ABSTRACT
Here, we report for the first time a waterborne outbreak of Shigella sonnei in China in 2015. Eleven multidrug-resistant (MDR) S. sonnei isolates were recovered, showing high resistance to azithromycin and third-generation cephalosporins in particular, due to an mph(A)- and blaCTX-M-14-harboring IncB/O/K/Z group transmissible plasmid of 104,285 kb in size. Our study highlights the potential prevalence of the MDR outbreak of S. sonnei in China and its further dissemination worldwide with the development of globalization.
KEYWORDS: Shigella sonnei, multidrug resistance, outbreak, waterborne
TEXT
Shigellosis is one of the most important causes of diarrhea worldwide (1). Shigellosis represents a significant public health burden in developing countries, with more than 160 million cases and 1.1 million deaths annually. Moreover, shigellosis has become one cause of disease-related death in children, particularly in children under 5 years of age (1, 2). Third-generation cephalosporins and fluoroquinolones are an effective treatment for adults. Azithromycin, a macrolide, is recommended by the American Academy of Pediatrics as the first-line empirical antimicrobial treatment for multidrug-resistant ([MDR] resistant to three or more classes of antimicrobials) Shigella spp. (3, 4) among children and as a second-line treatment among adults. Here, we report a waterborne outbreak of shigellosis caused by MDR Shigella sonnei showing particularly high resistance to azithromycin and third-generation cephalosporins.
On 9 September 2015, an outbreak of shigellosis occurred in a kindergarten class in Guangxi Province, China. We launched an epidemiological field investigation. This study was approved by the ethics committee of the Institute of Disease Control and Prevention, Academy of Military Medical Sciences. Cases were defined as students with two or more of the following symptoms of acute gastroenteritis between 30 August and 28 September 2015: diarrhea (more than 3 times per day), stool abnormalities (watery, mucus, bloody purulent, or bloody stools), nausea and/or vomiting, abdominal pain, and fever (>38°C). The first case was traced to a 5-year-old boy who presented with symptoms, including severe diarrhea (more than 10 watery stools/day), fever, tenesmus, and vomiting on 6 September. From 6 to 11 September, a total of 38 cases were identified among 285 children in the kindergarten class (attack rate, 13.33%). On 9 September, the incidence rate reached a peak (see Fig. S1 in the supplemental material). The male/female ratio was 1.53:1, and the median age was 4 years (range, 2 to 6 years) (see Table S1). Cases were observed from all eight kindergarten classes (see Fig. S2). All patients recovered, and no deaths were observed. Based on an environmental epidemiological investigation, we found that the canteen and water supply systems were in poor sanitary condition. After disinfection and other control measures, no new cases occurred.
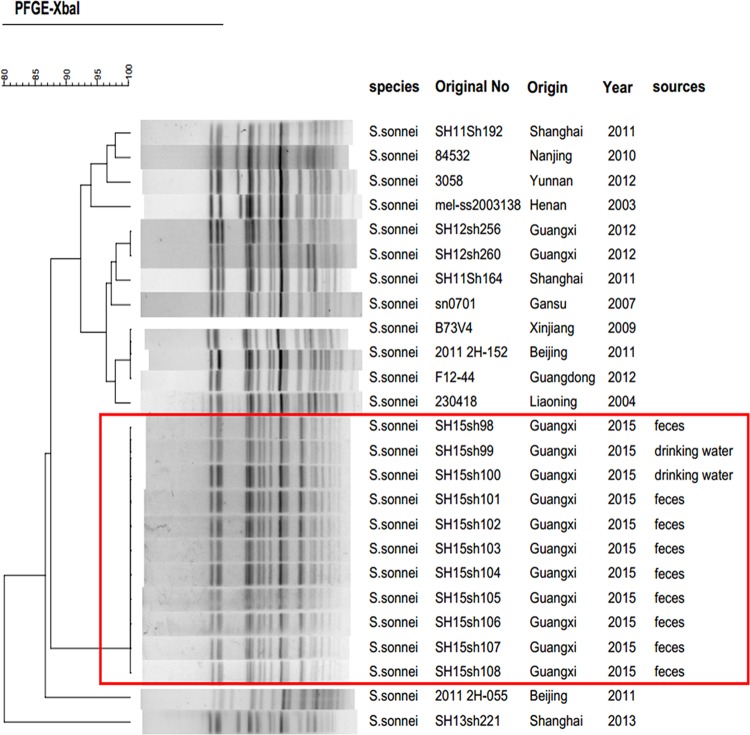
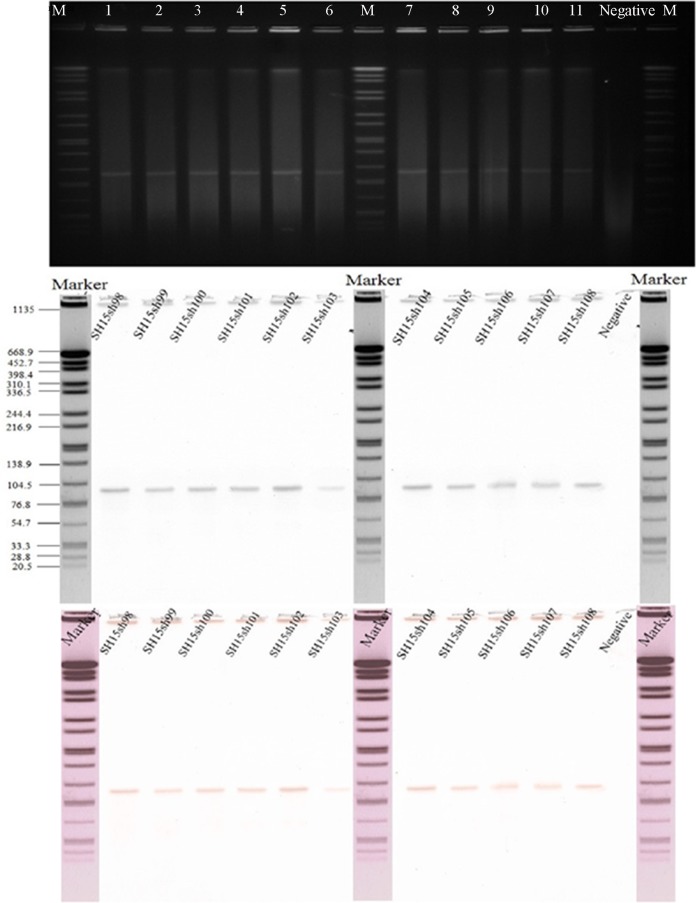
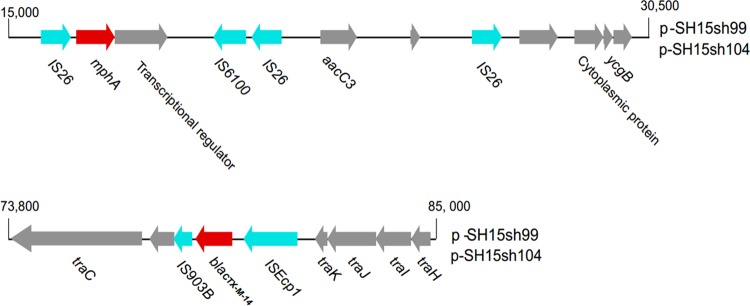
During the outbreak, we collected 35 fresh samples for laboratory identification of enteric pathogens, including 19 rectal swabs and 2 vomitus samples from 19 children with diarrhea, six drinking water samples, and eight suspicious residual foods from the kindergarten. All food and water samples were collected by sterile sampling according to the national food safety standards, such as GB 4789.1-2016, GB 4789.2-2016, GB 4789.3-2016, GB 4789.4-2016, GB 4789.6-2016, and GB 4789.10-2016, and standards for drinking water quality, GB 5749-2006. Suspected isolates recovered from the samples were identified using a commercial biochemical test kit (API 20E system; bioMérieux Vitek, France) according to the manufacturer's recommendations. The serotype of Shigella isolates was then determined using a serotyping kit (Denka Seiken, Tokyo, Japan) according to standard methods. We isolated 11 S. sonnei strains from the 35 samples, including nine in rectal swabs and two in drinking water. Antimicrobial susceptibility testing was performed by broth microdilution in 96-well microtiter plates (Sensititre, Trek Diagnostic Systems; Thermo Fisher Scientific, Inc., West Sussex, UK) according to the methods of the Clinical and Laboratory Standards Institute (5). The MICs of azithromycin and nalidixic acid were tested with concentrations of 4 to 256 μg/ml and 4 to 32 μg/ml, respectively. All 11 outbreak isolates displayed similar MDR profiles (Table 1). Moreover, high levels of resistance to azithromycin (MIC, ≥256 μg/ml) and nalidixic acid (MIC, >32 μg/ml) were observed. We used PCR to determine the presence of β-lactamase genes (6–8) and plasmid-mediated quinolone resistance (PMQR) determinants, such as qnrA, qnrB, qnrD, qnrS, and aac(6′)-Ib-cr (3, 9–11). The mph(A) gene (primers, mphA-F [5′-CGAAGACCTCCGAGTCCTGC-3′] and mphA-R [5′-CCGCCGATACCTCCCAACT-3′]) encoding macrolide 2-phosphotransferase (12) was also detected. Interestingly, all strains were negative for PMQR determinants but carried blaCTX-M-14 and mph(A). An analysis of point mutations in quinolone resistance-determining regions (QRDRs) of gyrA and parC (13, 14) showed that all isolates contained the gyrA(Ser83Leu) point mutation. Pulsed-field gel electrophoresis (PFGE) (15) showed that the outbreak isolates were genetically indistinguishable and were subtyped into one PFGE type, different from that of isolates from other regions of China (Fig. 1). The horizontal-transfer capability of blaCTX-M-14 and mph(A) was assessed by broth mating as reported previously (16) (see Table S2). Muller-Hinton agar (BD Biosciences, San Jose, CA, USA) plates containing 200 mg/liter sodium azide and 10 mg/liter cefotaxime were used as a selective medium for Escherichia coli J53 transconjugants. Putative transconjugants were confirmed by antimicrobial susceptibility testing and PCR detection of blaCTX-M-14 and mph(A) as described above. Plasmid conjugation transfer revealed that blaCTX-M-14 and mph(A) were successfully transferred into Escherichia coli J53. This result suggested that blaCTX-M-14 and mph(A) may be located on one plasmid. Plasmid profiling and Southern blot analysis were performed as reported previously (16) but with differences in switching time from 2.16 s to 29.27 s, as well as with using specific blaCTX-M-14 (probes, blaCTX-M-14-F [5′-AGCCTGCCGATCTGGTTAA-3′] and blaCTX-M-14-R [5′-CCGGTCGTATTGCCTTTG-3′]) and mph(A) (probes, mphA-F [5′-CGAAGACCTCCGAGTCCTGC-3′] and mphA-R [5′-CCGCCGATACCTCCCAACT-3′]) digoxigenin-labeled probes (Roche). It showed that each of the isolates harbored only one plasmid, with a size of approximately 90 to 100 kb. Notably, the plasmids were positive for both blaCTX-M-14 and mph(A) (Fig. 2), which is the primary cause of multidrug resistance in these S. sonnei strains. To identify the genes responsible for the multidrug resistance, the DNA of plasmids (pSH15sh99 and pSH15sh104) was extracted and subjected to high-throughput sequencing using Illumina MiSeq (San Diego, CA). The insertion sequences (ISs) were searched by using ISfinder (https://www-is.biotoul.fr/search.php), and the replicons genotypes were searched using PlasmidFinder (https://cge.cbs.dtu.dk/services/PlasmidFinder/). A structure analysis showed that the two plasmids were identical and belonged to incompatibility group IncB/O/K/Z with a size of 104,285 bp (Table 2). mph(A) and blaCTX-M-14 were located at 16,000 bp and 80,000 bp positions of the plasmid, respectively. In addition, IS26 and IS6100-IS26 transposon-like structures flanked mph(A) upstream and downstream. blaCTX-M-14 was found together with an upstream ISEcp1 and a downstream IS903B (Fig. 3). aacC3 was also found on the plasmid.
TABLE 1.
MIC results of 23 antibiotics for 11 outbreak S. sonnei strains
| Isolation no. | MICs (μg/ml)a |
||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CAZ | CRO | IMP | NIT | PIP | TET | FEP | CFP | CFZ | FOX | TOB | LVX | GEN | TIC | TIM | ATM | AMP | CHL | SXT | NOR | AMK | AZM | NAL | |
| SH15sh098 | <1 | >32 | <4 | <32 | >64 | >8 | <8 | >32 | >16 | <8 | >8 | <2 | >8 | >64 | 64 | 2 | >16 | <8 | >2 | <4 | <16 | >256 | >32 |
| SH15sh099 | <1 | >32 | <4 | <32 | >64 | >8 | <8 | >32 | >16 | <8 | >8 | <2 | >8 | >64 | 64 | 2 | >16 | <8 | >2 | <4 | <16 | >256 | >32 |
| SH15sh100 | <1 | >32 | <4 | <32 | >64 | >8 | <8 | >32 | >16 | <8 | >8 | <2 | >8 | >64 | <16 | 2 | >16 | <8 | >2 | <4 | <16 | >256 | >32 |
| SH15sh101 | <1 | >32 | <4 | <32 | >64 | >8 | <8 | >32 | >16 | <8 | >8 | <2 | >8 | >64 | 64 | 2 | >16 | <8 | >2 | <4 | <16 | >256 | >32 |
| SH15sh102 | <1 | >32 | <4 | <32 | >64 | >8 | <8 | >32 | >16 | <8 | >8 | <2 | >8 | >64 | <16 | 2 | >16 | <8 | >2 | <4 | <16 | >256 | >32 |
| SH15sh103 | <1 | >32 | <4 | <32 | >64 | >8 | <8 | >32 | >16 | <8 | >8 | <2 | >8 | >64 | 64 | 2 | >16 | <8 | >2 | <4 | <16 | >256 | >32 |
| SH15sh104 | <1 | >32 | <4 | <32 | >64 | >8 | <8 | >32 | >16 | <8 | >8 | <2 | >8 | >64 | <16 | 2 | >16 | <8 | >2 | <4 | <16 | >256 | >32 |
| SH15sh105 | <1 | >32 | <4 | <32 | >64 | >8 | <8 | >32 | >16 | <8 | >8 | <2 | >8 | >64 | 64 | 8 | >16 | <8 | >2 | <4 | <16 | >256 | >32 |
| SH15sh106 | <1 | >32 | <4 | <32 | >64 | >8 | <8 | >32 | >16 | <8 | >8 | <2 | >8 | >64 | <16 | 2 | >16 | <8 | >2 | <4 | <16 | >256 | >32 |
| SH15sh107 | <1 | >32 | <4 | <32 | >64 | >8 | <8 | >32 | >16 | <8 | >8 | <2 | >8 | >64 | <16 | 2 | >16 | <8 | >2 | <4 | <16 | >256 | >32 |
| SH15sh108 | <1 | >32 | <4 | <32 | >64 | >8 | <8 | >32 | >16 | <8 | >8 | <2 | >8 | >64 | 64 | 2 | >16 | <8 | >2 | <4 | <16 | >256 | >32 |
Cephalosporins: CAZ, ceftazidime; CRO, ceftriaxone; FEP, cefepime; CFP, cefoperazone; CFZ, cefazolin; and FOX, cefoxitin. Fluoroquinolones: LVX, levofloxacin; NOR, norfloxacin; and NAL, nalidixic acid. Penicillins: PIP, piperacillin; TIC, ticarcillin; AMP, ampicillin; TIM, ticarcillin-clavulanic acid; TOB, tobramycin; GEN, gentamicin; and AMK, amikacin. Phenicol: CHL, chloramphenicol. Sulfonamide: SXT, trimethoprim-sulfamethoxazole. Thienamycin: IPM, imipenem. Nitrofuran: NIT, nitrofurantoin. Tetracycline: TET, tetracycline. β-Lactam: ATM, aztreonam. Macrolide: AZM, azithromycin.
FIG 1.
Pulsed-field gel electrophoresis (PFGE) patterns created by digestion with the enzyme XbaI. Dendrogram showing the level of genetic relatedness based on the unweighted pair-group method using average linkages and the Dice coefficient for S. sonnei strains. The species, original number, origin, year, and sources of the stains are shown.
FIG 2.
Identification of mph(A)- and blaCTX-M-14-positive plasmids. (A) S1 nuclease plasmid pulsed-field electrophoresis profiles. (B) Southern blot hybridization for mph(A). Lanes: Marker, molecular size marker (strain H9812 digested with XbaI); lane 1 to lane 12 are the S. sonnei strains SH15sh98, SH15sh99, SH15sh100, SH15sh101, SH15sh102, SH15sh103, SH15sh104, SH15sh105, SH15sh106, SH15sh107, SH15sh108, and a negative-control strain, respectively. The molecular sizes on the left are in kilobases. (C) Southern blot hybridization for blaCTX-M-14.
TABLE 2.
Sequencing characteristics of the two plasmids of outbreak S. sonnei strains
| Name | Size (bp) | No. of total reads | Inca | No. of copiesb | IS types |
|---|---|---|---|---|---|
| pSH15sh099 | 104,285 | 313,326 | IncB/O/K/Z | 878 | ISEcp1, IS903B, IS6100, IS26 |
| pSH15sh104 | 104,285 | 366,962 | IncB/O/K/Z | 965 | ISEcp1, IS903B, IS6100, IS26 |
Inc, plasmid incompatibility group, as determined by PlasmidFinder version 1.2 (10).
Average copy number per cell, normalized to the chromosomal read coverage.
FIG 3.
The mosaic region of pSH15sh99 and pSH15sh104 with the surroundings of mph(A) and blaCTX-M-14. The resistance genes are denoted by arrows and colored based on gene function classification. mph(A) and blaCTX-M-14 are colored in red, IS elements are colored in light blue, and others are in gray.
Based on the above epidemiological and laboratory investigations, we concluded that this diarrhea outbreak was probably attributed to contamination of the water supply by MDR S. sonnei with high resistance to azithromycin and third-generation cephalosporins. Because of the low infectious dose and environmental persistence, S. sonnei can be spread by water (17–20), food (21, 22), and direct person-to-person transmission (23, 24), which can trigger sporadic cases or family outbreaks and may cause more extensive secondary transmission from a common source (25, 26). Therefore, S. sonnei outbreaks in childcare centers and schools can typically spread to the community or to other cities.
Concerns regarding S. sonnei infections with apparent high-level resistance to azithromycin and third-generation cephalosporins prompted this investigation. Previous outbreaks of S. sonnei with resistance to third-generation cephalosporins (27–30) or azithromycin (31–33) alone occurred elsewhere. However, to the best of our knowledge, this is the first report on the outbreak of S. sonnei with resistance to both third-generation cephalosporins and azithromycin, attributed to an mph(A)- and blaCTX-M-14-carrying IncB/O/K/Z group transmissible plasmid. In addition, the gyrA(Ser83Leu) single point mutation usually results in a high resistance to the narrow spectrum quinolone nalidixic acid and to reduced susceptibility to fluoroquinolones (13, 34). However, fluoroquinolone resistance is always due to multiple point mutations (such as gryA[Asp87Asn], gryA[Asp87Gly], gryA[His211Tyr], and parC[Ser80Ile]mutations) or to PMQR.
The mph(A) gene has been reported to be located on various plasmids, ranging from 10 to 208 kb (32, 35). Here, we found that the plasmid containing various mobile IS elements that carry multiple resistance determinants was 104,285 kb in size and could be successfully transferred into E. coli J53. The potentially widespread dissemination of pathogens with plasmid-mediated multidrug resistance, which is expedited by increasing globalization, may represent a major threat to global health.
In conclusion, we described a waterborne outbreak caused by S. sonnei with resistance to both azithromycin and third-generation cephalosporin in China, which was attributable to the presence of an IncB/O/K/Z plasmid coharboring mph(A) and blaCTX-M-14. The emergence and spread of MDR S. sonnei may necessitate improved strategies for the prevention and control of shigellosis. Moreover, our findings emphasize the importance of continuous surveillance of the prevalence of Shigella species and of changes in antibiotic resistance patterns on national and international scales. Whole-genome sequencing data are urgently needed to validate the detailed mechanisms of bacterial resistance, the true relatedness of strains, other resistance genes, and virulence factors.
Accession number(s).
The annotated plasmid sequences have been deposited in the GenBank database under accession numbers KY471628 (pSH15sh99) and KY471629 (pSH15sh104).
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the National Key Program for Infectious Diseases of China (no. 2012ZX10004215 and AWS15J006) and the National Nature Science Foundation of China (no. 81373053, 81473023, and 81371854).
We have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00308-17.
REFERENCES
- 1.Kotloff KL, Winickoff JP, Ivanoff B, Clemens JD, Swerdlow DL, Sansonetti PJ, Adak GK, Levine MM. 1999. Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull World Health Organ 77:651–666. [PMC free article] [PubMed] [Google Scholar]
- 2.Mathers CD, Boerma T, Ma Fat D. 2009. Global and regional causes of death. Br Med Bull 92:7–32. doi: 10.1093/bmb/ldp028. [DOI] [PubMed] [Google Scholar]
- 3.Cui X, Yang C, Wang J, Liang B, Yi S, Li H, Liu H, Li P, Wu Z, Xie J, Jia L, Hao R, Wang L, Hua Y, Qiu S, Song H. 2015. Antimicrobial resistance of Shigella flexneri serotype 1b isolates in China. PLoS One 10:e0129009. doi: 10.1371/journal.pone.0129009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui X, Wang J, Yang C, Liang B, Ma Q, Yi S, Li H, Liu H, Li P, Wu Z, Xie J, Jia L, Hao R, Wang L, Hua Y, Qiu S, Song H. 2015. Prevalence and antimicrobial resistance of Shigella flexneri serotype 2 variant in China. Front Microbiol 6:435. doi: 10.3389/fmicb.2015.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.CLSI. 2013. Performance standards for antimicrobial susceptibility testing, vol 33; 23rd informational supplement CLSI, Wayne, PA. [Google Scholar]
- 6.Ahmed AM, Furuta K, Shimomura K, Kasama Y, Shimamoto T. 2006. Genetic characterization of multidrug resistance in Shigella spp. from Japan. J Med Microbiol 55:1685–1691. doi: 10.1099/jmm.0.46725-0. [DOI] [PubMed] [Google Scholar]
- 7.Galani I, Souli M, Mitchell N, Chryssouli Z, Giamarellou H. 2010. Presence of plasmid-mediated quinolone resistance in Klebsiella pneumoniae and Escherichia coli isolates possessing blaVIM-1 in Greece. Int J Antimicrob Agents 36:252–254. doi: 10.1016/j.ijantimicag.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 8.Matar G, Jaafar R, Sabra A, Hart C, Corkill J, Dbaibo G, Araj G. 2007. First detection and sequence analysis of the blaCTX-M-15 gene in Lebanese isolates of extended-spectrum-β-lactamase-producing Shigella sonnei. Ann Trop Med Parasitol 101:511–517. doi: 10.1179/136485907X193860. [DOI] [PubMed] [Google Scholar]
- 9.Pan JC, Ye R, Meng DM, Zhang W, Wang HQ, Liu KZ. 2006. Molecular characteristics of class 1 and class 2 integrons and their relationships to antibiotic resistance in clinical isolates of Shigella sonnei and Shigella flexneri. J Antimicrob Chemother 58:288–296. doi: 10.1093/jac/dkl228. [DOI] [PubMed] [Google Scholar]
- 10.Tariq A, Haque A, Ali A, Bashir S, Habeeb MA, Salman M, Sarwar Y. 2012. Molecular profiling of antimicrobial resistance and integron association of multidrug-resistant clinical isolates of Shigella species from Faisalabad, Pakistan. Can J Microbiol 58:1047–1054. doi: 10.1139/w2012-085. [DOI] [PubMed] [Google Scholar]
- 11.Pu XY, Pan JC, Wang HQ, Zhang W, Huang ZC, Gu YM. 2009. Characterization of fluoroquinolone-resistant Shigella flexneri in Hangzhou area of China. J Antimicrob Chemother 63:917–920. doi: 10.1093/jac/dkp087. [DOI] [PubMed] [Google Scholar]
- 12.Ojo KK, Ulep C, Van Kirk N, Luis H, Bernardo M, Leitao J, Roberts MC. 2004. The mef(A) gene predominates among seven macrolide resistance genes identified in Gram-negative strains representing 13 genera, isolated from healthy Portuguese children. Antimicrob Agents Chemother 48:3451–3456. doi: 10.1128/AAC.48.9.3451-3456.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang H, Duan G, Zhu J, Zhang W, Xi Y, Fan Q. 2013. Prevalence and characterisation of plasmid-mediated quinolone resistance and mutations in the gyrase and topoisomerase IV genes among Shigella isolates from Henan, China, between 2001 and 2008. Int J Antimicrob Agents 42:173–177. doi: 10.1016/j.ijantimicag.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 14.Hu LF, Li JB, Ye Y, Li X. 2007. Mutations in the GyrA subunit of DNA gyrase and the ParC subunit of topoisomerase IV in clinical strains of fluoroquinolone-resistant Shigella in Anhui, China. J Microbiol 45:168–170. [PubMed] [Google Scholar]
- 15.Ribot EM, Fair M, Gautom R, Cameron D, Hunter S, Swaminathan B, Barrett TJ. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157: H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis 3:59–67. doi: 10.1089/fpd.2006.3.59. [DOI] [PubMed] [Google Scholar]
- 16.Zou D, Huang Y, Zhao X, Liu W, Dong D, Li H, Wang X, Huang S, Wei X, Yan X, Yang Z, Tong Y, Huang L, Yuan J. 2015. A novel New Delhi metallo-beta-lactamase variant, NDM-14, isolated in a Chinese hospital possesses increased enzymatic activity against carbapenems. Antimicrob Agents Chemother 59:2450–2453. doi: 10.1128/AAC.05168-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sezen F, Aval E, Agkurt T, Yilmaz S, Temel F, Gulesen R, Korukluoglu G, Sucakli MB, Torunoglu MA, Zhu BP. 2015. A large multi-pathogen gastroenteritis outbreak caused by drinking contaminated water from antique neighbourhood fountains, Erzurum city, Turkey, December 2012. Epidemiol Infect 143:704–710. doi: 10.1017/S0950268814001459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arias C, Sala MR, Dominguez A, Bartolome R, Benavente A, Veciana P, Pedrol A, Hoyo G. 2006. Waterborne epidemic outbreak of Shigella sonnei gastroenteritis in Santa Maria de Palautordera, Catalonia, Spain. Epidemiol Infect 134:598–604. doi: 10.1017/S0950268805005121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fleming CA, Caron D, Gunn JE, Horine MS, Matyas BT, Barry MA. 2000. An outbreak of Shigella sonnei associated with a recreational spray fountain. Am J Public Health 90:1641–1642. doi: 10.2105/AJPH.90.10.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Centers for Disease Control and Prevention. 1996. Shigella sonnei outbreak associated with contaminated drinking water–Island Park, Idaho, August 1995. MMWR Morb Mortal Wkly Rep 45:229–231. [PubMed] [Google Scholar]
- 21.Kuo H-W, Kasper S, Jelovcan S, Höger G, Lederer I, König C, Pridnig G, Luckner-Hornischer A, Allerberger F, Schmid D. 2009. A food-borne outbreak of Shigella sonnei gastroenteritis, Austria, 2008. Wien Klin Wochenschr 121:157–163. doi: 10.1007/s00508-008-1141-7. [DOI] [PubMed] [Google Scholar]
- 22.Gaynor K, Park SY, Kanenaka R, Colindres R, Mintz E, Ram PK, Kitsutani P, Nakata M, Wedel S, Boxrud D, Jennings D, Yoshida H, Tosaka N, He H, Ching-Lee M, Effler PV. 2009. International foodborne outbreak of Shigella sonnei infection in airline passengers. Epidemiol Infect 137:335–341. doi: 10.1017/S0950268807000064. [DOI] [PubMed] [Google Scholar]
- 23.Bennett JE, Dolin R, Blaser MJ. 2014. Principles and practice of infectious diseases, 8th ed, vol 1 Elsevier Health Sciences, Philadelphia, PA. [Google Scholar]
- 24.Qu F, Bao C, Chen S, Cui E, Guo T, Wang H, Zhang J, Wang H, Tang YW, Mao Y. 2012. Genotypes and antimicrobial profiles of Shigella sonnei isolates from diarrheal patients circulating in Beijing between 2002 and 2007. Diagn Microbiol Infect Dis 74:166–170. doi: 10.1016/j.diagmicrobio.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morera MA, Espejo E, Coll P, Simo M, Uriz MS, Llovet T, Rodriguez M, Martinez A, March F, Bella F. 1995. Epidemic outbreak of shigellosis following water intake. Enferm Infecc Microbiol Clin 13:160–165. (In Spanish.) [PubMed] [Google Scholar]
- 26.Masters BR. 2016. Mandell, Douglas, and Bennett's Principles and practice of infectious diseases, eighth edition (2015) eds: John E. Bennett, Raphael Dolin, Martin J. Blaser. ISBN: 13-978-1-4557-4801-3, Elsevier Saunders. Graefes Arch Clin Exp Ophthalmol 254:2285–2287. doi: 10.1007/s00417-015-2950-1. [DOI] [Google Scholar]
- 27.Centers for Disease Control and Prevention. 2006. Outbreaks of multidrug-resistant Shigella sonnei gastroenteritis associated with day care centers–Kansas, Kentucky, and Missouri, 2005. Morb Mortal Wkly Rep 55:1068–1071. [PubMed] [Google Scholar]
- 28.Kim JS, Kim JJ, Kim SJ, Jeon SE, Seo KY, Choi JK, Kim NO, Hong S, Chung GT, Yoo CK, Kim YT, Cheun HI, Bae GR, Yeo YH, Ha GJ, Choi MS, Kang SJ, Kim J. 2015. Outbreak of ciprofloxacin-resistant Shigella sonnei associated with travel to Vietnam, Republic of Korea. Emerg Infect Dis 21:1247–1250. doi: 10.3201/eid2107.150363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guzman-Herrador B, Vold L, Comelli H, MacDonald E, Heier BT, Wester AL, Stavnes TL, Jensvoll L, Lindegard Aanstad A, Severinsen G, Aasgaard Grini J, Werner Johansen O, Cudjoe K, Nygard K. 2011. Outbreak of Shigella sonnei infection in Norway linked to consumption of fresh basil, October 2011. Euro Surveill 16:pii=20007 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20007. [PubMed] [Google Scholar]
- 30.Huang IF, Chiu CH, Wang MH, Wu CY, Hsieh KS, Chiou CC. 2005. Outbreak of dysentery associated with ceftriaxone-resistant Shigella sonnei: first report of plasmid-mediated CMY-2-type AmpC beta-lactamase resistance in S. sonnei. J Clin Microbiol 43:2608–2612. doi: 10.1128/JCM.43.6.2608-2612.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heiman KE, Grass JE, Sjolund-Karlsson M, Bowen A. 2014. Shigellosis with decreased susceptibility to azithromycin. Pediatr Infect Dis J 33:1204–1205. doi: 10.1097/INF.0000000000000397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boumghar-Bourtchai L, Mariani-Kurkdjian P, Bingen E, Filliol I, Dhalluin A, Ifrane SA, Weill FX, Leclercq R. 2008. Macrolide-resistant Shigella sonnei. Emerg Infect Dis 14:1297–1299. doi: 10.3201/eid1408.080147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bowen A, Eikmeier D, Talley P, Siston A, Smith S, Hurd J, Smith K, Leano F, Bicknese A, Norton JC, Campbell D, Centers for Disease Control and Prevention (CDC). 2015. Notes from the field: outbreaks of Shigella sonnei infection with decreased susceptibility to azithromycin among men who have sex with men - Chicago and Metropolitan Minneapolis-St. Paul, 2014. MMWR Morb Mortal Wkly Rep 64:597–598. [PMC free article] [PubMed] [Google Scholar]
- 34.Ding J, Ma Y, Gong Z, Chen Y. 1999. A study on the mechanism of the resistance of Shigellae to fluoroquinolones. Zhonghua Nei Ke Za Zhi 38:550–553. (In Chinese.) [PubMed] [Google Scholar]
- 35.Howie RL, Folster JP, Bowen A, Barzilay EJ, Whichard JM. 2010. Reduced azithromycin susceptibility in Shigella sonnei, United States. Microb Drug Resist 16:245–248. doi: 10.1089/mdr.2010.0028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.