Abstract
Objective
Circadian Clock gene mutant mice show dampened 24-h feeding rhythms and an increased sensitivity to high-fat diet (HFD) feeding. Restricting HFD access to the dark phase counteracts its obesogenic effect in wild-type mice. The extent to which altered feeding rhythms are causative for the obesogenic phenotype of Clock mutant mice, however, remains unknown.
Methods
Metabolic parameters of wild-type (WT) and ClockΔ19 mutant mice (MT) were investigated under ad libitum and nighttime restricted HFD feeding. Liver circadian clock function was partially rescued by hydrodynamic tail vein delivery of WT-Clock DNA vectors in mutant mice and transcriptional, metabolic, endocrine and behavioral rhythms studied.
Results
Nighttime-restricted feeding restored food intake, but not body weight regulation in MT mice under HFD, suggesting Clock-dependent metabolic dysregulation downstream of circadian appetite control. Liver-directed Clock gene therapy partially restored liver circadian oscillator function and transcriptome regulation without affecting centrally controlled circadian behaviors. Under HFD, MT mice with partially restored liver clock function (MT-LR) showed normalized body weight gain, rescued 24-h food intake rhythms, and WT-like energy expenditure. This was associated with decreased nighttime leptin and daytime ghrelin levels, reduced hepatic lipid accumulation, and improved glucose tolerance. Transcriptome analysis revealed that hepatic Clock rescue in MT mice affected a range of metabolic pathways.
Conclusion
Liver Clock gene therapy improves resistance against HFD-induced metabolic impairments in mice with circadian clock disruption. Restoring or stabilizing liver clock function might be a promising target for therapeutic interventions in obesity and metabolic disorders.
Keywords: Circadian clock, Clock gene, High-fat diet, Liver, Transcription, Gene therapy
Graphical abstract
Highlights
-
•
Normalizing diurnal feeding rhythms does not rescue body weight regulation in Clock mutant mice.
-
•
Gene therapy restores liver clock function in Clock mutant mice.
-
•
Liver clock rescue improves metabolic homeostasis under high-fat diet conditions.
1. Introduction
In response to the Earth's rotation around its axis, most species have evolved endogenous circadian clocks, enabling them to adapt behavior and physiology to the 24-h rhythm of day and night [1], [2]. At the molecular level, these clocks are based on transcriptional-translational feedback loops built from a set of clock genes/proteins including the two transcription factors circadian locomotor output cycles kaput (CLOCK) and brain and muscle ANRT-like 1 (BMAL1 or ARNTL), which together drive rhythmic expression of three Period and two Cryptochrome genes through binding to E-box enhancer motifs. BMAL1 and CLOCK further regulate many other E-box-controlled genes in a tissue-specific manner, thereby translating the circadian clock rhythm into physiologically meaningful signals [3], [4]. Clocks are found in all body tissues and are synchronized via a master pacemaker located in the hypothalamic suprachiasmatic nucleus (SCN) [5], [6] that is entrained by the external light rhythm. Together, peripheral clocks and SCN-controlled sleep-wake and food intake rhythms regulate the expression of many metabolically relevant genes [7]. Interestingly, peripheral clocks not only respond to SCN signaling but also are reset by the timing of food intake. Therefore, mistimed feeding rhythms – as frequently occur in modern industrialized societies – can promote internal clock desynchrony and the development of metabolic disorders [8], [9], [10], [11], [12], [13].
The liver is the largest metabolic organ of the body. Transcriptome analyses have identified more than 3,000 rhythmic transcripts [14] and chromatin immunoprecipitation/DNA sequencing experiments revealed more than 2,000 DNA binding sites for BMAL1 in the murine liver [15]. Circadian regulation has been shown for several metabolic processes such as xenobiotic detoxification [16], [17], mitochondrial function [18], and lipid and glucose metabolism [14], [19], [20]. Mice with hepatocyte-specific abrogation of clock function through deletion of Bmal1 display impaired glucose homeostasis but normal body weight regulation [13].
In contrast, mice carrying a dominant negative mutation in the gene encoding the BMAL1 partner CLOCK (ClockΔ19) are overweight and, under high-fat diet conditions, develop symptoms of the metabolic syndrome [21]. This obesogenic phenotype is associated with deregulated feeding rhythms and over-eating during the normal rest phase. In addition, mistimed feeding is associated with the development of obesity in mice and humans [13], [22], [23], [24]. Moreover, in wild-type mice, restricting access to a HFD to the nighttime improves clock gene rhythms and normalizes body weight regulation [25], [26].
Appetite regulation and energy expenditure are centrally controlled and, thus, are difficult targets for clinical interventions [27], [28], [29]. On the other hand, metabolic feedback signals from the periphery such as leptin and ghrelin, but also liver-derived factors such as fibroblast growth factor 21 (FGF-21) [30] and ketone bodies [31], reach the brain and modulate neuronal circuits to adjust energy metabolism [32]. This bottom-up communication from peripheral metabolic tissues to central regulatory circuits is impaired during obesity [33], [34]. Thus, targeting the circadian regulation of metabolic feedback signals by manipulating peripheral clock function may provide a feasible means of restoring homeostatic set points in metabolic disorders. To test this hypothesis, we investigated the effects of liver Clock gene therapy on feeding behavior and energy metabolism in ClockΔ19 mutant mice.
2. Materials and methods
2.1. Mice
Adult male C57BL/6J (WT) and congenic homozygous ClockΔ19 (MT) [35] mice were bred under standard laboratory conditions with a temperature of 21 ± 2 °C, a relative humidity of 50 ± 5% and a 12-h:12-h light–dark cycle (LD; light phase: 200 ± 50 lux). Before mice were allocated to the experiments, they were group-housed with ad libitum access to standard rodent chow (58% carbohydrates, 33% protein, 9% fat; Ssniff, Germany) and water. After adaptation to single housing, 10-week old mice were assigned to the different experiments with either ad libitum access to standard rodent chow or regulated access (ad libitum vs. nighttime only) to HFD (45% kJ fat, Ssniff EF D12451) for 10 weeks. Food intake, body weight, locomotor activity, and energy expenditure were measured; then, animals were sacrificed by cervical dislocation for molecular characterization. All experiments were ethically assessed and licensed by the Office of Consumer Protection and Food Safety of the State of Lower Saxony and the Ministry of Energy Change, Agriculture, Environment and Rural Areas of the State of Schleswig–Holstein and performed according to international guidelines on the ethical use of experimental animals.
2.1.1. Feeding regimes
The effects of timed food access under HFD were analyzed in WT and MT mice (n = 12/group). Both genotypes received a HFD for 10 weeks either ad libitum (WT-ad libitum; MT-ad libitum) or restricted to the 12-h dark phase (restricted feeding: RF; WT-RF, MT-RF). Body weight and food intake were measured weekly. An additional cohort was analyzed for energy expenditure at 4 weeks into the feeding regime, followed by activity analysis one week later. In a second experiment, we restored the function of the liver Clock gene (liver rescue: LR; WT-LR, MT-LR) by hydrodynamic tail vein delivery (see below) and analyzed the HFD effects in these mice. 10-week old WT (n = 24), WT-LR (n = 13; data not shown), MT (n = 26) and MT-LR mice (n = 24) received a HFD ad libitum for ten weeks. Body weight and food intake were measured weekly; activity, energy expenditure, and other metabolic parameters (hormone levels, glucose tolerance) were investigated in an additional cohort in the middle of the feeding experiment after 5 weeks of HFD.
2.1.2. Activity analysis
Locomotor activity of single-housed mice was recorded either by custom-made infrared (IR) detectors placed on the cage lid or by running-wheels. IR recordings were performed for one week under standard LD conditions either in week 2 of the RF-paradigm (Figure 1: n = 12/group) or starting at week 5 after tail vein injection (Figure 4: n = 8/group). Wheel-running activity of adult ad libitum chow-fed WT, MT, and MT-LR was recorded for two weeks under standard LD conditions followed by a 4-week period in constant darkness (DD; Figure 3, n = 12 per group). IR and running-wheel measurements were analyzed using ClockLab software (Actimetrics, Evanston, USA).
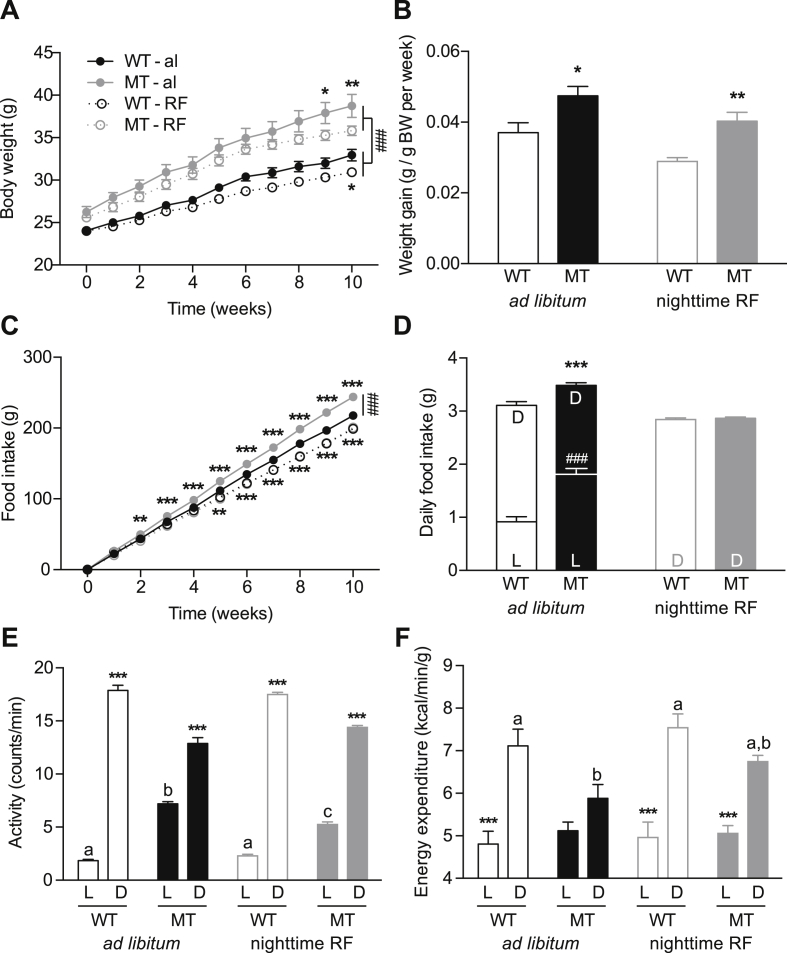
Figure 1.
Effect of nighttime restricted feeding on metabolic responses of WT and MT to HFD. A, B: Body weight development (A) and weekly weight gain (B) of WT and MT mice under ad libitum or nighttime-restricted (RF) access to HFD. C, D: Cumulative (C) and daily food intake (D) of ad libitum and nighttime-fed WT and MT mice. E, F: Daily activity (E) and energy expenditure (EE) (F) of ad libitum and nighttime-fed WT and MT mice. Data are shown as means ± SEM (n = 12 per group); 2-way ANOVA with Bonferroni post-test; *: p < 0.05; **: p < 0.01; ***/###: p < 0.001. L – light phase; D – dark phase. Different letters (a, b, …) depict statistically significant differences (p < 0.05).
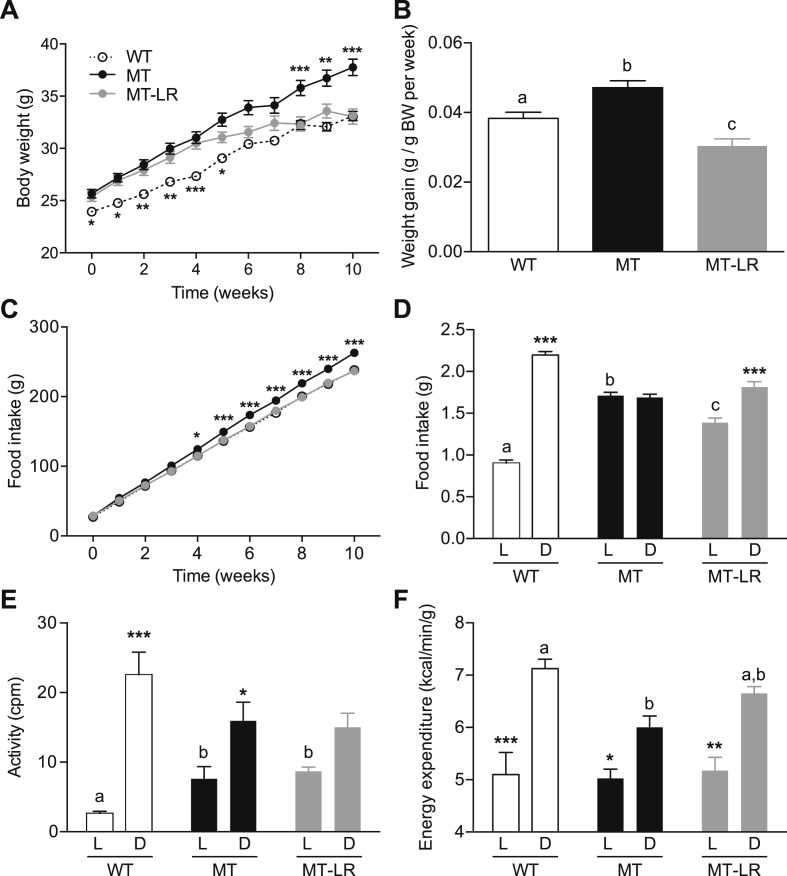
Figure 4.
Liver Clock gene therapy restores HFD responses in Clock mutant mice. A, B: Body weight development (A) and weekly weight gain (B) of WT, MT, and MT-LR mice under ad libitum access to HFD (n = 24–26). C, D: Cumulative (C) and daily food intake (D) of ad libitum HFD-fed WT, MT, and MT-LR mice (n = 24–26). E, F: Light:dark phase activity (E; n = 8) and energy expenditure (F; n = 12) and of ad libitum HFD-fed WT, MT, and MT-LR mice. Data are shown as means ± SEM; 1- and 2-way ANOVAs with Bonferroni post-test; different letters depict statistically different groups (p < 0.05); ***: p < 0.001 MT or WT vs. MT-LR (A, C) or light (L) vs. dark (D) groups (D–F).
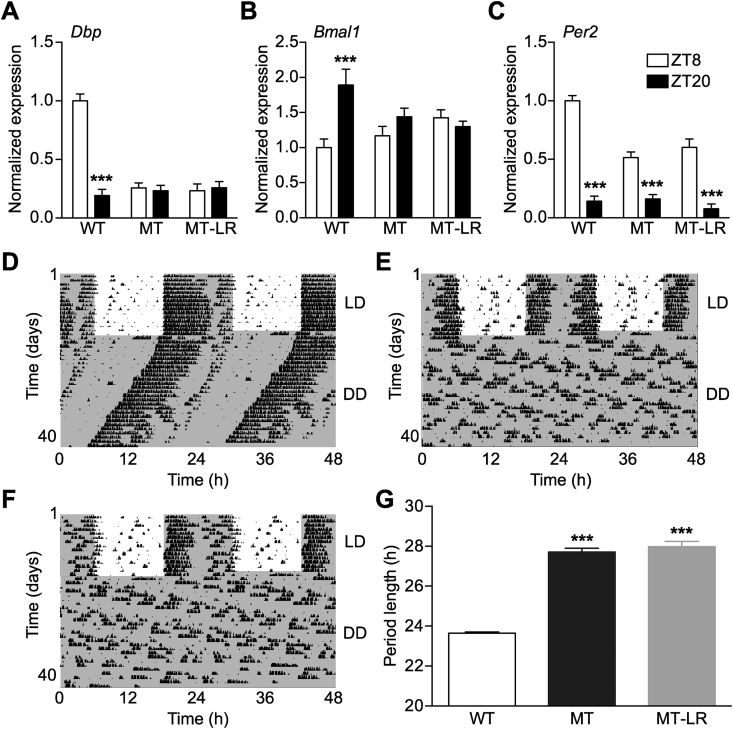
Figure 3.
Liver Clock gene therapy does not affect central pacemaker function. A–C: Day-night variations in SCN mRNA levels of Dbp (A), Bmal1 (B), and Per2 (C) analyzed by radioactive in situ hybridization (ZT: Zeitgeber time; means ± SEM (n = 8); 2-way ANOVA with Bonferroni post-test; ***: p < 0.001). D–F: Representative running-wheel recordings of WT (D), MT (E), and MT-LR mice (F) under 12-h:12-h light:dark (LD) and constant darkness conditions (DD). Actograms are double-plotted; dark phases are marked with grey shading. (F) Activity period length in DD (means ± SEM (n = 8); 1-way ANOVA with Bonferroni post-test; ***: p < 0.001).
2.1.3. Energy expenditure
Oxygen consumption and carbon dioxide production were investigated under standard housing conditions (LD, 21 °C) and the introduced feeding paradigm using an open-circuit indirect calorimetry system (TSE PhenoMaster Systems, Bad Homburg, Germany) after 2 weeks on RF-paradigm (Figure 1; n = 12 per group) or after 4 weeks on HFD (Figure 4; n = 24–26 per group). Mice were allowed to acclimatize to the system for 2 days, and then data were acquired in intervals of 15 min for 3–4 days. Energy expenditure was normalized to the individual body weight [36].
2.2. Liver Clock gene therapy
Hepatic Clock gene expression was manipulated by hydrodynamic tail vein injection [37]. In brief, either WT or MT mice were injected with a vector encoding the full-length, hemagglutinin (HA)-tagged wild-type Clock (GenBank AF000998), secreted embryonic alkaline phosphatase (SEAP), or lacZ cDNA under the control of a chimeric promoter composed of the mouse alpha fetoprotein enhancer II and a minimal mouse albumin (Alb1) promoter aimed at restricting expression specifically to hepatocytes (pLIVE; MIR 5420, Mirus Bio) [38]. Firefly Luciferase was under the control of the Bmal1 promoter [39]. All vectors were diluted in TransIT-EE Hydrodynamic Delivery Solution (Mirus Bio, Madison, USA) and injected into the tail vein at a constant rate over 5–7 s. The total volume was set to 10% of the total body weight. Pressing a moist paper towel on the injection point inhibited bleeding. Mice were returned to their home cage after observation for 5–10 min. Mouse Clock cDNA was cloned from a cDNA library of C57BL/6J mice into pLIVE vector followed by plasmid preparation (Endo-free Plasmid Maxi Kit, Qiagen, Hilden, Germany). The validation of delivery efficiency was performed using a liver-specific lacZ-expressing reporter plasmid (pLIVE-lacZ vector, MIR 5520, Mirus Bio). A working solution containing a total of 35 μg of Clock expressing plasmid per injection was used for all subsequent experiments. For the ex vivo visualization of liver clock rhythms, we co-injected the Bmal1-Luc plasmid together with the Clock plasmids. An Alb1-SEAP vector (pLIVE-SEAP; MIR 5620, Mirus Bio) was used for controls. Mice were allowed to recover for two days before the subsequent feeding experiments.
2.3. X-gal staining
Nine days after tail vein injection of the lacZ reporter plasmid, chow-fed mice were killed by cervical dislocation, and the medial liver lobe was removed and embedded in paraffin. 8-μm sections were prepared using a microtome (Leica, Eisfeld, Germany) and fixed in 0.2% glutaraldehyde (5 min at room temperature) followed by PBS washing and an overnight incubation in x-Gal staining solution at 37 °C. On the following day, sections were washed in PBS, dehydrated, mounted on slides, and analyzed using a camera-equipped microscope (Leica). Stained areas were calculated using NIH Image software (NIH, Bethesda, USA). Three sections of each animal were averaged (n = 7 per condition).
2.4. Luminescence explant recordings
Tissue explant clock rhythms were investigated by luminescence recording and imaging of 250-μm organotypic slice cultures. Three days after tail vein injections, tissues were dissected at 4–6 h after “lights on” and prepared for luminescence recordings as described [40]. For liver explants, median lobes were removed, embedded in low-melting agarose (Invitrogen, Carlsbad, USA), and sectioned with a vibratome (Campden Instruments, Loughborough, UK). Slices were transferred onto culture plate inserts (PICM01250; Millipore, Darmstadt, Germany) in 35-mm petri dishes filled with culture medium. Luminescence recordings were measured in a LumiCycle luminometer (Actimetrics, Evanston, USA) at 32.5 °C. Imaging was done using an Olympus LV-200 system (Olympus, Tokyo, Japan). Analyses were performed using the LumiCycle analysis (Actimetrics) and Prism software packages (GraphPad, La Jolla, USA). Raw data were baseline subtracted with running averages of 24 h (n = 8 per group; for each mouse 3 slices were averaged).
2.5. Quantitative real-time (q)PCR and in situ hybridization
mRNA expression was determined by quantitative real-time PCR (qPCR) in tissue samples (n = 3–5/group/time point) or by in situ hybridization of brain sections (n = 8 per group and time point) [41]. TRIzol extracted total RNA was transcribed to cDNA using random hexamer primers. qPCRs were run on a CFX96 thermocycler (Bio-Rad, Munich, Germany) with iQ-SYBR Green SuperMix (Bio-Rad). Eef1a was used as reference gene, and relative gene expression calculated with a modified ΔΔCT calculation as described [42]. Paraffin embedded 8-μm coronal brain sections were hybridized with 35S-UTP-labeled (PerkinElmer, Waltham, USA) antisense RNA probes as described [41]. Expression levels were determined by densitometric analysis of autoradiograph films. Three background corrected SCN sections per mouse were averaged. PCR primer and in situ probe sequences are provided upon request.
2.6. Protein expression
Expression of the wild-type CLOCK protein in MT-LR was determined by western blot analysis with antibodies directed against a C-terminal hemagglutinin (HA) tag (Cat # 9110, Abcam, Cambridge, UK). Liver tissue (n = 3–5) was homogenized in RIPA buffer (150 mM sodium chloride, 50 mM Tris, 0.1% SDS, 1% Triton X-100, 25 mM sodium deoxycholate, 5 mM ethylenediaminetetraacetic acid) with protease inhibitor cocktail (Roche, Grenzach-Wyhlen, Germany). Homogenates were boiled for 5 min followed by centrifugation. Protein concentrations were determined by BCA assay (Thermo Scientific, Waltham, USA). Total CLOCK protein was detected using a CLOCK antibody (Cat # CLO11-A, Alpha Diagnostics, Austin, USA). β-Actin (Cat # 3700, Cell Signaling Technology) was used as loading control. Densitometric analysis of band intensities was performed with Quantity One software (Bio-Rad).
2.7. Transcriptome analysis
After 5 weeks of HFD mice were sacrificed at ZT 10 (n = 3 per group). Livers were removed and total RNA isolated from TRIzol homogenized tissue and stored at −20 °C. cRNA synthesis, labeling, and hybridization (GeneChip Mouse Genome 430A 2.0, Affymetrix, Santa Clara, USA) was performed as described [43]. Microarray data are deposited in the GEO database (accession no. GSE94838). RMA normalization of gene expression was done using Affymetrix Expression Console (Affymetrix). Rank product tests (with a false discovery rate (FDR) cut-off of 0.05) were used to determine differentially expressed genes [44]. WebGestalt (Nucleic Acids Research, 2005. 33 p. W741-W748.) was used for gene enrichment analyses (significance level: p < 0.05; statistics test: hypergeometric with Benjamini-Hochberg correction for multiple testing; minimum group size: 5).
2.8. Lipid droplet staining
Hepatic lipid droplets were visualized by oil-red O staining of 8-μm sections as described [45]. In brief, livers were fixed overnight in 4% paraformaldehyde followed by cryo-protection in 30% sucrose and embedding in OCT. Liver sections were stained for 20 min with filtered oil-red O solution (Certistain Oil-red O, Merck, Darmstadt, Germany) and washed in 60% isopropyl alcohol and distilled water. Nuclei were counterstained with Mayer's hematoxylin for 2 min followed by incubation with tap water containing sodium borate until nuclei became blue. Covered sections were analyzed using a camera-equipped inverted microscope (Leica) and lipid accumulation quantified by densitometric analysis with NIH Image (National Institutes of Health, Bethesda, MA).
2.9. Glucose tolerance and hormone tests
Glucose clearance was investigated at ZT4-6 after 4 weeks on HFD [46]. Overnight fasted mice received an intraperitoneal (i.p.) injection of glucose (2 g/kg body weight) followed by repeated blood collection via the tail vein at defined time points (n = 8). Leptin and Acyl-ghrelin values where obtained from plasma samples collected in a 4-hour interval over 24 h. The hormone content was analyzed via ELISA (Crystal Chem, Downers Grove, USA and BioVendor, Brno, Czech Republic) and averaged values for all time points (n = 3) in the light and dark phases were used for the day/night concentration values.
2.10. Statistics
Statistical comparisons were performed with GraphPad Prism (GraphPad, La Jolla, USA). Time series were compared using 2-way ANOVAs with Bonferroni post-test. One condition between-genotype comparisons were done using 1-way ANOVAs with Bonferroni post-tests. Transcriptome data were analyzed as described above. 24-h rhythms were assessed by CircWave analysis [47].
3. Results
3.1. Restoration of daily food intake rhythms by restricted food access does not rescue the obese phenotype of Clock mutant mice
In line with previous reports [21], ten weeks of HFD feeding led to a significant increase in body weight in both WT and MT mice, but the effect was more pronounced in MTs (Figure 1A), in particular during the first weeks of the feeding paradigm. Over the whole 10 weeks, MTs gained 12.5 ± 0.9 g while WTs gained only 8.9 ± 0.7 g (WT vs. MT p < 0.01). Due to genotype differences in basal body weight [21] (WT: 24.02 ± 0.1; MT: 25.92 ± 0.4; p < 0.001), we calculated body weight gain relative to starting body weight. WTs increased their body weight by 3.7 ± 0.28% per week while MTs showed a weekly body weight gain of 4.7 ± 0.26% (WT vs. MT p < 0.05) under the same feeding conditions (Figure 1B). This increased susceptibility to the obesogenic effect of HFD in MT mice was associated with elevated cumulative (Figure 1C) and daily food intake, specifically during the light phase (Figure 1D) [21]. Despite equal total locomotor activity (Figure 1E), MT mice showed decreased energy expenditure specifically during the dark phase (Figure 1F). Disruptions in daily feeding rhythms were previously reported in Clock mutant [21] and HFD-fed WT mice [48]. Moreover, food access restricted to the normal active phase (i.e. night) leads to a weight gain resistance under HFD in WT mice [25], suggesting that the increase in light phase feeding may contribute to the obesity phenotype in Clock mutant mice [21]. To test this, we restricted the access to HFD to the dark phase in WT and MT mice. Restricted nighttime feeding (RF) reduced the anabolic properties of HFD in both genotypes (WT-RF: 6.9 ± 0.27 g; MT-RF: 10.2 ± 0–6 g; WT-RF vs. MT-RF p < 0.01). MT cumulative (Figure 1C) and daily food intake (WT-RF: 2.84 ± 0.03; MT-RF: 2.87 ± 0.03; Figure 1D) normalized to WT levels while activity remained largely unaltered (Figure 1E). Nevertheless, MT-RF showed a significantly higher weight gain compared to WT-RF (WT-RF: 2.9%; MT-RF 4.03%; WT-RF vs. MT-RF p < 0.001; Figure 1B). Restricted feeding did not significantly alter energy expenditure in either genotype, but overall energy expenditure remained lower in MT mice compared to WT controls in both conditions (ad libitum: WT: 51.00 ± 0.7 L/kg*h; MT: 47.28 ± 0.63 L/kg*h; p < 0.01; RF: WT: 53.15 ± 0.55 L/kg*h; MF: 50.57 ± 0.67 L/kg*h; p < 0.05). In summary, under HFD conditions metabolic effects of the Clock mutation were not rescued by nighttime restricted food access, suggesting that, besides mistimed feeding, other factors regulate metabolic homeostasis in these mice.
3.2. Partial restoration of liver circadian rhythms by Clock gene therapy
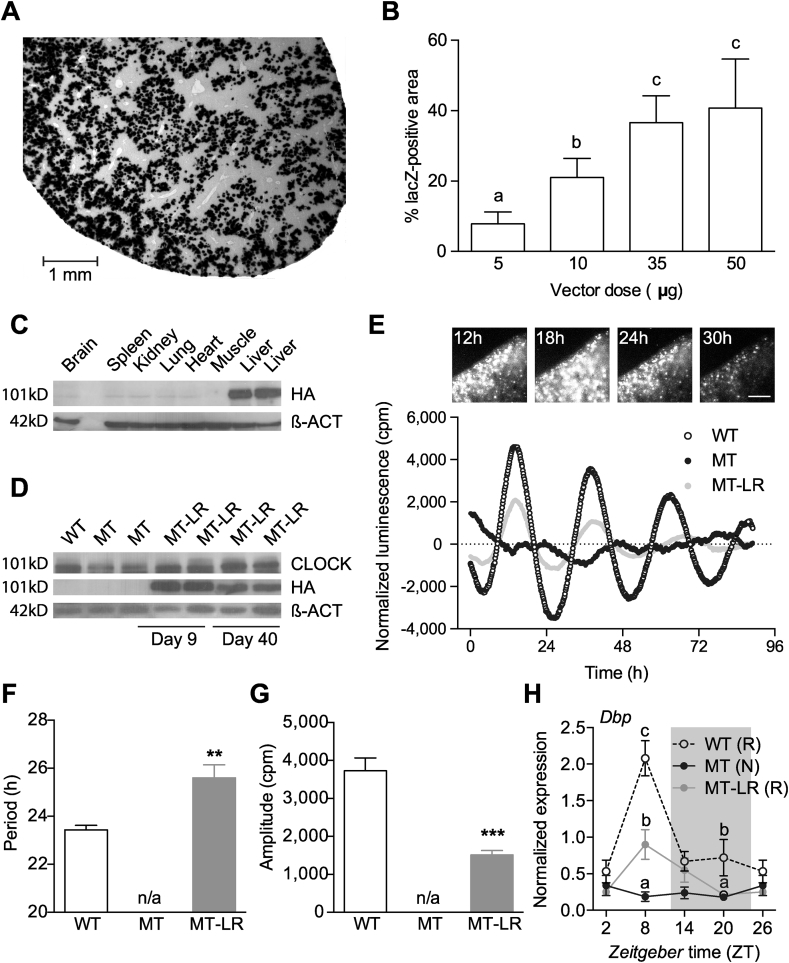
The liver is an important metabolic organ and the liver circadian clock has been shown to regulate glucose trafficking [13]. We therefore speculated that a restoration of liver clock rhythms might improve metabolic homeostasis in Clock mutant mice. To test this, we used hydrodynamic tail vein delivery of a hepatocyte-directed WT-CLOCK expression vector to restore liver clock function in MT mice (MT-LR). Injection of a lacZ reporter vector followed by x-Gal staining on liver sections was used to determine the optimal experimental parameters (Figure 2A). A maximal penetrance of 35–40% of the section surface was reached at a dose of 35–50 μg DNA per injection (Figure 2B). 35-μg doses were used for all following experiments. By western blotting, WT (HA-tagged) CLOCK protein was detected predominantly in liver, but faint bands were also seen in brain, spleen and kidney (Figure 2C). We further determined transgene expression in a number of metabolic tissues including white adipose tissue, pancreas and gut, and in the SCN. Compared to liver, HA-Clock expression was extremely low (below 2%) in all other tissues tested suggesting high tissue specificity (Suppl. Figure 2A). In MT-LR livers WT-CLOCK expression was stable for at least 40 (protein) to 80 days (mRNA) after injection (Figure 2D and Suppl. Figure 2B), making mice suitable for metabolic characterization.
Figure 2.
Liver Clock gene therapy restores rhythmic gene expression. A, B: lacZ-staining of sections of livers after treatment with different doses of an lacZ-expressing reporter vector. (A) micrograph of a liver section of mice treated with 35 μg of the vector. (B) dose response (means ± SEM (n = 7); 1-way ANOVA with Bonferroni post-test; different letters depict statistically different groups (p < 0.05)). (C) Western blot detecting HA-tagged CLOCK on day 9 after tail vein injection in different tissues. (D) Western blot detecting wild-type and HA-tagged CLOCK in livers of WT and MT mice at 9 and 40 days after treatment. E–G: Luminescence recordings of liver slices after co-injection of Bmal1-luc reporter w/o wild-type CLOCK. (E) Upper panel: luminescence micrographs taken at indicated time points after injection; lower panel: representative baseline-corrected luminescence traces of WT, MT and MT-LR liver slices. F, G: Luminescence rhythm period lengths (F) and amplitudes (G) of explant cultures (means ± SEM (n = 8); 1-way ANOVA with Bonferroni post-test; **: p < 0.01, ***: p < 0.001). (H) Diurnal liver gene expression profile for Dbp in WT, MT, and MT-LR mice (means ± SEM (n = 3); 2-way ANOVA with Bonferroni post-test; different letters depict statistically different groups (p < 0.05); letters in brackets indicate results from CircWave analysis for rhythmicity: R – rhythmic, N – non-rhythmic).
To see if WT-CLOCK expression in MT livers was sufficient to reinitiate liver circadian rhythms, we co-injected a circadian luciferase reporter construct and analyzed luciferase activity in liver slice cultures. As was seen for the lacZ vector, injection of WT-CLOCK and Bmal1-LUC vectors resulted in a patterned expression of LUCIFERASE in MT-LR liver slices (Figure 2E, upper panel). Extended cultivation revealed a partial rescue of luminescence rhythms in MT-LR compared to WT slices, while no endogenous rhythm was observed in untreated MT liver (Figure 2E) and explants of other tissues (kidney, lung, pancreas, epididymal and subcutaneous white adipose tissue (e/scWAT), spleen, heart, skeletal muscle, adrenal, thymus) of MT-LR mice (data not shown). Period length was increased by about 2 h (WT vs. MT-LR p < 0.01; Figure 2F) and amplitude dampened by around 2,000 cpm (WT vs. MT-LR p < 0.001; Figure 2G) in liver slices of MT-LR relative to WT mice. In line with this, liver Dbp mRNA expression MT-LR mice showed a partial rescue of the WT rhythm with high levels during the late light phase and minimal expression towards the end of the night (Figure 2H). Explant rhythms and Dbp profiles were also tested in WT mice receiving WT-CLOCK injections and in both genotypes after sham injection of a SEAP-expressing vector. In none of these mice, injections had significant effects compared to non-injected animals (data not shown). In summary WT-CLOCK overexpression in Clock mutant livers partially restores molecular clock rhythms.
3.3. The SCN clock is not affected by liver clock rescue
Since our western blot analysis revealed a small amount of the ectopically expressed WT-CLOCK in the MT-LR brain (Figure 2C), we characterized potential effects on SCN pacemaker function by clock gene in situ hybridization and running-wheel assays. Comparing the end of the light (ZT8) and the dark phase (ZT20), diurnal regulation of Dbp and Bmal1 was not affected by WT-CLOCK injection in MT SCNs (Figure 3A,B). In line with its direct light regulation [49], Per2 mRNA concentration in the SCN was elevated in the light phase in all genotypes, though expression levels were reduced in MT and MT-LR mice (Figure 3C). Consistently, running-wheel activity rhythms were also unaffected by liver gene therapy. Under standard light dark conditions, all three groups showed the typical activity rhythm of nocturnal species with wheel running predominantly confined to the dark phase (Figure 3D–F, representative actograms). Some MT-LR mice showed slightly more fragmented activity towards the end of the dark phase compared to untreated MT animals (compare 24–30 h in Figure 3E,F), but effects were minor compared to individual differences within each group. In constant darkness, WT mice showed a stable free-running period of 23.65 ± 0.06 h, while MT and MT-LR mice showed a lengthened period (MT: 27.71 ± 0.2 h; MT-LR: 27.98 ± 0.3 h; Figure 3G) with some mice losing stable circadian rhythms after extended time in DD (data not shown). WT-CLOCK injection did not affect free-running rhythmicity in WT mice, nor did SEAP expression affect rhythmicity and period in any genotype (data not shown). Together, these data suggest that liver Clock gene therapy had no functional impact on central pacemaker regulation and locomotor behavior.
3.4. Liver clock rescue reduces HFD sensitivity in Clock mutant mice
We next tested to what extent partial restoration of liver clock function would affect the increased HFD sensitivity in Clock mutant mice (Figure 1). In line with our previous experiment, all groups gained weight under HFD conditions, while MTs were more affected than WT mice (Figure 4A,B). Surprisingly, the increased initial weight of MT-LR adjusted to a final body weight development closer to that of WT than of MT mice during the second half of the experiment. On average, MT-LR mice compared to both non- and sham-treated genotypes showed a reduced weekly body weight gain (Figure 4B), indicating an increased resistance to the obesogenic effects of HFD (body weight gain, WT: 3.83 ± 0.17% per week, MT-LR: 3.04 ± 0.20%, MT: 4.71 ± 0.20%). Total food intake in MT-LR mice was restored to WT levels (Figure 4C). Interestingly, a partial restoration of diurnal feeding rhythms was observed in MT-LR mice, though day–night differences were dampened compared to WT mice (day: 1.39 ± 0.05 g; night: 1.82 ± 0.06 g; p < 0.001; Figure 4D). In WTs, WT-CLOCK gene therapy neither affected body weight gain or food intake behavior (Suppl. Figure 2). Similarly, body weight gain and food intake were unaffected by control (SEAP) vector injection in WT and MT mice (Suppl. Figure 2).
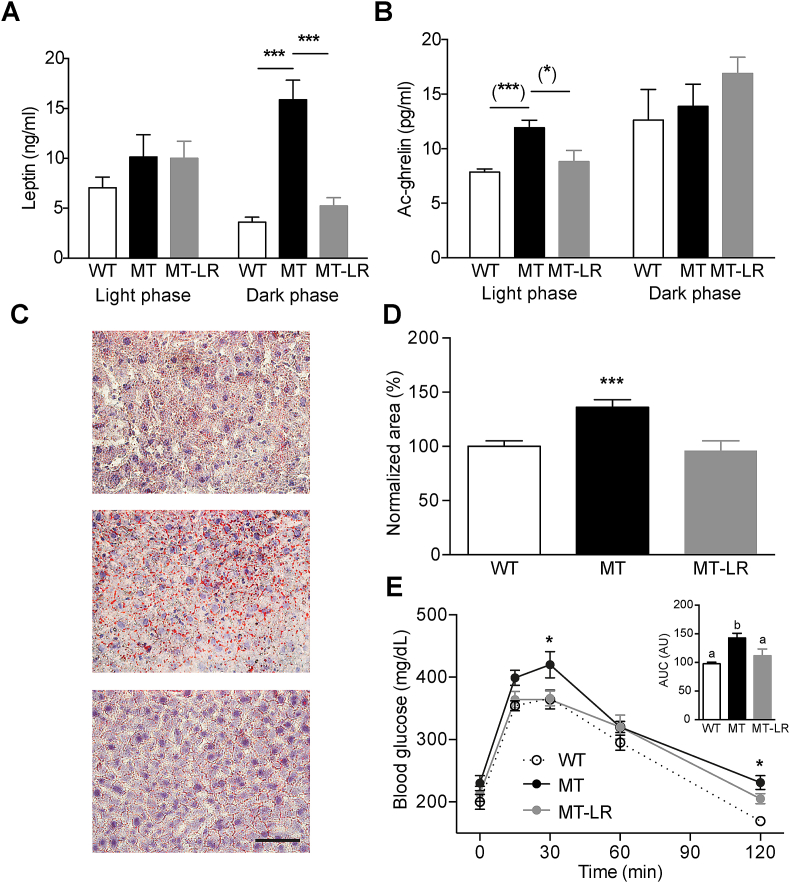
In week 5 of HFD, mice were tested for energy expenditure by indirect calorimetry. Liver Clock gene therapy restored energy expenditure to WT-like levels in MT mice (Figure 4E), without affecting MT-LR locomotor activity (Figure 4F), in line with comparable total activity observed under normal chow conditions (Figure 3) and HFD (Figure 1). Serum levels of the adipose tissue-derived satiety hormone leptin were largely unaltered in MT and MT-LR mice during the light phase but showed a profound upregulation in MT mice during the dark phase, which was reversed in MT-LR mice (Figure 5A). In contrast, the hunger signal Acyl-ghrelin was unaltered between genotypes during the dark phase, but Acyl-ghrelin levels were upregulated in MT mice during the light phase and this elevation was absent in MT-LR mice (Figure 5B). In line with partially restored energy homeostasis, liver clock rescue readjusted liver lipid content (Figure 5C,D) and glucose clearance (Figure 5E) in MT-LR mice.
Figure 5.
Liver Clock gene therapy partially restores metabolic parameters in Clock mutant mice. A, B: Light:dark phase leptin (A) and acylated (Ac-)ghrelin (B) serum concentrations in WT, MT, and MT-LR mice (means ± SEM (n = 9–11); 2-way ANOVAs with Bonferroni post-test; ***: p < 0.001; Student's t-test; (*): p < 0.05; (***): p < 0.001). C, D: Oil red-O staining of liver sections of WT, MT and MT-LR mice. (C) Representative micrographs; (D) Densitometric analysis (means ± SEM (n = 8); 1-way ANOVA with Bonferroni post-test; ***: p < 0.001). (D) blood glucose levels after intraperitoneal glucose injection in WT, MT, and MT-LR mice (means ± SEM (n = 8); 2-way ANOVA with Bonferroni post-test; *: p < 0.05 vs. WT). Inset shows area-under-the-curve analysis (letters depict significant differences (p < 0.05, 1-way ANOVA)).
3.5. Transcriptome analysis reveals liver clock rescue-associated changes in metabolic pathways
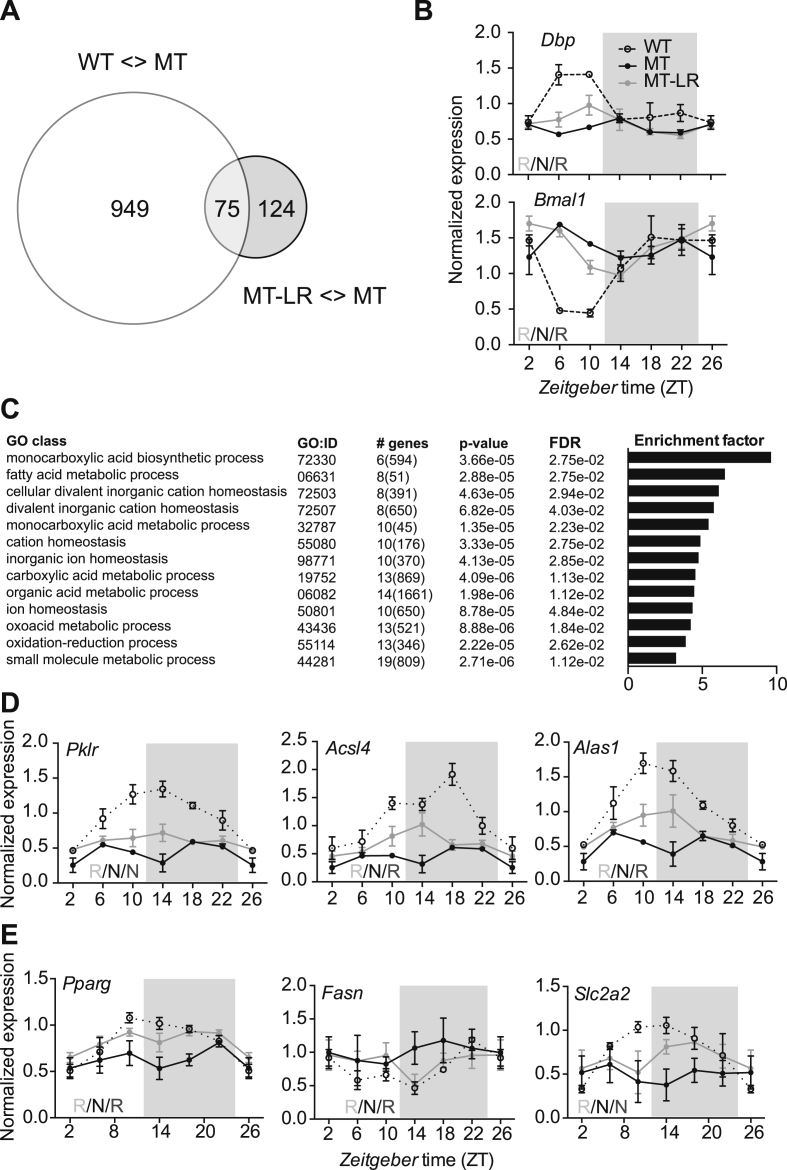
To gain some insight into which pathways are involved in conferring decreased HFD sensitivity in MT-LR mice, we compared the liver transcriptome between WT, MT, and MT-LR mice after 4 weeks of HFD at ZT10, the time of maximal CLOCK/BMAL1-mediated E-box transactivation in this tissue [15]. Mice were killed and livers isolated for total RNA extraction and samples prepared for microarray hybridization. Overall, we detected a significantly different (up- or down-) regulation of 1,024 probe sets between WT and MT tissue. A comparison of MT and MT-LR data yielded 199 differentially regulated transcripts (Figure 6A). 75 transcripts, which were differently expressed between WT and MT and at the same time between MT and MT-LR, were considered to be rescued by the LR treatment. (Figure 6A, Suppl. Table 1). Many of them were previously been identified as rhythmically expressed in mouse liver (Suppl. Table 1). To validate the microarray data, we analyzed 24-h expression profiles of selected clock genes by qPCR (Figure 6B). In MT livers, rhythmic expression of Dbp (as a representative E-box-regulated gene) and Bmal1 were abolished. In both cases, expression rhythms were partially restored in MT-LR (Figure 6B). Interestingly, while body weight was not significantly altered by the liver clock rescue during the first 4 weeks on diet, gene ontology enrichment analysis of the 75 rescued gene transcripts in MT-RL mice revealed that mainly pathways involved in metabolic function were affected, including oxidoreduction and fatty acid metabolism (Figure 6C). We analyzed diurnal transcription profiles of a set of metabolically relevant genes in all three cohorts. Similar to what was observed for clock genes, mRNA rhythms of these genes were non-rhythmic in MT livers, but partial restoration of rhythmicity was seen for some (acyl-CoA synthetase long-chain family member 4 – Acsl4, 5′-aminolevulinate synthase 1 – Alas1, fatty acid synthase – Fasn, peroxisome proliferator activated receptor gamma – Pparg, and glucose transporter 2 – Slc2a2) but not for all genes tested (Figure 6D,E). Together, our transcriptome data suggest a partial restoration of metabolic pathways mirroring the partial restoration of liver clock function in MT-LR mice. In this way, liver clock adjustment may promote the systemic rescue of metabolic and body weight homeostasis in MT-LR mice.
Figure 6.
Partial rescue of hepatic transcriptome regulation by liver Clock gene therapy in Clock mutant mice. (A) Venn diagram of differently regulated transcripts at ZT 10. White circle: number of transcripts with differential expression between WT and MT livers; grey circle: number of transcripts with differential expression between MT and MT-LR livers. (B) diurnal gene expression profiles for Dbp and Bmal1 in WT, MT, and MT-LR livers (means ± SEM; n = 3) determined by qPCR. Left bottom letters depict CircWave analysis (R – rhythmic; N – non-rhythmic). (C) gene enrichment analysis of genes with partially restored expression levels at ZT10 in MT-LR livers (GO: gene ontology; # genes: number of genes in group, total number of genes in GO category in brackets; FDR – false discovery rates). D, E: diurnal gene expression profiles for metabolically relevant genes taken from the microarray dataset (D) or from the literature (E) determined by qPCR (means ± SEM; n = 4). Left bottom letters depict CircWave analysis (R – rhythmic; N – non-rhythmic).
4. Discussion
Disrupted circadian rhythms are often associated with metabolic dysregulations altering glucose and energy homeostasis and promoting body weight gain [50]. Our results indicate that the circadian liver clock is a regulator of systemic energy homeostasis and targeting liver clock function may help to normalize body weight regulation.
In wild-type mice, HFD disrupts diurnal feeding rhythms and the obesogenic effects of HFD are inhibited by restricting food access to the active phase (i.e. the night) [25]. Disrupted diurnal food intake rhythms and obesity are similarly observed in Clock gene mutant mice [21]. Together, these data support the idea that the time of feeding is an important regulator of body weight gain [22], [25], [26]. However, in our experiments, restricting HFD access to the active nighttime reduced total food intake but did not normalize the energy balance of Clock mutant mice to that of WT controls. This clearly suggests that factors other than mistimed feeding regulate the increased sensitivity of these mice to high-fat food. The energy expenditure of MTs was significantly lower compared to WTs, explaining the higher anabolic impact and elevated body weight gain. In WTs, time-restricted access to a HFD improves circadian gene expression rhythms in tissues regulating energy expenditure [25], [51], [52], [53], [54]. In the absence of a functional clock machinery, however, restricted feeding can only partially recover rhythmic gene expression in peripheral metabolic tissues such as the liver [7], [55], which led us to speculate that liver clock function may be critical in defining the metabolic response to high-fat food.
Using a non-viral gene therapy approach, we overexpressed wild-type CLOCK protein in up to 40% of liver cells. This transduction rate was efficient enough to reestablish hepatic clock rhythmicity in vitro and in vivo, albeit both luciferase assay and gene expression analysis revealed reduced transcriptional amplitudes compared to WT livers. This was expected since Clock-Δ19 is a dominant negative mutation [35] and over-expressed WT proteins have to compete with mutant proteins for circadian DNA binding at E-boxes and activation of target genes such as Dbp [56], [57]. In line with this, liver explant rhythms of WT-Clock treated MT mice showed longer period lengths similar to what has been observed for heterozygous Clock mutant tissues [58]. Even though a liver-directed minimal Alb1 promoter was used to restrict transgene expression to hepatocytes, faint ectopic expression of tagged CLOCK protein and mRNA was detected in other tissues, in line with previous reports [38]. qPCR data showed low transcript levels in metabolic tissues such as WAT and pancreas (below 2% of what was seen in liver). Considering that the liver clock rescue was far from complete, one might assume that clock function in other tissues was likely unaffected. Still, restoration of clocks in some cells of other tissues cannot be excluded, and this may have impinged on metabolic regulation. Luciferase reporter explant, qPCR, and running-wheel analyses revealed that the functionality of the SCN pacemaker was not rescued by the treatment. In summary, we assume that metabolic effects are largely attributable to restoration of liver clock function in our mice.
The partial rescue of liver clock function in ClockΔ19 mice led to a restoration of the metabolic response to HFD with reduced food intake, partially restored diurnal feeding rhythms, and increased energy expenditure. Interestingly, in line with an absence of SCN pacemaker effects, locomotor activity rhythms remained dampened in MT-LR mice. Of note, feeding behavior as well as metabolic rate were improved before significant differences in body weight between MT and MT-LR mice were seen, indicating that the liver clock rescue per se altered gene expression and metabolic parameters independently of general metabolic state. The partially restored feeding rhythm may implicate further downstream effects on appetite-regulating centers in the brain. In a similar way, disruption of adipose tissue clocks has been shown to affect food intake rhythms and body weight [59].
In adipose clock-deficient mice, reduced secretion of poly-unsaturated fatty acid leads to a reduced satiety feedback to the brain, thus promoting over-eating [59]. In a similar way, liver derived factors such as ketone bodies [31] or FGF21 [60], [61] have been shown to affect appetite regulation and energy expenditure. Similar to the partial clock rescue, our transcriptome analysis reveals a partial restoration of metabolic transcript regulation in MT-LR mice. While the specific pathways mediating the restorative effect of liver Clock gene therapy on metabolic homeostasis remain to be defined, a normalization of peripheral metabolic feedback is further documented by restoration of major endocrine mediators of appetite regulation such as the stomach-derived ghrelin and the anorexigenic adipokine leptin. These findings suggest that diurnal regulation in other tissues may also benefit from liver clock rescue – though effects of ectopic clock rescue cannot not fully be excluded.
Many metabolic pathways in liver are under circadian control [62], [63], and hepatic BMAL1 was found to be a key regulator for metabolic fitness by regulating the hepatic mitochondrial dynamics [18]. In humans liver transplantation can result in reduced energy expenditure and consequential weight gain [52]. Along this line, Uno et al. have shown that hepatic overexpression of peroxisome proliferator-activated receptor (PPAR)-gamma (Pparg) increases energy expenditure [64]. In healthy subjects, PPARG is rhythmically expressed and links metabolism with the circadian system [51], [55], [65], [66]. In our mice, we also observed a partial rescue of Pparg mRNA expression rhythms after Clock gene therapy (Figure 6E). In this way, a recovery of hepatic metabolic pathway regulation in MT mice may reverse the obesogenic effects of clock disruption and restoring WT-like energy expenditure.
5. Conclusion
Our data show that liver gene therapy may partially restore metabolic responses to HFD in circadian Clock mutant mice, indicating an important role of liver clock function in energy homeostasis and appetite regulation. In modern societies, circadian rhythm disruptions, as observed in shift workers, increase the risk of developing metabolic disorders, imposing a huge burden on public health systems [67]. Another important obesogenic factor is the unlimited availability of energy-dense food. Animal data suggest that both aspects may reciprocally strengthen each other [21], [24], [48]. Therefore, it is of interest to develop therapeutic interventions reducing the obesogenic properties of high-energy food and circadian perturbation. Behavioral interventions such as temporally restricting food intake are often difficult to realize, e.g. in shift workers [68]. Moreover, central appetite centers are pharmacologically difficult to reach (though central application routes such as intranasal treatment may overcome this issue; [69]). Our data suggest that manipulation of liver clock function might be a promising and accessible target to stabilize metabolic homeostasis.
Acknowledgments
This study was supported by grants of the German Research Foundation (DFG; GRK-1957 & SFB-134) and a Lichtenberg Fellowship of the Volkswagen Foundation (HO). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molmet.2017.03.008.
Conflict of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
List of genes differentially expressed at ZT10 in MT-LR compared with WT and MT mice.
Tissue distribution and long-term expression of HA-Clock mRNA after tail vein injection. A: mRNA levels in different tissues at day 9 after injection determined by qPCR. B: mRNA levels in liver at different times after injection. All data are means ± SEM; n = 5.
Body weight development and food intake in control cohorts. A, B: body weight development (A) and weekly weight gain (B) of WT- and MT-CONTROL (injected with SEAP) mice and Clock-injected WT mice under ad libitum access to HFD (n = 12). C, D: Cumulative (C) and daily food intake (D) of WT- and MT-CONTROL (injected with SEAP) mice and Clock-injected WT mice (n = 12). Data are shown as means ± SEM; 1- and 2-way ANOVAs with Bonferroni post-test; different letters depict statistically different groups (p < 0.05); ***: p < 0.001 MT-CONTROL vs. WT groups (A, C) or light (L) vs. dark (D) groups (D). Letters in B depict significant differences (p < 0.05; 1-way ANOVA).
References
- 1.Tsang A.H., Barclay J.L., Oster H. Interactions between endocrine and circadian systems. Journal of Molecular Endocrinology. Feb 2014;52:R1–R16. doi: 10.1530/JME-13-0118. [DOI] [PubMed] [Google Scholar]
- 2.Bass J., Takahashi J.S. Circadian integration of metabolism and energetics. Science. Dec 3 2010;330:1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ueda H.R., Chen W., Adachi A., Wakamatsu H., Hayashi S., Takasugi T. A transcription factor response element for gene expression during circadian night. Nature. Aug 1 2002;418:534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- 4.Panda S., Antoch M.P., Miller B.H., Su A.I., Schook A.B., Straume M. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. May 3 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 5.Golombek D.A., Rosenstein R.E. Physiology of circadian entrainment. Physiological Reviews. Jul 2010;90:1063–1102. doi: 10.1152/physrev.00009.2009. [DOI] [PubMed] [Google Scholar]
- 6.Dibner C., Schibler U., Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annual Review of Physiology. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 7.Vollmers C., Gill S., DiTacchio L., Pulivarthy S.R., Le H.D., Panda S. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proceedings of the National Academy of Sciences of the United States of America. Dec 15 2009;106:21453–21458. doi: 10.1073/pnas.0909591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y., Sato Y., Yamaguchi N. Shift work and the risk of metabolic syndrome: a nested case-control study. International Journal of Occupational and Environmental Health. Apr-Jun 2011;17:154–160. doi: 10.1179/107735211799030960. [DOI] [PubMed] [Google Scholar]
- 9.Karlsson B., Knutsson A., Lindahl B. Is there an association between shift work and having a metabolic syndrome? Results from a population based study of 27,485 people. Occupational and Environmental Medicine. Nov 2001;58:747–752. doi: 10.1136/oem.58.11.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barclay J.L., Husse J., Bode B., Naujokat N., Meyer-Kovac J., Schmid S.M. Circadian desynchrony promotes metabolic disruption in a mouse model of shiftwork. PLoS One. 2012;7:e37150. doi: 10.1371/journal.pone.0037150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foster R.G., Kreitzman L. The rhythms of life: what your body clock means to you! Experimental Physiology. Apr 2014;99:599–606. doi: 10.1113/expphysiol.2012.071118. [DOI] [PubMed] [Google Scholar]
- 12.Jones S.G., Benca R.M. Circadian disruption in psychiatric disorders. Sleep Medicine Clinics. Dec 2015;10:481–493. doi: 10.1016/j.jsmc.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 13.Lamia K.A., Storch K.-F., Weitz C.J. Physiological significance of a peripheral tissue circadian clock. Proceedings of the National Academy of Sciences of the United States of America. September 30, 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang D., Tong X., Arthurs B., Guha A., Rui L., Kamath A. Liver clock protein BMAL1 promotes de novo lipogenesis through insulin-mTORC2-AKT signaling. Journal of Biological Chemistry. Jul 25 2014 doi: 10.1074/jbc.M114.567628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rey G., Cesbron F., Rougemont J., Reinke H., Brunner M., Naef F. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biology. Feb 2011;9:e1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gachon F., Olela F.F., Schaad O., Descombes P., Schibler U. The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metabolism. Jul 2006;4:25–36. doi: 10.1016/j.cmet.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 17.DeBruyne J.P., Weaver D.R., Dallmann R. The hepatic circadian clock modulates xenobiotic metabolism in mice. Journal of Biological Rhythms. Aug 2014;29:277–287. doi: 10.1177/0748730414544740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacobi D., Liu S., Burkewitz K., Kory N., Knudsen N.H., Alexander R.K. Hepatic Bmal1 regulates rhythmic mitochondrial dynamics and promotes metabolic fitness. Cell Metabolism. Oct 6 2015;22:709–720. doi: 10.1016/j.cmet.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukui K., Ferris H.A., Kahn C.R. Effect of cholesterol reduction on receptor signaling in neurons. Journal of Biological Chemistry. Oct 30 2015;290:26383–26392. doi: 10.1074/jbc.M115.664367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grechez-Cassiau A., Rayet B., Guillaumond F., Teboul M., Delaunay F. The circadian clock component BMAL1 is a critical regulator of p21WAF1/CIP1 expression and hepatocyte proliferation. Journal of Biological Chemistry. Feb 22 2008;283:4535–4542. doi: 10.1074/jbc.M705576200. [DOI] [PubMed] [Google Scholar]
- 21.Turek F.W., Joshu C., Kohsaka A., Lin E., Ivanova G., McDearmon E. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. May 13 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arble D.M., Sandoval D.A. CNS control of glucose metabolism: response to environmental challenges. Frontiers in Neuroscience. 2013;7:20. doi: 10.3389/fnins.2013.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rudic R.D., McNamara P., Curtis A.M., Boston R.C., Panda S., Hogenesch J.B. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biology. Nov 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barclay J.L., Shostak A., Leliavski A., Tsang A.H., Johren O., Muller-Fielitz H. High-fat diet-induced hyperinsulinemia and tissue-specific insulin resistance in cry-deficient mice. American Journal of Physiology, Endocrinology and Metabolism. May 15 2013;304:E1053–E1063. doi: 10.1152/ajpendo.00512.2012. [DOI] [PubMed] [Google Scholar]
- 25.Hatori M., Vollmers C., Zarrinpar A., DiTacchio L., Bushong E.A., Gill S. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metabolism. Jun 6 2012;15:848–860. doi: 10.1016/j.cmet.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sherman H., Genzer Y., Cohen R., Chapnik N., Madar Z., Froy O. Timed high-fat diet resets circadian metabolism and prevents obesity. FASEB Journal. Aug 2012;26:3493–3502. doi: 10.1096/fj.12-208868. [DOI] [PubMed] [Google Scholar]
- 27.Cornejo M.P., Hentges S.T., Maliqueo M., Coirini H., Becu-Villalobos D., Elias C.F. Neuroendocrine regulation of metabolism. Journal of Neuroendocrinology. Apr 26 2016 doi: 10.1111/jne.12395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waterson M.J., Horvath T.L. Neuronal regulation of energy homeostasis: beyond the hypothalamus and feeding. Cell Metabolism. Dec 1 2015;22:962–970. doi: 10.1016/j.cmet.2015.09.026. [DOI] [PubMed] [Google Scholar]
- 29.Horvath T.L., Diano S., Tschop M. Brain circuits regulating energy homeostasis. Neuroscientist. Jun 2004;10:235–246. doi: 10.1177/1073858403262151. [DOI] [PubMed] [Google Scholar]
- 30.Xu J., Lloyd D.J., Hale C., Stanislaus S., Chen M., Sivits G. Fibroblast growth factor 21 reverses hepatic steatosis, increases energy expenditure, and improves insulin sensitivity in diet-induced obese mice. Diabetes. Jan 2009;58:250–259. doi: 10.2337/db08-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chavan R., Feillet C., Costa S.S., Delorme J.E., Okabe T., Ripperger J.A. Liver-derived ketone bodies are necessary for food anticipation. Nature Communications. Feb 03 2016;7:10580. doi: 10.1038/ncomms10580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwartz M.W., Woods S.C., Porte D., Jr., Seeley R.J., Baskin D.G. Central nervous system control of food intake. Nature. Apr 6 2000;404:661–671. doi: 10.1038/35007534. [DOI] [PubMed] [Google Scholar]
- 33.Velloso L.A., Schwartz M.W. Altered hypothalamic function in diet-induced obesity. International Journal of Obesity (London) Dec 2011;35:1455–1465. doi: 10.1038/ijo.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yi C.X., Tschop M.H. Brain-gut-adipose-tissue communication pathways at a glance. Disease Models & Mechanisms. Sep 2012;5:583–587. doi: 10.1242/dmm.009902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vitaterna M.H., King D.P., Chang A.M., Kornhauser J.M., Lowrey P.L., McDonald J.D. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. Apr 29 1994;264:719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weir J.B. New methods for calculating metabolic rate with special reference to protein metabolism. Journal of Physiology. Aug 1949;109:1–9. doi: 10.1113/jphysiol.1949.sp004363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim M.J., Ahituv N. The hydrodynamic tail vein assay as a tool for the study of liver promoters and enhancers. Methods in Molecular Biology. 2013;1015:279–289. doi: 10.1007/978-1-62703-435-7_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma Y., Gao M., Sun H., Liu D. Interleukin-6 gene transfer reverses body weight gain and fatty liver in obese mice. Biochimica Biophysica Acta. May 2015;1852:1001–1011. doi: 10.1016/j.bbadis.2015.01.017. [DOI] [PubMed] [Google Scholar]
- 39.Nagoshi E., Saini C., Bauer C., Laroche T., Naef F., Schibler U. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. Nov 24 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 40.Landgraf D., Tsang A.H., Leliavski A., Koch C.E., Barclay J.L., Drucker D.J. Oxyntomodulin regulates resetting of the liver circadian clock by food. Elife. 2015;4:e06253. doi: 10.7554/eLife.06253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kiessling S., Eichele G., Oster H. Adrenal glucocorticoids have a key role in circadian resynchronization in a mouse model of jet lag. Journal of Clinical Investigation. Jul 2010;120:2600–2609. doi: 10.1172/JCI41192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. May 1 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Husse J., Hintze S.C., Eichele G., Lehnert H., Oster H. Circadian clock genes Per1 and Per2 regulate the response of metabolism-associated transcripts to sleep disruption. PLoS One. 2012;7:e52983. doi: 10.1371/journal.pone.0052983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laing E., Smith C.P. RankProdIt: a web-interactive rank products analysis tool. BMC Research Notes. Aug 06 2010;3:221. doi: 10.1186/1756-0500-3-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Churukian C.J. Lillie's oil red O method for neutral lipids. Journal of Histotechnology. 1999;22:309–311. [Google Scholar]
- 46.Koch C.E., Lowe C., Pretz D., Steger J., Williams L.M., Tups A. High-fat diet induces leptin resistance in leptin-deficient mice. Journal of Neuroendocrinology. Feb 2014;26:58–67. doi: 10.1111/jne.12131. [DOI] [PubMed] [Google Scholar]
- 47.Oster H., Damerow S., Hut R.A., Eichele G. Transcriptional profiling in the adrenal gland reveals circadian regulation of hormone biosynthesis genes and nucleosome assembly genes. Journal of Biological Rhythms. Oct 2006;21:350–361. doi: 10.1177/0748730406293053. [DOI] [PubMed] [Google Scholar]
- 48.Kohsaka A., Laposky A.D., Ramsey K.M., Estrada C., Joshu C., Kobayashi Y. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metabolism. Nov 2007;6:414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 49.Yan L., Takekida S., Shigeyoshi Y., Okamura H. Per1 and Per2 gene expression in the rat suprachiasmatic nucleus: circadian profile and the compartment-specific response to light. Neuroscience. 1999;94:141–150. doi: 10.1016/s0306-4522(99)00223-7. [DOI] [PubMed] [Google Scholar]
- 50.Marcheva B., Ramsey K.M., Peek C.B., Affinati A., Maury E., Bass J. Circadian clocks and metabolism. Handbook of Experimental Pharmacology. 2013:127–155. doi: 10.1007/978-3-642-25950-0_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen L., Yang G. PPARs integrate the mammalian clock and energy metabolism. PPAR Research. 2014;2014:653017. doi: 10.1155/2014/653017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Richardson R.A., Garden O.J., Davidson H.I. Reduction in energy expenditure after liver transplantation. Nutrition. Jul-Aug 2001;17:585–589. doi: 10.1016/s0899-9007(01)00571-8. [DOI] [PubMed] [Google Scholar]
- 53.Ayala J.E., Bracy D.P., Hansotia T., Flock G., Seino Y., Wasserman D.H. Insulin action in the double incretin receptor knockout mouse. Diabetes. Feb 2008;57:288–297. doi: 10.2337/db07-0704. [DOI] [PubMed] [Google Scholar]
- 54.Pazos P., Lima L., Tovar S., Gonzalez-Touceda D., Dieguez C., Garcia M.C. Divergent responses to thermogenic stimuli in BAT and subcutaneous adipose tissue from interleukin 18 and interleukin 18 receptor 1-deficient mice. Science Reports. 2015;5:17977. doi: 10.1038/srep17977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adamovich Y., Rousso-Noori L., Zwighaft Z., Neufeld-Cohen A., Golik M., Kraut-Cohen J. Circadian clocks and feeding time regulate the oscillations and levels of hepatic triglycerides. Cell Metabolism. Feb 4 2014;19:319–330. doi: 10.1016/j.cmet.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ripperger J.A., Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nature Genetics. Mar 2006;38:369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- 57.Doi M., Hirayama J., Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. May 5 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 58.Vitaterna M.H., Ko C.H., Chang A.M., Buhr E.D., Fruechte E.M., Schook A. The mouse clock mutation reduces circadian pacemaker amplitude and enhances efficacy of resetting stimuli and phase-response curve amplitude. Proceedings of the National Academy of Sciences of the United States of America. Jun 13 2006;103:9327–9332. doi: 10.1073/pnas.0603601103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paschos G.K., Ibrahim S., Song W.L., Kunieda T., Grant G., Reyes T.M. Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nature Medicine. Dec 2012;18:1768–1777. doi: 10.1038/nm.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Degirolamo C., Sabba C., Moschetta A. Therapeutic potential of the endocrine fibroblast growth factors FGF19, FGF21 and FGF23. Nature Reviews Drug Discovery. Jan 2016;15:51–69. doi: 10.1038/nrd.2015.9. [DOI] [PubMed] [Google Scholar]
- 61.Bookout A.L., de Groot M.H., Owen B.M., Lee S., Gautron L., Lawrence H.L. FGF21 regulates metabolism and circadian behavior by acting on the nervous system. Nature Medicine. Sep 2013;19:1147–1152. doi: 10.1038/nm.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miller B.H., McDearmon E.L., Panda S., Hayes K.R., Zhang J., Andrews J.L. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proceedings of the National Academy of Sciences of the United States of America. Feb 27 2007;104:3342–3347. doi: 10.1073/pnas.0611724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Akhtar R.A., Reddy A.B., Maywood E.S., Clayton J.D., King V.M., Smith A.G. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Current Biology. Apr 2 2002;12:540–550. doi: 10.1016/s0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- 64.Uno K., Katagiri H., Yamada T., Ishigaki Y., Ogihara T., Imai J. Neuronal pathway from the liver modulates energy expenditure and systemic insulin sensitivity. Science. Jun 16 2006;312:1656–1659. doi: 10.1126/science.1126010. [DOI] [PubMed] [Google Scholar]
- 65.Prime S.S., Nixon S.V., Crane I.J., Stone A., Matthews J.B., Maitland N.J. The behaviour of human oral squamous cell carcinoma in cell culture. The Journal of Pathology. Mar 1990;160:259–269. doi: 10.1002/path.1711600313. [DOI] [PubMed] [Google Scholar]
- 66.Inoue I., Shinoda Y., Ikeda M., Hayashi K., Kanazawa K., Nomura M. CLOCK/BMAL1 is involved in lipid metabolism via transactivation of the peroxisome proliferator-activated receptor (PPAR) response element. Journal of Atherosclerosis and Thrombosis. 2005;12:169–174. doi: 10.5551/jat.12.169. [DOI] [PubMed] [Google Scholar]
- 67.Vogel M., Braungardt T., Meyer W., Schneider W. The effects of shift work on physical and mental health. The Journal of Neural Transmission (Vienna) Oct 2012;119:1121–1132. doi: 10.1007/s00702-012-0800-4. [DOI] [PubMed] [Google Scholar]
- 68.Lowden A., Moreno C., Holmback U., Lennernas M., Tucker P. Eating and shift work – effects on habits, metabolism and performance. Scandinavian Journal of Work, Environment & Health. Mar 2010;36:150–162. doi: 10.5271/sjweh.2898. [DOI] [PubMed] [Google Scholar]
- 69.Jiang Y., Li Y., Liu X. Intranasal delivery: circumventing the iron curtain to treat neurological disorders. Expert Opinion on Drug Delivery. 2015;12:1717–1725. doi: 10.1517/17425247.2015.1065812. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of genes differentially expressed at ZT10 in MT-LR compared with WT and MT mice.
Tissue distribution and long-term expression of HA-Clock mRNA after tail vein injection. A: mRNA levels in different tissues at day 9 after injection determined by qPCR. B: mRNA levels in liver at different times after injection. All data are means ± SEM; n = 5.
Body weight development and food intake in control cohorts. A, B: body weight development (A) and weekly weight gain (B) of WT- and MT-CONTROL (injected with SEAP) mice and Clock-injected WT mice under ad libitum access to HFD (n = 12). C, D: Cumulative (C) and daily food intake (D) of WT- and MT-CONTROL (injected with SEAP) mice and Clock-injected WT mice (n = 12). Data are shown as means ± SEM; 1- and 2-way ANOVAs with Bonferroni post-test; different letters depict statistically different groups (p < 0.05); ***: p < 0.001 MT-CONTROL vs. WT groups (A, C) or light (L) vs. dark (D) groups (D). Letters in B depict significant differences (p < 0.05; 1-way ANOVA).