ABSTRACT
This study surveyed the prevalence of mcr-1 in extended-spectrum-β-lactamase (ESBL)-producing Escherichia coli strains of food origin in China and identified strains that carried mcr-1, fosA3, and ESBL genes, which were carried in various plasmids. The mcr-1 and ESBL genes could be cotransferred by one or more types of plasmids. The presence of these multidrug-resistant E. coli strains in food products might pose a huge threat to public health.
KEYWORDS: mcr-1, fosA3, ESBL-producing E. coli, transmission, circular intermediate, Tn6330, plasmids
TEXT
Clinical and public health problems due to multidrug-resistant (MDR) bacterial infections have been further aggravated in recent years following the emergence of blaNDM-1, a resistance gene that can mediate development of carbapenem resistance in the host strain and possesses the ability to disseminate rapidly among various species of bacterial pathogens worldwide (1, 2). Polymyxins have been regarded as the antibiotic of last resort to treat severe infections caused by carbapenem-resistant Enterobacteriaceae (CRE). Recently, a new plasmid-mediated colistin resistance mechanism, mediated by the MCR-1 protein, a phosphoethanolamine transferase that modifies bacterial lipid A through modification of its phosphoethanolamine moiety, was discovered (3). Since the initial discovery of mcr-1-positive Enterobacteriaceae strains in China in 2015, this resistance mechanism has been reported in various parts of the world (3, 4). However, there is still a lack of comprehensive information regarding the prevalence of mcr-1 in Escherichia coli of food origin. This study reports the isolation and characterization of foodborne E. coli strains that carried mcr-1, fosA3, and ESBL genes.
A total of 408 nonrepeated cefotaxime-resistant E. coli isolates were obtained from 828 retail food samples (484 pork, 76 beef, 143 chicken, and 125 shrimp) purchased from open-air markets and supermarkets in Shenzhen, China, during the period of 10 August 2015 to 22 February 2016. E. coli isolates were selected on MacConkey agar plates supplemented with 2 μg/ml cefotaxime and identified by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) using a Bruker MicroFlex LT mass spectrometer (Bruker Daltonics). E. coli isolates were further confirmed by API20E test strips (bioMérieux, Inc.). All the E. coli strains were subjected to antimicrobial susceptibility testing for 11 antimicrobials using the agar dilution method according to the CLSI guidelines (5). All isolates were shown to be resistant to ampicillin, cefotaxime, ceftriaxone, and sulfamethoxazole-trimethoprim. They also exhibited resistance to other antibiotics, such as tetracycline (96%), nalidixic acid (84%), chloramphenicol (86%), kanamycin (61%), and ciprofloxacin (59%). However, all isolates were susceptible to meropenem, and only 5% of these strains were resistant to amikacin.
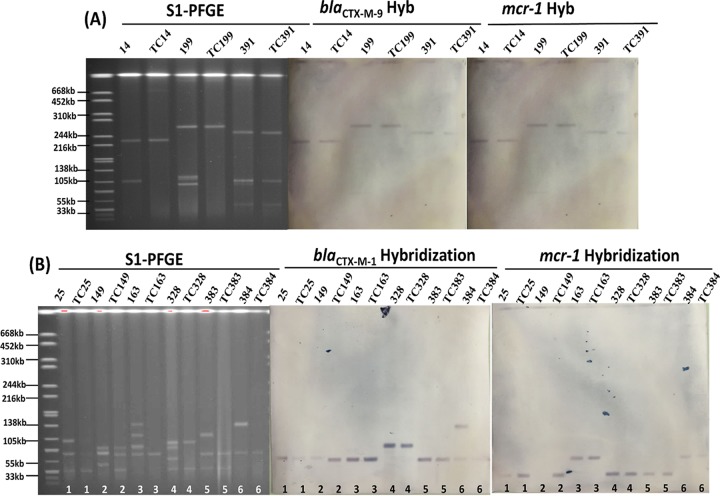
These E. coli isolates were subjected to screening for the presence of the mcr-1 gene by PCR assay as previously described (3). A total of 109 out of 408 (27%) cefotaxime-resistant E. coli strains were found to harbor the mcr-1 gene. Out of the 109 mcr-1-positive E. coli strains, 106 were resistant to colistin (MIC ≥ 4 μg/ml). Conjugation experiments were performed for all mcr-1-positive E. coli isolates as previously described (10) and showed that 35 out of the 109 isolates could successfully transfer their colistin resistance phenotypes to the recipient strain, E. coli J53. The MICs of colistin for these 35 transconjugants were mainly either 4 or 8 μg/ml, with the majority exhibiting resistance to multiple antimicrobial agents except for meropenem (Table 1). DNA linearization with S1 nuclease followed by pulsed-field gel electrophoresis (S1-PFGE) and Southern hybridization analyses was performed on these 35 E. coli strains and their transconjugants using the mcr-1 probe as previously described (6), with results confirming that the mcr-1 gene of 34 isolates was located on three major types of conjugative plasmids (∼33, ∼60, and ∼216 to 280 kb); interestingly, some isolates were found to harbor more than one mcr-1-bearing plasmid.
TABLE 1.
MICs, antimicrobial resistance genes, and plasmid profiles of E. coli strains and their corresponding transconjugantsd
| Strain | MIC (μg/ml) ofa: |
AMRb gene(s) | Plasmid(s) (kb) | mcr-1 plasmids (kb) | blaCTX-M plasmid (kb) | CIc form | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CLS | AMK | CTX | CIP | CRO | FOS | ||||||
| 14 | 8 | 4 | 8 | 0.12 | >16 | >512 | blaCTX-M-14, fosA3 | ∼104, ∼230 | ∼230, IncHI2 | ∼230 | + |
| TC14 | 4 | 0.5 | 8 | 0.06 | >16 | <4 | blaCTX-M-14 | ∼230 | ∼230, IncHI2 | ∼230 | + |
| 25 | 8 | 2 | >16 | >16 | >16 | >512 | blaCTX-M-55, fosA3 | ∼33, ∼60, ∼104 | ∼33, IncX4 | ∼60 | − |
| TC25 | 4 | 1 | 4 | 0.015 | >16 | <4 | blaCTX-M-55 | ∼33, ∼60 | ∼33, IncX4 | ∼60 | − |
| 149 | 8 | >128 | >16 | >16 | >16 | >512 | blaCTX-M-55, fosA3 | ∼33, ∼60, ∼90 | ∼33, IncX4 | ∼60 | − |
| TC149 | 8 | 0.5 | >16 | 0.015 | >16 | <4 | blaCTX-M-55 | ∼33, ∼60 | ∼33, IncX4 | ∼60 | − |
| 163 | 8 | 4 | >16 | >16 | >16 | >512 | blaCTX-M-55, blaCTX-M-65, fosA3 | ∼60, ∼78, ∼104, ∼138 | ∼60, IncI2 | ∼60 | − |
| TC163 | 8 | 0.5 | >16 | 0.015 | >16 | <4 | blaCTX-M-55 | ∼60 | ∼60, IncI2 | ∼60 | − |
| 199 | 8 | 1 | >16 | 0.5 | >16 | <4 | blaCTX-M-14 | ∼90, ∼120, ∼280 | ∼280, IncHI2 | ∼280 | + |
| TC199 | 8 | 0.5 | >16 | 0.015 | >16 | <4 | blaCTX-M-14 | ∼280 | ∼280, IncHI2 | ∼280 | + |
| 328 | 4 | 2 | >16 | 2 | >16 | >512 | blaCTX-M-55, fosA3 | ∼33, ∼90 | ∼33, IncX4 | ∼90 | − |
| TC328 | 4 | 0.5 | >16 | 0.0075 | >16 | <4 | blaCTX-M-55 | ∼33, ∼90 | ∼33, IncX4 | ∼90 | − |
| 383 | 8 | 4 | >16 | >16 | >16 | >512 | blaCTX-M-55, fosA3 | ∼33, ∼60, ∼78, ∼138, ∼200 | ∼33, IncX4 | ∼60 | − |
| TC383 | 4 | 0.5 | >16 | 0.0075 | >16 | <4 | blaCTX-M-55 | ∼33, ∼60 | ∼33, IncX4 | ∼60 | − |
| 384 | 4 | >128 | >16 | >16 | >16 | >512 | blaCTX-M-55, fosA3 | ∼60, ∼120 | ∼60, IncI2 | ∼60 | − |
| TC384 | 2 | 0.5 | >16 | 0.0075 | >16 | <4 | blaCTX-M-55 | ∼60 | ∼60, IncI2 | ∼60 | − |
| 391 | 4 | 2 | >16 | 0.5 | >16 | <4 | blaCTX-M-14 | ∼33, ∼104, ∼250 | ∼250, IncHI2 | ∼250 | + |
| TC391 | 4 | 0.5 | 4 | 0.015 | >16 | <4 | blaCTX-M-14 | ∼250 | ∼250, IncHI2 | ∼250 | + |
| J53 | 1 | 0.03 | 0.015 | 0.5 | 1 | <4 | − | − | − | − | − |
CLS, colistin; AMK, amikacin; CTX, cefotaxime; CIP, ciprofloxacin; CRO, ceftriaxone; FOS, fosfomycin.
AMR, antimicrobial resistance.
CI, circular intermediate.
−, negative; +, positive.
Among these 35 strains carrying conjugative mcr-1-encoding plasmids, 26 strains transferred only mcr-1-positive plasmids, while 9 were able to transfer both mcr-1-positive plasmids and ESBL-gene-encoding plasmids. These 9 E. coli strains were further characterized. Multilocus sequence typing (MLST) was performed for all these strains, and the results showed that strains 199 and 383 belonged to sequence type 10 (ST10), strain 25 belonged to ST641, and strain 328 belonged to ST4015, whereas the remaining 5 strains belonged to new different STs, suggesting the genetic diversity of these E. coli strains. β-Lactamase genes were screened in these 9 isolates and their transconjugants as previously described (7). The results showed that 6 out of 9 transconjugants were positive for the blaCTX-M-1 group gene and that the other three were positive for the blaCTX-M-9 group gene. Further sequencing of the full length of these β-lactamase genes confirmed that all blaCTX-M-1 group genes were blaCTX-M-55, with strain 149 also carrying a blaCTX-M-65, whereas the blaCTX-M-9 group genes were blaCTX-M-14. For blaCTX-M-55-positive transconjugants, the sizes of plasmids observed in these strains were ∼60 and ∼90 kb, with five plasmids being ∼60 kb (Table 1, Fig. 1). Plasmids harboring the mcr-1 gene were two sizes, ∼33 and ∼60 kb. Isolates 163 and 384 were found to harbor plasmids of ∼60 kb, which carried the mcr-1 and blaCTX-M-55 genes. For E. coli strains carrying the blaCTX-M-14 gene, the blaCTX-M-14 and mcr-1 genes were found to be located in plasmids of ∼230, ∼250, and ∼280 kb (Table 1, Fig. 1).
FIG 1.
S1-PFGE and Southern hybridization analyses of E. coli strains and their corresponding transconjugants carrying the mcr-1 and various blaCTX-M genes.
Recently, renewed attention has been paid to fosfomycin for the treatment of both urinary and systemic infections due to rapid dissemination of multidrug-resistant Gram-negative bacteria, especially strains of Enterobacteriaceae species that are resistant to traditionally used agents (8). Compared to other agents, fosfomycin seems to have retained antimicrobial activity against a substantial percentage of clinical isolates, particularly E. coli isolates. Therefore, the presence of a fosfomycin-resistant determinant commonly detected in China, the fosA3 gene, was screened for in these 9 E. coli isolates and their transconjugants as previously described. Surprisingly, 7 out of the 9 E. coli strains harbored the fosA3 gene, which was not cotransferred with the mcr-1 and ESBL genes. Plasmid profile analysis of both parental and transconjugants of these 9 E. coli strains suggests that the fosA3 gene might be located on additional plasmids of various sizes, with plasmids of ∼104 kb being the most dominant. Further research will be needed to understand more about the genetic features of the fosA3 gene in these E. coli strains.
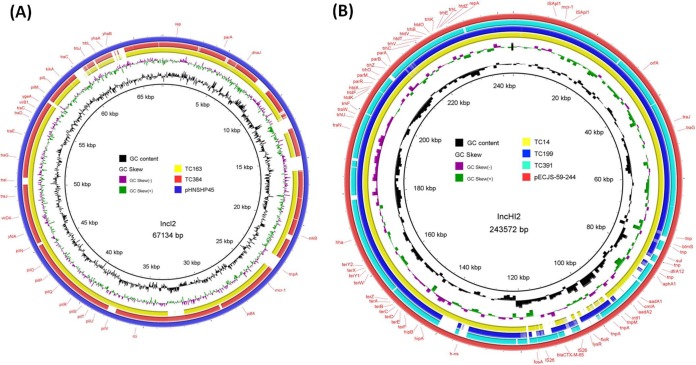
The total genomic DNA from the 9 transconjugants carrying the mcr-1 and blaCTX-Ms genes was extracted and sequenced using the Illumina platform as previously described (6). For the 6 E. coli transconjugants carrying blaCTX-M-55, two types of mcr-1-bearing plasmids were detected, the ∼30-kb IncX4 type and the ∼60-kb IncI2 type. Alignment of Illumina contigs of the four ∼30-kb plasmids harbored by E. coli strains TC25, TC149, TC383, and TC328 to several previously reported plasmids showed that they aligned very well to pOW3E1 (KX129783.1) (>99% in both identity and coverage; data not shown). Two transconjugants, TC163 and TC384, carried an ∼60-kb IncI2-type plasmid that carried the mcr-1 and blaCTX-M-55 genes. Illumina contigs of these two plasmids aligned well to IncI2 plasmid pA31-12 (KX034083), suggesting that these two plasmids were highly homologous to pA31-12 (9). Illumina contigs of plasmids carrying both mcr-1 and blaCTX-M-14 from 3 E. coli transconjugants, TC14, TC199, and TC391, exhibited a high degree of sequence homology with pECJS-59-244 (KX084394.1) and harbored a large MDR region which rendered IncHI2 the most genetically diverse plasmid type (Fig. 2). All three IncHI2 plasmids were found to harbor ISApI1–mcr-1–orf–ISApI1, the intact composite transposon Tn6330 (6). Tn6330 is known to form a circular intermediate comprising two direct repeats of ISApI1 and the mcr-1 gene, which may facilitate the transmission of mcr-1 in different plasmid backbones (10). All 9 transconjugants were then screened for the presence of the circular intermediate as previously described (10). Consistent with previous reports, circular intermediates could be detected only in E. coli strains that carried IncHI2 plasmids carrying intact Tn6330, not in strains carrying IncX4 or IncI2 plasmids. This confirmed that the circular intermediate can be generated only when the mcr-1-orf gene cassette is surrounded by two copies of ISApI1 to form intact Tn6330.
FIG 2.
Sequence alignment of IncI2- and IncHI2-type mcr-1-bearing plasmids. (A) The plasmid pA31-12 (KX034083) was used as a reference to compare with two mcr-1- and blaCTX-M-55-bearing plasmids which possess the IncI2 replicon. The outer circle with red arrows signifies annotation of the reference sequence. Gaps in the circle refer to plasmid regions which are missing compared to the reference. (B) The plasmid pECJS-59-244 (KX084394.1) was used as reference to compare with three mcr-1- and blaCTX-M-14-positive strains. The outer circle with red arrows denotes annotation of the reference plasmid. The three IncHI2 strains (TC14, TC199, and TC391) exhibited sequence similarity with the reference, but alignment was not successful because of the low overall sequence homology. The number of ISApI1 repeats is not depicted. One MDR region was observable in the backbone of all IncHI2 plasmids.
This study reports the comprehensive surveillance of mcr-1 in ESBL-producing E. coli strains isolated from food products in Shenzhen, China. Our data show that mcr-1 was very common in ESBL-producing E. coli strains from various food products, with some strains carrying mcr-1, fosA3, and ESBL genes. These E. coli isolates may pose a huge threat to public health and thus warrant further investigation.
ACKNOWLEDGMENTS
This work was supported by the Chinese National Key Basic Research and Development Program (2013CB127200).
We declare no conflicts of interest.
REFERENCES
- 1.Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, Chaudhary U, Doumith M, Giske CG, Irfan S, Krishnan P, Kumar AV, Maharjan S, Mushtaq S, Noorie T, Paterson DL, Pearson A, Perry C, Pike R, Rao B, Ray U, Sarma JB, Sharma M, Sheridan E, Thirunarayan MA, Turton J, Upadhyay S, Warner M, Welfare W, Livermore DM, Woodford N. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis 10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nordmann P, Dortet L, Poirel L. 2012. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol Med 18:263–272. doi: 10.1016/j.molmed.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 4.Skov RL, Monnet DL. 2016. Plasmid-mediated colistin resistance (mcr-1 gene): three months later, the story unfolds. Euro Surveill 21(9):pii=30155. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=21403. [DOI] [PubMed] [Google Scholar]
- 5.CLSI. 2016. Performance standards for antimicrobial susceptibility testing; 26th informational supplement. CLSI document M100-S26 CLSI, Wayne, PA. [Google Scholar]
- 6.Li R, Xie M, Zhang J, Yang Z, Liu L, Liu X, Zheng Z, Chan EW, Chen S. 2017. Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J Antimicrob Chemother 72:393–401. doi: 10.1093/jac/dkw411. [DOI] [PubMed] [Google Scholar]
- 7.Dallenne C, Da Costa A, Decre D, Favier C, Arlet G. 2010. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J Antimicrob Chemother 65:490–495. doi: 10.1093/jac/dkp498. [DOI] [PubMed] [Google Scholar]
- 8.Falagas ME, Kastoris AC, Kapaskelis AM, Karageorgopoulos DE. 2010. Fosfomycin for the treatment of multidrug-resistant, including extended-spectrum beta-lactamase producing, Enterobacteriaceae infections: a systematic review. Lancet Infect Dis 10:43–50. doi: 10.1016/S1473-3099(09)70325-1. [DOI] [PubMed] [Google Scholar]
- 9.Sun J, Li XP, Yang RS, Fang LX, Huo W, Li SM, Jiang P, Liao XP, Liu YH. 2016. Complete nucleotide sequence of an IncI2 plasmid coharboring blaCTX-M-55 and mcr-1. Antimicrob Agents Chemother 60:5014–5017. doi: 10.1128/AAC.00774-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li R, Xie M, Lv J, Wai-Chi Chan E, Chen S. 2017. Complete genetic analysis of plasmids carrying mcr-1 and other resistance genes in an Escherichia coli isolate of animal origin. J Antimicrob Chemother 72:696–699. doi: 10.1093/jac/dkw411. [DOI] [PubMed] [Google Scholar]