ABSTRACT
Type 3 secretion systems (T3SSs) are major virulence factors in Gram-negative bacteria. Pseudomonas aeruginosa expresses two T3SSs, namely, an injectisome (iT3SS) translocating effector proteins in the host cell cytosol and a flagellum (fT3SS) ensuring bacterial motility. Inhibiting these systems is an appealing therapeutic strategy for acute infections. This study examines the protective effects of the salicylidene acylhydrazide INP0341 and of the hydroxyquinoline INP1750 (previously described as T3SS inhibitors in other species) toward cytotoxic effects of P. aeruginosa in vitro. Both compounds reduced cell necrosis and inflammasome activation induced by reference strains or clinical isolates expressing T3SS toxins or only the translocation apparatus. INP0341 inhibited iT3SS transcriptional activation, including in strains with constitutive iT3SS expression, and reduced the total expression of toxins, suggesting it targets iT3SS gene transcription. INP1750 inhibited toxin secretion and flagellar motility and impaired the activity of the YscN ATPase from Yersinia pseudotuberculosis (homologous to the ATPase present in the basal body of P. aeruginosa iT3SS and fT3SS), suggesting that it rather targets a T3SS core constituent with high homology among iT3SS and fT3SS. This mode of action is similar to that previously described for INP1855, another hydroxyquinoline, against P. aeruginosa. Thus, although acting by different mechanisms, INP0341 and INP1750 appear as useful inhibitors of the virulence of P. aeruginosa. Hydroxyquinolines may have a broader spectrum of activity by the fact they act upon two virulence factors (iT3SS and fT3SS).
KEYWORDS: NLRC4 inflammasome, Pseudomonas aeruginosa, flagella, inhibitors, type three secretion system, virulence
INTRODUCTION
Increasing resistance of clinical isolates of Pseudomonas aeruginosa to current recommended antipseudomonal antibiotics is associated with poor clinical outcome and increased mortality (1–3), prompting the search for innovative therapeutic strategies. However, very few novel antipseudomonal agents are currently under development (4). This is partly due to difficulties in identifying and developing new molecules active on still unexploited targets, but it is also due to the low permeability of the outer membrane of P. aeruginosa (combining active efflux or poor porin permeability), which prevents many active molecules from reaching targets located in the bacterial cytosol or in the periplasmic space (5). In this context, innovative therapeutic strategies are needed (6), among which those aiming at reducing bacterial virulence are particularly appealing, because they reduce morbidity and improve antibacterial immune responses while preserving commensal flora (7). These strategies also remain compatible with antibiotic usage and are expected to have a lower risk of selecting resistance, since they only disarm bacteria, leaving the task of killing them to the immune system (8, 9).
Among other virulence factors, P. aeruginosa expresses two type 3 secretion systems (T3SSs), namely, an injectisome (referred to as iT3SS) translocating effector proteins directly in the host cell cytosol and a flagellum (designated fT3SS) ensuring bacterial motility (10). iT3SS expression is associated with poor clinical outcomes and high morbidity in acute infections (3, 11, 12). The system consists in a syringe-like machine injecting bacterial effector proteins from the bacterial cytosol directly into the host cell by the concerted action of a secretion apparatus (comprising among others the protein PscF forming the needle filament) and a translocon (proteins PopB, PopD, and PcrV; see Fig. S1 in the supplemental material for a schematic view). Among translocated proteins, effectors encoded by the iT3SS operon include ExoU (a phospholipase A2 causing rapid cell death by altering membrane integrity), ExoS and ExoT (causing disruption of actin filaments and subsequent prevention of phagocytosis, as well as cytotoxicity), and ExoY (contributing to actin cytoskeleton disorganization) (13). Importantly, other proteins can be translocated through the iT3SS, such as FliC (monomeric subunit of flagellum) (14, 15). FliC can cause host cell damage independently of effectors, notably by activating the intracellular cytosolic sensor NLRC4 (NLR family, CARD domain containing 4) inflammasome resulting in pyroptosis (16) and in liberation of active forms of interleukin-1β (IL-1β) and IL-18 (see reference 17 for a review). Animal models of acute pulmonary infections showed that the activation of this cascade contributes to increased lung injury and impaired P. aeruginosa clearance by reducing IL-17 signaling and antimicrobial peptide production (18, 19), the deleterious consequences of which for the host remain controversial (16). A third type of protein excreted by iT3SS is ExsE, a negative regulator of iT3SS transcription (20). It is secreted out of the bacteria under inducing conditions and liberates the transcription activator ExsA via a complex regulatory cascade, which then activates iT3SS gene transcription (21; see also reference 22 for a schematic view of this process). In vitro, iT3SS transcription is activated by contact with eukaryotic cells, low-Ca2+ medium, or the presence of serum (23–26).
fT3SS is also considered to be a virulence factor that is required for bacterial motility but also for adhesion to the surfaces of host cells, the dispersal of biofilms, or initiation of the inflammatory response (27). It consists of a basal body anchored in the membranes and comprises an export apparatus delivering flagellin subunits into a channel, from which they diffuse and assemble to form the flagellum. iT3SS and fT3SS share a similar general structure. Their assembly is dependent upon a T3SS located in the inner membrane and occurs according to a similar process. Proteins directly involved in the secretion activity of iT3SS and the export activity of fT3SS share a high degree of similarity (28).
On these bases, inhibiting T3SS appears as an attractive therapeutic strategy (29, 30). At this time, two main types of T3SS inhibitors have been investigated against P. aeruginosa (see reference 22 for a review). The first ones are therapeutic antibodies blocking PcrV protein or hybrid antibodies targeting both the PcrV protein and the Psl exopolysaccharide simultaneously. The second type of inhibitors consists of small molecules identified by in vitro screening of libraries of natural or synthetic compounds. However, biological evaluation of the pharmacological effect of these compounds is often limited to the demonstration of a reduction in cytotoxicity, while their effects on inflammasome activation, a key element in the host response to infection, have not been explored. Among them, two chemical classes have been identified as inhibitors of iT3SS transcription using screening-based approaches with Yersinia pseudotuberculosis, namely, the salicylidene acylhydrazides and the hydroxyquinolines. They have also proven active against other Gram-negative bacterial species such as Chlamydia, Shigella, and Salmonella spp. (31–34). Based on the high homology among iT3SS from Gram-negative bacteria, we undertook to investigate their activity against P. aeruginosa. In a previous report (35), we showed that the hydroxyquinoline INP1855 reduces the virulence of P. aeruginosa in vitro and in vivo (acute model of pulmonary infection) by inhibiting both the iT3SS and the fT3SS. This impairs the secretion of effectors and the flagellar motility, as well as the activity of a purified ATPase (homologous to that present in the basal core of these systems), protecting eukaryotic cells from necrosis and inflammasome activation and restoring bacterial phagocytosis.
In the present work, we selected a representative compound in each of these two chemical classes in order to compare their antipseudomonal activity. Among salicylidene acylhydrazides, we chose INP0341 (34, 36), which proved more potent against P. aeruginosa than other derivatives in the family (INP0010 and INP0406 [33, 37]) during a preliminary screening. Among hydroxyquinolines, we selected INP1750 (32), which differs from the previously studied INP1855 by the absence of fluorine (frequently used for pharmacomodulation) on the phenylpiperazine substituent of the hydroxyquinoline. In brief, we show that both INP0341 and INP1750 (Fig. 1) protect eukaryotic cells against cytotoxicity mediated by toxins and by inflammasome activation. These protective effects were observed against laboratory strains and clinical isolates. Interestingly, in spite of the similarity of their protective effects on the host cells, we show that they act by different mechanisms on bacteria.
FIG 1.
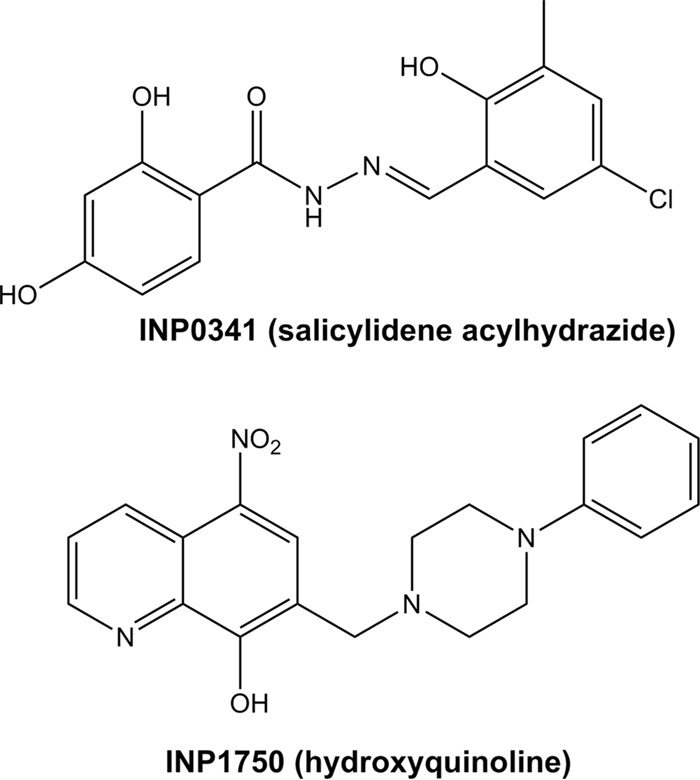
Chemical structure of the inhibitors used in this study.
RESULTS
Evaluation of INP0341 and INP1750 toxicity for bacteria and eukaryotic cells.
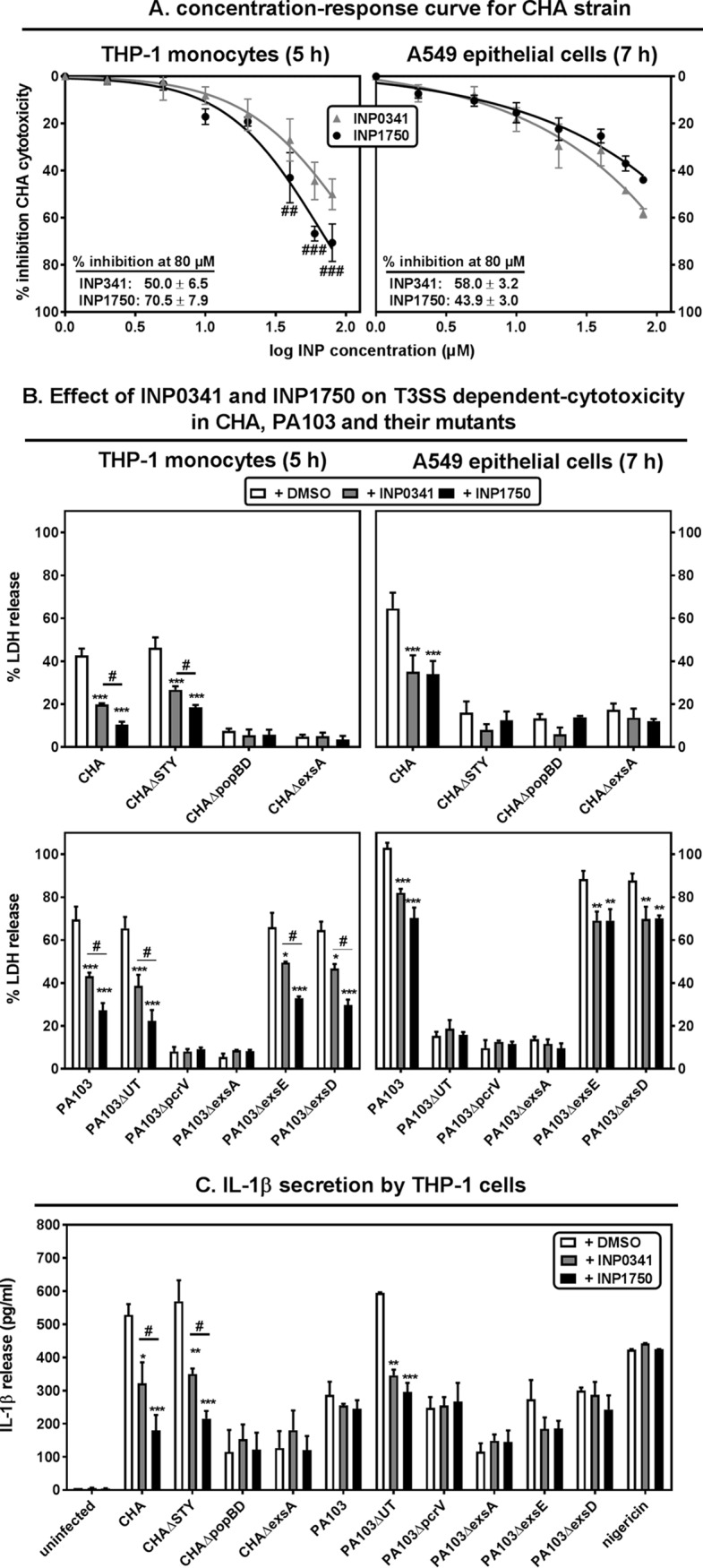
In preliminary experiments, we checked whether INPs exerted nonspecific toxic effects on bacteria and eukaryotic cells exposed to a broad range of INP concentrations (see Fig. S2 in the supplemental material). Toxicity toward bacteria was ruled out by demonstrating that INPs did not modify the growth rate of the clinical strain CHA at concentrations up to 250 μM. Toxicity toward eukaryotic cells was determined by measuring the release of the cytosolic enzyme lactate dehydrogenase in the culture medium of THP-1 cells and A549 lung epithelial cells incubated for 5 and 7 h, respectively, with increasing inhibitor concentrations. These incubation times were selected based on preliminary experiments showing that the cytotoxic effects of CHA were dependent on the iT3SS under these conditions (35). No significant release of lactate dehydrogenase was observed from both cell types for concentrations of ≤100 μM (INP0341) or ≤80 μM (INP1750). Further experiments were therefore performed with concentrations of ≤80 μM.
Influence of INP0341 and INP1750 on iT3SS-mediated cytotoxicity and bacterial internalization.
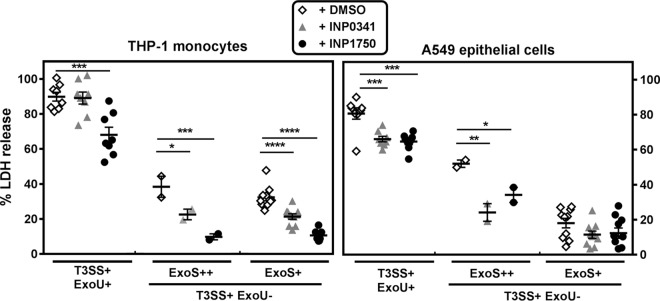
We then studied the effect of INP0341 and INP1750 on bacterial interactions with eukaryotic cells, focusing first on iT3SS-induced cytotoxicity. In a first set of experiments, the effect of INPs on CHA-induced cytotoxicity was evaluated after 5 h (THP-1 monocytes) or 7 h (A549 epithelial cells) of incubation with 10 bacteria/host cell and increasing, noncytotoxic concentrations of INPs (Fig. 2A). The two inhibitors decreased the cytotoxicity of CHA in a concentration-dependent manner in both cell types. INP0341 was equipotent toward both cell lines. INP1750 was more potent toward THP-1 cells than A549 cells at concentrations of ≥40 μM; it was also significantly more active than INP0341 over the same concentration range in THP-1 cells. The study was then extended to compare the effect of INP0341 and INP1750 at 80 μM on the cytotoxic effects of strains CHA, PA103, and selected mutants thereof (Fig. 2B). As previously described (35), CHA, PA103, and their mutants constitutively expressing the iT3SS (PA103ΔexsD and PA103ΔexsE) were toxic for both cell lines, with PA103 causing a higher lactate dehydrogenase (LDH) release than CHA. Mutants with no expression of toxins (CHAΔSTY and PA103ΔUT) were selectively toxic for THP-1 cells, while mutants with no expression of the translocation apparatus (CHAΔpopBD and PA103ΔpcrV) or of the iT3SS global regulator (CHAΔexsA and PA103ΔexsA) were not cytotoxic, demonstrating that the expression of iT3SS is responsible for the cytotoxicity observed in our experimental conditions. INP0341 and INP1750 impaired LDH release induced by all cytotoxic strains in both cell lines, with INP1750 being slightly more active than INP0341 in THP-1 cells.
FIG 2.
Influence of INP0341 and INP1750 on iT3SS-mediated effects in phagocytic cells and epithelial cells. (A) Inhibition of CHA cytotoxicity by INP0341 and INP1750. Cell viability was assessed by measuring the release of LDH in the culture medium after 5 h (THP-1) or 7 h (A549) of incubation with CHA (10 bacteria/cell) in the presence of increasing concentrations of INP0341 or INP1750. Values are expressed in percentage of inhibition of the cytotoxicity of CHA (as measured in the absence of inhibitors) and are the means ± the standard errors of the mean (SEM) of three experiments performed in quadruplicates. Statistical analyses were performed (two-way analysis of variance [ANOVA], Bonferroni posttest; comparison between cells incubated with INP0341 and cells incubated with INP1750 [###, P < 0.001; ##, P < 0.01]). (B) Cytotoxicity induced by different P. aeruginosa strains in THP-1 monocytes (left) or A549 cells (right) after, respectively, 5 and 7 h of incubation in the presence of 0.8% DMSO (vehicle) or 80 μM INP0341 or INP1750. Values are expressed as the percentage of LDH release and are means ± the SEM of three experiments performed in quadruplicates. Statistical analyses (two-way ANOVA with Bonferroni posttest comparing each of the inhibitors with cells incubated with 0.8% DMSO [***, P < 0.001; **, P < 0.01] and comparing the effect of INP0341 and of INP1750 on each strain [#, P < 0.05]). (C) IL-1β secretion in the culture medium of THP-1 monocytes preincubated during 4 h in the presence of LPS (100 ng/ml), centrifuged, resuspended in fresh medium, and incubated for 5 h with different bacterial strains (10 bacteria/cell) or nigericin (0.02 μM) in the presence of 0.8% DMSO (vehicle) or 80 μM INP0341 or INP1750. Values are means ± the SEM of two experiments performed in duplicates. Statistical analyses were performed (two-way ANOVA with Bonferroni posttest comparing each of the inhibitors with cells incubated with DMSO [***, P < 0.001; **, P < 0.01]; multiple t test comparing the effect of INP0341 and of INP1750 on each strain [#, P < 0.05]).
We and others previously showed that CHAΔSTY or PA103ΔUT (expressing the translocation apparatus but not the toxins) were selectively toxic for phagocytic cells by activating NLRC4 inflammasome cascade (16, 18, 35, 38). This effect follows the injection of flagellin (14, 15), rod or needle proteins (39, 40), or pilin (41) in the host cell cytosol via the iT3SS. NLRC4 inflammasome activation results in the cleavage of procaspase-1 into active caspase-1, which itself causes the maturation and release of IL-1β and IL-18, inducing cell death by pyroptosis (16). We therefore measured the release of IL-1β in the supernatant of THP-1 monocytes incubated for 5 h with different mutants of CHA and of PA103 in the presence of 80 μM INPs or their vehicle (Fig. 2C). Nigericin was used in parallel as an inducer of the NLRP3 inflammasome (42) to test for the specificity of the effects of INPs. CHA, CHAΔSTY, and PA103ΔUT strains caused a marked release of IL-1β that was significantly reduced by INP0341 and INP1750. Nigericin also increased IL-1β levels in the culture medium, but INPs did not reverse this effect. The other strains (including PA103) caused a smaller release of IL-1β (as previously described and discussed [35]) that was not prevented by INPs. Tumor necrosis factor alpha (TNF-α) release was measured in parallel as a control, its release being independent of inflammasome activation. All strains (whether expressing the iT3SS or not), as well as the combination of LPS and nigericin, induced the release of TNF-α release, but this effect was not prevented by INPs (see Fig. S3 in the supplemental material).
ExoS and ExoT inhibit bacterial internalization in eukaryotic cells by disorganizing actin microfilaments (43, 44). We therefore examined the influence of INPs on the internalization of CHA, using the aminoglycoside protection assay and focusing on phagocytic cells, which take up bacteria more avidly than epithelial cells. Bacteria were incubated during 3 h with 80 μM INP0341 or INP1750 in broth, pelleted, and incubated with THP-1 monocytes in the presence of inhibitors during 2 h; the phagocytosed bacteria were then quantified by CFU counting. INP0341 and INP1750 significantly increased (2.6- and 2.8-fold, respectively) CHA internalization within THP-1 monocytes (see Fig. S4 in the supplemental material).
Thus, the two inhibitors reduced the cytotoxicity of strains expressing toxins in both phagocytic and epithelial cells and reduced the toxin inhibitory effect on bacterial internalization by phagocytes. In addition, they prevented the development of toxicity mediated by inflammasome activation in THP-1 cells, including for strains expressing the translocation apparatus but no toxins.
Influence of INP0341 and INP1750 on iT3SS transcription.
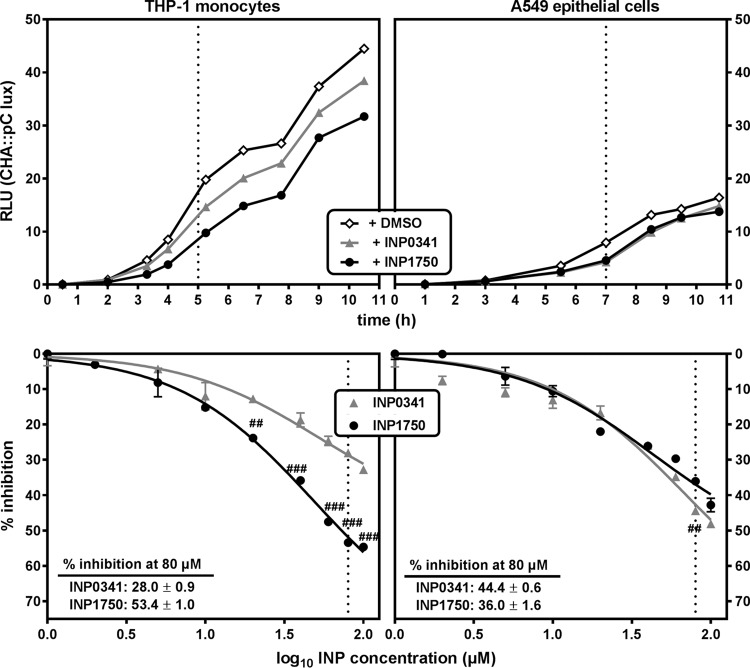
We then examined the effect of INP0341 and INP1750 at increasing concentrations on iT3SS transcription, using a bioluminescent reporter strain (CHA pC::lux). Bacteria were incubated in the presence of THP-1 monocytes or A549 epithelial cells, iT3SS transcription being activated by contact with the eukaryotic cells. As shown in Fig. 3, iT3SS transcription was greater in the presence of THP-1 cells than in the presence of A549 cells. In the presence of THP-1 cells, INP1750 was a more potent inhibitor of transcription than INP0341 (53 versus 28% inhibition after 5 h at 80 μM, respectively; there was a significant difference among them for concentrations ≥20 μM). Conversely, no statistically significant difference was observed between the two inhibitors in the presence of A549 cells (36 to 44% reduction in the bioluminescence signal after 7 h at a concentration of 80 μM).
FIG 3.
Influence of INP0341 and INP1750 on iT3SS transcription. (Top panels) iT3SS transcriptional activation of the exsCEBA operon, followed by measuring the bioluminescence signal of the reporter strain CHA pC::lux over time, under control conditions (0.8% DMSO) or in the presence of 80 μM INP0341 or INP1750. Bacteria (10/cell) were cultured in the presence of THP-1 monocytes in RPMI 1640 medium (left) or of A549 epithelial cells in DMEM (right). Bioluminescence is expressed in arbitrary units (RLU), which correspond to 10−4-fold the actual readings. The dotted lines indicate the time selected in the bottom panels. (Bottom panels) Inhibition of the transcriptional activation of exsCEBA operon by increasing concentrations of INP0341 or INP1750, as measured after 5 h of incubation of CHA pC::lux (10 bacteria/cell) in the presence of THP-1 monocytes (left) or after 7 h of incubation in the presence of A549 cells (right). Values are expressed in percentage of inhibition of the bioluminescence signal recorded in the absence of inhibitors. All values are means ± the SEM of two or three experiments. Statistical analyses were performed (two-way ANOVA, Bonferroni posttest; comparison between cells incubated with INP0341 and cells incubated with INP1750 [###, P < 0.001; ##, P < 0.01]).
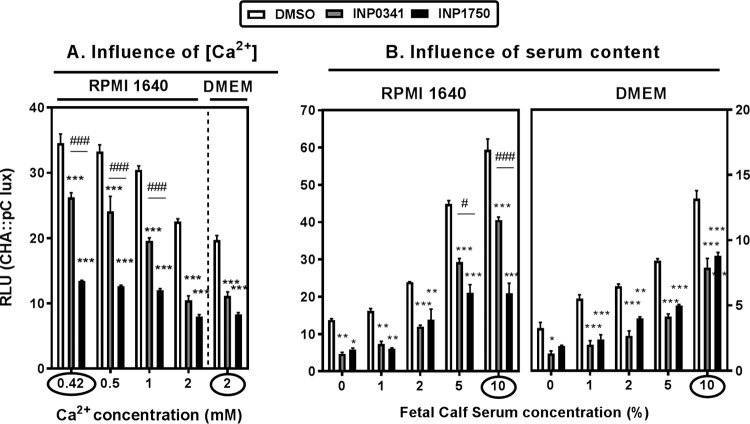
In addition to contact with eukaryotic cells, media depleted in Ca2+ (21) or enriched in serum (24, 25) also activate iT3SS transcription. Since THP-1 cells and A549 cells were cultivated in different media (RPMI 1640 and Dulbecco modified Eagle medium [DMEM], respectively), we examined the influence of the medium composition on iT3SS transcription and its inhibition by INPs used at a fixed concentration of 80 μM. The experiments illustrated in Fig. 3 were first performed in the absence of cells yielding similar results (see Fig. S5 in the supplemental material), suggesting that differences in culture media composition could by themselves explain the variation in iT3SS transcription levels observed in the absence of inhibitors between cultures in RPMI 1640 and in DMEM. Since RPMI 1640 contains less Ca2+ than DMEM, we supplemented RPMI 1640 with increasing amounts of Ca2+ until we reached the concentration found in DMEM (2 mM) (Fig. 4A). As anticipated, iT3SS transcription decreased when the Ca2+ content of RPMI 1640 was increased, reaching similar levels in the two media when their Ca2+ contents were similar. Interestingly, the inhibitory effect of INP0341 was much lower than that of INP1750 at low Ca2+ but similar at a high Ca2+ concentration. We then examined the influence of serum on iT3SS transcription and on INP inhibitory activity (Fig. 4B). Reducing the fetal calf serum (FCS) concentration in the medium caused an impairment of iT3SS transcription, as expected. Again, while both inhibitors were equipotent in media with a low FCS content, the inhibitory effect of INP0341 was less than that of INP1750 when the FCS content was increased. Finally, a previous work demonstrated iron-chelating properties for INP0341, leading to a reduction of its inhibitory effect toward intracellular multiplication of Chlamydia trachomatis in media enriched in ferric ions (45). We therefore also examined the effect of medium supplementation by ferric ions (over the range used in the Chlamydia model) on the inhibition of iT3SS transcription by INP0341 or INP1750, but we did not find evidence of any significant effect of the Fe3+ concentration on the inhibitory activity of INPs (see Fig. S6 in the supplemental material).
FIG 4.
Influence of culture conditions on iT3SS transcriptional activation and its inhibition by INP0341 and INP1750. The iT3SS transcriptional activation of the exsCEBA operon was monitored by measuring the bioluminescence signal of the reporter strain CHA pC::lux after 9 h of incubation in different media and in the presence of 0.8% DMSO (vehicle) or of 80 μM INP0341 or INP1750. (A) RPMI 1640 medium containing 10% FCS and supplemented with increasing concentrations of Ca2+ so as to reach the concentration present in DMEM (circled values correspond to the concentrations present in each medium). (B) RPMI 1640 or DMEM supplemented with increasing amount of FCS (circled values correspond to the concentrations used in the routine). Statistical analyses were performed (two-way ANOVA with Bonferroni posttest comparing each of the inhibitors with cells incubated with DMSO [***, P < 0.001; **, P < 0.01; *, P < 0.05]; multiple t test comparing the effect of INP0341 and of INP1750 on each strain [###, P < 0.001; **, P < 0.01; #, P < 0.05]).
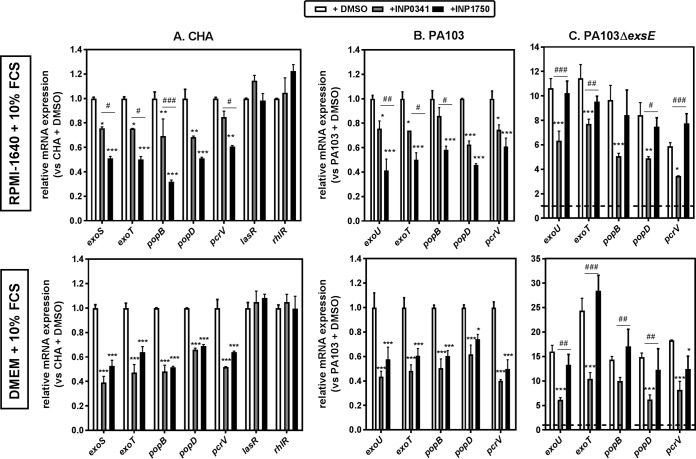
In parallel, using CHA and PA103, we examined the effects of INPs on mRNA levels for genes encoding the iT3SS effectors (exoS [CHA], exoU [PA103], and exoT [both strains]) and the translocation apparatus (popB, popD, and pcrV). Bacteria were incubated for 3 h (to reach an optical density at 620 nm [OD620] of 0.8) in the presence of 80 μM INP0341 or INP1750, or their vehicle, in RPMI 1640 or in DMEM supplemented with 10% FCS before mRNA isolation (Fig. 5A and B) or in Ca2+-depleted broth (see Fig. S7 in the supplemental material). Both compounds inhibited the expression of all tested genes. However, INP0341 was less effective in RPMI 1640 than in DMEM; it was also less effective than INP1750 in Ca2+-depleted broth. In contrast, INPs did not affect the expression of other genes such as lasR and rhlR (quorum-sensing regulators) in CHA (Fig. 5A) or the pyocyanin secretion (dependent on quorum sensing) in CHA (see Fig. S8 in the supplemental material), indicating that INPs are not general inhibitors of transcription. Importantly, Phe-Arg-β-naphthylamide (PAβN), a nonspecific efflux pump inhibitor (46), did not modify the inhibitory effects of INPs on gene transcription, suggesting that they are not substrates of the P. aeruginosa efflux pumps (data not shown).
FIG 5.
Influence of INP0341 and INP1750 on medium-dependent iT3SS mRNA expression levels. The influence of INP0341 and INP1750 on the expression levels of mRNA encoding iT3SS toxins (exoS and exoT), the iT3SS translocation apparatus (popB, popD, and pcrV), and the quorum sensing (QS) transcriptional activator (lasR and rhlR) was determined by real-time PCR. CHA, PA103, and PA103ΔexsE were grown from an OD620 of 0.1 to an OD620 of 0.8 with constant shaking in the presence of 0.8% DMSO (vehicle) or 80 μM INP0341 or INP1750, in eukaryotic cell culture medium (RPMI 1640 plus 10% FCS [top] or DMEM plus 10% FCS [bottom]). The results are expressed in mRNA relative expression compared to CHA (A) or PA103 (B and C) grown in the presence of DMSO. Dotted lines indicate the basal expression levels in PA103 in panel C. Values are means ± the SEM of two or three experiments performed in duplicates. Statistical analyses were performed (two-way ANOVA with Bonferroni posttest comparing each of the inhibitor with cells incubated with DMSO [***, P < 0.001; **, P < 0.01; *, P < 0.05] and comparing the effect of INP0341 and of INP1750 on each mRNA [###, P < 0.001; ##, P < 0.01; #, P < 0.05]).
The effects of INPs on iT3SS gene transcription were also examined using PA103ΔexsE and PA103ΔexsD strains, which constitutively express the iT3SS to high levels (see Fig. 5C for data with PA103ΔexsE; comparable results were obtained with PA103ΔexsD). Although INP0341 inhibited 30 to 40% of the expression of iT3SS genes, INP1750 was ineffective against this strain. These results suggest that INP0341 inhibits iT3SS gene expression, whereas INP1750 most probably acts indirectly by perturbing the ExsCEBA regulatory cascade (35).
Influence of INP0341 and INP1750 on iT3SS secretory activity and bacterial motility.
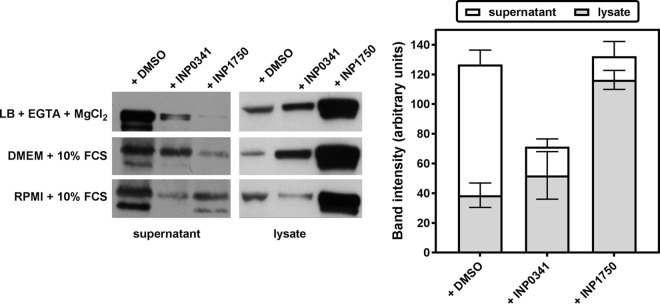
In a next step, we examined the influence of INPs on the secretion of the ExoS toxin by the iT3SS. To this effect, CHA was incubated for 3 h (to reach an OD620 of 0.8) in Ca2+-depleted broth or in eukaryotic cell culture media (RPMI 1640 plus 10% FCS or DMEM plus 10% FCS) in the presence of 80 μM INPs or their vehicle. ExoS was then detected by Western blot in the culture supernatant and in the bacterial lysate (Fig. 6). INP0341 reduced by ∼40% the total amount of toxin detected (lysate plus supernatant), suggesting an effect on toxin expression. At the same time, INP0341 also caused a marked decrease in the amount of ExoS detected in the supernatant, possibly due to a lower expression of the machinery responsible for its secretion. INP1750 reduced by ∼80% the amount of ExoS detected in the supernatant, with a commensurate increase in the amount detected in the lysate so that the total amount of toxin detected was not modified. This rather suggests an inhibition of the toxin secretion.
FIG 6.
Influence of INP0341 and INP1750 on ExoS secretion. CHA was grown from an OD620 of 0.1 to an OD620 of 0.8 with constant shaking in the presence of 0.8% DMSO (vehicle) or 80 μM INP0341 or INP1750 in low-calcium medium (LB broth, 5 mM EGTA, 20 mM MgCl2) or in eukaryotic cell culture medium (RPMI 1640 plus 10% FCS or DMEM plus 10% FCS). ExoS was detected by Western blotting after separation by SDS-PAGE of 13 μl (supernatants) or 5 μg (lysates) of proteins. The band intensity was quantified by using ImageJ software.
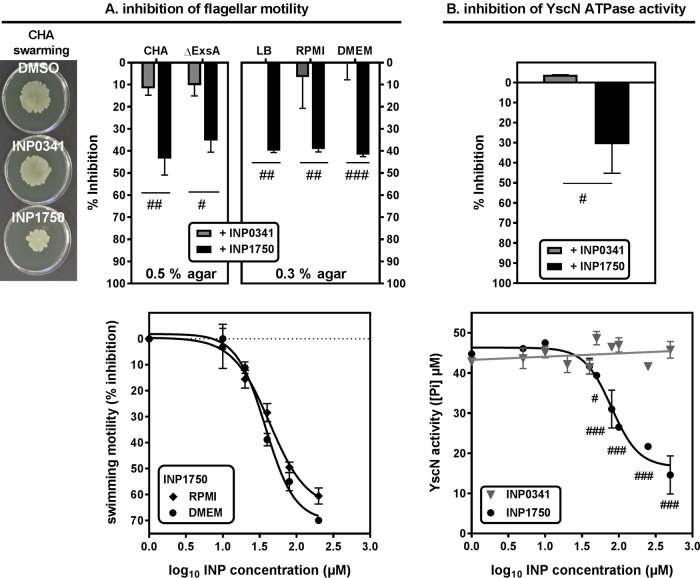
The assembly of the bacterial flagellum is also dependent upon a T3SS (fT3SS) distinct from the iT3SS (28). We therefore examined the effect of INP0341 and of INP1750 or their vehicle on bacterial flagellar motility. Bacteria were grown in the presence of 80 μM INPs in Ca2+-depleted broth and then placed in the center of a 0.5% agar plate and allowed to grow overnight (Fig. 7A). INP1750 reduced the swarming capacity of CHA and its iT3SS-negative mutant CHAΔexsA by ∼40 to 50%, but INP0341 had only a minimal effect (∼10% inhibition for both strains). Comparable results were obtained when examining swimming motility using 0.3% agar, as well as for bacteria precultivated in eukaryotic cell culture media (∼ 40% inhibition for INP1750 versus ∼1% inhibition for INP0341). The vehicle had no effect. The concentration dependency of the inhibitory effect of INP1750 on swimming motility is demonstrated in the lower panel for bacteria preincubated with the inhibitor in eukaryotic cell culture media.
FIG 7.
Influence of INP0341 and INP1750 on bacterial motility and YscN-ATPase activity. (A) Effect of INP0341 and INP1750 on flagellar motility. (Top) Swarming (left) and swimming (right) motility. Bacteria were grown from an OD620 of 0.1 to an OD620 of 0.8 with constant shaking in the presence of 0.8% DMSO (vehicle) or 80 μM INP0341 or INP1750; then, 3-μl portions of these cultures were placed in the center of agar LB plates and grown overnight at 37°C. The area covered by bacteria was evaluated using QuantityOne software (Bio-Rad). Swarming was evaluated using 0.5% agar for CHA (illustrated in the pictures) and CHAΔexsA strains that were precultivated with INPs in low-calcium medium (LB broth, 5 mM EGTA, 20 mM MgCl2) and swimming using 0.3% agar for CHA precultivated with INPs in low-calcium medium or in RPMI 1640 or DMEM added by 10% serum (right). (Bottom) Concentration effect relationship for swimming, with bacteria grown with increasing concentrations of INP1750 in RPMI 1640 or DMEM added by 10% serum, as described above. Values are means ± the SEM of two independent experiments performed in duplicate. Statistical analyses were performed (#, P < 0.05; ##, P < 0.01; ###, P < 0.001). A Student t test was used to compare values measured in the presence of INP0341 or in the presence of INP1750. (B) Effect of INP0341 and INP1750 on purified YscN T3SS ATPase. His-YscN (25 μg/ml) was incubated with INP0341 or INP1750 for 1 h; ATP (4 mM) was then added to the samples for 30 min, and the ATPase activity was measured as the amount of free phosphate liberated. Experiments were performed in duplicates. (Top) Percentage of inhibition of the enzymatic activity in the presence of 80 μM concentrations of each INP. (Bottom) Residual activity as a function of INP concentrations. Statistical analyes were performed (#, P < 0.05; ##, P < 0.01; ###, P < 0.001). A Student t test was used to compare values measured in the presence of INP0341 or in the presence of INP1750.
Taken together, these data showed that, in contrast to INP0341 that inhibits only iT3SS in our experimental setting, INP1750 also acts upon the fT3SS. We therefore wondered whether INP1750 could not act by inhibiting the ATPase activity of both T3SSs (that share a high degree of homology (47% identity between PscN [ATPase of iT3SS] and FliI [ATPase of fT3SS] based on a BLAST sequence alignment [28]). Since these enzymes had not been purified, we used the highly homologous iT3SS ATPase from Y. pseudotuberculosis (YscN) that shares 57 and 47% identity with P. aeruginosa PscN and FliI, respectively (BLAST sequence alignment). As shown in Fig. 7B, INP1750 caused a concentration-dependent inhibition of YscN ATPase activity, whereas INP0341 did not impair the enzymatic activity over the whole range of concentrations tested.
Influence of INP0341 and INP1750 on cytotoxicity of P. aeruginosa clinical isolates.
In order to better delineate the clinical potential of INP0341 and INP1750, we examined their effect on the cytotoxicity induced by clinical isolates collected from patients suffering from acute infections and expressing the iT3SS to variable levels (38). These isolates also showed resistance to one or several antibiotic classes (see Table S1 in the supplemental material). THP-1 monocytes or A549 epithelial cells were incubated in the presence of each of these isolates for 5 or 7 h, respectively, after which the cytotoxicity was evaluated by measuring the release of lactate dehydrogenase in the culture medium (Fig. 8). As previously observed, strains expressing ExoU were more cytotoxic than those expressing ExoS (38). In THP-1 monocytes, the cytotoxicity of ExoU-positive strains was not affected by INP0341 but was reduced by 23% by INP1750. The cytotoxicity of ExoS-positive strains was reduced by 33 and 51% by INP0341 and INP1750, respectively. In A549 epithelial cells, INP0341 and INP1750 reduced cytotoxicity by 18 to 20% for isolates expressing ExoU and by 54 and 35%, respectively, for isolates expressing ExoS (the difference in control values was statistically significant only for isolates expressing high levels of ExoS).
FIG 8.
Influence of INP0341 and INP1750 on the cytotoxicity of P. aeruginosa clinical isolates. The percentage of LDH release from THP-1 monocytes (left) or A549 cells (right) exposed for 5 h (left) or 7 h (right) to clinical isolates (10 bacteria/cell) in the presence of 0.8% DMSO (control; open symbol) or 80 μM INP0341 or INP1750 was determined. Strains were grouped according to the expression of ExoU toxin (T3SS+ ExoU+ versus T3SS+ ExoU−) or of the ExoS toxin (ExoS++, level of expression higher than that detected in CHA; ExoS+, level of expression lower than that detected in CHA). Values are means ± the SEM of three independent experiments performed in quadruplicate (LDH). Statistical analyses were performed (***, P < 0.001; **, P < 0.01; *, P < 0.05 [two-way ANOVA, Bonferroni posttest]) comparing control conditions with INP0341 or INP1750.
DISCUSSION
In this study, we compare the antivirulence properties of two molecules belonging to distinct chemical families (the salicylidene acylhydrazide INP0341 and the hydroxyquinoline INP1750) toward P. aeruginosa, which were originally described as inhibitors of iT3SS transcription in Y. pseudotuberculosis, Shigella, Salmonella, and Chlamydia spp. (31–33).
Both compounds protect eukaryotic cells from the toxic effects mediated by the iT3SS itself or by the toxins, which results in the release of mature IL-1β following inflammasome activation, the prevention of bacteria internalization, and the induction of cell necrosis. However, despite similarities in these protective effects, the two INPs differ in their direct effects on bacteria, suggesting that they act upon distinct pharmacological targets.
For the salicylidene acylhydrazide INP0341, we present two convergent pieces of evidence suggesting that it targets iT3SS gene transcription. First, INP0341 reduces the total amount of ExoS toxin detected by Western blot in treated cultures of CHA, a finding consistent with a reduction in its synthesis following impaired gene transcription. Second, INP0341 reduces iT3SS gene expression and iT3SS-mediated toxicity as efficiently in wild-type strains and in mutants that constitutively express iT3SS, indicating a direct effect on gene transcription. However, INP0341 does not affect the fT3SS (flagellum), as opposed to other salicylidene acylhydrazides that were shown to inhibit the flagellar motility of Salmonella enterica serovar Typhimurium (33). Although this difference may result from variations in the protocol used to assess flagellar motility (preincubation with INPs before inoculation in the agar here versus the addition of INPs in the agar in reference 33), INP0341 is nevertheless distinct from INP1750 in this context. The molecular mechanism by which inhibition of transcription by INP0341 takes place remains to be established and could be indirect. An interaction with specific proteins (namely, WrbA [NADP(H):quinone reductase], Tpx [thiol peroxidase], and FolX [epimerase]) has been demonstrated for other salicylidene acylhydrazides in Escherichia coli and Yersinia pseudotuberculosis (47, 48) and proposed to indirectly interfere with iT3SS regulation by altering cellular metabolism. Homologs of these enzymes are present in P. aeruginosa but could not be examined as potential targets for INP0341 in the present study.
For the hydroxyquinoline INP1750, our results point to a mechanism of action similar to what we previously described for INP1855, another hydroxyquinoline, consisting in an inhibition of a core component of T3SS sharing homology between iT3SS and fT3SS (35). Like INP1855, INP1750 indeed causes a drastic reduction of ExoS secretion (without reducing the total amount of toxin produced by the bacteria) and of flagellar motility, together with an inhibition of the activity of an enzyme homologous to the ATPases present in both T3SSs. As opposed to INP0341, it does not inhibit iT3SS transcription in constitutive mutants of PA103, while still decreasing their cytotoxicity. In the case of INP1750, inhibition of iT3SS transcription is probably not a primary effect but rather an indirect consequence of the inhibition of the secretory activity of iT3SS. In addition to ExoS, iT3SS indeed also secretes other proteins, including FliC and ExsE (the negative regulator of iT3SS gene expression). We previously showed that the hydroxyquinoline INP1855 impairs the secretion of these three proteins (ExoS, flagellin, and ExsE), leading us to conclude that iT3SS transcription by this compound is indirectly inhibited as a consequence of the increase in ExsE content inside the bacteria (35).
In addition to their mode of action, INP0341 and INP1750 also differ in their intrinsic activity, INP0341 being less potent than INP1750 through all the parameters investigated in this study. This difference could be due to the existence of distinct targets, but also to the differential influence exerted by the composition of the culture medium on their activity. We observe indeed that INP0341 is as active as INP1750 in DMEM but is less active in RPMI 1640. RPMI 1640 has a lower Ca2+ content than DMEM, which favors iT3SS expression (21, 23–25), and may consequently counteract the INP0341 activity on gene transcription. However, serum, which also increases iT3SS expression, affects to the same extent the activity of both INP0341 and INP1750 in Ca2+-rich DMEM. This therefore suggests another role of Ca2+ concentration, independent of its influence on iT3SS transcription level, in the modulation of INP0341 activity. A previous work with C. trachomatis failed to show an influence of Ca2+ concentration on INP0341 activity (45), but Ca2+ supplementation was performed in a medium as rich in Ca2+ as DMEM, in which INP0341 activity was already maximal in our setting. Conversely, that study also showed that the activity of INP0341 on C. trachomatis was impaired in Fe3+-enriched medium due to the iron-chelating properties of salicylidene hydrazides (45, 49). We did not observe any modification in the antipseudomonal activity of INP0341 upon medium supplementation in Fe3+, and we postulate therefore that its mode of action is probably different from that described against C. trachomatis. In addition to its effect on iT3SS transcription, serum also increases the outer membrane permeability of P. aeruginosa (50), which could modulate the activity of iT3SS, as well as its inhibition by INP0341, but this was not investigated here.
Moving now to INP1750, we see that its activity is only minimally influenced by the medium composition (RPMI 1640 or DMEM) whatever their Ca2 or serum content. INP1750 is also equipotent to INP1855 (35) when comparing their effects on bacteria (iT3SS secretory activity and transcription, flagellar motility, and inhibition of ATPase activity) and their consequences for the host cells (protection against necrosis and inflammasome activation).
Our work suffers from two main limitations. First, we did not identify at this stage the molecular target of these compounds, which would require additional experiments, including using global approaches such as transcriptomics or proteomics. The experimental approaches we set up could nevertheless represent a useful panel of techniques for the high-throughput screening of T3SS inhibitors in order to get a first insight of their main mechanism of action and also to screen larger series of compounds in order to discover more active derivatives based on refined structure-activity relationships. Applied to hydroxyquinolines and salicylidene acylhydrazides, these techniques allowed us to position them as a potentially useful addition to our future therapeutic armamentarium, since both show activity against clinical isolates, disregarding their resistance profile to currently used antipseudomonal antibiotics. Compared to previously described inhibitors of iT3SS in P. aeruginosa (see reference 22 for a review), they may offer a broader spectrum of activity than (i) compounds blocking toxins only (51–53), the activity of which will probably be restricted to toxin-producing strains, and (ii) antibodies for which activity can be variable due to the polymorphism of the targeted antigens (54). The present data, together with the results obtained previously with INP1855 (35), strongly suggest an advantage for hydroxyquinolines over salicylidene acylhydrazides. Hydroxyquinolines, indeed, show better effects on both iT3SS and the fT3SS, two key determinants in the pathogenicity of P. aeruginosa (3, 11, 12, 27, 55).
Second, we have not compared the activity of the two compounds in vivo. However, studies with other compounds in these families and/or with other bacterial species have demonstrated that they could be active in vivo (35, 36). Based on the in-depth pharmacological comparison performed here, future work is therefore warranted in order to characterize both their pharmacokinetics and their tolerance. This will allow us to define the best experimental conditions under which their efficacy could be studied in animal models of acute pseudomonal infection.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and susceptibility testing.
The strains and plasmids used here were described in a previous publication (35). PA103 is a cytotoxic strain expressing the phospholipase ExoU (56). CHA is a clinical strain isolated from a cystic fibrosis patient and expresses the ExoS/T toxins to a high level (57). The bioluminescent reporter strain CHA (pC::lux) was constructed by transcriptional fusion of Photorhabdus luminescens lux operon (luxCDABE) to the promoter of the iT3SS regulator operon exsCEBA (25). A series of strains derived from PA103 and CHA with deletions in the iT3SS regulon, the genes encoding iT3SS effector toxins or proteins from the translocon apparatus were also included in the study, together with 20 clinical isolates collected from acute infections in four Belgian hospitals (38). Unless stated otherwise, all strains were routinely grown overnight in Luria-Bertani (LB) broth (Becton Dickinson, Franklin Lakes, NJ) at 37°C with constant shaking (130 rpm) and under aerobic conditions. MICs were determined by geometric microdilution in cation-adjusted Mueller-Hinton broth (BD Diagnostics). Susceptibility was assessed according to the current interpretive criteria of both the European Committee on Antimicrobial Susceptibility Testing and the Clinical and Laboratory Standards Institute.
Cells and culture.
Human THP-1 cells (ATCC TIB-202) are a myelomonocytic cell line showing moderate phagocytic activity and low cell defense mechanisms (58). They were grown in suspension in RPMI 1640 supplemented with 10% fetal calf serum (FCS; Gibco/Life Technologies Corporation, Paisley, UK). Human alveolar epithelial A549 cells (ATCC CCL-185) are adherent cells derived from lung carcinoma (59). In contrast to THP-1 monocytes, these cells do not express NLRC4 (60). They were cultured in DMEM supplemented with 10% FCS and seeded in culture plates for 48 h prior being used for experiments. Cells were counted using a Burker hemocytometer to adjust their concentration prior infection.
T3SS inhibitors.
INP0341 and INP1750 were kindly provided by Creative Antibiotics Sweden AB (Umeå, Sweden). Stock solutions were prepared in dimethyl sulfoxide (DMSO) at a concentration of 10 mM and stored at room temperature in the dark for less than 1 month according to the manufacturer's instructions.
Cytotoxicity assay.
The cytotoxicity of INP0341 or INP1750 and of P. aeruginosa strains was determined by measuring the release of the cytoplasmic enzyme LDH in the culture medium, which occurs upon cell necrosis (61). Cells were seeded into 96-well plates (at a density of 2.5 × 105 cells/ml for phagocytic cells and of 8 × 104 cells/ml for epithelial cells) and incubated with INP0341 or INP1750 or/and with P. aeruginosa strains (10 bacteria/cell) for 5 h (THP-1 monocytes) or 7 h (A549 epithelial cells). The amount of LDH released was determined by using a cytotoxicity detection kit (Plus; Roche Diagnostics, Basel, Switzerland).
Internalization of P. aeruginosa in THP-1 cells.
Bacteria internalization was evaluated using a previously described protocol (aminoglycoside protection assay [62]) with minor adaptations. Overnight bacterial cultures were pretreated during 3 h with INP0341 or INP1750 in (0.8%) DMSO or with the vehicle, resuspended in eukaryotic cell medium with 10% human serum in the presence of INPs or DMSO, and incubated for 1 h at 37°C to allow for opsonization. Bacteria were then added to THP-1 monocytes (10 bacteria/cell; 7.5 × 105 cells/ml), again in the presence of INPs or DMSO. Infected cells were incubated at 37°C in a 5% CO2 atmosphere for 2 h to allow phagocytosis, washed in phosphate-buffered saline (PBS), and then exposed to tobramycin at 200 mg/liter for 1 h to kill extracellular bacteria. After three washes with PBS, the cells were lysed in 1 ml of sterile water, and aliquots were plated on tryptic soy agar plates after suitable dilution for the determination of CFU by colony counting after 24 h of incubation at 37°C. The results were normalized per mg of cell protein, as determined by Lowry's assay using a commercially available detection kit (Bio-Rad DC protein assay; Bio-Rad Laboratories, Hercules, CA).
Cytokine release assay.
THP-1 cells (2.5 × 105 cells/ml) were preincubated during 4 h with lipopolysaccharide (LPS; Sigma-Aldrich) at 100 ng/ml in order to stimulate the production of pro-IL-1β. The cells were then incubated for 5 h with INPs or DMSO alone or combined with (i) P. aeruginosa strains (10 bacteria/cell) or (ii) 0.02 μM nigericin, after which the cytokines released into the supernatant were measured using commercially available enzyme-linked immunosorbent assay kits (IL-1β [R&D Systems, Minneapolis, MN]; TNF-α [BD Biosciences, San Jose, CA]).
P. aeruginosa growth curves.
Overnight cultures were pelleted and resuspended in LB broth at an initial OD620 of 0.1; the increase in OD620 was then monitored over time during incubation at 37°C with aeration and constant shaking (300 rpm) in the presence of INP0341 and INP1750 in 2.5% DMSO or in 2.5% DMSO only (vehicle). Since some experiments were performed in the presence of eukaryotic cells, the growth rates were also measured in RPMI 1640 or DMEM supplemented by 10% FCS. No differences were observed compared to what was seen in LB broth (see Fig. S2 in the supplemental material).
Reporter genes assay.
An overnight culture of CHA (pC::lux) was centrifuged and resuspended in eukaryotic cell culture medium (supplemented by 10% of FCS) and added to THP-1 monocytes or A549 epithelial cells (5 × 105 cells/ml; 10 bacteria/cell) in the presence of increasing concentrations of INPs in DMSO (≤0.8% DMSO) or of DMSO only. Cultures were incubated without shaking at 37°C in a 5% CO2 atmosphere. Luminescence was measured in relative light units (RLU) over time using a SpectraMax spectrofluorometer (Molecular Devices LLC, Sunnyvale, CA). Concentration-response curves were also obtained after 5 h (THP-1) or 7 h (A549) of incubation (see reference 35 for a justification of these incubation times).
Real-time PCR.
Overnight cultures were resuspended in LB broth containing 5 mM EGTA and 20 mM MgCl2 or in eukaryotic cell medium (RPMI 1640 or DMEM) supplemented by 10% of FCS and grown from an OD620 of 0.1 to an OD620 of 0.8 with aeration and constant shaking (300 rpm) in the presence of INPs in 0.8% DMSO or 0.8% DMSO only. Total RNA was extracted by using an RNeasy minikit (Stratec Molecular GmbH, Berlin, Germany) and reverse transcribed using a Transcriptor first-strand kit (Roche Diagnostics) according to the manufacturer's instructions. RNA purity was checked for the absence of contaminating DNA prior to reverse transcription by PCR amplification of a fragment of the rpsL gene. Amplification reactions were performed in the presence of the Sybr green IQ Supermix (Bio-Rad Laboratories), using an iCycler iQ single-color real-time PCR detection system (MyiQreal Time PCR software; Bio-Rad). Relative quantification of the mRNA levels was obtained by the comparative CT (2−ΔΔCT) method (63), using rpsL as a housekeeping gene. The primers and conditions were as previously described (35, 38).
Pyocyanin production assay.
Bacteria were grown overnight in 5 ml of LB broth at 37°C with shaking at 130 rpm in the presence of INP0341 or INP1750 in DMSO or of DMSO only. After centrifugation at 4,000 rpm for 15 min, pyocyanin was extracted from the supernatant successively with 2.5 ml of chloroform and 0.2 N HCl and quantified by measuring the absorbance of the solution at 520 nm, based on a calibration curve established with purified pyocyanin (Sigma-Aldrich, St. Louis, MO) (64).
Detection of ExoS by Western blot.
Overnight cultures were resuspended in LB broth containing 5 mM EGTA and 20 mM MgCl2 or in eukaryotic cell medium (RPMI 1640 or DMEM) and grown from an OD620 of 0.1 to an OD620 of 0.8 with aeration and constant shaking (300 rpm) in the presence of INPs in 0.8% DMSO or of 0.8% DMSO only. Lysates were centrifuged (10 min at 4,000 rpm), and the pellet was collected, washed in ice-cold PBS, resuspended in radioimmunoprecipitation assay buffer (25 mM Tris-HCl [pH 7.6], 150 mM NaCl, 1% NP-40, 0.1% SDS, 1% sodium deoxycholate, and a cocktail of protease phosphatase inhibitors diluted according to the manufacturer's instructions [Sigma-Aldrich; catalog no. P8340]), and subjected to sonication. The gross debris were then eliminated by centrifugation for 15 min at 20,800 × g. Supernatants (5 ml) were concentrated by centrifugation (10 min; 4,200 × g) through Amicon Ultra 4 centrifugal filters (Millipore; Merck KGaA, Darmstadt, Germany) in order to reduce the volume to 100 μl. The protein content of both preparations was measured using a bicinchoninic acid protein assay (Bradford assay; BCA reagents; Pierce, Rockford, IL). This protein content was similar in all samples. Appropriate quantities of proteins (corresponding to the same volume for all samples) were mixed to 4× NuPAGE LDS sample buffer and 10× NuPAGE reducing agent and then heated for 10 min at 70°C. Samples were loaded on acrylamide gels (NuPAGE 10% Bis-Tris gel; Invitrogen, Carlsbad, CA). The proteins were electrotransferred after migration onto a polyvinylidene difluoride membrane, which was blocked by 1 h of incubation with 5% defatted milk in Tris-buffered saline (20 mM Tris-HCl, 500 mM NaCl [pH 7.5]) containing 0.1% Tween 20. The membranes were then incubated overnight with anti-ExoS antibody (Agrisera AB, Vännäs, Sweden; 1/5,000 dilution) and then with an appropriate horseradish peroxidase-coupled secondary antibody (1/10,000 dilution) for 1 h. Blots were revealed by chemiluminescence (SuperSignal West Pico; Pierce/Thermo Fisher Scientific, Inc., Rockford, IL). The band intensity was quantified using ImageJ software (v1.47; http://imagej.nih.gov/ij/).
Motility assay.
Overnight cultures were resuspended in LB broth or eukaryotic cell medium (RPMI 1640 or DMEM) with DMSO or INP0341 or INP1750 and grown from an OD620 of 0.1 to an OD620 of 0.8 with aeration and constant shaking (300 rpm) (65). Portions (3 μl) of these cultures were placed at the center of a 0.3% (swimming motility) or 0.5% (swarming motility) LB agar plate. Plates were incubated for 20 h at 37°C, and the dissemination area was evaluated using QuantityOne software (v4.5.0; Bio-Rad Laboratories).
ATPase purification and enzymatic activity assay.
Because the PscN ATPase of the iT3SS of P. aeruginosa has not been fully purified so far (35), we used the highly homologous YscN enzyme from Yersinia pseudotuberculosis, for which a vector encoding the His-tagged enzyme was available (57% identity; 72% positive substitutions with PscN by BLAST alignment). The enzyme was purified exactly as previously described (66), and its activity was assessed using a MAK113 kit (Sigma-Aldrich) according to the recommendations of the manufacturer.
Curve fittings and statistical analyses.
Curve fittings were performed with Prism (v7.02), and statistical analyses were completed using InStat (v3.1), both from GraphPad Prism Software, San Diego, CA.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to M. C. Cambier, V. Mohymont, and K. Santos for excellent technical assistance and to O. Denis (Hôpital Erasme, Université Libre de Bruxelles, Brussels, Belgium), Y. Glupczynski (CHU Dinant-Godinne UCL Namur, Université catholique de Louvain, Yvoir, Belgium), D. Pierard (Universitair Ziekenhuis Brussel, Vrije Universiteit Brussel, Brussels, Belgium), and H. Rodriguez-Villalobos (Cliniques universitaires Saint-Luc, Université catholique de Louvain, Brussels, Belgium) for collecting clinical strains. J. Mecsas (Tufts University School of Medicine, Boston, MA) kindly provided the plasmid encoding the YscN ATPase, and T. L. Yahr (University of Iowa, Iowa City, IA) provided the PA103 constitutive mutants. A.A. was successively Aspirant of the Belgian Fonds de la Recherche Scientifique (FRS-FNRS) and a research doctoral fellow of the Université Catholique de Louvain. F.V.B. is Maître de Recherches of the Belgian Fonds de la Recherche Scientifique.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02566-16.
REFERENCES
- 1.Dantas RC, Ferreira ML, Gontijo-Filho PP, Ribas RM. 2014. Pseudomonas aeruginosa bacteremia: independent risk factors for mortality and impact of resistance on outcome. J Med Microbiol 63(Pt 12):1679–1687. doi: 10.1099/jmm.0.073262-0. [DOI] [PubMed] [Google Scholar]
- 2.Hirsch EB, Tam VH. 2010. Impact of multidrug-resistant Pseudomonas aeruginosa infection on patient outcomes. Expert Rev Pharmacoecon Outcomes Res 10:441–451. doi: 10.1586/erp.10.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pena C, Cabot G, Gomez-Zorrilla S, Zamorano L, Ocampo-Sosa A, Murillas J, Almirante B, Pomar V, Aguilar M, Granados A, Calbo E, Rodriguez-Bano J, Rodriguez-Lopez F, Tubau F, Martinez-Martinez L, Oliver A. 2014. Influence of virulence genotype and resistance profile in the mortality of Pseudomonas aeruginosa bloodstream infections. Clin Infect Dis 60:539–548. doi: 10.1093/cid/ciu866. [DOI] [PubMed] [Google Scholar]
- 4.Page MGP, Heim J. 2009. Prospects for the next anti-Pseudomonas drug. Curr Opin Pharmacol 9:558–565. doi: 10.1016/j.coph.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 5.Lomovskaya O, Watkins WJ. 2001. Efflux pumps: their role in antibacterial drug discovery. Curr Med Chem 8:1699–1711. doi: 10.2174/0929867013371743. [DOI] [PubMed] [Google Scholar]
- 6.Czaplewski L, Bax R, Clokie M, Dawson M, Fairhead H, Fischetti VA, Foster S, Gilmore BF, Hancock RE, Harper D, Henderson IR, Hilpert K, Jones BV, Kadioglu A, Knowles D, Olafsdottir S, Payne D, Projan S, Shaunak S, Silverman J, Thomas CM, Trust TJ, Warn P, Rex JH. 2016. Alternatives to antibiotics: a pipeline portfolio review. Lancet Infect Dis 16:239–251. doi: 10.1016/S1473-3099(15)00466-1. [DOI] [PubMed] [Google Scholar]
- 7.Ramirez-Estrada S, Borgatta B, Rello J. 2016. Pseudomonas aeruginosa ventilator-associated pneumonia management. Infect Drug Resist 9:7–18. doi: 10.2147/IDR.S50669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maura D, Ballok AE, Rahme LG. 2016. Considerations and caveats in anti-virulence drug development. Curr Opin Microbiol 33:41–46. doi: 10.1016/j.mib.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rasko DA, Sperandio V. 2010. Anti-virulence strategies to combat bacteria-mediated disease. Nat Rev Drug Discov 9:117–128. doi: 10.1038/nrd3013. [DOI] [PubMed] [Google Scholar]
- 10.Diepold A, Armitage JP. 2015. Type III secretion systems: the bacterial flagellum and the injectisome. Philos Trans R Soc Lond B Biol Sci 370:pii20150020. doi: 10.1098/rstb.2015.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El-Solh AA, Hattemer A, Hauser AR, Alhajhusain A, Vora H. 2012. Clinical outcomes of type III Pseudomonas aeruginosa bacteremia. Crit Care Med 40:1157–1163. doi: 10.1097/CCM.0b013e3182377906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roy-Burman A, Savel RH, Racine S, Swanson BL, Revadigar NS, Fujimoto J, Sawa T, Frank DW, Wiener-Kronish JP. 2001. Type III protein secretion is associated with death in lower respiratory and systemic Pseudomonas aeruginosa infections. J Infect Dis 183:1767–1774. doi: 10.1086/320737. [DOI] [PubMed] [Google Scholar]
- 13.Hauser AR. 2009. The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat Rev Microbiol 7:654–665. doi: 10.1038/nrmicro2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miao EA, Ernst RK, Dors M, Mao DP, Aderem A. 2008. Pseudomonas aeruginosa activates caspase 1 through Ipaf. Proc Natl Acad Sci U S A 105:2562–2567. doi: 10.1073/pnas.0712183105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ince D, Sutterwala FS, Yahr TL. 2015. Secretion of flagellar proteins by the Pseudomonas aeruginosa type III secretion-injectisome system. J Bacteriol 197:2003–2011. doi: 10.1128/JB.00030-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sutterwala FS, Mijares LA, Li L, Ogura Y, Kazmierczak BI, Flavell RA. 2007. Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J Exp Med 204:3235–3245. doi: 10.1084/jem.20071239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von Moltke J, Ayres JS, Kofoed EM, Chavarria-Smith J, Vance RE. 2013. Recognition of bacteria by inflammasomes. Annu Rev Immunol 31:73–106. doi: 10.1146/annurev-immunol-032712-095944. [DOI] [PubMed] [Google Scholar]
- 18.Faure E, Mear JB, Faure K, Normand S, Couturier-Maillard A, Grandjean T, Balloy V, Ryffel B, Dessein R, Chignard M, Uyttenhove C, Guery B, Gosset P, Chamaillard M, Kipnis E. 2014. Pseudomonas aeruginosa type 3 secretion system dampens host defense by exploiting the NLRC4-coupled inflammasome. Am J Respir Crit Care Med 189:799–811. doi: 10.1164/rccm.201307-1358OC. [DOI] [PubMed] [Google Scholar]
- 19.Cohen TS, Prince AS. 2013. Activation of inflammasome signaling mediates pathology of acute Pseudomonas aeruginosa pneumonia. J Clin Invest 123:1630–1637. doi: 10.1172/JCI66142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Urbanowski ML, Brutinel ED, Yahr TL. 2007. Translocation of ExsE into Chinese hamster ovary cells is required for transcriptional induction of the Pseudomonas aeruginosa type III secretion system. Infect Immun 75:4432–4439. doi: 10.1128/IAI.00664-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yahr TL, Wolfgang MC. 2006. Transcriptional regulation of the Pseudomonas aeruginosa type III secretion system. Mol Microbiol 62:631–640. doi: 10.1111/j.1365-2958.2006.05412.x. [DOI] [PubMed] [Google Scholar]
- 22.Anantharajah A, Mingeot-Leclercq MP, Van Bambeke F. 2016. Targeting the type three secretion system in Pseudomonas aeruginosa. Trends Pharmacol Sci 37:734–749. doi: 10.1016/j.tips.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 23.Frank DW. 1997. The exoenzyme S regulon of Pseudomonas aeruginosa. Mol Microbiol 26:621–629. doi: 10.1046/j.1365-2958.1997.6251991.x. [DOI] [PubMed] [Google Scholar]
- 24.Kim J, Ahn K, Min S, Jia J, Ha U, Wu D, Jin S. 2005. Factors triggering type III secretion in Pseudomonas aeruginosa. Microbiology 151:3575–3587. doi: 10.1099/mic.0.28277-0. [DOI] [PubMed] [Google Scholar]
- 25.Shen DK, Filopon D, Kuhn L, Polack B, Toussaint B. 2006. PsrA is a positive transcriptional regulator of the type III secretion system in Pseudomonas aeruginosa. Infect Immun 74:1121–1129. doi: 10.1128/IAI.74.2.1121-1129.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sundin C, Thelaus J, Broms JE, Forsberg A. 2004. Polarisation of type III translocation by Pseudomonas aeruginosa requires PcrG, PcrV, and PopN. Microb Pathog 37:313–322. doi: 10.1016/j.micpath.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 27.Feldman M, Bryan R, Rajan S, Scheffler L, Brunnert S, Tang H, Prince A. 1998. Role of flagella in pathogenesis of Pseudomonas aeruginosa pulmonary infection. Infect Immun 66:43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blocker A, Komoriya K, Aizawa SI. 2003. Type III secretion systems and bacterial flagella: insights into their function from structural similarities. Proc Natl Acad Sci U S A 100:3027–3030. doi: 10.1073/pnas.0535335100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marshall NC, Finlay BB. 2014. Targeting the type III secretion system to treat bacterial infections. Expert Opin Ther Targets 18:137–152. doi: 10.1517/14728222.2014.855199. [DOI] [PubMed] [Google Scholar]
- 30.McShan AC, De Guzman RN. 2015. The bacterial type III secretion system as a target for developing new antibiotics. Chem Biol Drug Des 85:30–42. doi: 10.1111/cbdd.12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Veenendaal AK, Sundin C, Blocker AJ. 2009. Small-molecule type III secretion system inhibitors block assembly of the Shigella type III secreton. J Bacteriol 191:563–570. doi: 10.1128/JB.01004-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Enquist PA, Gylfe A, Hagglund U, Lindstrom P, Norberg-Scherman H, Sundin C, Elofsson M. 2012. Derivatives of 8-hydroxyquinoline–antibacterial agents that target intra- and extracellular Gram-negative pathogens. Bioorg Med Chem Lett 22:3550–3553. doi: 10.1016/j.bmcl.2012.03.096. [DOI] [PubMed] [Google Scholar]
- 33.Negrea A, Bjur E, Ygberg SE, Elofsson M, Wolf-Watz H, Rhen M. 2007. Salicylidene acylhydrazides that affect type III protein secretion in Salmonella enterica serovar Typhimurium. Antimicrob Agents Chemother 51:2867–2876. doi: 10.1128/AAC.00223-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muschiol S, Normark S, Henriques-Normark B, Subtil A. 2009. Small molecule inhibitors of the Yersinia type III secretion system impair the development of Chlamydia after entry into host cells. BMC Microbiol 9:75. doi: 10.1186/1471-2180-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anantharajah A, Faure E, Buyck JM, Sundin C, Lindmark T, Mecsas J, Yahr TL, Tulkens PM, Mingeot-Leclercq MP, Guery B, Van Bambeke F. 2016. Inhibition of the injectisome and flagellar type III secretion systems by INP1855 impairs Pseudomonas aeruginosa pathogenicity and inflammasome activation. J Infect Dis 214:1105–1116. doi: 10.1093/infdis/jiw295. [DOI] [PubMed] [Google Scholar]
- 36.Slepenkin A, Chu H, Elofsson M, Keyser P, Peterson EM. 2011. Protection of mice from a Chlamydia trachomatis vaginal infection using a salicylidene acylhydrazide, a potential microbicide. J Infect Dis 204:1313–1320. doi: 10.1093/infdis/jir552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bailey L, Gylfe A, Sundin C, Muschiol S, Elofsson M, Nordstrom P, Henriques-Normark B, Lugert R, Waldenstrom A, Wolf-Watz H, Bergstrom S. 2007. Small molecule inhibitors of type III secretion in Yersinia block the Chlamydia pneumoniae infection cycle. FEBS Lett 581:587–595. doi: 10.1016/j.febslet.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 38.Anantharajah A, Buyck JM, Faure E, Glupczynski Y, Rodriguez-Villalobos H, De Vos D, Pirnay JP, Bilocq F, Guery B, Tulkens PM, Mingeot-Leclercq MP, Van Bambeke F. 2015. Correlation between cytotoxicity induced by Pseudomonas aeruginosa clinical isolates from acute infections and IL-1β secretion in a model of human THP-1 monocytes. Pathog Dis 73:piiftv049. doi: 10.1093/femspd/ftv049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miao EA, Mao DP, Yudkovsky N, Bonneau R, Lorang CG, Warren SE, Leaf IA, Aderem A. 2010. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc Natl Acad Sci U S A 107:3076–3080. doi: 10.1073/pnas.0913087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang J, Zhao Y, Shi J, Shao F. 2013. Human NAIP and mouse NAIP1 recognize bacterial type III secretion needle protein for inflammasome activation. Proc Natl Acad Sci U S A 110:14408–14413. doi: 10.1073/pnas.1306376110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arlehamn CSL, Evans TJ. 2011. Pseudomonas aeruginosa pilin activates the inflammasome. Cell Microbiol 13:388–401. doi: 10.1111/j.1462-5822.2010.01541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petrilli V, Papin S, Dostert C, Mayor A, Martinon F, Tschopp J. 2007. Activation of the NALP3 inflammasome is triggered by low intracellular potassium concentration. Cell Death Differ 14:1583–1589. doi: 10.1038/sj.cdd.4402195. [DOI] [PubMed] [Google Scholar]
- 43.Frithz-Lindsten E, Du Y, Rosqvist R, Forsberg A. 1997. Intracellular targeting of exoenzyme S of Pseudomonas aeruginosa via type III-dependent translocation induces phagocytosis resistance, cytotoxicity and disruption of actin microfilaments. Mol Microbiol 25:1125–1139. doi: 10.1046/j.1365-2958.1997.5411905.x. [DOI] [PubMed] [Google Scholar]
- 44.Sundin C, Henriksson ML, Hallberg B, Forsberg A, Frithz-Lindsten E. 2001. Exoenzyme T of Pseudomonas aeruginosa elicits cytotoxicity without interfering with Ras signal transduction. Cell Microbiol 3:237–246. doi: 10.1046/j.1462-5822.2001.00108.x. [DOI] [PubMed] [Google Scholar]
- 45.Slepenkin A, Enquist PA, Hagglund U, de la Maza LM, Elofsson M, Peterson EM. 2007. Reversal of the antichlamydial activity of putative type III secretion inhibitors by iron. Infect Immun 75:3478–3489. doi: 10.1128/IAI.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lomovskaya O, Warren MS, Lee A, Galazzo J, Fronko R, Lee M, Blais J, Cho D, Chamberland S, Renau T, Leger R, Hecker S, Watkins W, Hoshino K, Ishida H, Lee VJ. 2001. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob Agents Chemother 45:105–116. doi: 10.1128/AAC.45.1.105-116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang D, Zetterstrom CE, Gabrielsen M, Beckham KSH, Tree JJ, Macdonald SE, Byron O, Mitchell TJ, Gally DL, Herzyk P, Mahajan A, Uvell H, Burchmore R, Smith BO, Elofsson M, Roe AJ. 2011. Identification of bacterial target proteins for the salicylidene acylhydrazide class of virulence-blocking compounds. J Biol Chem 286:29922–29931. doi: 10.1074/jbc.M111.233858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gabrielsen M, Beckham KSH, Feher VA, Zetterstrom CE, Wang D, Muller S, Elofsson M, Amaro RE, Byron O, Roe AJ. 2012. Structural characterisation of Tpx from Yersinia pseudotuberculosis reveals insights into the binding of salicylidene acylhydrazide compounds. PLoS One 7:e32217. doi: 10.1371/journal.pone.0032217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Engstrom P, Nguyen BD, Normark J, Nilsson I, Bastidas RJ, Gylfe A, Elofsson M, Fields KA, Valdivia RH, Wolf-Watz H, Bergstrom S. 2013. Mutations in hemG mediate resistance to salicylidene acylhydrazides, demonstrating a novel link between protoporphyrinogen oxidase (HemG) and Chlamydia trachomatis infectivity. J Bacteriol 195:4221–4230. doi: 10.1128/JB.00506-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buyck JM, Plesiat P, Traore H, Vanderbist F, Tulkens PM, Van Bambeke F. 2012. Increased susceptibility of Pseudomonas aeruginosa to macrolides and ketolides in eukaryotic cell culture media and biological fluids due to decreased expression of oprM and increased outer-membrane permeability. Clin Infect Dis 55:534–542. doi: 10.1093/cid/cis473. [DOI] [PubMed] [Google Scholar]
- 51.Kim D, Baek J, Song J, Byeon H, Min H, Min KH. 2014. Identification of arylsulfonamides as ExoU inhibitors. Bioorg Med Chem Lett 24:3823–3825. doi: 10.1016/j.bmcl.2014.06.064. [DOI] [PubMed] [Google Scholar]
- 52.Arnoldo A, Curak J, Kittanakom S, Chevelev I, Lee VT, Sahebol-Amri M, Koscik B, Ljuma L, Roy PJ, Bedalov A, Giaever G, Nislow C, Merrill AR, Lory S, Stagljar I. 2008. Identification of small molecule inhibitors of Pseudomonas aeruginosa exoenzyme S using a yeast phenotypic screen. PLoS Genet 4:e1000005. doi: 10.1371/journal.pgen.1000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee VT, Pukatzki S, Sato H, Kikawada E, Kazimirova AA, Huang J, Li X, Arm JP, Frank DW, Lory S. 2007. Pseudolipasin A is a specific inhibitor for phospholipase A2 activity of Pseudomonas aeruginosa cytotoxin ExoU. Infect Immun 75:1089–1098. doi: 10.1128/IAI.01184-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lynch SV, Flanagan JL, Sawa T, Fang A, Baek MS, Rubio-Mills A, Ajayi T, Yanagihara K, Hirakata Y, Kohno S, Misset B, Nguyen JC, Wiener-Kronish JP. 2010. Polymorphisms in the Pseudomonas aeruginosa type III secretion protein, PcrV: implications for anti-PcrV immunotherapy. Microb Pathog 48:197–204. doi: 10.1016/j.micpath.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Veesenmeyer JL, Hauser AR, Lisboa T, Rello J. 2009. Pseudomonas aeruginosa virulence and therapy: evolving translational strategies. Crit Care Med 37:1777–1786. doi: 10.1097/CCM.0b013e31819ff137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu PV. 1966. The roles of various fractions of Pseudomonas aeruginosa in its pathogenesis. 3. Identity of the lethal toxins produced in vitro and in vivo. J Infect Dis 116:481–489. [DOI] [PubMed] [Google Scholar]
- 57.Toussaint B, Delic-Attree I, Vignais PM. 1993. Pseudomonas aeruginosa contains an IHF-like protein that binds to the algD promoter. Biochem Biophys Res Commun 196:416–421. doi: 10.1006/bbrc.1993.2265. [DOI] [PubMed] [Google Scholar]
- 58.Tsuchiya S, Yamabe M, Yamaguchi Y, Kobayashi Y, Konno T, Tada K. 1980. Establishment and characterization of a human acute monocytic leukemia cell line (THP-1). Int J Cancer 26:171–176. doi: 10.1002/ijc.2910260208. [DOI] [PubMed] [Google Scholar]
- 59.Giard DJ, Aaronson SA, Todaro GJ, Arnstein P, Kersey JH, Dosik H, Parks WP. 1973. In vitro cultivation of human tumors: establishment of cell lines derived from a series of solid tumors. J Natl Cancer Inst 51:1417–1423. doi: 10.1093/jnci/51.5.1417. [DOI] [PubMed] [Google Scholar]
- 60.Vinzing M, Eitel J, Lippmann J, Hocke AC, Zahlten J, Slevogt H, N′guessan PD, Gunther S, Schmeck B, Hippenstiel S, Flieger A, Suttorp N, Opitz B. 2008. NAIP and Ipaf control Legionella pneumophila replication in human cells. J Immunol 180:6808–6815. doi: 10.4049/jimmunol.180.10.6808. [DOI] [PubMed] [Google Scholar]
- 61.Korzeniewski C, Callewaert DM. 1983. An enzyme-release assay for natural cytotoxicity. J Immunol Methods 64:313–320. doi: 10.1016/0022-1759(83)90438-6. [DOI] [PubMed] [Google Scholar]
- 62.Buyck JM, Tulkens PM, Van Bambeke F. 2013. Pharmacodynamic evaluation of the intracellular activity of antibiotics towards Pseudomonas aeruginosa PAO1 in a model of THP-1 human monocytes. Antimicrob Agents Chemother 57:2310–2318. doi: 10.1128/AAC.02609-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 64.Essar DW, Eberly L, Hadero A, Crawford IP. 1990. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J Bacteriol 172:884–900. doi: 10.1128/jb.172.2.884-900.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wei Q, Tarighi S, Dotsch A, Haussler S, Musken M, Wright VJ, Camara M, Williams P, Haenen S, Boerjan B, Bogaerts A, Vierstraete E, Verleyen P, Schoofs L, Willaert R, De Groote VN, Michiels J, Vercammen K, Crabbe A, Cornelis P. 2011. Phenotypic and genome-wide analysis of an antibiotic-resistant small colony variant (SCV) of Pseudomonas aeruginosa. PLoS One 6:e29276. doi: 10.1371/journal.pone.0029276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Davis AJ, Diaz DAD, Mecsas J. 2010. A dominant-negative needle mutant blocks type III secretion of early but not late substrates in Yersinia. Mol Microbiol 76:236–259. doi: 10.1111/j.1365-2958.2010.07096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.