Abstract
Intracerebral hemorrhage (ICH) is a subtype of stroke that is followed by primary and secondary brain injury. As a result of the injury, cell metabolism is disrupted and a series of stress responses are activated, such as endoplasmic reticulum (ER) stress and the unfolded protein response (UPR), leading to the re-establishment of cell homeostasis or cell death. As an important mechanism of cell homeostasis, autophagy has been widely studied, and the associations between autophagy, ER stress, and the UPR have also been demonstrated. Whether these mechanisms are beneficial or detrimental remains a matter of controversy, but there is no doubt as to their vital functions. An understanding of the mechanisms of injury and recovery after ICH is crucial to develop therapeutic strategies. In this review, we summarize the related studies and highlight the roles of autophagy, ER stress, and the UPR in disease, especially in ICH. We also provide an overview of therapeutic approaches that target autophagy, and we discuss the prospects for modulating autophagy, ER stress, and UPR mechanisms in ICH therapy.
Keywords: Intracerebral hemorrhage, unfolded protein response, ER stress, autophagy, therapy
Introduction
Autophagy plays an important role in cell metabolism and disease pathology [1-3]. This intracellular mechanism is essential for cell survival, facilitating the breakdown and eventual recycling of macromolecules during cellular adaptation to environmental changes. The endoplasmic reticulum (ER) is an organelle wherein proteins are synthesized, matured, and secreted. When a cell experiences stress conditions, such as starvation, redox imbalance, altered protein glycosylation, or protein folding defects, the normal functioning of the ER in protein synthesis is disrupted, and the ER switches to a stress state. As misfolded proteins accumulate, the unfolded protein response (UPR) is initiated to counter these stress effects [4,5]. The eventual outcome of ER stress determines whether a cell survives or undergoes programmed cell death [6]. The significant role of the ER stress-associated UPR in disease has been well studied [7-9]. It has been widely reported that autophagy is associated with several neurologic diseases, such as Alzheimer’s disease (AD) [10,11], Parkinson’s disease (PD) [12], amyotrophic lateral sclerosis (ALS) [2], subarachnoid hemorrhage and ICH [13,14]. As a subtype of stroke, intracerebral hemorrhage (ICH) is often associated with high mortality and morbidity [15], as well as with poor clinical outcomes. 20% of patients are functionally independent at 6 months after experiencing ICH [16], and survivors often suffer from serious neurologic impairments [17]. With the growing understanding of brain injury mechanisms after ICH, preventing brain injury and promoting neuronal survival have emerged as therapeutic goals. In this review, we provide an overview of the associations between autophagy, ER stress, and the UPR, as well as their roles in disease pathology, especially in ICH, and we highlight potential treatment strategies.
The mechanism of autophagy
In mammalian cells, there are three predominant subtypes of autophagy: macroautophagy, microautophagy, and chaperone-mediated autophagy (CMA). The term macroautophagy refers primarily to common autophagy. During macroautophagy, a double-membrane sequestering compartment is formed as phagophore and matures into an autophagosome. Following delivery to the vacuole or lysosome, the cargo is degraded and the resulting macromolecules are released back into the cytosol for reuse [18]. In microautophagy, the membrane of the lysosome invaginated and differentiates into an autophagic tube to enclose or engulf the cytosol directly. However, misfolded or unfolded proteins and other cytoplasmic material can also be imported by CMA directly into lysosomes, where they continue to be digested with the help of HSPA8/HSC70 (heat shock 70-kDa protein 8) [19]. Protein substrates containing a “KFERQ”-like motif are recognized by the constitutive heat shock cognate 70 (HSC70) chaperone and delivered to the lysosomes upon binding to lysosome-associated membrane protein (LAMP)2A receptors on the lysosomal membrane [20]. Another subtype of autophagy is noncanonical autophagy [21], in which phagocytes kill or digest extracellular pathogens directly.
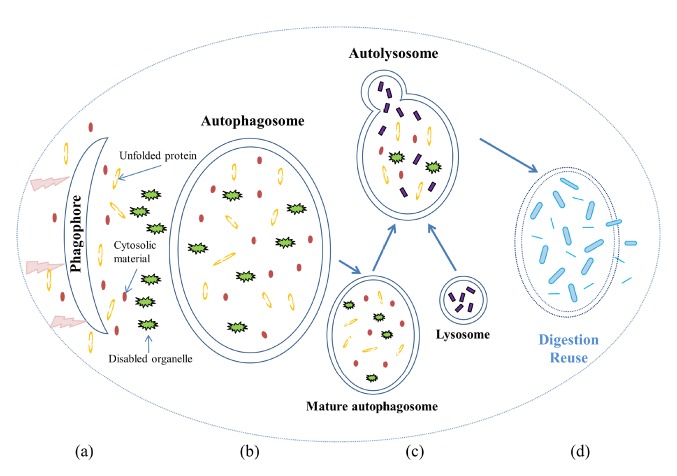
The activation of autophagy can be detected by transmission electron microscopy (TEM) during the development of phagophores (isolation membranes) and autophagosomes. The morphologic process of autophagy is shown in Figure 1. Based on our current knowledge, it appears that the formation of autophagosomes is regulated by a suite of proteins encoded by autophagy-related genes (ATGs). These genes, which are highly evolutionarily conserved, were originally discovered in yeast and have been systematically studied [22]. The initiation of autophagy could be induced by the dissociation of Beclin 1 (the mammalian ortholog of yeast autophagy-related gene 6, ATG6) from its inhibitors [23]. The extension of the isolation membrane is regulated by an ubiquitin-like conjugating system that converts LC3 (microtubule-associated protein 1 light chain 3, the mammalian homolog of yeast ATG8 ) from its free (LC3-I) to its lipidated, membrane bound (LC3-II) form [24]. The ULK1 (Serine/Threonine-Protein Kinase, also known as Unc-51 Like Autophagy Activating Kinase 1) complex [25] is also required during membrane initiation. The activation of the ULK complex is regulated by mTORC1 (mammalian target of rapamycin complex 1) and AMPK (AMP-activated protein kinase). The mTORC1 kinase complex consists of five subunits [26]: mTOR (mammalian target of rapamycin), RAPTOR (regulatory-associated protein of mTOR), mLST8 (mammalian lethal with SEC13 protein), DEPTOR (DEP domain-containing mTOR-interacting protein), and PRAS40 (40-kDa Pro-rich AKT substrate). mTOR is a central cell-growth regulator that integrates growth factors and nutrient signals. AMPK is a key energy sensor and regulates cellular metabolism to maintain energy homeostasis. Under nutrient sufficiency conditions, high mTOR activity prevents ULK1 activation by phosphorylating ULK1 Ser757 and disrupting ULK1 and AMPK interaction[27]. Conversely, under starvation or toxin accumulation, down-regulated mTORC1 leads to the activation of the ULK complex, resulting in the induction of autophagy [28]. AMPK also promotes autophagy by the direct activation of ULK1 through phosphorylation of Ser317 and Ser777 under glucose starvation[27]. During the process of autophagy induction, a mammalian-specific scaffold protein named AMBRA1 (activating Molecule in Beclin-1-Regulated Autophagy) is phosphorylated by ULK1 and translocates to the ER, where it can prime autophagosome formation[29]. These activations are also associated with the Beclin 1 complex (the autophagy-specific class III phosphoinositide-3-kinase complex, CIII PI3K) [30] and the ULK complex [31].
Figure 1.
Morphological process of autophagy. When a cell is experiencing starvation, the aggregation of unfolded proteins, a pathogen infection, or the accretion of any other cytotoxic factor, autophagy is initiated. (a) A flat bi-layer liposomal membrane known as the isolation membrane or phagophore forms in the cytosol. (b) As the membrane elongates, the phagophore seals itself to form an autophagosome that envelops the proteins, organelles, and other cytosolic material to be eliminated. The maturation of the autophagosome is coordinated with the endocytic system. (c) The mature autophagosome also fuses with the endosomal-lysosomal system, and lysosomal proteases are delivered to convert the autophagosome to an autolysosome. (d) The autolysosome digests the sequestered cytoplasmic material into amino acids and other molecules, which are reused after being transported across the membrane to the cytosol.
Some ULK complex and PI3K (phosphoinositide-3-kinase) complex phosphorylated proteins usually contribute to the closure of the isolation membrane and the nucleation of the autophagosome; examples of these proteins are PtdIns (3) (phosphorylatidylinositol-3-phosphate) [32] and Jumpy (a myotubularin-related phosphoinositol-3-phosphate phosphatase, also known as MTMR14) [33]. During the maturation of an autophagosome, LC3-II binds to the membrane and aids the expansion, making LC3 family protein a useful marker for identifying autophagosomes [34]. In contrast, the ATG proteins dissociate from the membrane before maturation, with the exception of ATG9, which participates in vesicles and lipid delivery and is involved in the entire process [35,36]. In response to starvation, phosphorylation of mATG9 by ULK1 promotes the interactions between mATG9 and the adaptor protein complex, leading to redistribution of mATG9 from the plasma membrane and juxta-nuclear region to the peripheral pool for autophagy initiation [36]. The origin and mechanism of autophagosome formation are not fully understood; however, the origin of the isolation membrane is considered to be associated with several cellular organelles, especially the ER, mitochondria, and Golgi apparatus [37].
Pathways associated with ER stress and the UPR
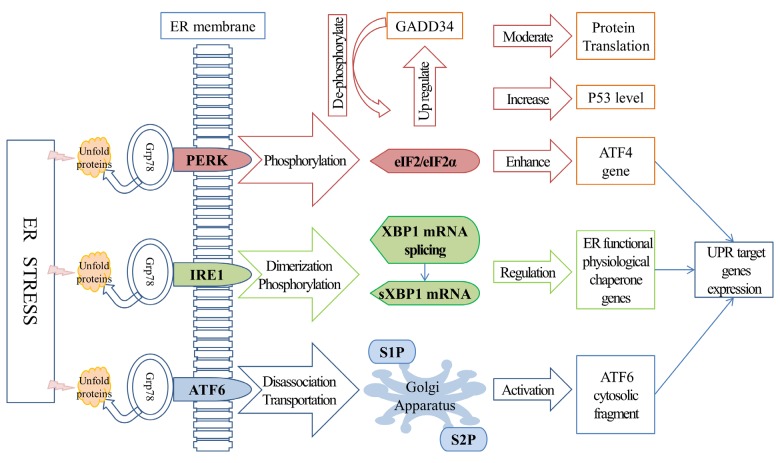
The hypothesis that misfolded or unfolded proteins and the UPR might be the primary promoters of cells surviving ER stress was first proposed in the 1980s [38] and led to the investigation of the influence of ER stress and the UPR on cell survival [39,40]. The UPR consists of three main signaling systems that are initiated by prototypical ER-localized stress sensors: PERK (protein kinase RNA-like endoplasmic reticulum kinase), IRE 1 (inositol-requiring enzyme 1), and ATF6 (activating transcription factor 6). These three sensors are all associated with Grp78 (glucose-regulated protein 78 kDa, a key ER chaperone that is also known as BiP), which controls their activation through a binding/release mechanism. Ordinarily, the activation of PERK, IRE1, and ATF6 is prevented by the binding of Grp78. However, when a cell is experiencing stress conditions, with misfolded or unfolded proteins accumulating in its ER, Grp78 switches its binding site to the hydrophobic domains and releases the sensors [41], thereby promoting the oligomerization and transphorylation of IRE1 and PERK [42] and revealing an ER export motif in ATF6 [43](as shown in Figure 2).
Figure 2.
UPR-associated pathways. UPR pathways play important roles in cell survival under stress conditions. IRE1, PERK, and ATF6 are three associated sensors that are normally inhibited by the binding of Grp78. However, under stress conditions, Grp78 dissociates from the sensors, thereby promoting the oligomerization and transphorylation of IRE1 and PERK, as well as the transportation of an ER export motif in ATF6a. The PERK downstream factor EIF2 attenuates translation or inhibits the cell cycle in order to maintain the coordination of protein synthesis and also enhances the translation of the ATF4 gene to support the recovery of translation. IRE1 dimerization and phosphorylation induced XBP1 mRNA splicing, resulting in the production of sXBP1. As a transcription factor, sXBP1 up regulates the expression of ER functional genes to promote the recovery of ER function. After its dissociation from Grp78, ATF6 undergoes trans-Golgi migration and releases a cytosolic fragment to activate the transcription of UPR target genes.
The PERK pathway
Under stress conditions, the protein kinase domain on PERK is activated, promoting its oligomerization and trans-auto-phosphorylation [44]. Phosphorylation of the α-subunit of EIF2 (eukaryotic initiation factor 2) subsequently inhibits EIF2β (a guanine nucleotide exchange factor), resulting in translation attenuation and reducing the excess of newly synthesized protein. Alternatively, phosphorylated EIF2 increases the translation of ATF4 (activating transcription factor 4) and induces the expression of ER-stress target genes to support the recovery of translation. A study in murine cells[45] found that the PERK-EIF2α signaling pathway was also involved in polyQ72-induced ATG12 upregulation and LC3 conversion, suggesting the pro-survival role that PERK-EIF2α pathway plays in autophagy. Another study [46] revealed that, in addition to reducing the protein overload in the ER, the inhibition of translation by PERK-EIF2α phosphorylation also increased the levels of p53 (transformation-related protein 53) through a PERK-dependent ribosomal-Hdm2 (human homolog of mouse double minute 2, also known as E3 ubiquitin-protein ligase MDM2) interaction, leading to cell cycle inhibition. Interestingly, the PERK-EIF2α pathway can also increase the expression of a protein phosphatase gene named GADD34 (DNA damage-inducible gene 34) and dephosphorylate EIF2 to restore global protein synthesis, which might be a feedback mechanism to maintain translation coordination [47].
The IRE1 pathway
As an ER-resident transmembrane kinase/RNase, IRE1 monitors the status of protein folding inside the ER in a similar manner to the PERK/EIF2α pathway. Grp78 dissociation initiates IRE1 dimerization and phosphorylation, inducing the splicing of XBP1 (X-box binding protein 1) mRNA [48], which is also activated by cytosolic kinase/endoribonuclease (RNase) domains [49]. Spliced XBP1 (sXBP1) mRNA is translated to a transcription factor that up-regulates the target genes via the ERSE (ER stress response element) promoter. sXBP1 is also involved in several aspects of ER function and physiology, such as protein folding, quality control, and the ERAD (ER-associated degeneration) system [50]. RNase substrate activation of IRE1α is restricted by its oligomerization status. Under conditions of chronic ER stress, the over-oligomerized IRE1a expands its substrates to include many ER-localized mRNAs, resulting in RIDD (regulated IRE1-dependent decay of mRNA) or apoptosis, whereas lower levels of oligomerization will block RIDD and maintain XBP1 splicing [48] to reduce cell degeneration.
The ATF6 pathway
As one of the stress response related transmembrane transcription factors, ATF6 has two isoforms in mammalian cells, ATF6α and ATF6β [51]. ATF6α and ATF6β are cleaved during the ER stress response. The resulting N-ATF6α and N-ATF6β have conserved DNA-binding domains and divergent transcriptional activation domains. N-ATF6β potentially acts as an endogenous transcriptional repressor of N-ATF6α. N-ATF6α and N-ATF6β translocate to the nucleus, bind to specific regulatory elements, and influence expression of ER stress response genes (e.g.GRP78) that contribute to resolving the ER stress response. Accordingly, cell viability are enhanced [52]. When a cell is under stress, ATF6 undergoes trans-Golgi migration instead of binding to the ER as usual, and an N-terminal DNA-binding transcription factor domain is released, catalyzed by two Golgi-resident enzymes named S1P and S2P proteases [44]. In addition to Grp78 dissociation, which is similar to that which occurs in PERK and IRE1 activation, the redox state of ATF6 is also involved in ER stress sensing and ATF6 activation [53]. Subsequently, the ATF6α cytosolic domain translocates to the nucleus and promotes adaptation processes, e.g., by activating the transcription of UPR target genes and AKT (also known as protein kinase B or PKB), resulting in negative regulation of mTORC1 and ULK1 activities [54]. A study conducted in ATF6α and ATF6β knockout mice showed that the maintenance of ER chaperones in mammalian cells depends on the function of ATF6, rather than that of IRE1 (as in worm and fly cells) [55]. However, the regulation of the ATF6 signal is highly dynamic and initially depends on the experimental system used, especially on the intensity of the stress induced by the different pharmacologic stressors [56]. Several regulators have been suggested to be involved in mediating ATF6, including WFS1 (Wolfram syndrome 1) [57] and PDIA5 (protein disulfide isomerase A5) [58], which have been implicated, respectively, in the degradation and activation of ATF6.
The activation of PERK, IRE1, and ATF6 produces several cytoprotective effects, such as reduced translation, enhanced ER protein-folding capacity, and clearance of misfolded ER proteins. UPR stress sensors are associated with many transcription factors and mediate the establishment of transcriptional patterns, indicating the functional role of the UPR in proteostasis.
Crosstalk between autophagy, ER stress, and the UPR
The associations between autophagy, ER stress, and the UPR have been demonstrated by several studies. As an essential organelle in which protein is processed and synthesized, the ER monitors protein folding, unpredicted errors and off-pathway intermediations during protein quality control. Under severe conditions, an overload of toxic polypeptides can challenge the capacity of the chaperone system, resulting in ER stress. The activation of the UPR promotes the refolding of non-native polypeptides or their elimination via ERAD and ER stress-activated autophagy [59]. It was initially thought that ER stress initiates autophagy only when aggregated proteins become excessive enough to overwhelm the canonical ubiquitin-proteasome-dependent ERAD. However, current findings suggest that ERAD-mediated proteins that are partially processed are also targets of autophagy. These encompass all other unfolded proteins [60]. ER stress-induced deactivation of mTOR contributes to the downregulation of AKT/TSC/mTOR pathway and promotes ER stress-induced autophagy [61]. Following stress conditions or nutrient deprivation, mTORC1 is quickly dissociated from the ULK1 complex. The decreased mTORC1 activity leads to dephosphorylation of ULK1, ULK2, and mAtg13, initiating autophagosome formation [62,63]. One link between autophagy and the UPR is the PERK/EIF2α pathway. This pathway is essential for autophagy induction after ER stress. Specifically required proteins during this process are the downstream ATF4 and CHOP (C/EBP homologous protein, a transcription factor induced by ATF4), which regulate several ATG genes [64]. It has been proven that autophagy acts as a cellular defense mechanism against polyQ72-induced ER-stress-mediated cell death. The PERK/eIF2α phosphorylation is involved in the polyQ72-induced LC3 conversion and autophagy induction [45]. Another associated pathway is the IRE1 pathway. It is proposed that the ATG16L1 (autophagy-related 16-like 1) gene inhibits IRE1α activity and that the escalation of autophagy reduces ER stress-induced inflammation, as well as cell death [65]. The IRE1-JNK (c-Jun N-terminal kinase) pathway is also demonstrated to be involved in activating the autophagy system [66]. TRAF2 (tumor necrosis factor receptor-associated factor 2)-dependent activation of IRE1 and JNK stimulated Bcl-2 (B-cell lymphoma 2) phosphorylation and Beclin 1 dissociation, as well as the activation of the PI3K complex, resulting in the activation of autophagy [67].
The interconnections between ER stress and autophagy have recently been reviewed [68], and a series of ER stress-regulated autophagy adaptors, such as mTORC1, AMPK (AMP-activated protein kinase), Beclin 1, and the ATG genes, have been highlighted to demonstrate the associations between ER stress (UPR) pathways and autophagy.
Intracerebral Hemorrhage
ICH is a subtype of stroke and often associated with high mortality and morbidity. The most common cause of spontaneous ICH is hypertension; other risk factors include amyloid angiopathy, arteriovenous malformation, intracranial aneurysm, angioma, neoplasm, coagulopathy, vasculitis, and the use of anticoagulants [69]. The pathology of ICH is very complex and several cell signaling pathways are involved in the neuron loss process. Brain injuries that occur after hemorrhage can be classified as primary or secondary injuries. Primary injuries usually occur within minutes to hours of the initial bleeding as a result of the mechanical effects and physical disruption caused by the hemorrhage. The mass effect of a hematoma usually results in the compression of some brain regions, increased intracranial pressure, and changes in blood flow. The usual strategies for alleviating the primary injury include surgically removing the hematoma [70], preventing the mass effect [71], and ameliorating the perihematomal blood flow [72]. Secondary injuries arise from the physiologic responses to hematoma and perihematomal edema (PHE) [73,74]. The resulting accumulations of blood components, overproduced iron complexes, dysfunctional organelles, and other cytokines can disrupt normal protein folding leading to activation of UPR/ER stress, which plays a critical role in the ICH brain injury and contributes to neuropathological responses [75]. These injuries eventually lead to the irreversible disruption of brain parenchyma and massive cell death. However, some protective responses and mechanisms are also activated in an effort to overcome the effects of the injury or to remove the cellular debris in order to restore cell homeostasis, such as autophagy, ER stress, and the UPR. The processes leading to neuronal death or survival represent a critical phase after brain injury and determine the subsequent neurological deficits and quality of life of the patient.
Autophagy in ICH and other diseases
Autophagy has been implicated in the pathophysiology of many neurological disorders [76,77], such as PD [12], AD [10], ALS and other neurodegenerative disorders [78,79]. The involvement of autophagy in ALS has been recently confirmed by whole-exome gene sequencing of a large cohort of ALS patients and controls [2]. The activation of autophagy is also involved in several brain injury associated disorders. Previous studies [80-82] provided evidences that autophagy pathways were initiated after cerebral ischemia, and functional impairment of autophagy resulted in the accumulation of protein aggregates, damaged organelles, and ultimately neuronal death. A recent study [83] on traumatic brain injury (TBI) examined the levels of autophagy and autophagic flux at different time points following TBI, confirming the accumulation of LC3 and autophagosomes in the ipsilateral cortex and hippocampus within hours of brain injury. The results also indicated that autophagic clearance was impaired as a result of lysosomal dysfunction and correlated with neuronal cell death. With similar mechanisms, brain injury usually occurs immediately after ICH, resulting in cellular stress or neuron death. Autophagy pathway activation was detected in neurons after experimentally induced ICH [84], and subarachnoid hemorrhage [85] in rat models.
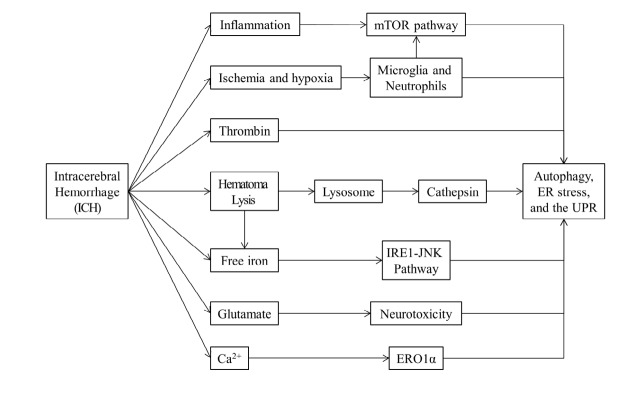
Although therapeutic targeting of autophagy to limit brain injury or facilitate recovery after ICH still requires further exploration, several studies have highlighted the roles and mechanisms of autophagy in ICH (Figure 3). For instance, autophagic vacuoles have been detected by electron microscopy in several ICH studies, and astrocytes are demonstrated as the most autophagic brain cells after ICH [3,84,86,87]. Another notable consequence of ICH is inflammation, which correlates with activation of resident microglia as well as migration of neutrophils into brain [88,89]. Moreover, the initial hemorrhage increases intracranial pressure and decreases blood flow, leading to perihematomal ischemia and hypoxia. Mediated by HIF-1α (hypoxia-inducible factor 1α), hypoxia-induced autophagy promotes microglia cell death after ICH [90]. The mTOR pathway integrates diverse environmental signals to regulate autophagy, and mTOR upregulation attenuates microglial autophagy and inflammation after ICH [91]. Neutrophil extracellular traps (NETs) are extracellular chromatin structures that trap and degrade microbes [92]. It is reported that neutrophil depletion reduces blood-brain barrier breakdown and inflammation after ICH, and autophagy and superoxide production also occur following intracellular chromatin decondensation and NET formation [93]. These results indicate that inflammation-associated autophagy is involved in ICH-induced brain injury. Thrombin, a serine protease, is produced immediately after ICH to induce hemostasis. Researchers find that ICH-induced autophagic activation is associated with thrombin. Inhibition of autophagy aggravates thrombin-induced cell death after ICH [87]. Lysosome is an essential organelle for execution of autophagy activity. Lysosomal enzymes are required for degradation of the contents in autophagic vacuoles [94]. As a hydrolytic enzyme in lysosomes, cathepsin D acts as an autophagic mediator and the inhibition of cathepsin D prevents the formation of autophagic vacuoles [95]. Following ICH, cathepsin D is induced in neurons and astrocytes within 1 day, reaches peak levels after 1 week, and remains up-regulated for at least 4 weeks [84], suggesting elevated lysosomal activity and enhanced autophagy activation after ICH. In summary, multiple layers of evidence have linked autophagy and ICH-associated events (Figure 3), highlighting the crucial role autophagy in ICH pathology.
Figure 3.
Autophagy, ER Stress, and the UPR related ICH events and components. Several events arising after ICH and the components releasing after ICH are associated with Autophagy, ER Stress and the UPR, such as Inflammation and perihematomal ischemia and hypoxia after ICH, and thrombin, free iron and glutamate released after ICH
Following primary brain injuries, secondary injuries often occur through many parallel pathological pathways, such as the cytotoxicity of blood, hypermetabolism, and excitotoxity. Meanwhile, the disturbances in cell homeostasis and the accumulations of misfolded/unfolded proteins gradually prolong ER stress, leading to sustained ER dysfunction and autophagy (Figure 2 and Figure 3). Iron is an important promoter of ICH-induced autophagic cell death and also contributes to ICH-induced brain injury [96]. As a component from plasma and hemoglobin after erythrocyte lysis, the overloaded free irons can cause free radical formation and brain damage [69]. JNK pathway activation may influence cell survival via transcriptional and post-translational regulation of proteins [97]. The fusion of irons trigger JNK pathway activation, and conversely the application of iron chelates reduce free iron levels and attenuates activation of JNK [98]. With the accumulation of free iron, ER stress is triggered, and autophagosome formation is also increased via IRE1-JNK signaling pathway [66]. These results indicate that JNK pathway is involved in iron triggered ER stress and autophagy activation. Glutamate-induced neuron toxicity is an ideal model for studying neurological diseases. The increased glutamate level in perihematomal is a major metabolic effect of hemorrhage [99,100]. The stimulated glutamate signaling contributes to neuronal excitation and increases intracellular concentrations of calcium and sodium ions, leading to the hyper-perfusion and hyper-metabolism after ICH. The activity of autophagy can be enhanced during the process of glutamate-induced neurotoxicity, and excessive glutamate further results in ER dysfunction and initiates ER stress [101]. The disruption of Ca2+ homeostasis also plays an important role in neuronal function and survival after ICH [102]. ER is the major storage for cellular Ca2+, energy failure after ICH releases intracellular Ca2+ and disrupts ER-associated Ca2+ channels, leading to Ca2+ loss in ER and ER-associated Ca2+-ATPase activation. Meanwhile, the failure of EROlα (ER oxidoreductin-1 alpha) disrupts protein disulfide bond formation and decreases protein folding, resulting in accumulation of unfolded proteins and activation of the UPR under ER stress [94]. COL4 (collagen, type IV) A1 and COL4A2 are the most abundant type IV collagens that are basement membrane proteins [103]. It is recently reported that COL4A1 and its mutations are associated with cerebrovascular disease pathology, including ICH [13,14]. The COL4A2 mutations that perturb collagen biosynthesis can increase the risk of sporadic ICH in humans [104]. Additionally, Mutations on COL4A1 and COL4A2 often lead to their intracellular accumulation, enhancing their cytotoxic effects. A recent study identified a dominant COL4A mutation in the collagen domain of COL4A2 which caused ER stress and UPR activation in primary fibroblasts from a hemorrhagic stroke patient, and COL4A mutant was shown to be degraded via the proteasome[105]. The COL4A1 and COL4A2 mutation-associated cellular pathophysiology continually triggers ER stress and the UPR, leading to irreversible cell death. These results indicate the potential role of COL4A mutation in ER stress and ICH pathology.
Numerous hypotheses have been proposed regarding the role of autophagy in other diseases. The results of studies employing autophagy inducers, autophagy inhibitors, and ATG7 knockout mice indicated that autophagy may play a role in metabolic diseases, such as obesity and diabetes, and even in the neuroendocrine system [106-109]. Autophagy is also involved in both innate and adaptive immunity, which is attributable to autophagy protein-dependent or -independent functions [110]. With regard to innate immune signaling molecules, the upregulation of autophagy enhanced immune signaling, as well as pathogen degradation and antigen presentation [111]; however, in the adaptive immune response, autophagy primarily affected the regulation of the inflammatory transcriptional response, in addition to negatively regulating inflammasome activation [112]. The role of autophagy in cancer has been well studied [113,114], but the effect of autophagy on cancer is largely unknown. While most evidences support a role of autophagy in sustaining cell survival, cell death resulting from progressive cellular consumption has been attributed to unrestrained autophagy [114]. The loss of the pro-survival function of autophagy promotes tumorigenesis. For instance, ATG5 is required for maintaining T-cell survival and proliferation. Mice with the ATG5 deletion and liver-specific ATG7 knockout were prone to develop liver adenomas, and this predisposition was attenuated by the deletion of p62 (an autophagy adaptor protein) [115]. Autophagy is also required for the survival of tumor cells to metabolic stress. In cancer cells, metabolic stress robustly induces autophagy, which is sustained when apoptosis is blocked. It is reported that autophagy may function in tumor suppression by mitigating metabolic stress. In concert with apoptosis, autophagy can also suppress tumor by preventing necrosis-induced cell death [116]. Meanwhile, in some preclinical studies, inhibition of pro-survival autophagy was shown to kill tumor cells and trigger apoptosis [116-118]. Several oncosuppressor proteins and oncoproteins, such as PTEN (phosphatase and tensin homolog), TSC1 (tuberous sclerosis 1), and RAS (a family of proteins involved in cellular signal transduction), have also been reported to stimulate or obstruct autophagy flow through mTORC1 restraint or ULK1 activation [119,120]. These results highlight the complex interactions between autophagy and cancer.
Autophagy-associated potential therapy strategies in ICH
Investigation of the autophagy mechanism is crucial for understanding the development of disease and exploring therapeutic strategies. Some reported autophagy-targeting approaches highlight the potential of autophagy modulation as a therapeutic strategy. For medical purposes, several categories of inducers and inhibitors of autophagy have been identified [121], characterized by their functional pathways, such as mTOR complex 1 targeting AMPK activity, the phosphatidylinositol signaling pathway, the cyclic AMP-associated pathway, and unforeseen autophagy-blocking effects. Other reasons for considering autophagy modulation as a possible treatment are its influence on the immune response [122,123] and its association with apoptosis and necroptosis [124,125].
Strategies to modulate the autophagy mechanism for disease treatment have been widely evaluated [113,126,127]. For now, whether inducing autophagy and ER stress after brain injury or inhibiting these processes protects neurons is still under debating. However several encouraging progressions have been made in targeting autophagy in ICH. A rat model study [128] proved that autophagy activation induced by ischemic preconditioning may ameliorate brain damage by constraining excessive ER stress, whereas inhibiting autophagy with the inhibitor 3-MA (3-methyladenine) may result in excessive ER stress and the aggravation of ischemic neuronal damage. The results of another study in a rat middle cerebral artery occlusion (MCAO) model[129] suggested that ganglioside (GM1) inhibited autophagy and protected neurons after an experimentally induced stroke. However, this biological function could be abolished by Tat-Beclin 1, a recently identified autophagy-inducing agent [130]. Researchers investigating ICH [131,132] have proposed that autophagy contributes to microglial activation and that the inhibition of autophagy partly reduces brain damage after ICH. The effect of age was also evaluated in a rat model of ICH [86], with more severe autophagy and neurologic deficits being seen in aged rats after ICH than in younger animals. A sex-specific study[3] in a rat model of ICH demonstrated that the suppression of ferrous citrate-induced autophagy contributes to the less severe brain injury caused by iron overload in female rats as compared to that in male rats. It was recently demonstrated in rat models that many secondary injuries caused by ICH could be substantially reduced by injecting the animals with minocycline and that the medicinal effect of minocycline was exerted through both anti-autophagy and anti-apoptosis pathways [133]. Clonidine and rilmenidine have also been reported to be pharmacologically protective against neurologic disease through their positive effect on autophagy induction [134]. Although there is still controversy as to whether the mechanisms of autophagy, ER stress, and the UPR are beneficial or detrimental to injured neurons after ICH, the above results highlight the applications and therapeutic potential of autophagy pathway modulators for the treatment of ICH.
Conclusions
For many years, there has been increasing interest in the function of autophagy in neurodegenerative disease [77,135] and cerebral ischemic injury[128,136,137]; however, ICH, despite having similar pathology and a more severe outcome, has attracted less attention. A comprehensive understanding of the roles played by autophagy in ICH and the appropriate implementation of autophagy in ICH therapy are essential. Nevertheless, there remain several challenges to exploring therapeutic applications of autophagy. For instance, although there is confidence in the potential benefits of therapeutic applications of autophagy in light of recent studies, further investigations are needed to determine whether autophagy upregulation or downregulation is the best approach, as well as to determine the appropriate time for such an intervention. Meanwhile, a better understanding of the autophagy mechanism in ICH pathophysiology and a means of determining which of the competing hypotheses is correct are both desirable. The integration of multi-omics studies, such as proteomics, genomics, and epigenetics, is also highly recommended. Furthermore, there are well-developed large-scale analysis methods and technologies that could be applied to the treatment-targeting study [129,136,138-141]. Finally, the prominent roles of autophagy and the ER stress-associated UPR in cell survival, as revealed in recent studies, have highlighted a new prospect for researchers investigating potential ICH therapies.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (grant no. 81473764 to Wei Zou) and the China Scholarship Council (certification no. 201408230060 to Mingming Niu). The authors thank Keith A. Laycock, PhD, ELS, for scientific editing of the manuscript.
Footnotes
Conflict of interest statement: The authors declare that they have no conflict of interest
References
- [1].Yorimitsu T., Klionsky D. J.. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12:1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Cirulli E. T., Lasseigne B. N., Petrovski S., Sapp P. C., Dion P. A., Leblond C. S., Science. New York, N.Y.: Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Chen C. W., Chen T. Y., Tsai K. L., Lin C. L., Yokoyama K. K., Lee W. S.. et al. Inhibition of autophagy as a therapeutic strategy of iron-induced brain injury after hemorrhage. Autophagy. 2012;8:1510–1520. doi: 10.4161/auto.21289. [DOI] [PubMed] [Google Scholar]
- [4].Hetz C.. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- [5].Rutkowski D. T., Hegde R. S.. Regulation of basal cellular physiology by the homeostatic unfolded protein response. J. Cell Biol. 2010;189:783–794. doi: 10.1083/jcb.201003138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Senft D., Ronai Z. A.. UPR, autophagy, and mitochondria crosstalk underlies the ER stress response. Trends Biochem. Sci. 2015;40:141–148. doi: 10.1016/j.tibs.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hollien J., Weissman J. S. Science. Vol. 313. New York, N.Y.: 2006. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response; pp. 104–107. [DOI] [PubMed] [Google Scholar]
- [8].Lin J. H., Li H., Yasumura D., Cohen H. R., Zhang C., Panning B., Science. Vol. 318. New York, N.Y.: 2007. IRE1 signaling affects cell fate during the unfolded protein response; pp. 944–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Aragon T., van Anken E., Pincus D., Serafimova I. M., Korennykh A. V., Rubio C. A.. et al. Messenger RNA targeting to endoplasmic reticulum stress signalling sites. Nature. 2009;457:736–740. doi: 10.1038/nature07641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Salminen A., Kaarniranta K., Kauppinen A., Ojala J., Haapasalo A., Soininen H.. et al. Impaired autophagy and APP processing in Alzheimer’s disease: The potential role of Beclin 1 interactome. Prog Neurobiol. 2013;106 doi: 10.1016/j.pneurobio.2013.06.002. [DOI] [PubMed] [Google Scholar]
- [11].Nijholt D. A., de Graaf T. R., van Haastert E. S., Oliveira A. O., Berkers C. R., Zwart R.. et al. Endoplasmic reticulum stress activates autophagy but not the proteasome in neuronal cells: implications for Alzheimer’s disease. Cell Death Differ. 2011;18:1071–1081. doi: 10.1038/cdd.2010.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Gan Or Z., Dion P. A., Rouleau G. A.. Genetic perspective on the role of the Autophagy-Lysosome Pathway in Parkinson disease. Autophagy. 2015;0 doi: 10.1080/15548627.2015.1067364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gould D. B., Phalan F. C., Breedveld G. J., van Mil S. E., Smith R. S., Schimenti J. C., Science. Vol. 308. (New York, N.Y.): 2005. Mutations in Col4a1 cause perinatal cerebral hemorrhage and porencephaly; pp. 1167–1171. [DOI] [PubMed] [Google Scholar]
- [14].Vahedi K., Kubis N., Boukobza M., Arnoult M., Massin P., Tournier-Lasserve E.. et al. COL4A1 mutation in a patient with sporadic, recurrent intracerebral hemorrhage. Stroke; a journal of cerebral circulation. 2007;38:1461–1464. doi: 10.1161/STROKEAHA.106.475194. [DOI] [PubMed] [Google Scholar]
- [15].Feigin V. L., Lawes C. M., Bennett D. A., Barker-Collo S. L., Parag V.. Worldwide stroke incidence and early case fatality reported in 56 population-based studies: a systematic review. Lancet Neurol. 2009;8:355–369. doi: 10.1016/S1474-4422(09)70025-0. [DOI] [PubMed] [Google Scholar]
- [16].Van Asch C. J., Luitse M. J., Rinkel G. J., van der Tweel I., Algra A., Klijn C. J.. Incidence, case fatality, and functional outcome of intracerebral haemorrhage over time, according to age, sex, and ethnic origin: a systematic review and meta-analysis. Lancet Neurol. 2010;9:167–176. doi: 10.1016/S1474-4422(09)70340-0. [DOI] [PubMed] [Google Scholar]
- [17].Keep R. F., Hua Y., Xi G.. Intracerebral haemorrhage: mechanisms of injury and therapeutic targets. Lancet Neurol. 2012;11:720–731. doi: 10.1016/S1474-4422(12)70104-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Feng Y., He D., Yao Z., Klionsky D. J.. The machinery of macroautophagy. Cell Res. 2014;24:24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Stricher F., Macri C., Ruff M., Muller S.. HSPA8/HSC70 chaperone protein: structure, function, and chemical targeting. Autophagy. 2013;9:1937–1954. doi: 10.4161/auto.26448. [DOI] [PubMed] [Google Scholar]
- [20].Kaushik S., Cuervo A. M.. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol. 2012;22:407–417. doi: 10.1016/j.tcb.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Mehta P., Henault J., Kolbeck R., Sanjuan M. A.. Noncanonical autophagy: one small step for LC3, one giant leap for immunity. Curr. Opin. Immunol. 2014;26:69–75. doi: 10.1016/j.coi.2013.10.012. [DOI] [PubMed] [Google Scholar]
- [22].Mizushima N., Yoshimori T., Ohsumi Y.. The role of Atg proteins in autophagosome formation. Annu. Rev Cell Dev. Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- [23].Furuya N., Yu J., Byfield M., Pattingre S., Levine B.. The Evolutionarily Conserved Domain of Beclin 1 is Required for Vps34 Binding, Autophagy, and Tumor Suppressor Function. Autophagy. 2005;1:46–52. doi: 10.4161/auto.1.1.1542. [DOI] [PubMed] [Google Scholar]
- [24].Tanida I., Minematsu-Ikeguchi N., Ueno T., Kominami E.. Lysosomal Turnover, but Not a Cellular Level, of Endogenous LC3 is a Marker for Autophagy. Autophagy. 2005;1:84–91. doi: 10.4161/auto.1.2.1697. [DOI] [PubMed] [Google Scholar]
- [25].Wong P.-M., Puente C., Ganley I. G., Jiang X.. The ULK1 complex. Autophagy. 2013;9:124–137. doi: 10.4161/auto.23323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Laplante M., Sabatini D. M.. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kim J., Kundu M., Viollet B., Guan K.-L.. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Alers S., Loffler A. S., Wesselborg S., Stork B.. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol Cell Biol. 2012;32:2–11. doi: 10.1128/MCB.06159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Nazio F., Strappazzon F., Antonioli M., Bielli P., Cianfanelli V., Bordi M.. et al. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol. 2013;15:406–416. doi: 10.1038/ncb2708. [DOI] [PubMed] [Google Scholar]
- [30].Strappazzon F., Vietri-Rudan M., Campello S., Nazio F., Florenzano F., Fimia G. M.. et al. Mitochondrial BCL-2 inhibits AMBRA1-induced autophagy. EMBO J. 2011;30:1195–1208. doi: 10.1038/emboj.2011.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ragusa M. J., Stanley R. E., Hurley J. H.. Architecture of the Atg17 complex as a scaffold for autophagosome biogenesis. Cell. 2012;151:1501–1512. doi: 10.1016/j.cell.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lu Q., Yang P., Huang X., Hu W., Guo B., Wu F.. et al. The WD40 repeat PtdIns(3)P-binding protein EPG-6 regulates progression of omegasomes to autophagosomes. Dev Cell. 2011;21:343–357. doi: 10.1016/j.devcel.2011.06.024. [DOI] [PubMed] [Google Scholar]
- [33].Vergne I., Roberts E., Elmaoued R. A., Tosch V., Delgado M. A., Proikas-Cezanne T.. et al. Control of autophagy initiation by phosphoinositide 3-phosphatase Jumpy. EMBO J. 2009;28:2244–2258. doi: 10.1038/emboj.2009.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Klionsky D. J.. The molecular machinery of autophagy: unanswered questions. J. Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Orsi A., Razi M., Dooley H. C., Robinson D., Weston A. E., Collinson L.M.. et al. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol. Biol Cell. 2012;23:1860–1873. doi: 10.1091/mbc.E11-09-0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zhou C., Ma K., Gao R., Mu C., Chen L., Liu Q.. et al. Regulation of mATG9 trafficking by Src- and ULK1-mediated phosphorylation in basal and starvation-induced autophagy. Cell Res. 2017;27:184–201. doi: 10.1038/cr.2016.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lamb C. A., Yoshimori T., Tooze S. A.. The autophagosome: origins unknown, biogenesis complex. Nat. Rev. Mol Cell Biol. 2013;14:759–774. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- [38].Kozutsumi Y., Segal M., Normington K., Gething M. J., Sambrook J.. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature. 1988;332:462–464. doi: 10.1038/332462a0. [DOI] [PubMed] [Google Scholar]
- [39].Cox J. S., Walter P.. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- [40].Rosbash M.. Mixed mechanisms in yeast pre-mRNA splicing? Cell. 1996;87:357–359. doi: 10.1016/s0092-8674(00)81355-0. [DOI] [PubMed] [Google Scholar]
- [41].Condello M., Caraglia M., Castellano M., Arancia G., Meschini S.. Structural and functional alterations of cellular components as revealed by electron microscopy. Microsc Res. Tech. 2013;76:1057–1069. doi: 10.1002/jemt.22266. [DOI] [PubMed] [Google Scholar]
- [42].Bertolotti A., Zhang Y., Hendershot L. M., Harding H. P., Ron D.. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- [43].Shen J., Snapp E. L., Lippincott-Schwartz J., Prywes R.. Stable binding of ATF6 to BiP in the endoplasmic reticulum stress response. Molecular and cellular biology. 2005;25:921–932. doi: 10.1128/MCB.25.3.921-932.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ron D., Walter P.. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev. Mol. Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- [45].Kouroku Y., Fujita E., Tanida I., Ueno T., Isoai A., Kumagai H.. et al. ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007;14:230–239. doi: 10.1038/sj.cdd.4401984. [DOI] [PubMed] [Google Scholar]
- [46].Zhang F., Hamanaka R. B., Bobrovnikova-Marjon E., Gordan J. D., Dai M.S., Lu H.. et al. Ribosomal stress couples the unfolded protein response to p53-dependent cell cycle arrest. J. Biol. Chem. 2006;281:30036–30045. doi: 10.1074/jbc.M604674200. [DOI] [PubMed] [Google Scholar]
- [47].Harding H. P., Zhang Y., Zeng H., Novoa I., Lu P. D., Calfon M.. et al. An Integrated Stress Response Regulates Amino Acid Metabolism and Resistance to Oxidative Stress. Mol Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- [48].Ghosh R., Wang L., Wang E. S., Perera B. G., Igbaria A., Morita S.. et al. Allosteric inhibition of the IRE1alpha RNase preserves cell viability and function during endoplasmic reticulum stress. Cell. 2014;158:534–548. doi: 10.1016/j.cell.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Calfon M., Zeng H., Urano F., Till J. H., Hubbard S. R., Harding H. P.. et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- [50].Hetz C., Martinon F., Rodriguez D., Glimcher L. H.. The unfolded protein response: integrating stress signals through the stress sensor IRE1alpha. Physiol. Rev. 2011;91:1219–1243. doi: 10.1152/physrev.00001.2011. [DOI] [PubMed] [Google Scholar]
- [51].Haze K., Okada T., Yoshida H., Yanagi H., Yura T., Negishi M.. et al. Identification of the G13 (cAMP-response-element-binding protein-related protein) gene product related to activating transcription factor 6 as a transcriptional activator of the mammalian unfolded protein response. Biochem. J. 2001;355:19–28. doi: 10.1042/0264-6021:3550019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Thuerauf D. J., Marcinko M., Belmont P. J., Glembotski C. C.. Effects of the isoform-specific characteristics of ATF6 alpha and ATF6 beta on endoplasmic reticulum stress response gene expression and cell viability. J. Biol. Chem. 2007;282:22865–22878. doi: 10.1074/jbc.M701213200. [DOI] [PubMed] [Google Scholar]
- [53].Nadanaka S., Okada T., Yoshida H., Mori K.. Role of disulfide bridges formed in the luminal domain of ATF6 in sensing endoplasmic reticulum stress. Mol Cell Biol. 2007;27:1027–1043. doi: 10.1128/MCB.00408-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Appenzeller-Herzog C., Hall M. N.. Bidirectional crosstalk between endoplasmic reticulum stress and mTOR signaling. Trends Cell Biol. 2012;22:274–282. doi: 10.1016/j.tcb.2012.02.006. [DOI] [PubMed] [Google Scholar]
- [55].Yamamoto K., Sato T., Matsui T., Sato M., Okada T., Yoshida H.. et al. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell. 2007;13:365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- [56].Hetz C., Chevet E., Oakes S. A.. Proteostasis control by the unfolded protein response. Nat Cell Biol. 2015;17:829–838. doi: 10.1038/ncb3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Fonseca S. G., Ishigaki S., Oslowski C. M., Lu S., Lipson K. L., Ghosh R.. et al. Wolfram syndrome 1 gene negatively regulates ER stress signaling in rodent and human cells. J. Clin. Invest. 2010;120:744–755. doi: 10.1172/JCI39678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Higa A., Taouji S., Lhomond S., Jensen D., Fernandez-Zapico M. E., Simpson J. C.. et al. Endoplasmic reticulum stress-activated transcription factor ATF6alpha requires the disulfide isomerase PDIA5 to modulate chemoresistance. Mol Cell Biol. 2014;34:1839–1849. doi: 10.1128/MCB.01484-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Digaleh H., Kiaei M., Khodagholi F.. Nrf2 and Nrf 1 signaling and ER stress crosstalk: implication for proteasomal degradation and autophagy. Cell Mol Life Sci. 2013;70:4681–4694. doi: 10.1007/s00018-013-1409-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Houck S. A., Ren H. Y., Madden V. J., Bonner J. N., Conlin M. P., Janovick J. A.. et al. Quality control autophagy degrades soluble ERAD-resistant conformers of the misfolded membrane protein GnRHR. Mol Cell. 2014;54:166–179. doi: 10.1016/j.molcel.2014.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Qin L., Wang Z., Tao L., Wang Y.. ER stress negatively regulates AKT/TSC/mTOR pathway to enhance autophagy. Autophagy. 2010;6:239–247. doi: 10.4161/auto.6.2.11062. [DOI] [PubMed] [Google Scholar]
- [62].Yang Z., Klionsky D. J.. Mammalian autophagy: core molecular machinery and signaling regulation. Current Opinion in Cell Biology. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Hosokawa N., Hara T., Kaizuka T., Kishi C., Takamura A., Miura Y.. et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol. Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].B’Chir W., Maurin A. C., Carraro V., Averous J., Jousse C., Muranishi Y.. et al. The eIF2alpha/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 2013;41:7683–7699. doi: 10.1093/nar/gkt563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Adolph T. E., Tomczak M. F., Niederreiter L., Ko H. J., Bock J., Martinez-Naves E.. et al. Paneth cells as a site of origin for intestinal inflammation. Nature. 2013;503:272–276. doi: 10.1038/nature12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Ogata M., Hino S., Saito A., Morikawa K., Kondo S., Kanemoto S.. et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Deegan S., Saveljeva S., Gorman A. M., Samali A.. Stress-induced self-cannibalism: on the regulation of autophagy by endoplasmic reticulum stress. Cell Mol Life Sci. 2013;70:2425–2441. doi: 10.1007/s00018-012-1173-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Rashid H. O., Yadav R. K., Kim H. R., Chae H. J.. ER stress: Autophagy induction, inhibition and selection. Autophagy. 2015;11:1956–1977. doi: 10.1080/15548627.2015.1091141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Xi G., Keep R. F., Hoff J. T.. Mechanisms of brain injury after intracerebral haemorrhage. Lancet Neurol. 2006;5:53–63. doi: 10.1016/S1474-4422(05)70283-0. [DOI] [PubMed] [Google Scholar]
- [70].Morgenstern L. B., Hemphill J. C., Anderson C., Becker K., Broderick J. P., Connolly E. S. Jr.. et al. uidelines for the management of spontaneous intracerebral hemorrhage: a guideline for healthcare professionals from the American Heart Association/American Stroke Association. Stroke; a journal of cerebral circulation. 2010;41:2108–2129. doi: 10.1161/STR.0b013e3181ec611b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Liu J., Gao B. B., Clermont A. C., Blair P., Chilcote T. J., Sinha S.. et al. Hyperglycemia-induced cerebral hematoma expansion is mediated by plasma kallikrein. Nat Med. 2011;17:206–210. doi: 10.1038/nm.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Herweh C., Juttler E., Schellinger P. D., Klotz E., Jenetzky E., Orakcioglu B.. et al. Evidence against a perihemorrhagic penumbra provided by perfusion computed tomography Stroke. a journal of cerebral circulation. 2007;38:2941–2947. doi: 10.1161/STROKEAHA.107.486977. [DOI] [PubMed] [Google Scholar]
- [73].Urday S., Beslow L. A., Goldstein D. W., Vashkevich A., Ayres A. M., Battey T. W.. et al. Measurement of perihematomal edema in intracerebral hemorrhage Stroke. a journal of cerebral circulation. 2015;46:1116–1119. doi: 10.1161/STROKEAHA.114.007565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Aronowski J., Zhao X.. Molecular pathophysiology of cerebral hemorrhage: secondary brain injury Stroke. a journal of cerebral circulation. 2011;42:1781–1786. doi: 10.1161/STROKEAHA.110.596718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Guo F., Hua Y., Wang J., Keep R. F., Xi G.. Inhibition of carbonic anhydrase reduces brain injury after intracerebral hemorrhage. Transl Stroke Res. 2012;3:130–137. doi: 10.1007/s12975-011-0106-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Mizushima N., Levine B., Cuervo A. M., Klionsky D. J.. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Nixon R. A.. The role of autophagy in neurodegenerative disease. Nat. Med. 2013;19:983–997. doi: 10.1038/nm.3232. [DOI] [PubMed] [Google Scholar]
- [78].Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I.. et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- [79].Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R.. et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- [80].Liu C., Gao Y., Barrett J., Hu B.. Autophagy and protein aggregation after brain ischemia. J. Neurochem. 2010;115:68–78. doi: 10.1111/j.1471-4159.2010.06905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Adhami F., Schloemer A., Kuan C. Y.. The roles of autophagy in cerebral ischemia. Autophagy. 2007;3:42–44. doi: 10.4161/auto.3412. [DOI] [PubMed] [Google Scholar]
- [82].Zhu C., Wang X., Xu F., Bahr B. A., Shibata M., Uchiyama Y.. et al. The influence of age on apoptotic and other mechanisms of cell death after cerebral hypoxia-ischemia. Cell Death Differ. 2005;12:162–176. doi: 10.1038/sj.cdd.4401545. [DOI] [PubMed] [Google Scholar]
- [83].Sarkar C., Zhao Z., Aungst S., Sabirzhanov B., Faden A. I., Lipinski M. M.. Impaired autophagy flux is associated with neuronal cell death after traumatic brain injury. Autophagy. 2014;10:2208–2222. doi: 10.4161/15548627.2014.981787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].He Y., Wan S., Hua Y., Keep R. F., Xi G.. Autophagy after experimental intracerebral hemorrhage. J. Cereb. Blood Flow Metab. 2008;28:897–905. doi: 10.1038/sj.jcbfm.9600578. [DOI] [PubMed] [Google Scholar]
- [85].Wang Z., Shi X.-Y., Yin J., Zuo G., Zhang J., Chen G.. Role of Autophagy in Early Brain Injury after Experimental Subarachnoid Hemorrhage. J. Mol. Neurosci. 2012;46:192–202. doi: 10.1007/s12031-011-9575-6. [DOI] [PubMed] [Google Scholar]
- [86].Gong Y., He Y., Gu Y., Keep R. F., Xi G., Hua Y.. Effects of aging on autophagy after experimental intracerebral hemorrhage. Acta Neurochir. Suppl. 2011;111:113–117. doi: 10.1007/978-3-7091-0693-8_18. [DOI] [PubMed] [Google Scholar]
- [87].Hu S., Xi G., Jin H., He Y., Keep R. F., Hua Y.. Thrombin-induced autophagy: A potential role in intracerebral hemorrhage. Brain Research. 2011;1424:60–66. doi: 10.1016/j.brainres.2011.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Hwang B. Y., Appelboom G., Ayer A., Kellner C. P., Kotchetkov I. S., Gigante P. R.. et al. Advances in neuroprotective strategies: potential therapies for intracerebral hemorrhage. Cerebrovasc. Dis. 2011;31:211–222. doi: 10.1159/000321870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Wang J., Dore S.. Inflammation after intracerebral hemorrhage. J. Cereb. Blood Flow Metab. 2007;27:894–908. doi: 10.1038/sj.jcbfm.9600403. [DOI] [PubMed] [Google Scholar]
- [90].Yang Z., Zhao T.-z., Zou Y.-j., Zhang J. H., Feng H.. Hypoxia Induces Autophagic Cell Death through Hypoxia-Inducible Factor 1a in Microglia. PLoS ONE. 2014;9:e96509. doi: 10.1371/journal.pone.0096509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Yu A., Zhang T., Zhong W., Duan H., Wang S., Ye P.. et al. miRNA-144 induces microglial autophagy and inflammation following intracerebral hemorrhage. Immunol. Lett. 2017;182:18–23. doi: 10.1016/j.imlet.2017.01.002. [DOI] [PubMed] [Google Scholar]
- [92].Remijsen Q., Berghe T. V., Wirawan E., Asselbergh B., Parthoens E., De Rycke R.. et al. Neutrophil extracellular trap cell death requires both autophagy and superoxide generation. Cell Res. 2011;21:290–304. doi: 10.1038/cr.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Moxon-Emre I., Schlichter L. C.. Neutrophil Depletion Reduces Blood-Brain Barrier Breakdown, Axon Injury, and Inflammation After Intracerebral Hemorrhage. J. Neuropathol. Exp. Neurol. 2011;70:218–235. doi: 10.1097/NEN.0b013e31820d94a5. [DOI] [PubMed] [Google Scholar]
- [94].Wen Y.-D., Sheng R., Zhang L. S., Han R., Zhang X., Zhang X.-D.. et al. Neuronal injury in rat model of permanent focal cerebral ischemia is associated with activation of autophagic and lysosomal pathways. Autophagy. 2008;4:762–769. doi: 10.4161/auto.6412. [DOI] [PubMed] [Google Scholar]
- [95].Araki T., Hayashi M., Saruta T.. Anion-exchange blocker enhances cytoplasmic vacuole formation and cell death in serum-deprived mouse kidney epithelial cells in mice. Cell Biol. Int. 2006;30:93–100. doi: 10.1016/j.cellbi.2005.10.020. [DOI] [PubMed] [Google Scholar]
- [96].Hua Y., Keep R. F., Hoff J. T., Xi G.. Brain injury after intracerebral hemorrhage: the role of thrombin and iron Stroke. a journal of cerebral circulation. 2007;38:759–762. doi: 10.1161/01.STR.0000247868.97078.10. [DOI] [PubMed] [Google Scholar]
- [97].Manning A. M., Davis R. J.. Targeting JNK for therapeutic benefit: from junk to gold? Nat. Rev. Drug Discov. 2003;2:554–565. doi: 10.1038/nrd1132. [DOI] [PubMed] [Google Scholar]
- [98].Wan S., Zhan R., Zheng S., Hua Y., Xi G.. Activation of c-Jun-N-terminal kinase in a rat model of intracerebral hemorrhage: the role of iron. Neurosci. Res. 2009;63:100–105. doi: 10.1016/j.neures.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Sharp F., Liu D.-Z., Zhan X., Ander B. P. In Cerebral Hemorrhage. Springer Vienna; Vienna: 2008. [Google Scholar]
- [100].Wagner K. R.. Modeling intracerebral hemorrhage: glutamate, nuclear factor-kappa B signaling and cytokines Stroke. a journal of cerebral circulation. 2007;38:753–758. doi: 10.1161/01.STR.0000255033.02904.db. [DOI] [PubMed] [Google Scholar]
- [101].Chen Z., Lu T., Yue X., Wei N., Jiang Y., Chen M.. et al. Neuroprotective effect of ginsenoside Rb1 on glutamate-induced neurotoxicity: With emphasis on autophagy. Neurosci. Lett. 2010;482:264–268. doi: 10.1016/j.neulet.2010.07.052. [DOI] [PubMed] [Google Scholar]
- [102].Xu C., Bailly-Maitre B., Reed J. C.. Endoplasmic reticulum stress: cell life and death decisions. J. Clin. Invest. 2005;115:2656–2664. doi: 10.1172/JCI26373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Urabe N., Naito I., Saito K., Yonezawa T., Sado Y., Yoshioka H.. et al. Basement membrane type IV collagen molecules in the choroid plexus, pia mater and capillaries in the mouse brain. Arch. Histol. Cytol. 2002;65:133–143. doi: 10.1679/aohc.65.133. [DOI] [PubMed] [Google Scholar]
- [104].Jeanne M., Labelle-Dumais C., Jorgensen J., Kauffman W. B., Mancini G. M., Favor J.. et al. COL4A2 mutations impair COL4A1 and COL4A2 secretion and cause hemorrhagic stroke. Am. J. Hum. Genet. 2012;90:91–101. doi: 10.1016/j.ajhg.2011.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Murray L. S., Lu Y., Taggart A., Van Regemorter N., Vilain C., Abramowicz M.. et al. Chemical chaperone treatment reduces intracellular accumulation of mutant collagen IV and ameliorates the cellular phenotype of a COL4A2 mutation that causes haemorrhagic stroke. Hum. Mol. Genet. 2014;23:283–292. doi: 10.1093/hmg/ddt418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Yang L., Li P., Fu S., Calay E. S., Hotamisligil G. S.. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Codogno P., Meijer A. J.. Autophagy: a potential link between obesity and insulin resistance. Cell Metab. 2010;11:449–451. doi: 10.1016/j.cmet.2010.05.006. [DOI] [PubMed] [Google Scholar]
- [108].Kaushik S., Rodriguez-Navarro J. A., Arias E., Kiffin R., Sahu S., Schwartz G. J.. et al. Autophagy in hypothalamic AgRP neurons regulates food intake and energy balance. Cell Metab. 2011;14:173–183. doi: 10.1016/j.cmet.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].He C., Bassik M. C., Moresi V., Sun K., Wei Y., Zou Z.. et al. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature. 2012;481:511–515. doi: 10.1038/nature10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Levine B., Mizushima N., Virgin H. W.. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Meissner F., Molawi K., Zychlinsky A. Proc. Natl. Acad. Sci. Vol. 107. U. S. A.: 2010. Mutant superoxide dismutase 1-induced IL-1beta accelerates ALS pathogenesis; pp. 13046–13050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Saitoh T., Fujita N., Jang M. H., Uematsu S., Yang B. G., Satoh T.. et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- [113].Yang Z. J., Chee C. E., Huang S., Sinicrope F. A.. The role of autophagy in cancer: therapeutic implications. Mol Cancer Ther. 2011;10:1533–1541. doi: 10.1158/1535-7163.MCT-11-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Mathew R., Karantza-Wadsworth V., White E.. Role of autophagy in cancer. Nat. Rev. Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Takamura A., Komatsu M., Hara T., Sakamoto A., Kishi C., Waguri S.. et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Degenhardt K., Mathew R., Beaudoin B., Bray K., Anderson D., Chen G.. et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].White E., DiPaola R. S.. The double-edged sword of autophagy modulation in cancer. Clin. Cancer Res. 2009;15:5308–5316. doi: 10.1158/1078-0432.CCR-07-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Ding W. X., Ni H. M., Gao W., Chen X., Kang J. H., Stolz D. B.. et al. Oncogenic transformation confers a selective susceptibility to the combined suppression of the proteasome and autophagy. Mol Cancer Ther. 2009;8:2036–2045. doi: 10.1158/1535-7163.MCT-08-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Morselli E., Galluzzi L., Kepp O., Marino G., Michaud M., Vitale I.. et al. Oncosuppressive functions of autophagy. Antioxid Redox Signal. 2011;14:2251–2269. doi: 10.1089/ars.2010.3478. [DOI] [PubMed] [Google Scholar]
- [120].Yin D, Xia X, Zhang J, Zhang S, Liao F, Zhang G. et al. Impact of NUDT15 polymorphisms on thiopurines-induced myelotoxicity and thiopurines tolerance dose. Oncotarget. 2017 doi: 10.18632/oncotarget.14594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Rubinsztein D. C, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat. Rev. Drug Discov. 2012;11:709–730. doi: 10.1038/nrd3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Michaud M, Martins I, Sukkurwala A. Q, Adjemian S, Ma Y, Pellegatti P, Science. Vol. 334. New York, N.Y.: 2011. Autophagy-dependent anticancer immune responses induced by chemotherapeutic agents in mice; pp. 1573–1577. [DOI] [PubMed] [Google Scholar]
- [123].Rao S, Tortola L, Perlot T, Wirnsberger G, Novatchkova M, Nitsch R. et al. A dual role for autophagy in a murine model of lung cancer. Nat. Commun. 2014;5:3056. doi: 10.1038/ncomms4056. [DOI] [PubMed] [Google Scholar]
- [124].Jin S. M, Jang H. W, Sohn S. Y, Kim N. K, Joung J. Y, Cho Y. Y. et al. Role of autophagy in the resistance to tumour necrosis factor-related apoptosis-inducing ligand-induced apoptosis in papillary and anaplastic thyroid cancer cells. Endocrine. 2014;45:256–262. doi: 10.1007/s12020-013-9997-8. [DOI] [PubMed] [Google Scholar]
- [125].Marino G, Niso-Santano M, Baehrecke E. H, Kroemer G. Self-consumption: the interplay of autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 2014;15:81–94. doi: 10.1038/nrm3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Janku F, McConkey D. J, Hong D. S, Kurzrock R. Autophagy as a target for anticancer therapy. Nat. Rev. Clin. Oncol. 2011;8:528–539. doi: 10.1038/nrclinonc.2011.71. [DOI] [PubMed] [Google Scholar]
- [127].Pal I, Parida S, Prashanth Kumar B. N, Banik P, Kumar Dey K, Chakraborty S. et al. Blockade of autophagy enhances proapoptotic potential of BI-69A11, a novel Akt inhibitor, in colon carcinoma. Eur. J. Pharmacol. 2015;765:217–227. doi: 10.1016/j.ejphar.2015.08.039. [DOI] [PubMed] [Google Scholar]
- [128].Sheng R, Liu X. Q, Zhang L. S, Gao B, Han R, Wu Y. Q. et al. Autophagy regulates endoplasmic reticulum stress in ischemic preconditioning. Autophagy. 2012;8:310–325. doi: 10.4161/auto.18673. [DOI] [PubMed] [Google Scholar]
- [129].Li L, Tian J, Long M K.-W, Chen Y, Lu J, Zhou C. et al. Protection against Experimental Stroke by Ganglioside GM1 Is Associated with the Inhibition of Autophagy. PLoS ONE. 2016;11:e0144219. doi: 10.1371/journal.pone.0144219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Shoji-Kawata S, Sumpter R, Leveno M, Campbell G. R, Zou Z, Kinch L. et al. Identification of a candidate therapeutic autophagy-inducing peptide. Nature. 2013;494:201–206. doi: 10.1038/nature11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Yuan B, Shen H, Lin L, Su T, Zhong L, Yang Z. Autophagy Promotes Microglia Activation Through Beclin-1-Atg5 Pathway in Intracerebral Hemorrhage. Mol. Neurobiol. 2016:1–10. doi: 10.1007/s12035-015-9642-z. [DOI] [PubMed] [Google Scholar]
- [132].Yang Z, Liu B, Zhong L, Shen H, Lin C, Lin L. et al. Toll-like receptor-4-mediated autophagy contributes to microglial activation and inflammatory injury in mouse models of intracerebral haemorrhage. Neuropath. Appl. Neuro. 2015;41:e95–e106. doi: 10.1111/nan.12177. [DOI] [PubMed] [Google Scholar]
- [133].Wu Z, Zou X, Zhu W, Mao Y, Chen L, Zhao F. Minocycline is effective in intracerebral hemorrhage by inhibition of apoptosis and autophagy. J. Neurol. Sci. 2016;371:88–95. doi: 10.1016/j.jns.2016.10.025. [DOI] [PubMed] [Google Scholar]
- [134].Rose C, Menzies F. M, Renna M, Acevedo-Arozena A, Corrochano S, Sadiq O. et al. Rilmenidine attenuates toxicity of polyglutamine expansions in a mouse model of Huntington’s disease. Hum. Mol. Genet. 1992;39:292–313. doi: 10.1093/hmg/ddq093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Rubinsztein D. C, Marino G, Kroemer G. Autophagy and agin. Cell. 2011;146:682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- [136].Wang P, Xu T.-Y, Wei K, Guan Y.-F, Wang X, Xu H. et al. ARRB1/β-arrestin-1 mediates neuroprotection through coordination of BECN1-dependent autophagy in cerebral ischemia. Autophagy. 2014;10:1535–1548. doi: 10.4161/auto.29203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Zhang X, Yuan Y, Jiang L, Zhang J, Gao J, Shen Z. et al. Endoplasmic reticulum stress induced by tunicamycin and thapsigargin protects against transient ischemic brain injury: Involvement of PARK2-dependent mitophagy. Autophagy. 2014;10:1801–1813. doi: 10.4161/auto.32136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Tan H, Wu Z, Wang H, Bai B, Li Y, Wang X. et al. Refined phosphopeptide enrichment by phosphate additive and the analysis of human brain phosphoproteome. Proteomics. 2014;15:500–507. doi: 10.1002/pmic.201400171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Wang H, Yang Y, Li Y, Bai B, Wang X, Tan H. et al. Systematic optimization of long gradient chromatography mass spectrometry for deep analysis of brain proteome. J. Proteome Res. 2015;14:829–838. doi: 10.1021/pr500882h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Wang X, Li Y, Wu Z, Wang H, Tan H, Peng J. JUMP: A Tag-based Database Search Tool for Peptide Identification with High Sensitivity and Accuracy. Mol. Cell Proteomics. 2014;13:3663–3673. doi: 10.1074/mcp.O114.039586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Li Y, Wang X, Cho J. H, Shaw T. I, Wu Z, Bai B. et al. JUMPg: An Integrative Proteogenomics Pipeline Identifying Unannotated Proteins in Human Brain and Cancer Cells. J. Proteome Res. 2016;15:2309–2320. doi: 10.1021/acs.jproteome.6b00344. [DOI] [PMC free article] [PubMed] [Google Scholar]