyacrylamide gel in a 44.5 mM Tris-borate buffer (pH 8.3) containing 1 mM EDTA at 10 V/cm at 4°C for 3 h. The gels were dried and exposed to a phosphorimager screen (Typhoon FLA-9000; GE Healthcare), and the bands were visualized using the software supplied by the manufacturer. The data were quantified using the software UVIbandmap (Uvitec Cambridge; http://www.uvitec.co.uk/) and plotted in the software GraphPad Prism (GraphPad Software, https://www.graphpad.com/).
DNA pairing assays
The reaction mixtures (20 μL) contained 2 nM of the specified 32P-labeled duplex DNA substrate comprising a G/C-rich sequence, 10 mM Tris-HCl (pH 7.5), and the indicated concentrations of Hop1 or Rad17. After incubation at 30°C for 30 min, these were deproteinized by the addition of proteinase K (0.2 mg/mL) and SDS (0.2%) and were further incubated at 37°C for 20 min. The reaction was terminated by the addition of 2 μL of a gel loading dye (0.25% xylene cyanol, 0.25% bromophenol blue in 20% glycerol). The reaction mixtures were subjected to electrophoresis on a 10% native polyacrylamide gel (PAGE) in a 44.5 mM Tris-borate buffer (pH 8.3) containing 1 mM EDTA at 10 V/cm at 4°C for 5 h. The gels were dried and visualized by a Typhoon FLA-9000 phosphorimager. The data was quantified using the software UVIbandmap and plotted in GraphPad Prism.
Taq polymerase stop assay
The polymerase stop assay was carried out as described in Han et al. (87). Briefly, the annealing reactions (20 μL) contained 10 mM Tris-HCl (pH 8), 10 nM of an ssDNA template bearing G-quadruplex (WT), or the corresponding mutant template, 15 nM of a 32P-labeled primer, and specified concentrations of KCl. The reaction mixture was first heated at 95°C for 5 min, and followed by slow cooling at room temperature over a period of 24 h. A primer extension reaction was performed by mixing annealed substrates with an assay buffer (2 μL) containing 5 mM MgCl2, 1.5 mg/mL BSA, and 0.2 mM dNTPs. After incubation with Taq DNA polymerase for 30 min at 37°C, the reaction was stopped by the addition of a loading dye (95% formamide, 10 mM EDTA, 10 mM NaOH, 0.1% xylene cyanol, 0.1% bromophenol blue). The samples were separated on 12% denaturing PAGE. The gels were dried and exposed to a phosphorimager screen (Typhoon FLA-9000), and the bands were visualized using the software supplied by the manufacturer.
DNA polymerase stop assay with i-motif and its mutant templates
The annealing mixtures (19 μL) contained the i-motif template or its mutant template (10 nM), a 32P-labeled primer (15 nM), and 5 mM MgCl2 in a 50 mM sodium phosphate buffer of pH 4.5, 5.5, 6.5, or 7.5. The reaction mixtures were heated at 95°C for 5 min and then allowed to cool slowly at room temperature before the assay. The annealing mixture was mixed with a reaction buffer (1 μL) containing 1.5 mg/mL bovine serum albumin, 0.2 mM dNTPs, and 0.5 U of KOD-Plus DNA polymerase (Toyobo, Osaka, Japan). After incubation for 30 min at 37°C, the reaction was stopped by the addition of a loading dye (95% formamide, 10 mM EDTA, 10 mM NaOH, 0.1% xylene cyanol, 0.1% bromophenol blue). The samples were separated on 12% denaturing PAGE. The gels were dried and exposed to a phosphorimager screen (Typhoon FLA-9000), and the bands were visualized using the software supplied by the manufacturer.
Construction of GFP reporter plasmids
The gene sequence corresponding to the GFP was amplified by PCR using gene-specific primers. The PCR product was gel purified and digested with EcoRI and XhoI. The PCR product containing the GFP cassette was inserted into a centromeric plasmid pRS416, in which the expression of GFP is under the control of a TEF promoter. The resulting plasmid was designated “pGFP”. To clone GQ or i-motif-forming sequences upstream of the GFP coding sequence, the pGFP was digested with BamHI and EcoRI. The double-stranded DNA inserts (Table S2) containing GQ or i-motif-forming sequences were directionally cloned into the pGFP vector. Plasmids with a G-rich sequence or a C-rich sequence in the sense strand were designated “G-plasmid” and “C-plasmid”, respectively. Similarly, mutant plasmids were generated using sequences that had G-T or C-T substitutions. The names of the constructs used in this study are listed in Table S3.
Confocal microscopy
The recombinant plasmids listed in Table S3 were transformed into the S. cerevisiae strain W303 and selected on Ura− plates. One colony from the transformation plate was inoculated into the SC-Ura− broth. Aliquots of 2 mL from the exponentially growing cultures were centrifuged and washed three times with 1× phosphate-buffered saline (PBS) and resuspended in 200 μL PBS. Aliquots of 10 μL were spotted on a glass slide. The cells were imaged using a confocal microscope (Carl Zeiss, Oberkochen, Germany).
GFP measurement assay
S. cerevisiae W303 cells were transformed with the specified recombinant GFP construct and selected on Ura− plates. The transformation efficiency of WT constructs was similar to those of mutant and control plasmids (∼3.9 × 105 to 4.2 × 105 CFU/μg plasmid DNA). One colony from the transformation plate was inoculated into the SC-Ura− broth. Aliquots of 2 mL from the exponentially growing cultures were centrifuged and washed three times with 1× PBS. The GFP levels were determined with an Infinite 200 PRO Multimode Reader (Tecan, Männedorf, Switzerland) with serially diluted cells (0.2, 0.4, and 0.6 OD) in a 96-well black plate.
Results
The G-rich ssDNA from S. cerevisiae meiosis-specific DSB folds into intramolecular G-quadruplex DNA
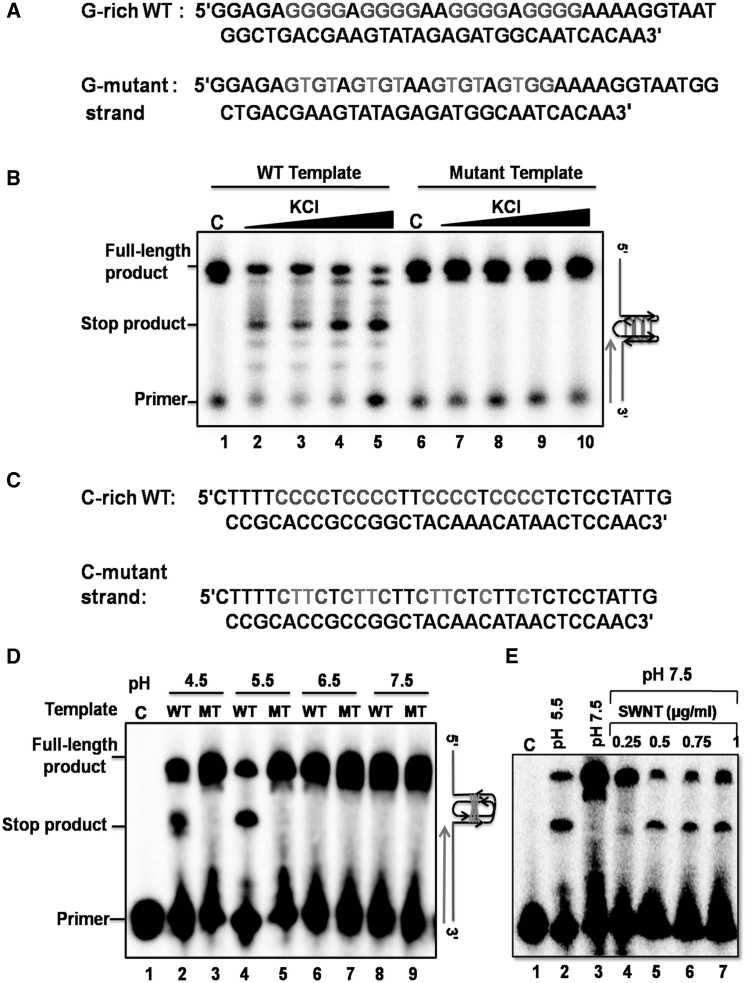
In this study, to determine the structure and functional significance of the G/C-rich motif associated with a meiosis-specific DSB in S. cerevisiae (6, 88), we chose a meiosis-specific G/C-rich motif on chromosome IV (coordinates 1242544–1242567) (6). We designed a 24-mer ODN corresponding to the motif above, which contained four repeats of four contiguous guanine residues. We also designed a modified ODN of the same length bearing G to T base substitutions (Fig. 1 A). Similarly, we used a complementary 24-mer ODN containing four repeats of four contiguous cytosine residues and its corresponding modified 24-mer ODN containing the C to T base substitutions (Fig. 2 A). An oligo(dT) homopolymer of identical length served as an additional negative control for G-quadruplex and i-motif formation.
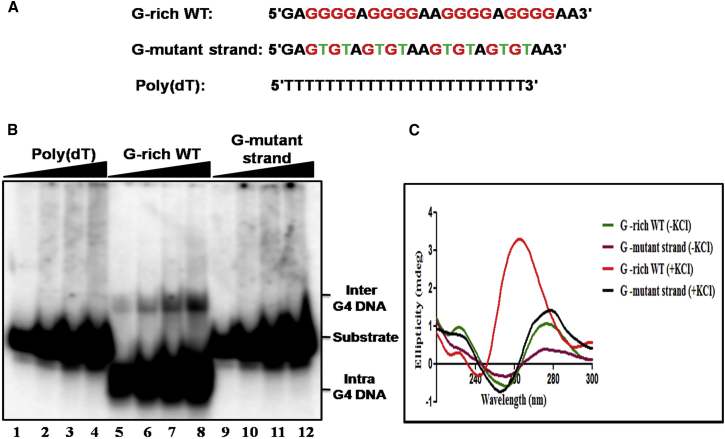
Figure 1.
The G-rich motif associated with a meiosis-specific DSB on chromosome IV of S. cerevisiae folds into a G-quadruplex structure. (A) Given here are nucleotide sequences of G-rich WT, G-MT, and poly (dT). The guanine residues are highlighted in red. The G to T substitution in the mutant strand is denoted by green. (B) Given here is the EMSA analysis of the G-quadruplex structures formed by increasing concentrations of a 32P-labeled G-rich WT, G-mutant and poly(dT) ODNs in the presence of 120 mM KCl. The solid triangle on top of the gel image denotes increasing concentrations of the indicated ODN. Lanes 1–4 contained 50, 100, 150, and 200 nM poly(dT), lanes 5–8 contain G-rich WT, and lanes 9–12 G-mutant ODN, respectively. The positions of inter- and intramolecular G-quadruplex and the substrate is indicated on the right-hand side of the image. (C) Effect of KCl on the formation of G-quadruplex DNA. The CD spectral tracings of the indicated ODNs were obtained in the absence or presence of 120 mM KCl.
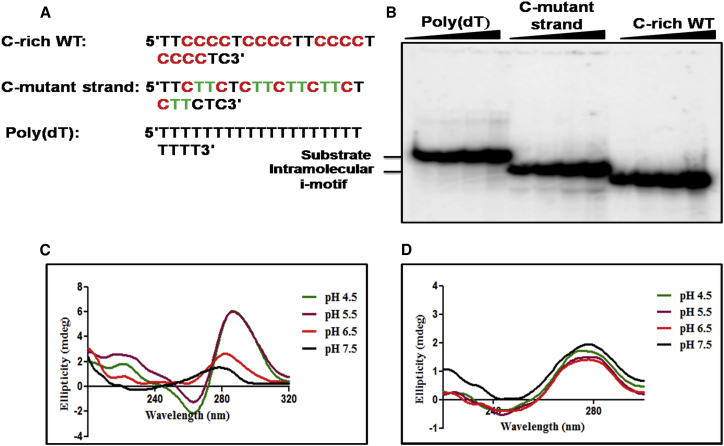
Figure 2.
The C-rich sequence associated with a meiosis-specific DSB on chromosome IV of S. cerevisiae folds into an intramolecular i-motif. (A) Shown here are the nucleotide sequences of C-rich WT, C-MT, and poly (dT). The C-residues are highlighted in red. The C to T substitution in the mutant sequence is denoted by green. (B) Shown here is the EMSA analysis of i-motif structures formed by increasing concentrations of a 32P-labeled poly(dT), C-mutant, and C-rich WT ODNs in a 50 mM sodium phosphate buffer (pH 5.5). The solid triangle on top of the gel image denotes increasing concentrations of the indicated ODN. Lanes 1–4 contain 50, 100, 150, and 200 nM poly(dT), lanes 5–8 contain 50, 100, 150, and 200 nM C-mutant, and lanes 9–12 contain 50, 100, 150, and 200 nM C-rich WT ODN, respectively. (C) Shown here is the effect of pH on the formation of i-motif by C-rich WT ODN. (D) Shown here is the effect of pH on the formation of i-motif by C-mutant ODN.
To investigate whether the 24-mer G-rich ODN can fold into a GQ, increasing concentrations of 32P-labeled ODNs were incubated in the presence of 120 mM KCl. The reaction products were resolved by native PAGE. Under nondenaturing conditions, we found the coexistence of two, to our knowledge, new species of DNA in a reaction mixture containing G-rich ODNs, albeit at a varying relative abundance: a major species migrating faster than the 24-mer ODN and a minor species migrating more slowly. These correspond to intra- and interstrand GQs, respectively (Fig. 1 B, lanes 5–8). The G-mutant strand (MT) and the poly(dT) ODNs failed to show new bands in the gel, indicating the involvement of contiguous G residues in quadruplex formation (Fig. 1 B). Next, we used CD spectroscopy to determine quantitatively the formation of G-quadruplex DNA. An analysis of CD spectra revealed that 24-mer G-rich ODN showed an intense positive peak around 265 nm and a small negative peak at 240 nm in the presence of KCl (Fig. 1 C). These results are in agreement with those reported previously for the G-rich hTERT promoter, suggesting the coexistence of two different GQ conformations (89). Furthermore, we found that the characteristic CD spectroscopic signatures of GQ DNA were absent in reaction mixtures containing mutant 24-mer ODN or lacking KCl (Fig. 1 C). Additionally, DMS footprinting assays confirmed the formation of G-quadruplex DNA (Fig. S1). Together, these results support the idea that the G-rich ssDNA associated with meiosis-specific DSB folds predominantly into an intramolecular GQ in the presence of KCl.
The C-rich ssDNA from S. cerevisiae meiosis-specific DSB adopts i-motif conformation
Multiple studies have shown that the formation of i-motif structures by C-rich ODNs result in a characteristic retarded mobility of DNA in polyacrylamide gels, and a specific CD spectroscopic signature with a maximum positive peak between 280 and 288 nm and a negative peak within 260–267 nm (63, 64, 65, 90). After having shown that the G-rich ssDNA derived from a meiosis-specific DSB folds into intramolecular GQs, we asked whether the C-rich strand complementary to the G-rich sequence can adopt i-motif conformation. Increasing concentrations of 32P-labeled WT and mutant ODNs were incubated in a 50 mM sodium phosphate buffer (pH 5.5). As above, a poly(dT) ODN of equivalent length was used as a control. The formation of i-motif DNA was monitored by analyzing the reaction mixtures by PAGE. Under nondenaturing conditions, the C-rich ODN exhibited faster mobility with respect to C mutant and poly(dT) ODNs (Fig. 2 B, lanes 9–12), indicating the formation of intramolecularly folded i-motif structures (63, 64). In contrast, C-mutant and poly(dT) ODNs displayed relatively slow-moving bands under identical conditions (Fig. 2 B, lanes 1–8).
We used CD spectroscopy to determine the secondary structure in 24-mer C-rich ODN derived from a meiosis-specific DSB. Fig. 2 C shows the CD spectra obtained at four different pH values, ranging from pH 4.5 to 7.5. It is evident that the spectra exhibited significant changes concomitant with pH change from 7.5 to 4.5. The CD spectra showed a large increase in ellipticity with a positive band near 288 nm and a negative peak at 262 nm at pH 4.5 and 5.5, which are the hallmark characteristics of i-motif structure (91, 92, 93). At pH 7.5, the positive maxima shifted to 278–280 nm with a drastic decrease in molar ellipticity, consistent with an unordered random-coiled structure. The observed red shifts of the CD bands with pH change from pH 7.5 to 5.5; this is in agreement with the involvement of intercalated C-C+ basepairs in the formation of an i-motif conformation (94).
For further characterization of the i-motif structure, we used a modified 24-mer ODN containing C to T base substitutions. The CD spectra obtained under similar conditions showed the absence of positive and negative peaks at 288 and 262 nm, respectively, consistent with the notion that contiguous C residues are essential for the formation of an i-motif structure (Fig. 2 D). We next sought to determine if 24-mer C-rich ODN can adopt an i-motif conformation under cell-mimicking molecular crowding conditions. To this end, we measured and analyzed the CD spectra in the presence of 20% PEG 3350. The CD data show that PEG induces the formation of an i-motif conformation at pH 6.5 (compare Fig. 3 A with Fig. 2 C). These results support the idea that crowding agents under cellular conditions can induce the formation of i-motif conformations at a near-neutral pH (53, 95, 96).
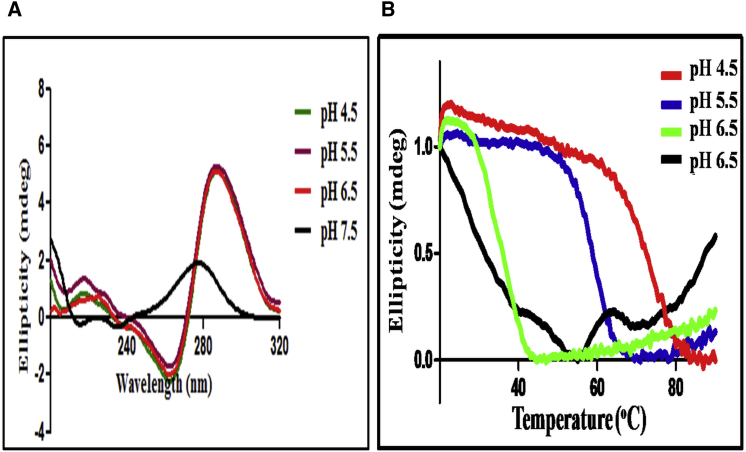
Figure 3.
Biophysical characterization of i-motif formed by a meiosis-specific DSB from chromosome IV of S. cerevisiae. (A) A molecular crowding agent facilitates the folding of C-rich sequence into i-motif at a near-physiological pH. Reaction mixtures contained 5 μM C-rich WT ODN (Fig. 3A) in 50 mM sodium phosphate buffer at different pH values. CD spectra were obtained in the presence of 20% PEG 3350 at several pH values: green, pH 4.5; magenta, pH 5.5; red, pH 6.5; and black, pH 7.5. (B) Shown here are denaturation profiles of i-motif structures at different pH values. Molar ellipticity changes in the far UV CD range plotted against increase in temperature.
We next assessed the thermal stability of the i-motif by following changes in the CD spectra with increasing temperatures and at various pH values. Fig. 3 B shows the thermal denaturation profiles of the i-motif generated by plotting the fraction of folded i-motif DNA versus temperature. Overall, these results suggest that i-motif DNA has a significantly higher melting temperature under acidic pH conditions with a distinct inverse sigmoidal melting curve at 286 nm. The higher TM at an acidic pH is due to the presence of additional CH+:C bp that stabilizes the i-motif structure (53).
NMR evidence suggests that the C-rich ssDNA associated with a meiosis-specific DSB folds into the i-motif structure
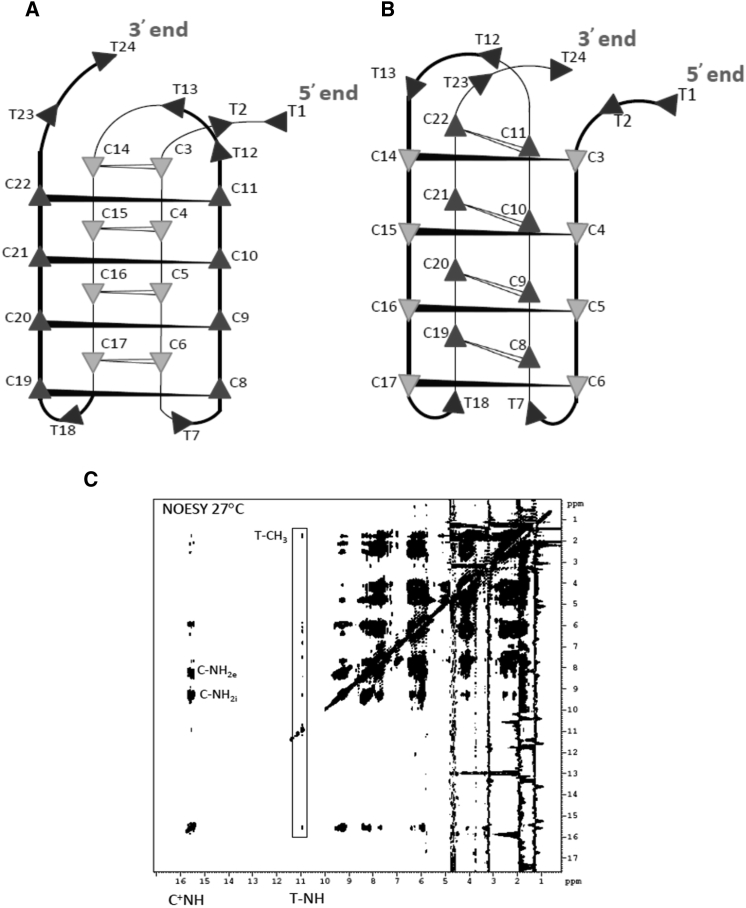
We used 2D solution-state NMR spectroscopy as a probe to characterize the i-motif structure. Fig. 4 iA shows the 14.5–16.5 ppm motif of the 1H NMR spectrum at 27°C of 24-mer C-rich ssDNA recorded on an 800-MHz spectrometer. The spectrum shows peaks between 14.8 and 16.1 ppm, which is a characteristic feature of C-C+ pairing via protonation of one of the C-residues at the N3 position (97). The DNA sequence, which is rich in C-residues, can pair, either via inter- or intramolecular folding, wherein the C residues come closer to form C-C+ pairs. To assess whether both occur, we diluted the sample 10-fold. The spectrum in Fig. 4 iB shows the identical motif as depicted in Fig. 4 iA. We therefore conclude that the spectrum in Fig. 4 i is indicative of a structure formed by an intramolecular folding of the DNA sequence. After dilution, no pH changes were observed. However, the slight shuffling in the central peaks on dilution could be attributed to minor changes in solution conditions after dilution. An examination of the DNA sequence reveals that C-rich ODN can fold several times, with thymine residues occupying the loop positions, so that the four arrays of 4 C residues can be brought in proximity to form C-C+ basepairs.
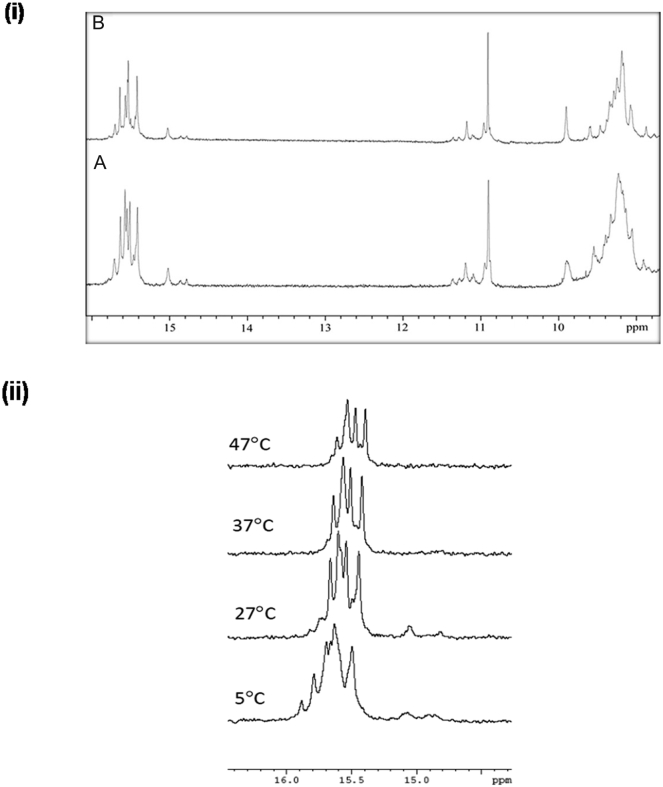
Figure 4.
(i) Given here is the 1D 800 MHz 1H NMR spectrum of 1 mM 24-mer C-rich ODN (Fig.2A) derived from meiosis-specific DSB on chromosome IV of S. cerevisiae. (A) Given here is the spectrum at 1 mM 24-mer C-rich ODN (Fig. 2A) at 27°C showing the imino region and (B) the spectrum recorded after 10 times dilution with water. (ii) Shown here are the 500 MHz 1D NMR spectra of 1 mM 24-mer C-rich ODN (Fig. 2A) showing the imino region (14.5–16.0 ppm) at different temperatures, 5°C, 27°C, 37°C, and 47°C.
Fig. 4 ii shows the temperature dependence of the spectral motif above recorded at 500 MHz. The data suggest that the structure is highly stable even at 47°C. Approximately four sharp peaks could be visualized. Thus, the folding of the DNA sequence leads to a very stable structure, and this seems to implicate the formation of the i-motif wherein one C-C+ duplex formed by two parallel strands interdigitates with another similar duplex running in the opposite direction. Each of these duplexes contains 4 C-C+ basepairs. In a structure, there will be eight nonequivalent C+ imino protons, and, therefore, one would expect to observe eight peaks. However, we observed a large number of peaks (Fig. 4 i), which seems to imply that the chain can fold into the i-motif in more ways than one. Indeed, as can be seen in Fig. 5, A and B, one could readily conceive of two ways that differ in the order of interdigitation of the two C-C+ duplex molecules. In Fig. 4 ii, we observed that the different peaks have different line widths and they melt at different rates as the temperature is raised. We can readily see that the outer peaks around 14.8 ppm are the first to vanish and these must, therefore, belong to the outermost C-C+ pairs in the structure. The fact that there are at least three (if not four) peaks in this region seems to suggest that there are perhaps two i-motif structures that coexist in solution. The central peaks that are retained at higher temperatures must correspond to the interior C-C+ pairs in i-motif structures.
Figure 5.
(A and B) Given here is the schematic illustration of two folding topologies. The intercalation topology of the cytidine core is identical in both the cases, although the arrangement of the CC stretches (folding topology) is not. (C) Given here is the complete 800 MHz 2D NOESY spectrum (200 ms) of 1 mM 24-mer C-rich ODN (Fig. 2A) recorded at 27°C. External and internal imino protons involved in C-C+ pairing are marked NH2e and NH2i, respectively.
We also recorded TOCSY and NOESY spectra of the sample with a view to characterizing the i-motif structure (Fig. 5 C). The spectrum does, indeed, display a large number of peaks typical of a well-formed stable structure, but an extensive overlap even at 800 MHz renders a detailed analysis and sequence-specific assignments impossible. Evidently, there are peaks at the expected spectral regions, reflecting imino-imino, imino-amino proton correlations, and between nonexchangeable protons in the bases and the sugar rings. Importantly, the set of nuclear Overhauser effects (NOEs) enclosed in the strip around 11 ppm includes NOEs to protons at ∼1.5 ppm, which can be readily attributed to T methyls. Thus, a natural deduction would be that the peak at 11 ppm belongs to the T-imino proton. Interestingly, this proton also shows NOEs to C+-imino and amino protons. Clearly, this imino proton is sufficiently stable even at 27°C. This is significant considering that the T-imino is not involved in any kind of basepairing. Thus, the high stability of the T-imino must be attributed to its poor accessibility to the solvent in the structure. Such a feature must be expected for the structural models in Fig. 5 C, especially for the loops involving a single T nucleotide. The NOE patterns from this proton would provide strong support for such structural models. Altogether, these results support the idea that the C-rich ssDNA corresponding to a DSB site on chromosome IV adopts an i-motif structure.
G-quadruplex blocks primer extension by DNA polymerase
Our analyses, thus far, have relied on techniques such as electrophoretic mobility shift assays, CD, and NMR spectroscopy, which are similar to previous studies that probed the formation of quadruplex structures (1, 2, 3). Although our results revealed the formation of i-motif and G-quadruplex structures by ssDNA associated with meiosis-specific DSB, we sought to determine the functional relevance of these structures. A DNA polymerase stop assay has been used to examine the effect of quadruplex structures on DNA replication in vitro (98). Accordingly, to determine the DNA replication-blocking potential of the sequences at meiosis-specific DSB on chromosome IV, we designed 66-mer WT and mutant ssDNA templates, placing the G-rich motifs at the center (Fig. 6 A). Also, we used a primer sequence complementary to the sequence located at the 3′ end of the template. The template and 32P-labeled primer (at a template/primer ratio 1:1.5) were annealed in the assay buffer containing increasing concentrations of KCl. In the absence of KCl, both the WT and mutant templates supported the synthesis of a 66-mer product by Taq DNA polymerase (Fig. 6 B, lanes 1 and 6). In the case of WT template, we found that the readthrough by DNA polymerase was blocked at a site corresponding to the site of G-quadruplex formation. Indeed, with the addition of increasing concentrations of KCl, the yield of the full-length product decreased with a concomitant increase in the amount of truncated product (Fig. 6 B, lanes 2–5). In contrast, the template containing the mutant G-rich sequence allowed polymerase to pass without being blocked (Fig. 6 B, lanes 7–10).
Figure 6.
The G/C-rich sequence associated with a meiosis-specific DSB on chromosome IV of S. cerevisiae folds into intramolecular G-quadruplex or i-motif structures and blocks DNA synthesis. (A) Here is a schematic diagram showing the nucleotide sequence of G-rich (WT) and its corresponding mutant template. (B) The Taq polymerase stop assay was performed in the absence (lanes 1 and 6) or presence of increasing concentrations of KCl (10, 25, 50, and 100 mM; lanes 2–5 and lanes 7–10, respectively). The solid triangle on top of the gel denotes increasing concentrations of KCl. (C) Here we show a diagram of C-rich (WT) and its corresponding mutant DNA template. (D) Given here is the KOD-Plus DNA polymerase (from Thermococcus kodakaraensis) stop assay performed with WT i-motif and its corresponding mutant template at the indicated pH values. Lane 1 is the primer used in this study. (E) SWNTs stabilize the i-motif DNA structure at neutral pH, which, in turn, blocks primer extension by DNA polymerase.
The i-motif blocks primer extension by DNA polymerase
We next asked whether the presence of an i-motif structure in a template would obstruct DNA polymerase to read through and synthesize full-length ssDNA. To investigate this premise, we annealed a 32P-labeled primer to the 3′ end of the 66-mer i-motif forming ODN or to its corresponding mutant template. The primer extension reactions were performed with KOD-Plus DNA polymerase, which is active under acidic conditions (99). Consistent with the results shown in Fig. 6 D, primer extension was blocked in the case of the WT template at a site coinciding with the i-motif formation in the pH range of 4.5–5.5, but not at higher pH values (Fig. 6 D, lanes 2 and 4). In contrast, a full-length product was produced with the template containing the mutant sequence (Fig. 6 D, lanes 3 and 5). However, there was no blocking with the same WT template when the reactions were performed in pH values from 6.5 to 7.5 (Fig. 6 D, lanes 6 and 8). There was no change in the magnitude of primer extension arrest with time, suggesting that KOD-Plus DNA polymerase was blocked rather than pausing at the site of i-motif structure, and we did not observe any shorter products than the full-length product (data not shown). Collectively, these results support the idea that the G/C-rich motif derived from the meiosis-specific DSB folds into G-quadruplex and i-motif structures, which, in turn, block DNA replication.
Several lines of evidence suggest that G-quadruplexes are stable under physiological conditions (1, 2, 3). On the other hand, i-motif structures are stable only in a pH range of 4.5–6 because of the need for protonation of the cytosine residues. We therefore reasoned that the loss of DNA polymerase arrest at the site of i-motif formation under neutral pH could be due to the instability of these structures. Previous studies have shown that single-walled carbon nanotubes (SWNTs) can induce human telomeric i-motif formation under physiological conditions (100). We, therefore, carried out a polymerase arrest assay at pH 7.5 in the presence of increasing concentrations of SWNTs. We observed that reaction mixtures containing the WT template blocked primer extension in the presence of SWNTs exactly at the site coinciding with the formation of the i-motif structure (Fig. 6 E). Thus, these results indicate that factors such as the superhelical density of the DNA, the binding of specific proteins, and macromolecular crowding conditions inside the cell could facilitate the formation of i-motif structures under physiological conditions (101, 102).
The G/C-rich motif folds into GQ DNA and i-motif structures in S. cerevisiae cells
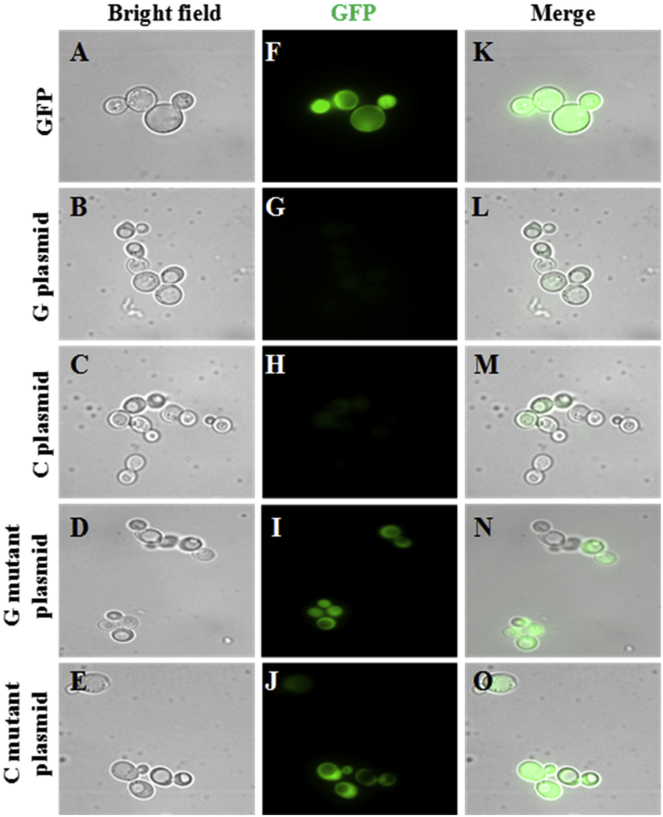
After having established that the meiosis-specific G/C-rich DSB from chromosome IV folds into GQ DNA and i-motif structures in vitro, we wished to examine if this motif could fold into quadruplex structures in vivo as well. The in vivo existence of GQ has been demonstrated using specific antibodies (8, 10). However, the in vivo existence of the i-motif structures and their ability to act as cis-regulatory elements remains to be elucidated. To examine the effects of a meiosis-specific G/C-rich DSB on gene expression, we developed a GFP-based reporter assay. The GQ DNA or i-motif-forming sequences associated with the G/C-rich motif along with its 6 bp upstream and downstream motifs were placed upstream of the GFP coding sequence in plasmids optimized for protein expression in S. cerevisiae. In these constructs, the correct reading frame of mRNA is maintained during translation. The plasmids with G-rich or C-rich sequences placed on the sense strand were termed G-plasmids or C-plasmids, respectively. For comparison, mutant constructs (GM and CM plasmids) were generated harboring the corresponding mutations in the G- or C-rich sequence. Duplex DNA containing a random sequence of equivalent length, cloned upstream of the GFP coding motif, served as a control plasmid. Fig. S2 shows a schematic depicting the potential formation of quadruplex structures by WT or mutant sequences on sense or antisense strands. In principle, the folding of the G/C-motif into the GQ or an i-motif structure should impair RNA polymerase activity on the DNA template, therefore reducing the expression of the fused GFP. The constructs were transformed into S. cerevisiae W303 cells and selected on Ura− plates. Midexponentially grown cells were examined by fluorescence confocal microscopy. The results revealed a significant decrease in the expression of GFP in cells containing the G-plasmid or C-plasmid (Fig. 7, L and M), but not those that contained the corresponding mutant sequences (Fig. 7, N and O). Further, GFP expression in cells containing mutant plasmids was similar to the vector control (Fig. 7, compare K with N and O).
Figure 7.
The G-rich or C-rich sequences associated with a meiosis-specific DSB on chromosome IV of S. cerevisiae fold into G-quadruplex and i-motif structures, respectively. The S. cerevisiae strain W303 was transformed individually with a centromeric pRS416 plasmid expressing GFP or GFP tagged to a G-rich sequence (G plasmid) or GFP tagged to a C-rich sequence (C plasmid) or GFP tagged to their corresponding mutant sequences (G mutant or C mutant plasmid). Fluorescence and bright-field images were acquired with a confocal laser scan microscope. The panels show bright-field images (A)–(E), GFP fluorescence images (F)–(J), and merged fluorescence-bright field images (K)–(O) of the constructs above.
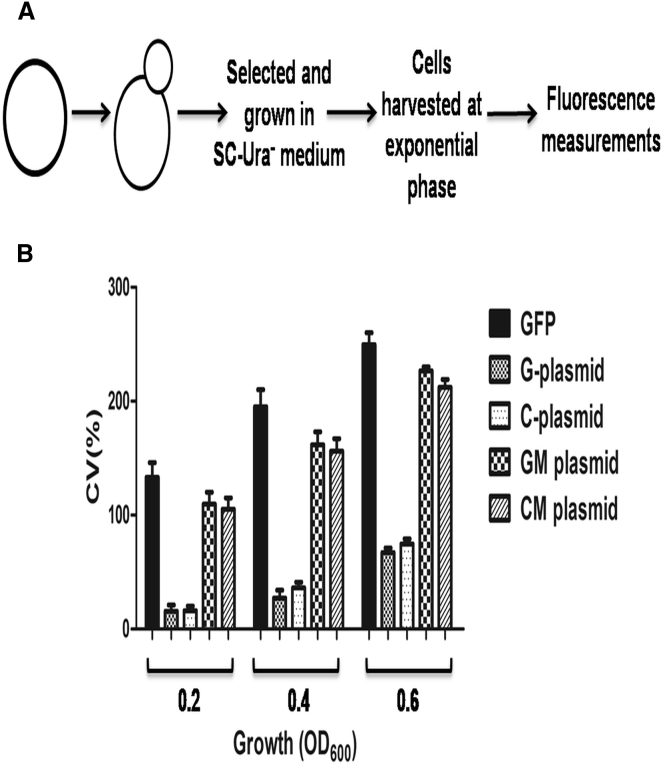
Next, we quantified the levels of GFP expression in midexponentially grown cells containing the recombinant plasmids described above by flow cytometry. The cultures were harvested, centrifuged, and the cell pellet was diluted in PBS to A600 = 0.2, 0.4, and 0.6. From the data obtained from flow cytometry, we calculated the average mean GFP expression as well as the coefficient of variation. Fig. 8 shows the relative differences in GFP expression among the various recombinant plasmids. At all cell densities, the highest expression was observed in cells containing the GFP-expressing control plasmid. The GFP fluorescence markedly reduced (75–85%) in cells containing WT G-rich or C-rich sequences upstream of the GFP coding sequence, compared with the GFP-expressing control plasmid. Interestingly, in cells containing plasmids with mutated G-rich or C-rich sequences, GFP expression was restored to the levels seen in cells containing the positive control plasmid. Altogether, these results support the idea that both G-rich and C-rich sequences derived from a meiosis-specific DSB can fold into GQ DNA and i-motif structures in vivo, respectively, and independent of each other, thus leading to a substantial decrease in GFP expression.
Figure 8.
The formation of G-quadruplex and i-motif structures in vivo lead to decreased GFP expression. (A) Given here is a schematic representation of the experimental design. (B) Histograms depict the coefficient of variation in the amount of GFP expression in S. cerevisiae cells containing the indicated recombinant constructs. Exponentially growing cells of S. cerevisiae strain W303 were serially diluted (OD600 = 0.2, 0.4, and 0.6) and used to measure GFP fluorescence.
To further confirm if changes in GFP fluorescence reflect underlying changes in the GFP reporter gene mRNA abundance, we measured the relative levels of gfp transcripts after the isolation of total RNA from the strains containing the WT and mutant constructs. The gfp mRNA yield obtained from a construct lacking the GQ DNA and i-motif-forming sequences served as a positive control. The relative gfp mRNA levels significantly decreased in cells bearing WT GQ DNA and i-motif-forming constructs (i.e., a reduction of ∼60% in the case of GQ DNA and ∼40% with i-motif-forming sequences, respectively) compared to GFP-positive cells (Fig. S3). The reason for these differences is not clear; however, it could be due to differences in the relative stability of GQ DNA and i-motif structures in vivo that need further investigation. Nevertheless, it seems that quadruplex formation in vivo efficiently inhibits gfp mRNA transcription.
Full-length Hop1 and Hop1CTD associate preferentially with G-quadruplex DNA formed by the G/C-rich motif associated with meiosis-specific DSB
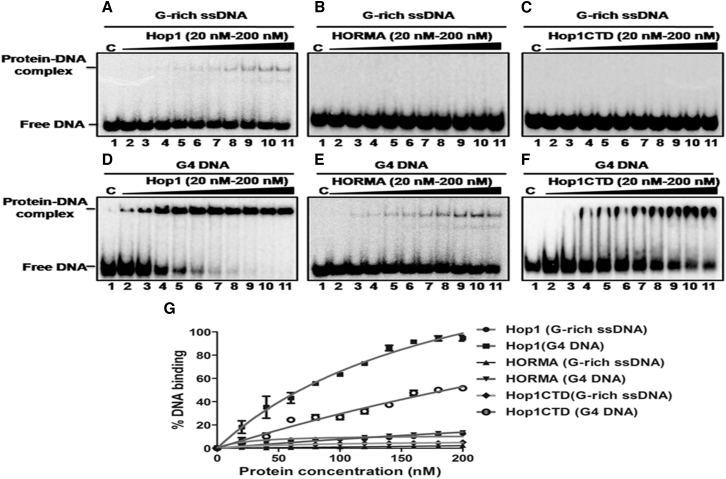
Our previous studies have shown that Hop1 possesses a modular organization with distinct N- and C-terminal domains, called “HORMA” and “Hop1CTD”, respectively (103). Further, the full-length Hop1 and Hop1CTD associate with linear duplex DNA as well as synthetic G4 DNA with high affinity, in the low nanomolar range (13, 103, 104, 105). Using purified preparations of full-length Hop1 and its truncated derivatives (Fig. S4), we carried out electrophoretic mobility shift assays to assess their ability to bind the G4 DNA motif derived from a meiosis-specific DSB. The assay was performed with a constant amount of 32P-labeled GQ DNA or ssDNA and various concentrations of full-length Hop1, HORMA, or Hop1CTD. Although the full-length Hop1 and its truncated derivatives exhibited little or no G-rich ssDNA binding activity (Fig. 9, A–C), full-length Hop1 formed a discrete band with GQ DNA (Fig. 9 D), and the Hop1CTD bound weakly to GQ DNA producing a diffuse smear after gel electrophoresis (Fig. 9 F). Under these conditions, the HORMA domain showed weaker GQ DNA binding activity even at high protein concentrations used in the assay (Fig. 9 E).
Figure 9.
Full-length Hop1 and Hop1CTD, but not the N-terminal fragment of Hop1 (HORMA), exhibit efficient binding to the G-quadruplex formed by a meiosis-specific DSB from chromosome IV of S. cerevisiae. The reaction mixtures (20 μL) contained 0.5 nM 32P-labeled G-rich ssDNA (A–C) or G4 DNA (D–F). The reaction mixtures were incubated in the absence (lane 1) or presence of (lanes 2–11) 20, 40, 60, 80, 100, 120, 140, 160, 180, and 200 nM of full-length Hop1, HORMA, or Hop1CTD, respectively. In (D)–(F), the reaction mixtures also contained 50 mM KCl. The solid triangle on top of the gel denotes increasing concentrations of the specified protein. (G) Shown here is a graphical representation of the extent of protein binding to G-rich ssDNA or G4 DNA. The extent of formation of a protein–DNA complex in (A)–(F) is plotted versus varying concentrations of the specified protein. Error bars indicate mean ± SE.
To evaluate the substrate preference, we quantitatively determined the apparent dissociation constants for full-length Hop1 and its truncated derivatives. The results shown in Fig. 9 G suggest that Hop1 associates preferentially with GQ DNA formed by the G-rich motif associated with a meiosis-specific DSB, and this preference was reflected by a Kd value of 68.45 nM, which was approximately threefold lower than that obtained for GQ DNA-Hop1CTD interaction. The Kd values determined for full-length Hop1 and Hop1CTD are in agreement with the previously determined values (13, 103, 105). Interestingly, the full-length Hop1 was devoid of significant i-motif binding activity, but Hop1CTD bound the i-motif with a reduced affinity (Kd = 412 nM) compared to GQ DNA (Table 1).
Table 1.
Kd Values for the Binding of Full-length Hop1 and Its Truncated Derivatives to GQ DNA and i-Motif
| DNA Substrate | Protein | Kd (nM) |
|---|---|---|
| G-quadruplex | Hopl | 68.45 ± 1.5 |
| i-motif | Hopl | not determined |
| G-quadruplex | HORMA | not determined |
| i-motif | HORMA | not determined |
| G-quadruplex | HoplCTD | 186.57 ± 2.8 |
| i-motif | HoplCTD | 412.75 ± 2.3 |
The Hop1CTD binds efficiently to i-motif DNA
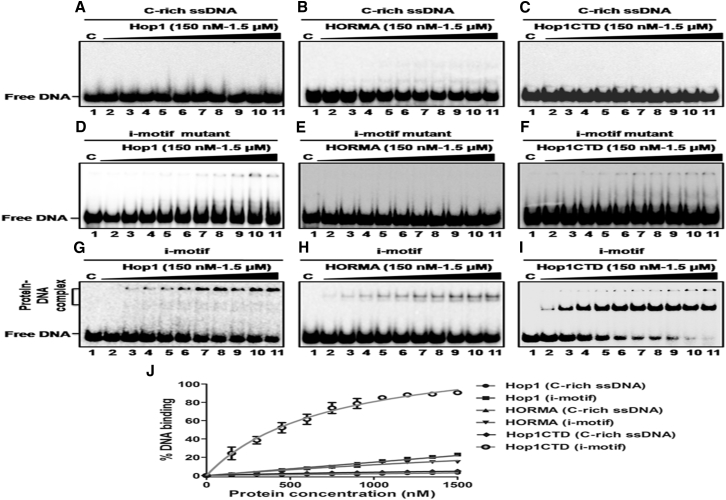
Given the evidence that the C-rich motif associated with a meiosis-specific DSB folds into an i-motif structure, we investigated whether full-length Hop1 or its truncated derivatives exhibit i-motif binding activity. Using electrophoretic mobility shift assays, we explored the ability of full-length Hop1 and its derivatives to bind i-motif, its constituent WT 24-mer ODN, or a mutant form of the i-motif structure. The full-length Hop1 or its truncated derivatives did not show measurable binding activity to either C-rich ODN or the mutant i-motif structure (Fig. 10, A–F). However, at high concentrations of full-length Hop1, we noticed only a small amount of i-motif-protein complex, although these complexes failed to enter the gel indicating possible aggregation (Fig. 10 G). An estimate of the formation of DNA-protein complexes revealed that i-motif binding required more than sixfold-higher concentrations of full-length Hop1 to reach 50% binding relative to GQ DNA (Fig. 10 J). Like full-length Hop1, HORMA was able to bind weakly to the i-motif albeit at high concentrations. However, unlike full-length Hop1, HORMA formed a discrete band on the gel (Fig. 10 H). Strikingly, Hop1CTD exhibited strong i-motif binding activity and generated a discrete band in the gel: its intensity increased with the addition of increasing protein concentrations and reached a plateau at 1.5 μM (Fig. 10 I). Under these conditions, and at high concentrations of Hop1CTD, a second band of much weaker intensity could be seen in the wells of the gel (Fig. 10 I) whose structure remain to be investigated. Nevertheless, the specific association of Hop1CTD with i-motif supports the idea that this activity has some biological relevance to the synapsis of meiotic chromosomes.
Figure 10.
Hop1CTD, but not full-length Hop1 or HORMA, exhibits a high affinity binding to the i-motif formed by meiosis-specific DSB from chromosome IV of S. cerevisiae. Reaction mixtures (20 μL) containing 0.5 nM 32P-labeled C-rich ssDNA (A–C) or i-motif mutant DNA (D–F) or i-motif DNA (G–I) were incubated in the absence (lane 1) or the presence of (lanes 2–11) 150, 300, 450, 600, 750, 900, 1050, 1200, 1350, and 1500 nM of full-length Hop1, HORMA, or Hop1CTD, respectively. The solid triangle on top of the gel denotes increasing concentrations of the specified protein. (J) Given here is a graphical representation of the extent of protein binding to C-rich ssDNA, i-motif mutant, or i-motif structures. The extent of the formation of the protein–DNA complex in (A)–(I) is plotted versus varying concentrations of specified protein. Error bars indicate mean ± SE.
Hop1 promotes the pairing of duplex DNA segments containing the G/C-rich motif associated with a meiosis-specific DSB
We have previously demonstrated that full-length Hop1 or Hop1CTD promotes an intra- and intermolecular synapsis between linear or circular duplex DNA molecules (84, 103, 104). We now asked whether Hop1 can promote pairing between linear duplex DNA segments having the G/C motif associated with a meiosis-specific DSB. For this purpose, we designed a 52-bp duplex DNA fragment with a 24-bp G/C-rich motif placed at the center of the molecule (Fig. 11 A). The sequences flanking the 24-bp G/C-rich motif represent the sequences in the natural context of the meiosis-specific DSB on chromosome IV. Likewise, we designed a 52-bp duplex fragment with a mixed DNA sequence (Fig. 11 B). Using mobility shift assays, we first examined the ability of full-length Hop1 to bind 2 nM 32P-labeled 52-bp WT and mutant DNA fragments. Consistent with our previous data (84, 103, 104), Hop1 bound efficiently to both WT and mutant 52-bp DNA fragments in a concentration-dependent manner (Fig. S5, C and D). To further confirm the specificity, we used the S. cerevisiae Rad17 protein. We found that Rad17 bound weakly to the WT 52-bp duplex DNA fragment, generating diffuse bands after gel electrophoresis (Fig. S5 E).
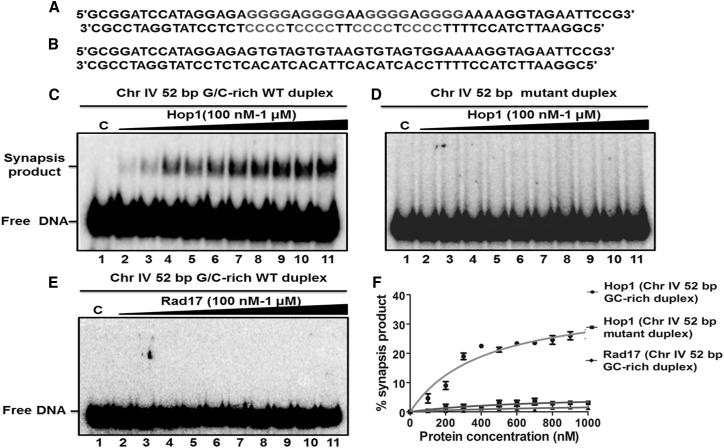
Figure 11.
The pairing of double-stranded DNA by Hop1 is dependent on the presence of a G/C-rich sequence. (A) Given here is a schematic representation of the 52-bp duplex DNA containing the G/C-rich motif embedded at the center. (B) Given here is a schematic representation of duplex DNA with a mixed sequence. (C) DNA pairing assays were carried out with a 52-bp G/C-rich duplex DNA with increasing concentrations of Hop1. (D) The pairing of duplex DNA molecules with a mixed sequence in the presence of increasing concentrations of Hop1 is shown. Reaction mixtures (20 μL) contained 2 nM of the indicated 32P-labeled duplex DNA in the absence (lane 1) or presence of 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, and 1 μM of Hop1(lanes 2–11), respectively. (E) DNA pairing assays were performed with a 52-bp duplex DNA in the absence (lane 1) and in the presence of 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, and 1 μM Rad17 (lanes 2–11), respectively. The samples were incubated with proteinase K and SDS and analyzed as described under Materials and Methods. The positions of free DNA and products are indicated on the left-hand side of the image. Lane 1 was the reaction performed in the absence of protein. (F) Given here is a graphical representation of the extent of the generation of a synapsis product with G/C-rich duplex DNA or mutant duplex DNA. The extent of the product formed in (C)–(E) is plotted versus varying concentrations of the specified protein. Error bars indicate mean ± SE.
Previous studies have shown that full-length Hop1 or Hop1CTD promote intermolecular synapsis between synthetic DNA segments containing centrally positioned G/C-rich sequences via Hoogsteen basepairing (Fig. S6). To determine the ability of full-length Hop1 to promote an intermolecular synapsis between two linear duplex DNA segments having a G/C motif derived from a meiosis-specific DSB, a constant amount of 32P-labeled 52-bp WT DNA fragment (Fig. S7 A) was incubated with a saturating amount of full-length Hop1 for the indicated time periods (Fig. S7 B). The reaction mixtures were deproteinized and the reaction products were separated by electrophoresis on a 10% native polyacrylamide gel. In good agreement with previous data (84, 103), we noticed two bands: a lower band corresponding to the free DNA substrate, and an upper band to a, to our knowledge, new DNA species whose migration is consistent with the formation of a side-by-side synapsis between two 52-bp linear duplex DNA fragments (Fig. S7 B). A quantification of the newly formed DNA species revealed that its abundance increased in a time-dependent manner, reaching a maximum of 30% in ∼10 min, and plateaued thereafter (Fig. S7 C).
We then sought to determine the effect of various amounts of full-length Hop1 on the formation of a synapsis product between 52-bp duplex DNA fragments containing a centrally positioned 24-bp G/C-rich motif and a similar fragment with a mixed DNA sequence. It is evident that the synapsis between duplex molecules (Fig. 11 A) increased with the addition of increasing concentrations of full-length Hop1 (Fig. 11 C). As previously reported, however, the formation of synapsis product did not increase above 30%, even with high levels of Hop1 (Fig. 11 F). Although full-length Hop1 bound efficiently to the fragment containing a mixed DNA sequence, it did not promote synapsis between the duplex DNA molecules (Fig. 11 D). Similarly, although S. cerevisiae Rad17 bound efficiently to the fragment containing the centrally positioned 24-bp G/C-rich motif, it failed to promote synapsis between the duplex DNA segments (Fig. 11 E). Altogether, these results support the idea that Hop1 promotes pairing between duplex DNA molecules containing G/C-rich motifs associated with a meiosis-specific DSB.
Discussion
In this study, we investigated the ability of a G/C-rich motif associated with a meiosis-specific DSB to fold into GQ and i-motif structures. Numerous studies have demonstrated that G/C-rich motifs of genomic DNA fold into GQs and impact various processes associated with nucleic acid metabolism (2, 3). We reveal, in this study, using a multitude of approaches, that a G-rich motif associated with a meiosis-specific DSB from S. cerevisiae chromosome IV folds into GQ, and the C-rich sequence complementary to the G-rich sequence forms an i-motif. Further, we found that the presence of G-quadruplex or i-motif forming sequences upstream of the GFP-coding sequence drastically reduce GFP expression, thereby indicating the existence and in vivo functional importance of these structures. Surprisingly, the Hop1 C-terminal but not the N-terminal domain exhibited strong i-motif binding activity, implying that the C-terminal domain has a distinct substrate specificity. Consistent with a proof-of-concept study using synthetic substrates (84), we found that Hop1 promotes intermolecular synapsis between G/C-rich DNA molecules derived from a meiosis-specific DSB site. Overall similar results were observed with G/C motifs derived from chromosome IX and X (data not shown). Altogether, these results reveal that the G/C-rich motif associated with a meiosis-specific DSB folds into mutually exclusive G-quadruplex and i-motif structures, both in vitro and in S. cerevisiae cells. Additionally, our data provide, to our knowledge, new insights into the activities of S. cerevisiae Hop1 that further illuminate the complexity of its role in meiotic chromosome synapsis and recombination.
As noted in the Introduction, tetramolecular and bimolecular GQs have been extensively studied using a wide variety of experimental approaches compared to i-motif structures (2, 3). The biological significance of the ubiquitous GQs has been established in different cellular contexts and in different organisms (1, 31, 105, 106, 107). The formation of GQ DNA and GQ RNA have been shown to modulate a variety of cellular functions such as replication, transcription, translation, and recombination (4, 30, 34, 108). One caveat of interpreting some of these studies is that the occurrence of GQs in cells employed GQ-specific antibodies, fragmented genomic DNA, or the quadruplex-specific ligands. Therefore, it is necessary to provide direct in vivo evidence of the existence and regulatory role(s) of GQs in processes associated with DNA or RNA. We began this study with the goal of determining the ability of a G/C-rich motif associated with a meiosis-specific DSB from S. cerevisiae chromosome IV to fold into quadruplex structures. Our data, obtained through a number of biochemical and biophysical methods, show that the G/C-rich motif folds into GQ, and the C-rich motif complementary to the G-rich motif folds into the i-motif. Furthermore, our studies reveal that the C-rich sequence folds into i-motif at a near neutral pH in the presence of a crowding agent. More importantly, we show that the WT G-rich or C-rich motifs, but not their corresponding mutated sequences, placed upstream of the GFP coding sequence attenuate its expression, and impede the progression of a replicative DNA polymerase.
Recent advances in the field of nucleic acid structure have generated much interest regarding the ability of C-rich strands to adopt i-motif conformation (53). Despite great interest and recent progress in the field of nucleic acid structure, very little is known about the biological significance of the i-motif structure. Here, we have characterized the formation of i-motif through a variety of biochemical, biophysical, and molecular and cell biological techniques. First, we show that the C-rich motif of a meiosis-specific DSB assumes an i-motif DNA conformation under mild acidic conditions. Second, the formation of an i-motif conformation occurs under physiological conditions in the presence of crowding agents. Third, CD melting experiments suggest that i-motif DNA has a significantly higher stability and that the Tm values decrease with an increase in the pH value. Finally, the analysis of NMR data is consistent with the view that the C-rich ssDNA corresponding to a DSB site on chromosome IV folds into an intramolecular i-motif structure.
The functional significance of the i-motif structure, currently, is less clear compared to the GQ because of its instability at a neutral pH (2, 3). However, it is possible that specific nuclear proteins could promote and/or stabilize i-motif structures under physiological conditions in vivo (56, 67, 68, 69, 70). The formation of an i-motif structure by a C-rich motif associated with a meiosis-specific DSB at a near-neutral pH supports the notion that this structure is physiologically relevant. We ourselves found unexpectedly, that Hop1CTD, a known GQ-binding domain, showed a binding preference for the i-motif structure. Given the fact that Hop1 is a structural component of SC, its binding to the i-motif implies that it might play a role during chromosome synapsis and meiotic recombination. More importantly, the results of primer extension and GFP expression with WT sequences are consistent with the notion that the i-motif structure formed by the C-rich sequence of a meiosis-specific DSB is functionally significant. Previous studies have shown that three closely related proteins of S. cerevisiae, i.e., Imd2, Imd3, and Imd4, possess C-rich telomeric DNA binding activity (109).
The precise molecular mechanism(s) underlying the alignment of homologous chromosomes and recombination during meiosis remain to be fully elucidated. In addition, the most poorly understood step in the DSB repair pathway is the functional significance of G/C-rich motifs at meiotic DSB sites. Several lines of evidence have provided a strong correlation between the G/C-motif at DSB sites and the high recombination rates in S. cerevisiae. This was further supported by the fact that the G/C-rich motifs derived from bacterial plasmids display a high frequency of recombination in meiotic cells (110, 111, 112, 113). Previous studies have also demonstrated that transcription factors play an important role in the formation of meiosis-specific DSB ends (75, 114, 115). However, less is known regarding the occurrence of GQ motifs at meiosis-specific DSB ends: if they exist, what role do they play in meiotic chromosome synapsis and recombination? Consistent with our previous work (84), full-length Hop1 associates preferentially with GQs as well as mediates the GQ-dependent synapsis between duplex molecules derived from meiosis-specific DSB sites. On the other hand, the binding of Hop1 to the i-motif produces a complex that is retained in the well of the gel. Previously, we showed that Hop1CTD interacts with GQ (103), and herein we show that the same domain binds to the i-motif. Although full-length Hop1 did not produce a specific complex with the i-motif, it is possible that the Hop1 structure could change after its interaction with GQ and unmask i-motif binding activity.
The ability of chromosomal G/C-rich sequences to form G-quadruplexes and their widespread occurrence has led to the question of whether these structures contribute to the regulation of cellular processes involving DNA or RNA. A number of studies have established that more than 20 helicases, including human BLM, WRN, FANCJ, and Pif1, can bind and/or unwind GQs in vitro, and mutations in these helicases cause genome instability, genomic instability-associated diseases in humans, and an increased risk of cancer (2, 3, 14). Although much is known concerning the formation and putative roles of GQs, relatively less is understood about the physical identity and functional properties of i-motif binding proteins. What could be the role of Hop1 in the processing of meiosis-specific DSB ends? In principle, there are several possibilities. Genetic and biochemical studies have shown that Hop1 protects DSB ends and recombination intermediates from exonucleolytic degradation (106, 116, 117, 118). Given the fact that both the i-motif and GQ form simultaneously within the same G/C-rich motif (62), and Hop1 binds as a dimer to DNA (86), we hypothesize that the C-terminal domain from each monomer might interact with GQ and i-motif, respectively. In continuation of this premise, Hop1 binding to GQ and i-motif structures, which exist at the DSB sites or are produced from the ssDNA generated after meiotic DSB resection, could prevent the loss of genomic integrity. In summary, the findings presented in this report, on the one hand, provide important mechanistic and functional insights into the connection between meiotic DSBs and the formation of GQ and i-motif structures and, on the other, Hop1 binding to these structures.
Author Contributions
K.M. conceived and coordinated the study. K.M., R.V.H., and R.K. designed the study and analyzed the data. R.K. conducted all the experiments (except NMR) with assistance from K.K. at the initial stages of the study. M.V.J. conducted NMR experiments under R.V.H.’s supervision. K.M. and R.V.H. wrote the article with assistance from all co-authors. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Dr. Virginia Zakian (Princeton University, Princeton) for providing the chromosome coordinates of S. cerevisiae meiosis-specific DSB sites. We thank the anonymous reviewers for their constructive comments and suggestions, which helped us to improve the manuscript.
This work was supported by a grant from the Department of Science and Technology, New Delhi and from the DBT-IISc Partnership program of the Department of Biotechnology, New Delhi. K.M. is the recipient of the J. C. Bose National Fellowship from the Department of Science and Technology, New Delhi.
Editor: Jason Kahn.
Footnotes
Seven figures and three tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30444-7.
Supporting Material
References
- 1.Bochman M.L., Paeschke K., Zakian V.A. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012;13:770–780. doi: 10.1038/nrg3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rhodes D., Lipps H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015;43:8627–8637. doi: 10.1093/nar/gkv862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mendoza O., Bourdoncle A., Mergny J.L. G-quadruplexes and helicases. Nucleic Acids Res. 2016;44:1989–2006. doi: 10.1093/nar/gkw079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schaffitzel C., Berger I., Plückthun A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl. Acad. Sci. USA. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paeschke K., Simonsson T., Lipps H.J. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat. Struct. Mol. Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 6.Capra J.A., Paeschke K., Zakian V.A. G-quadruplex DNA sequences are evolutionarily conserved and associated with distinct genomic features in Saccharomyces cerevisiae. PLOS Comput. Biol. 2010;6:e1000861. doi: 10.1371/journal.pcbi.1000861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith J.S., Chen Q., Johnson F.B. Rudimentary G-quadruplex-based telomere capping in Saccharomyces cerevisiae. Nat. Struct. Mol. Biol. 2011;18:478–485. doi: 10.1038/nsmb.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biffi G., Tannahill D., Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lam E.Y., Beraldi D., Balasubramanian S. G-quadruplex structures are stable and detectable in human genomic DNA. Nat. Commun. 2013;4:1796. doi: 10.1038/ncomms2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henderson A., Wu Y., Lansdorp P.M. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 2014;42:860–869. doi: 10.1093/nar/gkt957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giraldo R., Rhodes D. The yeast telomere-binding protein RAP1 binds to and promotes the formation of DNA quadruplexes in telomeric DNA. EMBO J. 1994;13:2411–2420. doi: 10.1002/j.1460-2075.1994.tb06526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghosal G., Muniyappa K. Saccharomyces cerevisiae Mre11 is a high-affinity G4 DNA-binding protein and a G-rich DNA-specific endonuclease: implications for replication of telomeric DNA. Nucleic Acids Res. 2005;33:4692–4703. doi: 10.1093/nar/gki777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muniyappa K., Anuradha S., Byers B. Yeast meiosis-specific protein Hop1 binds to G4 DNA and promotes its formation. Mol. Cell. Biol. 2000;20:1361–1369. doi: 10.1128/mcb.20.4.1361-1369.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat. Struct. Mol. Biol. 2006;13:1055–1059. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- 15.London T.B., Barber L.J., Hiom K. FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J. Biol. Chem. 2008;283:36132–36139. doi: 10.1074/jbc.M808152200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biffi G., Tannahill D., Balasubramanian S. An intramolecular G-quadruplex structure is required for binding of telomeric repeat-containing RNA to the telomeric protein TRF2. J. Am. Chem. Soc. 2012;134:11974–11976. doi: 10.1021/ja305734x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paeschke K., Bochman M.L., Zakian V.A. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013;497:458–462. doi: 10.1038/nature12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paeschke K., Capra J.A., Zakian V.A. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell. 2011;145:678–691. doi: 10.1016/j.cell.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vannier J.B., Pavicic-Kaltenbrunner V., Boulton S.J. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell. 2012;149:795–806. doi: 10.1016/j.cell.2012.03.030. [DOI] [PubMed] [Google Scholar]
- 20.Brázda V., Hároníková L., Fojta M. DNA and RNA quadruplex-binding proteins. Int. J. Mol. Sci. 2014;15:17493–17517. doi: 10.3390/ijms151017493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brosh R.M., Jr. DNA helicases involved in DNA repair and their roles in cancer. Nat. Rev. Cancer. 2013;13:542–558. doi: 10.1038/nrc3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zahler A.M., Williamson J.R., Prescott D.M. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 23.Liu Z., Gilbert W. The yeast KEM1 gene encodes a nuclease specific for G4 tetraplex DNA: implication of in vivo functions for this novel DNA structure. Cell. 1994;77:1083–1092. doi: 10.1016/0092-8674(94)90447-2. [DOI] [PubMed] [Google Scholar]
- 24.Larson E.D., Duquette M.L., Maizels N. MutSα binds to and promotes synapsis of transcriptionally activated immunoglobulin switch regions. Curr. Biol. 2005;15:470–474. doi: 10.1016/j.cub.2004.12.077. [DOI] [PubMed] [Google Scholar]
- 25.Sun D., Thompson B., Hurley L.H. Inhibition of human telomerase by a G-quadruplex-interactive compound. J. Med. Chem. 1997;40:2113–2116. doi: 10.1021/jm970199z. [DOI] [PubMed] [Google Scholar]
- 26.Siddiqui-Jain A., Grand C.L., Hurley L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marian C.O., Cho S.K., Bachoo R.M. The telomerase antagonist, imetelstat, efficiently targets glioblastoma tumor-initiating cells leading to decreased proliferation and tumor growth. Clin. Cancer Res. 2010;16:154–163. doi: 10.1158/1078-0432.CCR-09-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maji B., Kumar K., Bhattacharya S. Design and synthesis of new benzimidazole-carbazole conjugates for the stabilization of human telomeric DNA, telomerase inhibition, and their selective action on cancer cells. J. Med. Chem. 2014;57:6973–6988. doi: 10.1021/jm500427n. [DOI] [PubMed] [Google Scholar]
- 29.Kaulage M., Maji B., Muniyappa K. Discovery and structural characterization of G-quadruplex DNA in human acetyl-CoA carboxylase gene promoters: its role in transcriptional regulation and as a therapeutic target for human disease. J. Med. Chem. 2016;59:5035–5050. doi: 10.1021/acs.jmedchem.6b00453. [DOI] [PubMed] [Google Scholar]
- 30.Balasubramanian S., Hurley L.H., Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat. Rev. Drug Discov. 2011;10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kudlicki A.S. G-Quadruplexes involving both strands of genomic DNA are highly abundant and co-localize with functional sites in the human genome. PLoS One. 2016;11:e0146174. doi: 10.1371/journal.pone.0146174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cox R., Mirkin S.M. Characteristic enrichment of DNA repeats in different genomes. Proc. Natl. Acad. Sci. USA. 1997;94:5237–5242. doi: 10.1073/pnas.94.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rawal P., Kummarasetti V.B., Chowdhury S. Genome-wide prediction of G4 DNA as regulatory motifs: role in Escherichia coli global regulation. Genome Res. 2006;16:644–655. doi: 10.1101/gr.4508806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cahoon L.A., Seifert H.S. An alternative DNA structure is necessary for pilin antigenic variation in Neisseria gonorrhoeae. Science. 2009;325:764–767. doi: 10.1126/science.1175653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maizels N., Gray L.T. The G4 genome. PLoS Genet. 2013;9:e1003468. doi: 10.1371/journal.pgen.1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Todd A.K.M.J., Johnston M., Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sen D., Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 38.Dunnick W., Hertz G.Z., Gritzmacher C. DNA sequences at immunoglobulin switch region recombination sites. Nucleic Acids Res. 1993;21:365–372. doi: 10.1093/nar/21.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williamson J.R., Raghuraman M.K., Cech T.R. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 40.Parkinson G.N., Lee M.P., Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 41.Hanakahi L.A.H.S., Sun H., Maizels N. High affinity interactions of nucleolin with G-G-paired rDNA. J. Biol. Chem. 1999;274:15908–15912. doi: 10.1074/jbc.274.22.15908. [DOI] [PubMed] [Google Scholar]
- 42.Connor A.C., Frederick K.A., McGown L.B. Insulin capture by an insulin-linked polymorphic region G-quadruplex DNA oligonucleotide. J. Am. Chem. Soc. 2006;128:4986–4991. doi: 10.1021/ja056097c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gomez D., Lemarteleur T., Riou J.F. Telomerase downregulation induced by the G-quadruplex ligand 12459 in A549 cells is mediated by hTERT RNA alternative splicing. Nucleic Acids Res. 2004;32:371–379. doi: 10.1093/nar/gkh181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seenisamy J., Rezler E.M., Hurley L.H. The dynamic character of the G-quadruplex element in the c-MYC promoter and modification by TMPyP4. J. Am. Chem. Soc. 2004;126:8702–8709. doi: 10.1021/ja040022b. [DOI] [PubMed] [Google Scholar]
- 45.Onyshchenko M.I., Gaynutdinov T.I., Panyutin I.G. Stabilization of G-quadruplex in the BCL2 promoter region in double-stranded DNA by invading short PNAs. Nucleic Acids Res. 2009;37:7570–7580. doi: 10.1093/nar/gkp840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hershman S.G., Chen Q., Johnson F.B. Genomic distribution and functional analyses of potential G-quadruplex-forming sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 2008;36:144–156. doi: 10.1093/nar/gkm986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fullerton S.M., Bernardo Carvalho A., Clark A.G. Local rates of recombination are positively correlated with GC content in the human genome. Mol. Biol. Evol. 2001;18:1139–1142. doi: 10.1093/oxfordjournals.molbev.a003886. [DOI] [PubMed] [Google Scholar]
- 48.Gerton J.L., DeRisi J., Petes T.D. Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2000;97:11383–11390. doi: 10.1073/pnas.97.21.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gehring K., Leroy J.L., Guéron M. A tetrameric DNA structure with protonated cytosine–cytosine base pairs. Nature. 1993;363:561–565. doi: 10.1038/363561a0. [DOI] [PubMed] [Google Scholar]
- 50.Leroy J.L., Guéron M., Hélène C. Intramolecular folding of a fragment of the cytosine-rich strand of telomeric DNA into an i-motif. Nucleic Acids Res. 1994;22:1600–1606. doi: 10.1093/nar/22.9.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Diederichsen U. Oligomers with intercalating cytosine–cytosine+ base pairs and peptide backbone: DNA i-motif analogues. Angew. Chem. 1998;37:2273–2276. doi: 10.1002/(SICI)1521-3773(19980904)37:16<2273::AID-ANIE2273>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 52.Brooks T.A., Kendrick S., Hurley L. Making sense of G-quadruplex and i-motif functions in oncogene promoters. FEBS J. 2010;277:3459–3469. doi: 10.1111/j.1742-4658.2010.07759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dembska A. The analytical and biomedical potential of cytosine-rich oligonucleotides: a review. Anal. Chim. Acta. 2016;930:1–12. doi: 10.1016/j.aca.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 54.Gallego J., Chou S.H., Reid B.R. Centromeric pyrimidine strands fold into an intercalated motif by forming a double hairpin with a novel T:G:G:T tetrad: solution structure of the d(TCCCGTTTCCA) dimer. J. Mol. Biol. 1997;273:840–856. doi: 10.1006/jmbi.1997.1361. [DOI] [PubMed] [Google Scholar]
- 55.Xu Y., Sugiyama H. Structural and functional characterizations of the G-quartet and i-motif elements in retinoblastoma susceptibility genes (Rb) Nucleic Acids Symp. Ser. (Oxf.) 2005;49:177–178. doi: 10.1093/nass/49.1.177. [DOI] [PubMed] [Google Scholar]
- 56.Kendrick S., Kang H.J., Hurley L.H. The dynamic character of the BCL2 promoter i-motif provides a mechanism for modulation of gene expression by compounds that bind selectively to the alternative DNA hairpin structure. J. Am. Chem. Soc. 2014;136:4161–4171. doi: 10.1021/ja410934b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khan N., Aviñó A., Gargallo R. Solution equilibria of the i-motif-forming region upstream of the B-cell lymphoma-2 P1 promoter. Biochimie. 2007;89:1562–1572. doi: 10.1016/j.biochi.2007.07.026. [DOI] [PubMed] [Google Scholar]
- 58.Sun D., Hurley L.H. The importance of negative superhelicity in inducing the formation of G-quadruplex and i-motif structures in the c-Myc promoter: implications for drug targeting and control of gene expression. J. Med. Chem. 2009;52:2863–2874. doi: 10.1021/jm900055s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simonsson T., Pribylova M., Vorlickova M. A nuclease hypersensitive element in the human c-myc promoter adopts several distinct i-tetraplex structures. Biochem. Biophys. Res. Commun. 2000;278:158–166. doi: 10.1006/bbrc.2000.3783. [DOI] [PubMed] [Google Scholar]
- 60.Brazier J.A., Shah A., Brown G.D. I-motif formation in gene promoters: unusually stable formation in sequences complementary to known G-quadruplexes. Chem. Commun. (Camb.) 2012;48:10739–10741. doi: 10.1039/c2cc30863k. [DOI] [PubMed] [Google Scholar]
- 61.Bucek P., Gargallo R., Kudrev A. Spectrometric study of the folding process of i-motif-forming DNA sequences upstream of the c-kit transcription initiation site. Anal. Chim. Acta. 2010;683:69–77. doi: 10.1016/j.aca.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 62.Cui Y., Kong D., Mao H. Mutually exclusive formation of G-quadruplex and i-motif is a general phenomenon governed by steric hindrance in duplex DNA. Biochemistry. 2016;55:2291–2299. doi: 10.1021/acs.biochem.6b00016. [DOI] [PubMed] [Google Scholar]
- 63.Dhakal S., Yu Z., Mao H. G-quadruplex and i-motif are mutually exclusive in ILPR double-stranded DNA. Biophys. J. 2012;102:2575–2584. doi: 10.1016/j.bpj.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kendrick S., Akiyama Y., Hurley L.H. The i-motif in the bcl-2 P1 promoter forms an unexpectedly stable structure with a unique 8:5:7 loop folding pattern. J. Am. Chem. Soc. 2009;131:17667–17676. doi: 10.1021/ja9076292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kang H.J., Kendrick S., Hurley L.H. The transcriptional complex between the BCL2 i-motif and hnRNP LL is a molecular switch for control of gene expression that can be modulated by small molecules. J. Am. Chem. Soc. 2014;136:4172–4185. doi: 10.1021/ja4109352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tomonaga T., Levens D. Activating transcription from single stranded DNA. Proc. Natl. Acad. Sci. USA. 1996;93:5830–5835. doi: 10.1073/pnas.93.12.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miglietta G., Cogoi S., Xodo L.E. GC-elements controlling HRAS transcription form i-motif structures unfolded by heterogeneous ribonucleoprotein particle A1. Sci. Rep. 2015;5:18097. doi: 10.1038/srep18097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Paramasivam M., Membrino A., Xodo L.E. Protein hnRNP A1 and its derivative Up1 unfold quadruplex DNA in the human KRAS promoter: implications for transcription. Nucleic Acids Res. 2009;37:2841–2853. doi: 10.1093/nar/gkp138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Michelotti E.F., Michelotti G.A., Levens D. Heterogeneous nuclear ribonucleoprotein K is a transcription factor. Mol. Cell. Biol. 1996;16:2350–2360. doi: 10.1128/mcb.16.5.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Uribe D.J., Guo K., Sun D. Heterogeneous nuclear ribonucleoprotein K and nucleolin as transcriptional activators of the vascular endothelial growth factor promoter through interaction with secondary DNA structures. Biochemistry. 2011;50:3796–3806. doi: 10.1021/bi101633b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eid J.E., Sollner-Webb B. ST-1, a 39-kiloDalton protein in Trypanosoma brucei, exhibits a dual affinity for the duplex form of the 29-base-pair subtelomeric repeat and its C-rich strand. Mol. Cell. Biol. 1995;15:389–397. doi: 10.1128/mcb.15.1.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sarig G., Weisman-Shomer P., Fry M. Purification and characterization of qTBP42, a new single-stranded and quadruplex telomeric DNA-binding protein from rat hepatocytes. J. Biol. Chem. 1997;272:4474–4482. doi: 10.1074/jbc.272.7.4474. [DOI] [PubMed] [Google Scholar]
- 73.Amato J., Iaccarino N., Pagano B. Noncanonical DNA secondary structures as drug targets: the prospect of the i-motif. ChemMedChem. 2014;9:2026–2030. doi: 10.1002/cmdc.201402153. [DOI] [PubMed] [Google Scholar]
- 74.Day H.A., Pavlou P., Waller Z.A. i-Motif DNA: structure, stability and targeting with ligands. Bioorg. Med. Chem. 2014;22:4407–4418. doi: 10.1016/j.bmc.2014.05.047. [DOI] [PubMed] [Google Scholar]
- 75.Petes T.D. Meiotic recombination hot spots and cold spots. Nat. Rev. Genet. 2001;2:360–369. doi: 10.1038/35072078. [DOI] [PubMed] [Google Scholar]
- 76.Zickler D., Kleckner N. Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb. Perspect. Biol. 2015;7:a016626. doi: 10.1101/cshperspect.a016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kauppi L., Jeffreys A.J., Keeney S. Where the crossovers are: recombination distributions in mammals. Nat. Rev. Genet. 2004;5:413–424. doi: 10.1038/nrg1346. [DOI] [PubMed] [Google Scholar]
- 78.Baudat F., Nicolas A. Clustering of meiotic double-strand breaks on yeast chromosome III. Proc. Natl. Acad. Sci. USA. 1997;94:5213–5218. doi: 10.1073/pnas.94.10.5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Petes T.D., Merker J.D. Context dependence of meiotic recombination hotspots in yeast: the relationship between recombination activity of a reporter construct and base composition. Genetics. 2002;162:2049–2052. doi: 10.1093/genetics/162.4.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hansen L., Kim N.K., Landsman D. Analysis of biological features associated with meiotic recombination hot and cold spots in Saccharomyces cerevisiae. PLoS One. 2011;6:e29711. doi: 10.1371/journal.pone.0029711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Eisenbarth I., Vogel G., Assum G. An isochore transition in the NF1 gene region coincides with a switch in the extent of linkage disequilibrium. Am. J. Hum. Genet. 2000;67:873–880. doi: 10.1086/303085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bagshaw A.T., Pitt J.P., Gemmell N.J. Association of poly-purine/poly-pyrimidine sequences with meiotic recombination hot spots. BMC Genomics. 2006;7:179. doi: 10.1186/1471-2164-7-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hollingsworth N.M., Goetsch L., Byers B. The HOP1 gene encodes a meiosis-specific component of yeast chromosomes. Cell. 1990;61:73–84. doi: 10.1016/0092-8674(90)90216-2. [DOI] [PubMed] [Google Scholar]
- 84.Anuradha S., Muniyappa K. Meiosis-specific yeast Hop1 protein promotes synapsis of double-stranded DNA helices via the formation of guanine quartets. Nucleic Acids Res. 2004;32:2378–2385. doi: 10.1093/nar/gkh559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sambrook J., Fritsch E.F., Maniatis T. 2nd Ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 86.Kironmai K.M., Muniyappa K., Byers B. DNA-binding activities of Hop1 protein, a synaptonemal complex component from Saccharomyces cerevisiae. Mol. Cell. Biol. 1998;18:1424–1435. doi: 10.1128/mcb.18.3.1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Han H., Hurley L.H., Salazar M. A DNA polymerase stop assay for G-quadruplex-interactive compounds. Nucleic Acids Res. 1999;27:537–542. doi: 10.1093/nar/27.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jacq C., Alt-Mörbe J. The nucleotide sequence of Saccharomyces cerevisiae chromosome IV. Nature. 1997;387(6632, Suppl):75–78. [PubMed] [Google Scholar]
- 89.Lim K.W., Amrane S., Phan A.T. Structure of the human telomere in K+ solution: a stable basket-type G-quadruplex with only two G-tetrad layers. J. Am. Chem. Soc. 2009;131:4301–4309. doi: 10.1021/ja807503g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Reilly S.M., Morgan R.K., Wadkins R.M. Effect of interior loop length on the thermal stability and pKa of i-motif DNA. Biochemistry. 2015;54:1364–1370. doi: 10.1021/bi5014722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Manzini G., Yathindra N., Xodo L.E. Evidence for intramolecularly folded i-DNA structures in biologically relevant CCC-repeat sequences. Nucleic Acids Res. 1994;22:4634–4640. doi: 10.1093/nar/22.22.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Guo K., Gokhale V., Sun D. Intramolecularly folded G-quadruplex and i-motif structures in the proximal promoter of the vascular endothelial growth factor gene. Nucleic Acids Res. 2008;36:4598–4608. doi: 10.1093/nar/gkn380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li W., Wu P., Sugimoto N. Characterization and thermodynamic properties of quadruplex/duplex competition. FEBS Lett. 2002;526:77–81. doi: 10.1016/s0014-5793(02)03118-6. [DOI] [PubMed] [Google Scholar]
- 94.Mathur V., Verma A., Chowdhury S. Thermodynamics of i-tetraplex formation in the nuclease hypersensitive element of human c-myc promoter. Biochem. Biophys. Res. Commun. 2004;320:1220–1227. doi: 10.1016/j.bbrc.2004.06.074. [DOI] [PubMed] [Google Scholar]
- 95.Mergny J.L., Lacroix L., Helene C. Intramolecular folding of pyrimidine oligodeoxynucleotides into an i-DNA motif. J. Am. Chem. Soc. 1995;117:8887–8898. [Google Scholar]
- 96.Fojtík P., Vorlícková M. The fragile X chromosome (GCC) repeat folds into a DNA tetraplex at neutral pH. Nucleic Acids Res. 2001;29:4684–4690. doi: 10.1093/nar/29.22.4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kallenbach N.R., Daniel W.E., Jr., Kaminker M.A. Nuclear magnetic resonance study of hydrogen-bonded ring protons in oligonucleotide helices involving classical and nonclassical base pairs. Biochemistry. 1976;15:1218–1224. doi: 10.1021/bi00651a007. [DOI] [PubMed] [Google Scholar]
- 98.Weitzmann M.N., Woodford K.J., Usdin K. The development and use of a DNA polymerase arrest assay for the evaluation of parameters affecting intrastrand tetraplex formation. J. Biol. Chem. 1996;271:20958–20964. doi: 10.1074/jbc.271.34.20958. [DOI] [PubMed] [Google Scholar]
- 99.Takagi M., Nishioka M., Imanaka T. Characterization of DNA polymerase from Pyrococcus sp. strain KOD1 and its application to PCR. Appl. Environ. Microbiol. 1997;63:4504–4510. doi: 10.1128/aem.63.11.4504-4510.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li X., Peng Y., Qu X. Carboxyl-modified single-walled carbon nanotubes selectively induce human telomeric i-motif formation. Proc. Natl. Acad. Sci. USA. 2006;103:19658–19663. doi: 10.1073/pnas.0607245103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dettler J.M., Buscaglia R., Lewis E.A. Biophysical characterization of an ensemble of intramolecular i-motifs formed by the human c-MYC NHE III1 P1 promoter mutant sequence. Biophys. J. 2010;99:561–567. doi: 10.1016/j.bpj.2010.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rajendran A., Nakano S., Sugimoto N. Molecular crowding of the cosolutes induces an intramolecular i-motif structure of triplet repeat DNA oligomers at neutral pH. Chem. Commun. (Camb.) 2010;46:1299–1301. doi: 10.1039/b922050j. [DOI] [PubMed] [Google Scholar]
- 103.Khan K., Madhavan T.P., Muniyappa K. N-terminal disordered domain of Saccharomyces cerevisiae Hop1 protein is dispensable for DNA binding, bridging, and synapsis of double-stranded DNA molecules but is necessary for spore formation. Biochemistry. 2013;52:5265–5279. doi: 10.1021/bi4005528. [DOI] [PubMed] [Google Scholar]
- 104.Khan K., Karthikeyan U., Muniyappa K. Single-molecule DNA analysis reveals that yeast Hop1 protein promotes DNA folding and synapsis: implications for condensation of meiotic chromosomes. ACS Nano. 2012;6:10658–10666. doi: 10.1021/nn3038258. [DOI] [PubMed] [Google Scholar]
- 105.Anuradha S., Muniyappa K. Molecular aspects of meiotic chromosome synapsis and recombination. Prog. Nucleic Acid Res. Mol. Biol. 2005;79:49–132. doi: 10.1016/S0079-6603(04)79002-9. [DOI] [PubMed] [Google Scholar]
- 106.Anuradha S., Tripathi P., Muniyappa K. Meiosis-specific yeast Hop1 protein promotes pairing of double-stranded DNA helices via G/C isochores. Biochem. Biophys. Res. Commun. 2005;336:934–941. doi: 10.1016/j.bbrc.2005.08.196. [DOI] [PubMed] [Google Scholar]
- 107.Ghosal G., Muniyappa K. Hoogsteen base-pairing revisited: resolving a role in normal biological processes and human diseases. Biochem. Biophys. Res. Commun. 2006;343:1–7. doi: 10.1016/j.bbrc.2006.02.148. [DOI] [PubMed] [Google Scholar]
- 108.Kumari S., Bugaut A., Balasubramanian S. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cornuel J.F., Moraillon A., Guéron M. Participation of yeast inosine 5′-monophosphate dehydrogenase in an in vitro complex with a fragment of the C-rich telomeric strand. Biochimie. 2002;84:279–289. doi: 10.1016/s0300-9084(02)01400-1. [DOI] [PubMed] [Google Scholar]
- 110.Stapleton A., Petes T.D. The Tn3 β-lactamase gene acts as a hotspot for meiotic recombination in yeast. Genetics. 1991;127:39–51. doi: 10.1093/genetics/127.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wu T.C., Lichten M. Factors that affect the location and frequency of meiosis-induced double-strand breaks in Saccharomyces cerevisiae. Genetics. 1995;140:55–66. doi: 10.1093/genetics/140.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Birdsell J.A. Integrating genomics, bioinformatics, and classical genetics to study the effects of recombination on genome evolution. Mol. Biol. Evol. 2002;19:1181–1197. doi: 10.1093/oxfordjournals.molbev.a004176. [DOI] [PubMed] [Google Scholar]
- 113.Borde V., Wu T.C., Lichten M. Use of a recombination reporter insert to define meiotic recombination domains on chromosome III of Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:4832–4842. doi: 10.1128/mcb.19.7.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fan Q., Xu F., Petes T.D. Meiosis-specific double-strand DNA breaks at the HIS4 recombination hot spot in the yeast Saccharomyces cerevisiae: control in cis and trans. Mol. Cell. Biol. 1995;15:1679–1688. doi: 10.1128/mcb.15.3.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mieczkowski P.A., Lemoine F.J., Petes T.D. Recombination between retrotransposons as a source of chromosome rearrangements in the yeast Saccharomyces cerevisiae. DNA Repair (Amst.) 2006;5:1010–1020. doi: 10.1016/j.dnarep.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 116.Schwacha A., Kleckner N. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell. 1994;76:51–63. doi: 10.1016/0092-8674(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 117.Mao-Draayer Y., Galbraith A.M., Malone R.E. Analysis of meiotic recombination pathways in the yeast Saccharomyces cerevisiae. Genetics. 1996;144:71–86. doi: 10.1093/genetics/144.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Woltering D., Baumgartner B., Hollingsworth N.M. Meiotic segregation, synapsis, and recombination checkpoint functions require physical interaction between the chromosomal proteins Red1p and Hop1p. Mol. Cell. Biol. 2000;20:6646–6658. doi: 10.1128/mcb.20.18.6646-6658.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.