Summary
Background
Rare highly penetrant gain of function mutations in caspase recruitment domain family, member 14 (CARD14) can lead to psoriasis, a chronic inflammatory disease of the skin and other organs.
Objectives
To investigate the contribution of rare CARD14 variants to psoriasis in the Tunisian population and expand knowledge of CARD14 variants in the European population.
Methods
CARD14 coding exons were re-sequenced in psoriasis cases and controls from Tunisia and Europe including included sixteen European cases with generalized pustular psoriasis (GPP). Novel variants seen in cases were evaluated for their effect upon NF-kb signalling.
Results
Rare variants in CARD14 were significantly enriched in Tunisian cases compared to controls. Three were collectively found in 5% of Tunisian cases and all affected the N terminal region of the protein harbouring its CARD or coiled-coil domain. These variants were: c.349G>A (p.Gly117Ser), c.205C>T (p.Arg69Trp) and c.589G>A (p.Glu197Lys). c.589G>A (p.Glu197Lys) led to upregulation of NF-kb activity in a similar manner to previously described psoriasis-associated mutations. p.Arg69Trp led to seven fold down-regulation of NF-kb activity. One Tunisian case harboured a c.1356+5G>A splice alteration that is predicted to lead to loss of exon 9 which encodes part of the coiled-coil domain. No GPP cases harboured an IL36RN mutation, but one out of 16 GPP cases with a family history of PV harboured a c.1805C>T (p.Ser602Leu) mutation.
Conclusions
These observations provide further insights into the genetic basis of psoriasis in the Tunisian population and provide functional information on novel CARD14 variants seen in cases from Tunisia and other populations.
Psoriasis is a chronic inflammatory disease of the skin and other organs affecting ~2–3% of the population of European ancestry but is less common in other populations1. Psoriasis consists of a number of distinct clinical phenotypes with plaque psoriasis accounting for 80% of cases2. To identify common and rare variants leading to psoriasis, genetic studies have been performed with linkage analysis and genome-wide association studies1,3. Rare highly penetrant mutations in caspase recruitment domain family, member 14 (CARD14) can lead to psoriasis and accounts for PSORS2 (psoriasis susceptibility locus 2). These mutations can also lead to psoriatic arthritis (PA)4, generalized pustular psoriasis (GPP)4,5 and pityriasis rubra pilaris (PRP)5. A common mis-sense variant in CARD14 also increases psoriasis and PA risk 1.15 fold in European and Asian populations4,6,7. Familial GPP can be caused by recessively inherited mutations in the IL36RN gene, which encodes interleukin 36 receptor antagonist (IL-36Ra)8,9.
The contribution of rare CARD14 variants to psoriasis in other populations is not well understood. To investigate this in the Tunisian population we directly sequenced its coding exons in a cohort of Tunisian psoriasis cases and controls. We also expanded our sequencing efforts of CARD14 in the European population. Rare novel CARD14 variants were identified in the Tunisian population and an excess of rare variants was detected in Tunisian psoriasis cases. We also re-sequenced 16 cases of generalized pustular psoriasis in psoriasis sib pair families, and did not detect any IL13RN mutations, but identified one case with a family history of psoriasis and a rare CARD14 mutation leading to a c.1805C>T (p.Ser602Leu) alteration in the PDZ domain. Sequencing of exon 4 of CARD14 which is a hotspot for mutations in additional European psoriasis cases revealed additional mutations. CARD14 is a scaffold protein that regulates NF-kb (nuclear factor of kappa light chain enhancer in B-cells) pathway signalling10, and we determined the effects of the novel CARD14 variants detected in cases on its activity. We identified likely pathogenic variants with effects in both directions and this is discussed. Our studies enhance our knowledge of CARD14 variants contributing to psoriasis in different human populations and an examination of their effect on NF-kB signaling provides insights into their likely pathogenicity.
Materials and methods
Subjects
Tunisian cases consisted of 282 (104 women and 178 men, mean age 41.9). Patients were recruited between 2008 and 2012 from the outpatient clinics and hospital wards of two participating dermatology departments: La Rabta Hospital of Tunis and the Military Hospital of Tunis. Each patient was examined by a dermatologist who evaluated disease type and severity. A detailed questionnaire concerning age of diagnosis and family history of psoriasis was collected. The psoriasis cases included seventy-five PA cases with comitant psoriasis who had been recruited from the Rheumatology Department of La Rabta Hospital and the Military Hospital. All cases of PA were confirmed by a rheumatologist. While the majority of the psoriasis cases were diagnosed with classic plaque-type psoriasis vulgaris, (PV) 20% had other forms of psoriasis that included guttate psoriasis (5%), Generalized Pustular Psoriasis (GPP) (2%), localized palmo plantar psoriasis (PP) (9%) and Erythrodermic Psoriasis (4%). A sample of 192 healthy ethnically, sex and age matched individuals (mean age 43) was also recruited. Individuals with previously diagnosed inflammatory disease were excluded from the control group. Informed written consent was obtained from all individuals participating in this study prior to blood sampling. Cases and controls for the European cohorts are described elsewhere11 and included 242 affected sib pair families and their parents, and cases and controls from the National Psoriasis Foundation Victor Henschel Tissue Repository (NPF). This study was approved by the ethics committees of participating institutions in accordance with the Declaration of Helsinki.
Sanger resequencing
All coding exons of full-length CARD14 (CARD14fl) were re-sequenced in the 272 Tunisian sporadic psoriasis cases, 192 Tunisian controls, and at least one case from 10 families with multiple psoriasis-affected members and described elsewhere12. Sixteen GPP cases of European ancestry from nuclear families with affected sibling pairs were also re-sequenced for all coding exons of CARD14 and IL36RN. Having previously identified exon 4 as a mutation hotspot in CARD144, we re-sequenced that exon in one affected case from each of the additional European families (240 total). We also re-sequenced 944 psoriasis cases, 675 controls, and 17 samples of unknown status from NPF for exon 4. Of those samples, 486 cases and 152 controls were Caucasian. Demographic information was not available for most of the other NPF cases and controls. Primers are available upon request. Mutations in cDNA shown in Tables 1A and 1B refer to CARD14fl sequence NM_001257970.
Table 1.
| A. Rare CARD14 variants identified in the Tunisian population by resequencing. | |||||||
|---|---|---|---|---|---|---|---|
|
CARD14 Exon |
cDNA Mutation and Corresponding Protein Change |
SNP ID | Protein Domain |
Observed in Tunisian Cases/Controls |
Consensus prediction on protein function |
Effect on NF-kb Activation (FC versus Wild-type CARD14sh) (significant at P < 0.05*; P < 0.01** or P < 0.001***) |
Previously Published? |
| 2 | c.G185A (p.Arg62Gln) | rs115582620 | 2PV (1 homozygote) | Benign | 1.06 | Yes | |
| 2 | c.205C>T (p.Arg69Trp) | rs375624435 | CARD | 4 PV, 1 PA | Damaging | 0.144*** | No |
| 3 | c.349G>A (p.Gly117Ser) | rs281875215 | Between CARD and coiled-coil | 1 PV (familial), 1PA | Damaging | 3.714 | Yes4,12 |
| 4 | c.T449G (p.Leu150Arg) | rs146214639 | Coiled-coil | 1 PV | Neutral | 1.794 | Yes11 |
| 4 | c.452G>A (p.Arg151Gln) | rs200731780 | Coiled-coil | 1 PV | Benign | 0.576** | No |
| 4 | c.589G>A (p.Glu197Lys) | rs200790561 | Coiled-coil | 4 PV, 3 PA, 1 C | Damaging | 1.667* | Yes17 |
| 4 | c.599G>A (p.Ser200Asn) | rs114688446 | Coiled-coild | 2 PV | Benign | 0.6711 | Yes11 |
| 7 | c.1012A>G (p.Met338Val) (homozygote) | rs200132496 | Coiled-coile | 1 PV | Benign | 0.914 | No |
| 7 | c.1049T>C (p.Leu350Pro) | none | Coiled-coile | 1C | Damaging | ND | No |
| 7 | c.1079T>C (p.Leu357Pro) | none | Coiled-coil | 1C | Damaging | ND | No |
| 9 | c.1258A>G (p.Thr420Ala) | none | none | 1 PV | Benign | 0.663*** | No |
| 9 | c.1356+5G>A | rs376524884 | NA | 1 PV | Damaging | NA | No |
| B. Novel rare CARD14 variants identified in Europeans. | |||||||
|---|---|---|---|---|---|---|---|
|
CARD14 Exon |
cDNA Mutation and Corresponding Protein Change |
SNP ID | Protein Domain |
Observed in European (and other)a Cases/Controls |
Predicted Effect on Protein Function |
Effect on NF-kb Activation (FC versus Wild-type CARD14sh) (significant at P < 0.05*; P < 0.01** or P < 0.001***) |
Previously Published? |
| 4 | c.451C>T (p.Arg151Trp) | none | Coiled-coil | 1 unknown (NPF) | Probably damaging | 1.766*** | No |
| 4 | c.452G>A (p.Arg151Gln) | rs200731780 | Coiled-coil | 1 PV (NPF); 1C (NPF) | Benign | 0.576** | No |
| 4 | c.589G>A (p.Glu197Lys) | rs200790561 | Coiled-coil | 1 C (NPF) | Probably damaging | 1.667* | Yes17 |
| 4 | c.626T>C (p.Leu209Pro) | none | Coiled-coil | 1 PV (PSSP128-1) | Probably damaging | 0.785 | No |
| 4 | c.646G>A (p.Ala216Thr) | rs574982768 | Coiled-coil | 1 PV (NPF) | Benign | 0.575* | Yes18 |
| 4 | c.652C>T (p.Arg218Cys) | none | Coiled-coil | 1 C (NPF) | Damaging | 0.658* | No |
| 13 | c.1805C>T (p.Ser602Leu) | rs201285077 | PDZ | GPP, PPP, PV (siblings, parent) | Damaging | 1.196 | No |
| 14 | c.1916C>G (p.Ala639Gly) | none | PDZ | 1 atopic dermatitis case | Damaging | 0.917 | No |
CARD14 missense variants are listed with details on their locations in critical CARD14 protein domains, their predicted effect on protein function (where predicted to be damaging by four out of five prediction algorithms they have been deemed damaging, and where predicted to be damaging by three out of five prediction algorithms they have been deemed to be probably damaging) (See also Supplementary Table 1 for a complete description of variants characterized by ANNOVAR) and their effect on NF-kb activation (fold change compared to un-stimulated wild-type CARD14sh; see also Figure 2).
The following abbreviations are used: FC, fold change; NA, not applicable; ND, not done; NPF, National Psoriasis Foundation; PSSP, psoriasis case from the St.Louis/Dallas/UCSF cohort of Northern European ancestry23,24. PV, psoriasis vulgaris; PA, psoriatic arthritis; PP, palmar plantar psoriasis; C, control.
CARD14 missense variants are listed with details on their locations in critical CARD14 protein domains, their predicted effect on protein function (See also Supplementary Table 1 for a complete description of variants characterized by ANNOVAR) and their effect on NF-kb activation (fold change compared to un-stimulated wild-type CARD14sh; see also Figure 2).
Abbreviations are described in the legend of Table 1A.
Statistical Analysis
The burden test for association of the selected rare variants with the disease was performed with the SKAT package in R version 3.0.113. The logistic weights were calculated from the minor allele frequencies and were applied to compute the p-values. The following 12 variants were included in the test: c.G185A, c.C205T, c.G349A, c.T449G, c.G452A, c.G589A, c.G599A, c.A301G, c.A547G, c.T338C, c.T359C and rs376524884.
NF-kb luciferase reporter assay for CARD14 variants
Expression plasmids for CARD14 are described elsewhere4,11. Briefly, full length CARD14sh and CARD14cl (Genebank BC018142 and NM_052819) coding for 740 and 434 amino acids respectively were cloned into pReceiver-M11 (Capital Biosciences). The CARD14sh constructs were subjected to in vitro mutagenesis with the QuikChange Site-Directed Mutagenesis Kit (Stratagene). Numbering of all CARD14 variants in this manuscript is based on RefSeq NM_024110.3. For novel missense variants, constructs with the mutant allele were generated. The assay for NF-kb activation was performed as we have described4,11. HEK293 (human embryonic kidney) cells were cultured under standard conditions. Cells were co-transfected with (1) constructs encoding wild-type CARD14sh, mutant CARD14sh, or the wild type CARD14cl isoform which lacks the CARD domain, (2) pTAL-luc or pNF-kb-luc plasmid, and (3) pGL4.70 Renilla reporter plasmid (Promega). All transfections were performed in triplicate. Cells were harvested twenty-four hours after transfection and firefly luciferase activity was determined with the Dual-Luciferase Reporter Assay System (Promega). Relative NF-kb luciferase activity was calculated by background subtraction of pTAL luciferase activity followed by division of the normalized NF-kb luciferase value by the background pTAL luciferase value.
Results
We re-sequenced all coding exons of CARD14 in 272 Tunisian psoriasis cases, seventy five of whom had also been diagnosed with PA, at least one member of 10 multiply affected Tunisian families and 192 Tunisian controls. We also re-sequenced all coding exons of CARD14 and IL36RN in 16 GPP cases from affected sibling pair/nuclear European families and all coding exons of CARD14 in 14 atopic dermatitis (AD) European cases. We had previously identified exon 4 of CARD14 as a mutation hotspot, so we also re-sequenced this exon from DNA from one affected member from the remaining affected sibling-pair/nuclear families of European ancestry (240 in total) and from DNA of 1164 psoriasis cases, 675 controls, and 17 samples of unknown status from the National Psoriasis Victor Henschel Tissue Repository (NPF)
Among all these samples, we identified twelve rare variants in CARD14 in Tunisians (Table 1B) and eight in Europeans (Table 1B). Additional information on variants including allele frequencies in public databases and prediction of protein function from a variety of computational algorithms provided through ANNOVAR14 is provided in Supplementary Table 1. Specific alterations are discussed further below.
CARD14 Mutations in Tunisians
Three variants predicted to be damaging were collectively found in 5% of Tunisian PV cases and 0.5% of Tunisian controls: c.349G>A (p.Gly117Ser), c.205C>T (p.Arg69Trp) and c.589G>A (p.Glu197Lys) (9 PV cases, 5 PA and 1 control). All of these variants were rare or absent in other populations (frequencies in ExAC were 0, 0.0002 and 0.0008 respectively). The c.589G>A (p.Glu197Lys) mutation was seen in a total of 4 Tunisian PV and 3 PA cases and one control. It was also identified in a female control of unknown ancestry from the NPF. The c.205C>T (p.Arg69Trp) variant was seen in four PV and one PA case and no controls. None of the Tunisian cases with other forms of psoriasis harboured CARD14 mutations.
A c.G185A (p.Arg62Gln) variant previously reported by us in 0.15% of European cases and 0.8% of controls11, but predicted to be benign was seen in two Tunisian PV cases and no controls. One of the Tunisian PV cases was homozygous for the allele encoding the altered variant. The c.599G>A (p.Ser200Asn) variant was seen in three Tunisian cases and no controls. We identified a c.452G>A (p.Arg151Gln) substitution in a Tunisian case. This individual also harboured the c.589G>A (p.Glu197Lys) variant. We detected the same c.452G>A (p.Arg151Gln) mutation in a single case and a single control from the NPF suggesting that it is unrelated to disease and that it is the c.589G>A (p.Glu197Lys) variant that is pathogenic in this individual. One Tunisian case was homozygous for a c.1012A>G (p.Met338Val) alteration. This lies in the coiled-coil domain of the CARD14 peptide but the alteration was predicted to be benign. We also detected a c.1258A>G (p.Thr420Ala) alteration in a single case with PV, but this mutation is predicted to be benign. We identified a single case with a c.T449G (p.Leu150Arg) alteration that we have described elsewhere (allele frequency = 0.25% in cases and 0.16% in controls)11. It is predicted to be damaging and leads to 1.79 fold upregulation in NF-kB signaling. We also identified a single case with a c.1258A>G alteration in the consensus splice donor sequence of exon 9 in a single Tunisian case with PV. This mutation is predicted to affect splicing and led to skipping of exon 9 which encodes a portion of the CARD14 coiled-coil domain. Two controls harboured c.1049T>C (p.Leu350Pro) and c.1079T>C (p.Leu357Pro) alterations that interestingly are also predicted to be damaging.
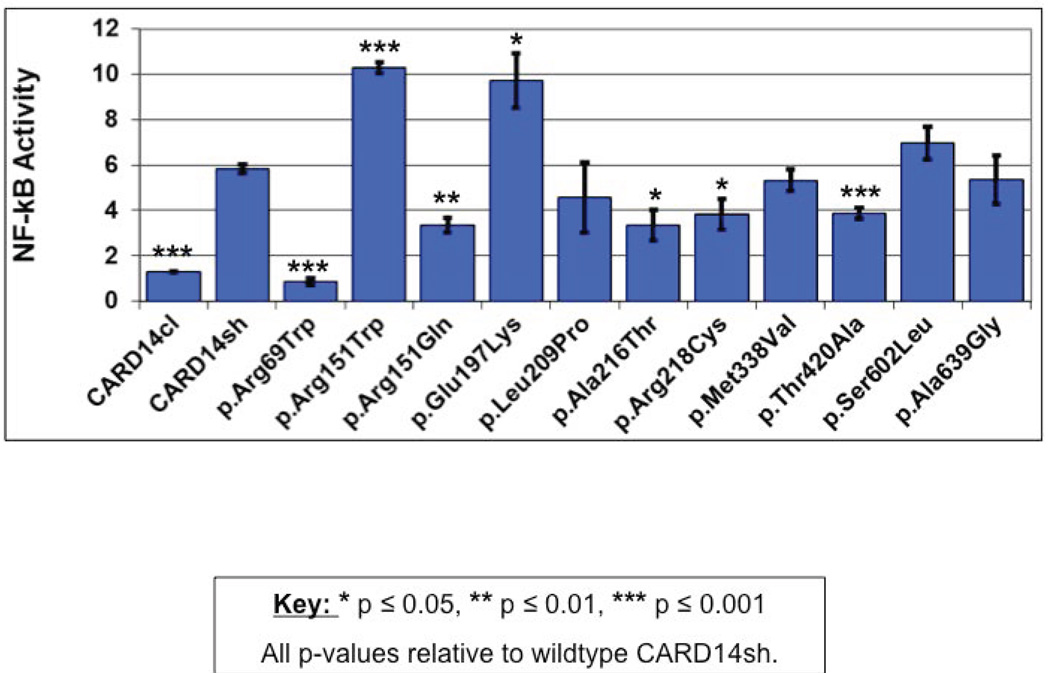
SKAT (SNP-set (Sequence) Kernel Association test15 was performed to compare the distribution of rare CARD14 variants in cases and controls in Tunisians and yielded a Burden test P value=1.420078e-19. We also tested the effects of up-regulated and down-regulated variants on NF-kB activation. The 3 down-regulated variants (c.205C>T (p.Arg69Trp), c.452G>A (p.Arg151Gln), c.1258A>G (p.Thr420Ala)) and two up regulated variants (p c.349G>A (p.Gly117Ser) and c.589G>A (p.Glu197Lys) showed significant differences between the case and controls (up-regulation p-value=0.001 and down-regulation p-value=6.8e-08). While suggesting that there is excess of rare CARD14 variants in cases versus controls, and that variants leading to both enhanced and decreased levels of NF-kB activation compared to wild type CARD14 can contribute to psoriasis, these results should be viewed with caution in light of small sample size and unadjusted for any covariates.
Novel European variants
We detected eight rare CARD14 variants in Europeans: four variants in psoriasis cases. A c.626T>C (p.Leu209Pro) was detected in a female of Northern European ancestry with PV (PSSP128-1). This alteration, is novel and not found in any public database (Supplementary Table 2), but was also present in the patient’s affected sibling who also had PV. Both siblings were diagnosed with PV between the ages of 21 and 30.The mother of these two girls had also been diagnosed with PV but her sequence, and that of their father was not available to determine from which parent the variant had been inherited. The c.626T>C (p.Leu209Pro) alteration lies in the coiled-coil region of CARD14 and is predicted to be damaging by a number of prediction programs. A c.646G>A (p.Ala216Thr) mutation was observed in a male of Northern European ancestry from the NPF who was diagnosed with psoriasis at 19 years of age and a c.652C>T (p.Arg218Cys) variant was identified in a male control of unknown ancestry from the NPF. An atopic dermatitis case harboured a c.1916C>G (p.Ala639Gly) mutation. One NPF case and one control harboured an c.452G>A (p.Arg151Gln) variant that had also seen in a Tunisian case, but this is predicted to be benign.
We did not identify any mutations in IL36RN in the sixteen sequenced pustular cases, but we did detect a mutation in CARD14 in one case with GPP which had developed during her teenage years. This mutation (c.1805C>T; p.Ser602Leu) alters the PDZ domain of CARD14 and is predicted to be damaging by a number of prediction programs (Supplementary Table 1). This mutation had been inherited from the patient’s mother who had PV and who had also transmitted this mutation to a male sibling with PV. The p.Ser602Leu was not present in the unaffected father. This patient also had a male sibling with palmar plantar hyperkertotic psoriasis but DNA was unavailable for sequencing.
We also detected two other variants in European cases, a c.451C>T (p.Arg151Trp) alteration in an individual from the NPF of unknown clinical status, and a c.589G>A (p.Glu197Lys) variant in a control (discussed below).
Effect of mutations on NF-kb activation
The identification of genes for complex traits is a challenge. While rare damaging variants can be identified in many genes, their effect on disease can still be hard to gauge. Rare variants do not necessarily lead to disease and programs predicting pathogenicity of alterations in proteins are not always reliable. Hence, functional assays that determine function of protein variants are important in establishing their pathogenicity. We have previously shown that significant up-regulation of NF-kB signaling can be a predictor of psoriasis causing variants of CARD144. We therefore tested the effect of novel CARD14 variants found in cases on NF-kB signalling using a previously described luciferase reporter assay4. Results are shown in Figure 2. Previous studies on p.G117S and p.S200N alterations showed that these alterations lead to significant changes in NF-kB signaling (3.71 and 0.67 fold respectively compared to wild type CARD14). In the case of the newly described alterations, p.Arg151Trp and p.Glu197Lys induced significantly more activation of NF-kB compared to that induced by the wild-type CARD14sh isoform (1.8-fold (P<0.001) and 1.7-fold (P < 0.05) respectively). The p c.205C>T (p.Arg69Trp), c.452G>A (p.Arg151Gln), c.646G>A (p.Ala216Thr), c.652C>T (p.Arg218Cys), and c.1258A>G (p.Thr420Ala) variants induced significantly less NF-kB activation than wild-type CARD14. Other variants had no significant effect.
Figure 2. Effect of wild-type and novel CARD14 variants on NF-kb activation.
HEK293 cells were transfected with the construct that codes for wild type CARD14sh, the same construct harbouring one of the rare variants shown, or a construct that codes for CARD14cl and lacks the CARD domain. NF-kb activity was determined by measuring relative luciferase activity. All values were first normalized to Renilla expression to control for transfection efficiency and then adjusted to control for activity of the empty background vector, pTAL-luc. Change in NF-kb activity relative to background vector was determined for each variant (y-axis, NF-kb activity). Every data point represents the average of three replicates. Asterisks show results from two-tailed, unpaired student’s t tests comparing NF-kb activation induced by the indicated variant to that of unstimulated cells with CARD14sh. * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001.
Discussion
Gain of function mutations in CARD14 can lead to psoriasis4. These mutations included familial mutations: a.Gly117Ser seen in a North American family of European origin with PV and PA and a splicing mutation at the acceptor site of the same exon in a Taiwanese family with PV. A de-novo p.Glu138Ala mutation led to GPP4. Further studies in other populations have revealed association of a p.Asp176His alteration with GPP with PV in Japan16. This variant had been described earlier11but did not provide evidence for association with PV. However, this alteration increased NF-kB activation ±2.8 fold compared to WT CARD14, suggesting that in some circumstances it is pathogenic. An p.Asp176His variant was also found in 2/251 PPP German cases compared to 1/1049 controls17 although no association with this variant and GPP was detected in Han Chinese18. All of these variants affect the coiled-coil region of CARD14, and most occur in exon 4. A common variant in CARD14 (Trp820Arg;rs11652075) that lies in the MAGUK domain at the C terminus was also shown to be associated with psoriasis11and achieved genome-wide significance in larger cohorts from Europe and China6,19. CARD14 mutations can also lead to pityriasis rubra pilaris5. The variants identified in the current study are discussed below.
The pathogenic CARD14 c.349G>A (p.Gly117Ser) mutation previously shown to be associated in rare familial forms of psoriasis and PA4 was found in a sporadic PA Tunisian case. We previously demonstrated that it segregates with PV in a multiplex Tunisian family12. Its frequency of ±0.3% in Tunisian cases is higher than the estimate of 0.02% in psoriasis cases from the northern European population11. The sporadic Tunisian case with this mutation had also developed PA, similar to 30% of the European family harboring this change4. We also identified a novel c.589G>A (p.Glu197Lys) alteration in CARD14 in ±2.5% of Tunisian cases with PV. Three out of seven of these cases had also been diagnosed with PA. The c.589G>A (p.Glu197Lys) variant is predicted to be damaging by a number of prediction algorithms (Table 1A, Supplementary Table 1) and is rare, with a frequency in the ExAC database of 0.0008. It led to 1.7 fold upregulation of NF-B signalling. In our previous studies, we suggested that variants in CARD14 had to induce at least a 2.5-fold increase in NF-kb activation relative to wild-type CARD14sh in order to lead to psoriasis4. However, in those studies, few variants induced a fold increase in NF-kb relative to wild-type CARD14sh in the lower 1.7–2.4-fold range that are described for several variants in the current study. The results obtained with these new variants suggest that the threshold for NF-kB might be lower than that we originally proposed. It is important to note that our assay for NF-kb activation levels corresponds very well with other assays we previously used to investigate the pathogenic effects of mutant CARD14, including upregulation of NF-kb targets such as IL8 by keratinocytes11. Consistent with this hypothesis, levels of NF-kb activation of less than 2.5 fold were also recently described for several PRP CARD14 mutations20. However, the c.589G>A (p.Glu197Lys) alteration was recently reported in three out of over 1000 German controls17, illustrating the problem of determining pathogenicity of rare variants, where numbers of affected cases are small, and functional assays are not available or indeterminate.
We also identified a novel c.205C>T (p.Arg69Trp) alteration in five PV (one with PA) cases and no controls (ExAC frequency = 0.0002). This variant was also predicted to be damaging by a number of algorithms (Supplementary Table 1) but unlike other pathogenic psoriasis mutations its effect on NF-kB activation was in the opposite direction since it led to a seven fold decrease. However, we previously described an p.Arg38Cys alteration in CARD14 in a single psoriasis case that leads to a similar decrease in NF-kB activation levels11. It is possible that disruption of NF-kb signalling can lead to disease, regardless of its direction since a critical level of NF-kb signalling is required to maintain skin homeostasis21. Alternatively, some CARD14 variants could be affecting signalling pathways other than NF-kb. Additional support for the importance of this p.Arg69 residue comes from a German study which revealed one GPP patient with an p.Arg69Gln mutation that was also predicted to be damaging22.
We saw an c.G185A (p.Arg62Gln) variant previously reported in 0.15% of European cases and 0.8% of controls11 in two Tunisian PV cases and no controls. One of the Tunisian PV cases was homozygous for the p.Glu62 allele. We had previously seen an p.Arg62Glu (CGG to CAG) variant in a child with GPP who harboured the allele encoding p.Glu138Ala4. The p.Arg62Glu variant had been inherited from her unaffected father and it is predicted to be benign. Hence, its role in psoriasis pathogenesis is inconclusive.
A single Tunisian case with PV was homozygous for a novel p.Met338Val (c.1012A>G) mutation but this was predicted to be benign. One case with PV harboured a mutation (c.1356+5G>A) in the splice donor of exon 9 which is likely to affect protein function by disrupting the coiled-coil domain of CARD14.
The c.599G>A (p.Ser200Asn) alteration was seen in 3 Tunisian cases and no controls. In our earlier study its association with psoriasis achieved borderline significance (P = 0.05 in a meta-analysis)11 but this variant is predicted to be benign and leads to a 0.67 fold down-regulation in NF-kB signalling. This variant was recently shown to be more common in German PPP cases than in controls where allele frequencies were 0.012 versus 0.005 respectively17. However, control frequencies in this population were less than those reported previously in the cohort of North American controls of European origin where they were estimated to be 0.008411. Hence, its relationship to psoriasis susceptibility is not clear.
Despite the ambiguous effects of some CARD14 alterations, we detected an excess of rare variants in cases versus controls indicating that as in the European population, rare variants in CARD14 can play an important role in triggering PV and PA in the Tunisian population.
In the European population we identified a c.626T>C (p.Leu209Pro) mutation in two siblings with psoriasis. This variant is not reported in any public database and is predicted to be damaging. A c.646G>A (p.Ala216Thr) variant was seen in a PV case from the NPF tissue bank. This mutation was previously described in a PV patient from the Chinese Han population18. This alteration lies in the coiled-coil region of CARD14 and led to a significant down-regulation in NF-kB activation (±0.6 fold)
We also identified a c.1805C>T (p.Ser602Leu) mutation in a GPP case whose mother and brother with PV also harboured the mutation. This case had a second brother with palmoplantar hyperkeratotic psoriasis but unfortunately his DNA was not available for sequencing. This alteration led to 1.2 fold upregulation of NF-kB which is not significantly different from wild type CARD14sh, so that at this stage, its effect upon this pathway is unclear. However, this variant is very rare (freq = 0.0002 in ExAC (Supplementary Table 1). The altered amino acid lies within the PDZ domain and has a strong likelihood of being damaging. Hence, if this mutation is indeed responsible for the different forms of psoriasis in this family, it could be acting through a different pathway, or could be responding to an external signal relayed through the MAGUK domain of CARD14. An Asp176His alteration in CARD14, originally described by Jordan11 was recently described in two European patients with palmar plantar pustulosis (PPP) who were negative for mutations in IL36RN17. Hence, a further evaluation of CARD14 in additional cases of PPP is worthwhile.
Finally, we were interested in determining if rare CARD14 variants are ever seen in atopic dermatitis, an inflammatory skin disorder leading to an altered skin barrier. Resequencing of 16 AD cases revealed one with a p.Ala639Gly alteration that lies within the PDZ domain. This is predicted to be damaging, although it did not affect NF-kB signaling. Hence, further analysis of CARD14 in additional inflammatory skin diseases is warranted.
Overall, our observations increase the number of highly penetrant psoriasis mutations, help to clarify the functions of some of the recently described CARD14 variants, and provide further insights into the genetic basis of different forms of psoriasis in the Tunisian population.
Supplementary Material
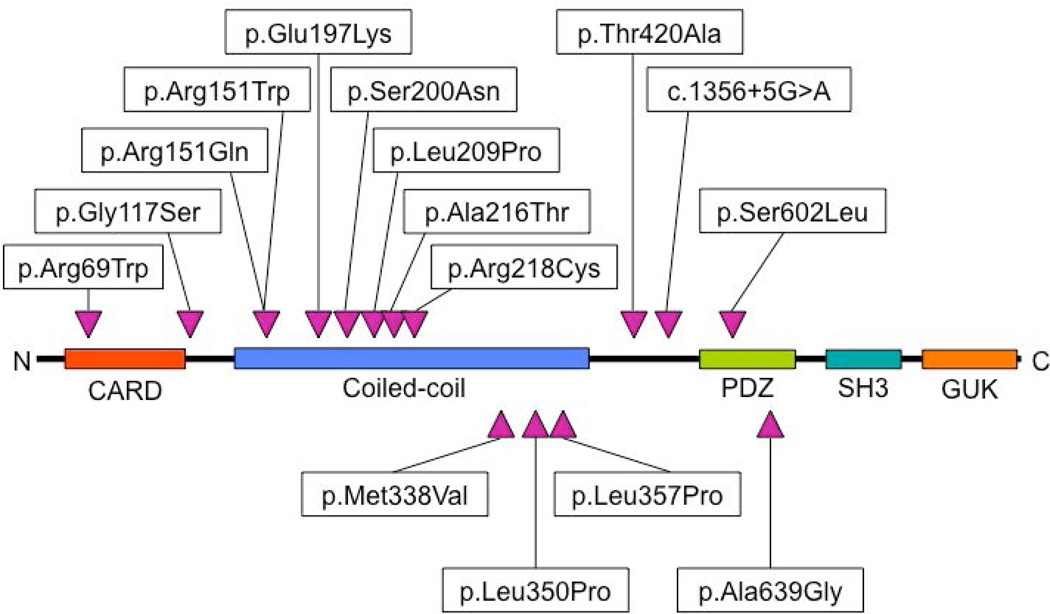
Figure 1. CARD14 protein domains and locations of amino acid substitutions.
Novel missense variants identified in CARD14 by re-sequencing are shown relative to key protein domains.
What's already known about this topic?
Mutations in CARD14 lead to psoriasis
What does this study add?
Knowledge of CARD14 mutations in Tunisian cases and further information in European psoriasis cases and functional consequences of mutations.
Acknowledgements
This study was supported in part by NIH grant AR050266 to AMB.
References
- 1.Lowes MA, Bowcock AM, Krueger JG. Pathogenesis and therapy of psoriasis. Nature. 2007;445:866–873. doi: 10.1038/nature05663. [DOI] [PubMed] [Google Scholar]
- 2.Griffiths CE, Christophers E, Barker JN, et al. A classification of psoriasis vulgaris according to phenotype. The British journal of dermatology. 2007;156:258–262. doi: 10.1111/j.1365-2133.2006.07675.x. [DOI] [PubMed] [Google Scholar]
- 3.Rahman P, Elder JT. Genetic epidemiology of psoriasis and psoriatic arthritis. Annals of the rheumatic diseases. 2005;64(Suppl 2):ii37–ii39. doi: 10.1136/ard.2004.030775. discussion ii40-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jordan CT, Cao L, Roberson ED, et al. PSORS2 is Due to Mutations in CARD14. American journal of human genetics. 2012 doi: 10.1016/j.ajhg.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuchs-Telem D, Sarig O, van Steensel MA, et al. Familial pityriasis rubra pilaris is caused by mutations in CARD14. American journal of human genetics. 2012;91:163–170. doi: 10.1016/j.ajhg.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsoi LC, Spain SL, Knight J, et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nature genetics. 2012;44:1341–1348. doi: 10.1038/ng.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowes J, Budu-Aggrey A, Huffmeier U, et al. Dense genotyping of immune-related susceptibility loci reveals new insights into the genetics of psoriatic arthritis. Nature communications. 2015;6:6046. doi: 10.1038/ncomms7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marrakchi S, Guigue P, Renshaw BR, et al. Interleukin-36-receptor antagonist deficiency and generalized pustular psoriasis. The New England journal of medicine. 2011;365:620–628. doi: 10.1056/NEJMoa1013068. [DOI] [PubMed] [Google Scholar]
- 9.Onoufriadis A, Simpson MA, Pink AE, et al. Mutations in IL36RN/IL1F5 are associated with the severe episodic inflammatory skin disease known as generalized pustular psoriasis. American journal of human genetics. 2011;89:432–437. doi: 10.1016/j.ajhg.2011.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bertin J, Wang L, Guo Y, et al. CARD11 and CARD14 are novel caspase recruitment domain (CARD)/membrane-associated guanylate kinase (MAGUK) family members that interact with BCL10 and activate NF-kappa B. The Journal of biological chemistry. 2001;276:11877–11882. doi: 10.1074/jbc.M010512200. [DOI] [PubMed] [Google Scholar]
- 11.Jordan CT, Cao L, Roberson ED, et al. Rare and common variants in CARD14, encoding an epidermal regulator of NF-kappaB, in psoriasis. American journal of human genetics. 2012;90:796–808. doi: 10.1016/j.ajhg.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ammar M, Bouchlaka-Souissi C, Helms CA, et al. Genome-wide linkage scan for psoriasis susceptibility loci in multiplex Tunisian families. The British journal of dermatology. 2013;168:583–587. doi: 10.1111/bjd.12050. [DOI] [PubMed] [Google Scholar]
- 13.Li D. SKAT: SNP-set (Sequence) Kernel Association Test. R package version 1.0.1 2014 [Google Scholar]
- 14.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei C, Li M, He Z, et al. A weighted U-statistic for genetic association analyses of sequencing data. Genet Epidemiol. 2014;38:699–708. doi: 10.1002/gepi.21864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sugiura K, Muto M, Akiyama M. CARD14 c.526G>C (p.Asp176His) is a significant risk factor for generalized pustular psoriasis with psoriasis vulgaris in the Japanese cohort. The Journal of investigative dermatology. 2014;134:1755–1757. doi: 10.1038/jid.2014.46. [DOI] [PubMed] [Google Scholar]
- 17.Mossner R, Frambach Y, Wilsmann-Theis D, et al. Palmoplantar Pustular Psoriasis is Associated with Missense Variants in CARD14, but not with loSs-of-Function Mutations in IL36RN in European Patients. The Journal of investigative dermatology. 2015 doi: 10.1038/jid.2015.186. [DOI] [PubMed] [Google Scholar]
- 18.Qin P, Zhang Q, Chen M, et al. Variant analysis of CARD14 in a Chinese Han population with psoriasis vulgaris and generalized pustular psoriasis. The Journal of investigative dermatology. 2014;134:2994–2996. doi: 10.1038/jid.2014.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang H, Jin X, Li Y, et al. A large-scale screen for coding variants predisposing to psoriasis. Nature genetics. 2014;46:45–50. doi: 10.1038/ng.2827. [DOI] [PubMed] [Google Scholar]
- 20.Li Q, Jin Chung H, Ross N, et al. Analysis of CARD14 Polymorphisms in Pityriasis Rubra Pilaris: Activation of NF-kappaB. The Journal of investigative dermatology. 2015 doi: 10.1038/jid.2015.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pasparakis M. Regulation of tissue homeostasis by NF-kappaB signalling: implications for inflammatory diseases. Nat Rev Immunol. 2009;9:778–788. doi: 10.1038/nri2655. [DOI] [PubMed] [Google Scholar]
- 22.Korber A, Mossner R, Renner R, et al. Mutations in IL36RN in patients with generalized pustular psoriasis. The Journal of investigative dermatology. 2013;133:2634–2637. doi: 10.1038/jid.2013.214. [DOI] [PubMed] [Google Scholar]
- 23.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nature methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature protocols. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.