ABSTRACT
The Klebsiella pneumoniae carbapenemase gene (blaKPC) is typically located within mobile transposon Tn4401. Enhanced KPC expression has been associated with deletions in the putative promoter region upstream of blaKPC. Illumina sequences from blaKPC-positive clinical isolates from a single institution were mapped to a Tn4401b reference sequence, which carries no deletions. The novel isoform Tn4401h (188-bp deletion [between istB and blaKPC]) was present in 14% (39/281) of clinical isolates. MICs showed that Escherichia coli strains containing plasmids with Tn4401a and Tn4401h were more resistant to meropenem (≥16 and ≥16, respectively), ertapenem (≥8 and 4, respectively), and cefepime (≥64 and 4, respectively) than E. coli strains with Tn4401b (0.5, ≤0.5, and ≤1, respectively). Quantitative real-time PCR (qRT-PCR) demonstrated that Tn4401a had a 16-fold increase and Tn4401h a 4-fold increase in blaKPC mRNA levels compared to the reference Tn4401b. A lacZ reporter plasmid was used to test the activity of the promoter regions from the different variants, and the results showed that the Tn4401a and Tn4401h promoter sequences generated higher β-galactosidase activity than the corresponding Tn4401b sequence. Further dissection of the promoter region demonstrated that putative promoter P1 was not functional. The activity of the isolated P2 promoter was greatly enhanced by inclusion of the P1-P2 intervening sequence. These studies indicated that gene expression could be an important consideration in understanding resistance phenotypes predicted by genetic signatures in the context of sequencing-based rapid diagnostics.
KEYWORDS: KPC, Tn4401, carbapenem-resistant Enterobacteriaceae, gene expression, meropenem, promoters
INTRODUCTION
Carbapenem-resistant Enterobacteriaceae (CRE) are an urgent public health threat because of their increasing incidence and the high mortality seen with infection (1). Currently, the most commonly reported mechanism of carbapenem resistance in clinical Enterobacteriaceae is Klebsiella pneumoniae carbapenemase (KPC), which has disseminated globally over the last decade, with infections becoming endemic in several geographic locations such as Italy, Greece, and the United States (2). KPC is a class A serine beta-lactamase which can hydrolyze penicillins, cephalosporins, aztreonam, and carbapenems, limiting treatment options in infected patients. The KPC gene (blaKPC) is typically located within a 10-kb mobile transposon (Tn4401), which is most often situated on conjugative plasmids (3, 4). The association of blaKPC with mobile genetic elements such as plasmids and transposons contributes to intraspecies gene transfer and dissemination of carbapenem resistance (2, 5–8).
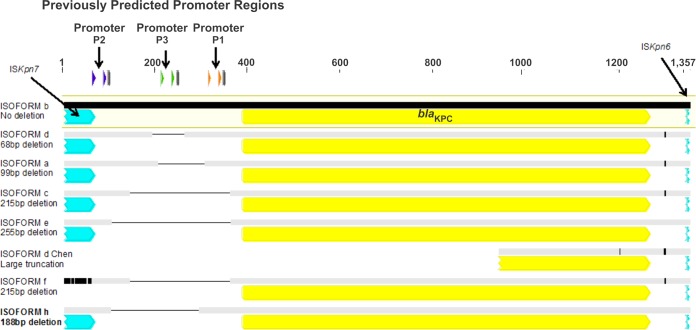
Tn4401 is composed of blaKPC, a transposase gene (tnpA), a resolvase gene (tnpR), and two insertion sequences, ISKpn6 and ISKpn7, all flanked by two 32-bp inverted repeat sequences (3). To date, seven unique Tn4401 isoforms (Tn4401a to Tn4401g) have been identified (9–12), with Tn4401a and Tn4401b being the most widespread (13). The original characterization of these isoforms demonstrated deletions in the noncoding region upstream of blaKPC or lacking genes or both as follows: a, −99 bp (14, 15); b, no deletion (3); c, −215 bp (16); d, −68 bp (9) and −5.3 kb (9, 17, 18); e, −255 bp (17); f, truncated tnpA and lacking tnpR, ISKpn7 left, and Tn4401 IRL-1 (10); g, lacking tnpA, tnpR, and ISKpn7 left, with a −215-bp deletion in the noncoding region (Fig. 1). Most recently, a truncated version of Tn4401e, lacking tnpA and tnpR, was also described (19).
FIG 1.
Structural variation in published isoforms of the Tn4401 transposons. The sequence highlighted in yellow denotes the reference Tn4401b sequence (EU176013). Gray sequence bars denote areas of 100% identity to the reference; black sequence bars denote areas of nucleotide variation.
Previously, the noncoding region between ISKpn7 and blaKPC was proposed to contain three putative promoter regions (P1, P2, and P3); P1 and P2 were demonstrated to affect expression, while P3 was shown to not be a real promoter (5, 8, 20). Deletions in the noncoding region upstream of blaKPC have demonstrated variable expression of KPC in vitro, with the highest levels of expression observed with Tn4401a (presence of P1 and P2). However, the relative contributions of P1 and P2 and regions of associated flanking sequences to KPC expression have not been fully characterized.
Here, we describe a novel Tn4401 transposon variant (Tn4401h) which has a unique 188-bp deletion in the noncoding region upstream of blaKPC and, similarly to Tn4401a, retains promoters P1 and P2. We explore the effect of this 188-bp deletion on the level of KPC expression and the degree of phenotypic resistance compared to the most frequently seen and best characterized isoforms, Tn4401a and Tn4401b (4), and further assess promoter activity using fusions to a lacZ reporter. Understanding the impact of genetic variability in noncoding and/or promoter regions on resistance gene expression and its correlation with MICs will be important for the use of sequencing-based approaches in clinical diagnostics and antimicrobial susceptibility prediction (8, 21).
RESULTS
The molecular epidemiology of Tn4401 in the study isolates.
The Tn4401 sequence was identified in 281 isolates across eight genera, mainly represented by Klebsiella spp. (n = 129), Enterobacter spp. (n = 101), and Citrobacter spp. (n = 32), with blaKPC-2 found in the majority (230/281; 82%) of the isolates as previously characterized (6). The same work also reported that the most common Tn4401 variant was Tn4401b (n = 230); Tn4401a was seen in 8 isolates (all K. pneumoniae), and a novel variant (Tn4401h) (all with blaKPC-2 [Fig. 1]) with a 188-bp deletion was identified in 39 isolates from 28 patients. Tn4401h was first identified in a K. pneumoniae strain (February 2009); all other Tn4401h variants were identified in highly genetically related Enterobacter cloacae isolates from all 28 patients (including the patient with the initial K. pneumoniae isolate with Tn4401h) over 3 years (May 2009 to December 2012), consistent with the idea of local interspecies and interpatient transmission (6).
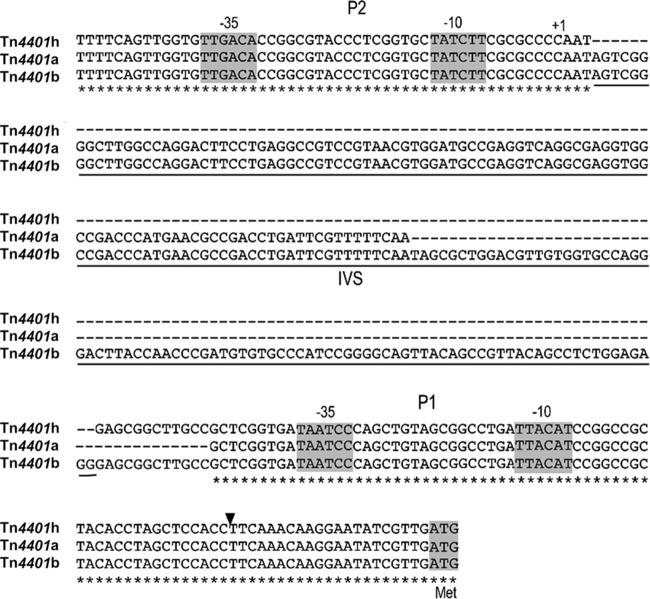
Structural variation in published isoforms of Tn4401 and the novel Tn4401h isoform, including variation in ISKpn6, ISKpn7, and the putative promoter (P1, P2, and P3) regions, is shown in Fig. 1 (9). Notably, isoforms a and h do not contain putative promoter P3 and have nucleotide variation in the intervening sequence (IVS) between P2 and P1 (Fig. 2).
FIG 2.
Alignment of the promoter regions of blaKPC isoforms with putative promoters as defined by Naas et al (20). −35 regions and −10 regions are highlighted in gray. +1 indicates the P2 transcription start site previously identified by RACE (rapid amplification of cDNA ends). Isoforms a and h have deletion differences in the intervening sequence between P2 and P1. The extent of the intervening sequence that is deleted in isoform h is underlined and denoted the IVS. The start codon of blaKPC is highlighted in gray. The arrowhead indicates the site of fusions to the lacZ reporter.
Susceptibility testing.
Susceptibility results for selected parent strains and Escherichia coli transformants from CAV1016, CAV1746, and CAV1438 containing Tn4401b, Tn4401a, and Tn4401h, respectively, are presented in Table 1. Parent strains CAV1746 (Tn4401a) and CAV1438 (Tn4401h) were resistant to cefepime and meropenem by broth dilution and VITEK2 (Table 1). CAV1016 (Tn4401b) was resistant to meropenem by broth dilution and VITEK2 but was susceptible to cefepime by VITEK2.
TABLE 1.
MICs for parent strains CAV1016, CAV1746, and CAV1438 and E. coli transformants AMGH-1, ACGH, and ACGH2 containing plasmids bearing Tn4401b, Tn4401a, and Tn4401h, respectively, from the parent strainsa
| Strain | Tn4401 variant | Plasmid size, PCR replicon type, putative plasmid backbone (reference) | Species | MIC (μg/ml) |
||||
|---|---|---|---|---|---|---|---|---|
| VITEK2 |
Broth microdilution |
|||||||
| Meropenem | Ertapenem | Cefepime | Meropenem | Cefepime | ||||
| CAV1016 | Tn4401b | K. pneumoniae | ≥16 | 4 | 2 | 256 | 64 | |
| CAV1746 | Tn4401a | K. pneumoniae | ≥16 | ≥8 | ≥64 | 1,024 | 512 | |
| CAV1438 | Tn4401h | E. cloacae | ≥16 | ≥8 | ≥64 | 1,024 | 512 | |
| E. coli transformed strain (parent strain) | ||||||||
| AMGH-1 (CAV1016) | Tn4401b | 43 kb, nontypeable, confirmed pKPC_UVA01 (CP017937.1) (22) | E. coli | 0.5 | ≤0.5 | ≤1 | 1 | 8 |
| ACGH (CAV1746) | Tn4401a | >60 kb, IncR, putatively resembles pKPC-484 (CP008798.1) | E. coli | ≥16 | ≥8 | ≥64 | 16 | 128 |
| ACGH2 (CAV1438) | Tn4401h | >70 kb, IncR, putatively resembles pKPC_CAV1176 (CP011661.1) (6) | E. coli | ≥16 | 4 | 4 | 4 | 64 |
MICs were determined using the VITEK2 automated susceptibility testing platform and broth microdilution.
Testing of susceptibility to cefepime and meropenem of E. coli transformants containing Tn4401a or Tn4401h by VITEK2 and broth dilution demonstrated MICs that were all in the resistant range. For the Tn4401b E. coli transformant, cefepime results were in the susceptible dose-dependent (SDD) range and meropenem results were in the susceptible range by broth dilution. By VITEK2, both cefepime and meropenem demonstrated MICs which were in the susceptible range (2015 CLSI breakpoints; 23) (Table 1). The plasmid backbones for each transformant were unique, but both Tn4401h and Tn4401a were on large IncR plasmids whereas Tn4401b was on a nontypeable 43-kb plasmid (pKPC_UVA01) (Table 1).
For parent strains CAV1016, CAV1746, and CAV1438 and for the E. coli transformants, carbapenemase production was indicated by the results of both the modified Hodge test and the indirect carbapenemase test (24).
Expression assays.
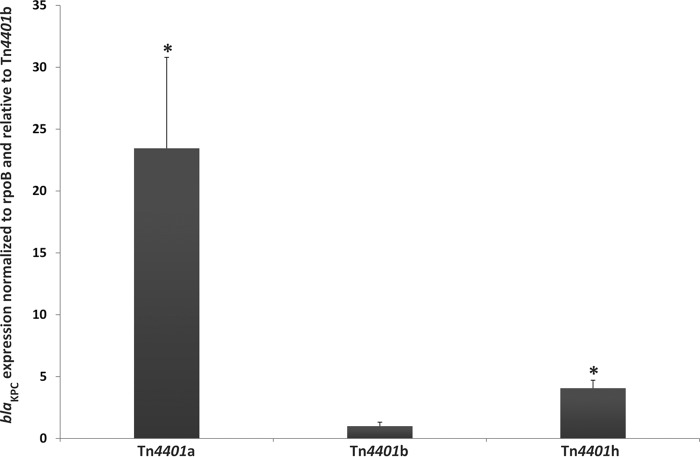
Two-step quantitative real-time PCR (qRT-PCR) was performed on all three E. coli transformants using primers as noted in Table S1 in the supplemental material. Fold differences in blaKPC expression (normalized to rpoB) were quantified relative to Tn4401b. Tn4401a had 23-fold-greater blaKPC expression than Tn4401b (Student t test, P = 0.011) (Fig. 3). Tn4401h had 4-fold-greater blaKPC expression than Tn4401b (Student t test, P = 0.03) (Fig. 3).
FIG 3.
Differences in blaKPC expression normalized to rpoB and relative to Tn4401b. Tn4401a had significantly higher β-galactosidase activity than Tn4401b and Tn4401h (P < 0.05), and the Tn4401h putative promoter sequence had significantly higher β-galactosidase activity than Tn4401b (P < 0.05).
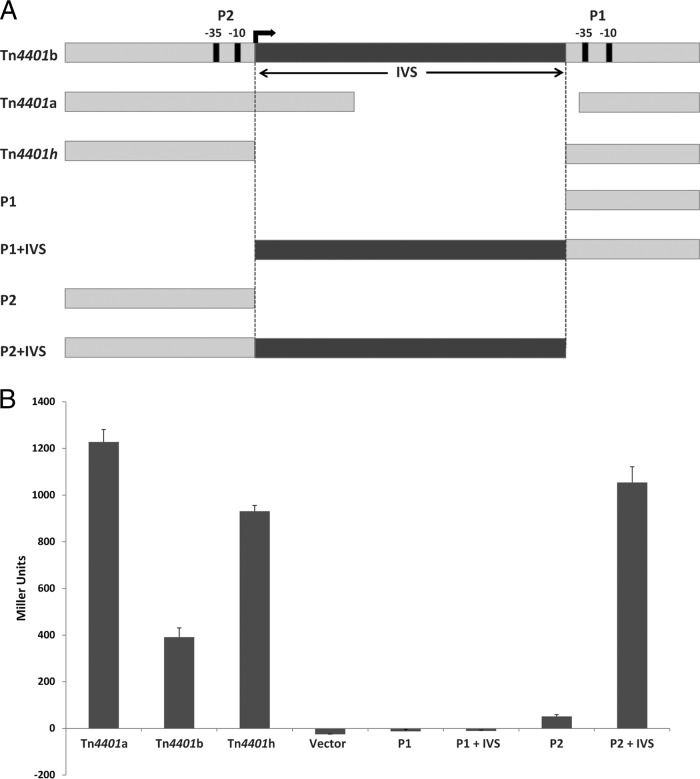
Transcription fusions to the lacZ reporter in plasmid pRS551 (25) of the putative promoter sequences from plasmids carrying Tn4401a, Tn4401b, and Tn4401h (Table S1) (Fig. 4) were used to quantify β-galactosidase activity associated with each Tn4401 variant. The promoter sequence of Tn4401a had significantly higher β-galactosidase activity than Tn4401b (Student t test, P = 0.0001) and Tn4401h (Student t test, P = 0.0002), and the Tn4401h putative promoter sequence had significantly higher β-galactosidase activity than Tn4401b (Student t test, P < 0.0001) (Fig. 4). These results were consistent with the effect that each deletion had on the level of blaKPC mRNA in the transformants (Fig. 3).
FIG 4.
(A) Schematic to explain the constructs, showing the promoter regions cloned in lacZ fusions. The arrow indicates the P2 transcription start site. (B) β-Galactosidase activity level associated with putative promoter regions of Tn4401a, Tn4401b, and Tn4401h isoforms (putative promoters P1, P1+IVS, P2, and P+IVS). lacZ reporter plasmid pRS551 (vector) served as a negative control. Among the reporter constructs, P2 had significantly higher β-galactosidase activity than vector, P1, and P1+IVS (P < 0.05); P2+IVS had significantly higher β-galactosidase activity than P2, P1, and P1+IVS (Student t test, P < 0.05); and promoter sequence Tn4401a had significantly higher β-galactosidase activity than P2+IVS (P < 0.05).
Tn4401h is missing an intervening sequence (IVS) of 188 nucleotides between P1 and P2, whereas Tn4401a carries a smaller deletion in this region (Fig. 2 and 4A). To better discern the contributions of the different sequence domains to gene expression, lacZ fusions were constructed of the isolated P1 and P2 promoters as well as of these promoter regions carrying the IVS. Reporter constructs containing P1 or P1 plus IVS (P1+IVS) showed no β-galactosidase activity. Reporter constructs containing P2 showed significantly higher β-galactosidase activity than those containing vector (Student t test, P = 0.00025) or P1 (Student t test, P = 0.0034) and P1+IVS (Student-t test, P = 0.0020). Reporter constructs containing P2+IVS had significantly higher β-galactosidase activity than those containing P2 alone (Student t test, P = 0.015), P1 (Student t test, P < 0.001), and P1+IVS (Student t test, P < 0.001) (Fig. 4). Additionally, reporter constructs containing promoter sequence Tn4401a had significantly higher β-galactosidase activity than reporter constructs containing P2+IVS (Student t test; P = 0.006).
DISCUSSION
As KPC-producing Enterobacteriaceae are seen in increasing numbers around the globe, understanding expression of blaKPC and its effect on the degree of associated carbapenem resistance could have significant clinical consequences. The Tn4401 transposon has been the most consistently observed unit of blaKPC transfer, and the noncoding region upstream of blaKPC has been described as a regulatory region (3, 22, 26). We describe a novel isoform of Tn4401, Tn4401h, which was first identified in a K. pneumoniae strain in a patient who also had an E. cloacae strain that was subsequently transmitted between several patients over a period of 3 years in our institution.
The 188-bp deletion of Tn4401h, encompassing the region between P1 and P2, demonstrated increased blaKPC expression compared with Tn4401b (no deletions), although to a lesser degree than Tn4401a (99-bp deletion; with a smaller deletion between P1 and P2). To exclude the impact of variability of gene copy number and plasmid background on these results, we directly investigated the impact of promoter regions and intervening sequences on gene expression in a model system by inserting these promoter regions and promoter/intervening sequence combinations into a lacZ reporter.
The Tn4401h promoter region showed significantly higher β-galactosidase activity than Tn4401b, again to a lesser degree than the Tn4401a promoter region and consistent with the blaKPC expression results (Tn4401a → Tn4401h → Tn4401b). Given that both Tn4401a and Tn4401h have deletions in the intervening sequence between P1 and P2, with P1 and P2 intact, the differential levels of gene expression and promoter activity remain incompletely explained. One possible explanation for this variability is that the differing lengths of the intervening sequences between P2 and P1 for Tn4401a (99-bp deletion) and Tn4401h (188-bp deletion) may result in a more stable RNA structure for Tn4401a.
Previously, Naas et al. reported only P1 and P2 as true promoters involved in blaKPC expression. A review of the −10 and −35 regions of the promoter P1 and P2 sequences shows that the P2 promoter (initially identified by Roth [27]) has a strong −35 consensus sequence (TTGACA) and only two minor differences (indicated with lowercase characters) from the canonical −10 sequence (TATctT), while the P1 promoter has a poor consensus sequence, with 4 differences from the canonical −35 sequence (TaatCc) and 3 differences from the canonical −10 sequence (TtacAT). However, the relative contributions of putative promoter P1 by itself and in relation to the IVS remained previously unknown. Hence, promoter sequences for P1 alone and in combination with IVS were cloned into a lacZ reporter plasmid. Surprisingly, the reporter constructs containing P1 and P1+IVS showed no β-galactosidase activity, indicating that this promoter was not active under our assay conditions. We additionally attempted to characterize the relative contributions of promoter P2 alone and in combination with IVS. Higher β-galactosidase activity was observed with the promoter P2 lacZ construct than with P1, and this was further enhanced when IVS was included. These results are consistent with findings of Naas et al. (20). The exact role of IVS in the enhanced activity of P2+IVS compared to P2 will require future evaluation, and the data would ideally reflect variations in IVS length.
E. coli transformants containing Tn4401a and Tn4401h variants on native parent plasmid backgrounds showed meropenem and cefepime resistance MICs compared to E. coli transformants containing Tn4401b, consistent with the variations in blaKPC expression (i.e., Tn4401a → Tn4401h → Tn4401b). Previous studies have suggested that another mechanism, in addition to gene expression and copy number, namely, production of porin channel defects, is a primary contributor to the variability in susceptibility in testing KPC-producing isolates (3, 11, 20, 28–30). Here we excluded the impact of porin channel variation on susceptibility by evaluating plasmids bearing the Tn4401-blaKPC units in a fixed E. coli background. Although we did not exclude gene copy number as a factor contributing to expression, as the different Tn4401 variants were located on unique plasmid backbones, the differences in the levels of promoter activity seen in the β-galactosidase assay align with the differences seen in the E. coli transformants. In addition, Roth et al. previously showed that in blaKPC-2 transformants, blaKPC gene copy number did not correlate with expression and increases in β-lactam MICs; similarly, Kitchel et al. demonstrated that factors other than blaKPC copy number likely contribute to elevated KPC production and high-level carbapenem resistance in certain isolates (26, 27). Taken together, these data suggest that the promoter region upstream of blaKPC appears to have the most substantial effect on expression of carbapenem resistance in the transformants. Future studies utilizing methods such as chromatin immunoprecipitation with sequencing (ChIP-sequencing) could be used to examine the interaction of transcription factors and thus offer further insight into regulation of blaKPC expression.
By characterizing the Tn4401h isoform, we have highlighted that the promoter region may be more complex than previously described. Correctly ascertaining the role of promoter regions and 5′ untranslated regions in resistance gene expression is of particular relevance to clinical diagnostic microbiology given the development of an increasing number of genotype-based approaches (e.g., microarray, multiplex, or whole-genome sequencing) to resistance prediction (31–35). There is a trend toward incorporating rapid genotypic methods into routine laboratory diagnostics, and yet there is increasing recognition of the importance of pharmacokinetics/pharmacodynamics in relation to the MIC. Therefore, understanding the predicted impact of promoter regions on the MIC may be of increasing importance to clinical care.
In summary, we have found a novel isoform, Tn4401h, with a 188-bp deletion in the upstream noncoding region between ISKpn7 and blaKPC. We have shown that this deletion has a differential effect on carbapenem MICs compared with other common Tn4401 variants. This work increases our understanding of the complexity of blaKPC expression and has implications for predicting the degree of resistance based solely on the presence or absence of a particular resistance gene.
MATERIALS AND METHODS
Sampling.
The sampling of blaKPC-positive Enterobacteriaceae clinical and surveillance isolates from the University of Virginia Health System (UVaHS) between August 2007 and December 2012 has been previously published (6). Sheppard et al. performed Illumina sequencing on the collected isolates and determined the Tn4401 isoform of each isolate using BLASTn comparisons between the isolate's de novo assembly and the Tn4401b reference sequence (EU176013.1) (3, 6). Where available, ertapenem MIC results from a VITEK2 AST GN-70 test kit (bioMérieux, Durham, NC) were obtained retrospectively from laboratory records.
Purified plasmid DNA was electroporated into electrocompetent E. coli Genehog cells (Invitrogen, Carlsbad, CA) as previously described (22). To examine the plasmid background in the newly generated transformant incompatibility group, PCR typing was performed (36). For CAV1746 and CAV1438 parent strains (those with unknown blaKPC plasmids), the Illumina contigs containing the corresponding replicon sequences (1,960 bp and 15,241 bp, respectively) and the contigs containing Tn4401 (18,813 bp and 10,011 bp, respectively) were used as BLASTn queries to determine similarity to previously described plasmids in GenBank. For CAV1016, the blaKPC plasmid background has been previously described (22). For plasmid size estimation, BamHI and EcoRI (New England BioLabs, Ipswich, MA) digested and undigested plasmid extractions were run on a 0.8% agarose gel over 8 h at 70 V with V517 and Hyperladder I (Bioline, Taunton, MA) to estimate plasmid size as previously described (37).
Quantitative real-time PCR (qRT-PCR) experiments.
Total RNA was extracted from E. coli transformants containing parent plasmid Tn4401a, Tn4401b, and Tn4401h using an RNeasy minikit (Qiagen, GmBH, Hilden, Germany). RNA products underwent column DNase I digestion (Qiagen) and were quantified by measuring the optical density at 260 nm using a NanoDrop 1000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). cDNA was synthesized from RNA using a commercial method (qScript cDNA Supermix; Quanta Biosciences, Gaithersburg, MD) in accordance with the manufacturer's instructions.
blaKPC gene expression was examined in E. coli transformants utilizing qRT-PCR and the synthesized cDNA (primers are listed in Table S1 in the supplemental material). A 25-ng volume of RNA-equivalent DNA was used in triplicate. qRT-PCR was performed using SsoFast EvaGreen Supermix (Bio-Rad Laboratories, Hercules, CA), template DNA, and 500 nM (each) primer RT-KPC-F/RT-KPC-R or EcoliRPOB-F/EcoliRPOB-R (Table S1). Cycling conditions included a 3-min enzyme activation step at 95°C followed by 40 cycles of melting (95°C for 10 s) and annealing/extension (56.3°C for 30 s/72°C for 30 s) followed by a final extension for 72°C for 1 min. Standard curves were generated for both the target (blaKPC) and the endogenous control (rpoB) using 4-fold dilutions of template DNA at known concentrations (from 0.39 ng to 100 ng) and by plotting the logarithm of the initial quantity of template (along the x axis) versus the respective cycle threshold (CT) values (along the y axis). Thereafter, the amount of blaKPC and rpoB was calculated from the appropriate standard curve for each experimental sample. Expression levels of blaKPC were first normalized relative to the rpoB gene in E. coli, and fold difference in blaKPC expression was calculated as follows: (normalized blaKPC expression in E. coli transformant Tn4401a or Tn4401h)/(normalized blaKPC expression in E. coli transformant Tn4401b).
PCR and cloning.
Online ClustalW software (http://www.ebi.ac.uk/Tools/msa/clustalw2/) was utilized to align promoter regions of isoforms Tn4401a, Tn4401b, and Tn4401h. Primers (F-ISKpn7, R-prom) containing restriction sites for BamHI and EcoRI (New England BioLabs, Ipswich, MA) were used to amplify putative promoter sequences from purified plasmids carrying Tn4401a, Tn4401b, and Tn4401h (Table S1). Primers were also used to amplify putative promoters P1 (P1Eco-F, R-prom), P1 plus the intervening sequence (IVS; i.e., the sequence intervening between P1 and P2 that is missing in Tn4401h) (P1+IVS Eco-F, R-prom), P2 (F-ISKpn7; P2 Bam –R), and P2+IVS (which is in F-ISKpn7; P2+IVS Bam –R), in order to assess the relative activities of putative promoters P1 and P2 alone and in combination with the IVS (Fig. 2 and 4; see also Table S1). All amplified PCR products were ligated into lacZ reporter plasmid pRS551 (25) after restriction digestion was performed using BamHI and EcoRI. The ligated constructs and lacZ reporter plasmid pRS551 were transformed into E. coli DH10B calcium-competent cells and then grown on ampicillin (100 μg/ml)-selective LB plates. Plasmids isolated from transformants were screened by PCR, and all 7 ligated constructs were sequenced using primer (pRS551-R sequencing; Table S1) to confirm the proper orientation and nucleotide sequence of the inserted regions.
β-Galactosidase assay.
The protocol for the assay of β-galactosidase activity was adapted from Griffith and Wolf (38). A 5-μl volume of the overnight culture of the DH10B E. coli transformants was added to 3 ml LB broth containing ampicillin (50 μg/ml), and the cultures were incubated on a platform shaker at 37°C until the culture reached an optical density at 600 nm of 0.4 to 0.5.
For cell permeabilization to release the enzyme, 1 ml of Z buffer (60 mM Na2HPO4 · 7H2O, 40 mM NaH2PO4 · H2O, 10 mM KCl, 1 mM MgSO44 · 7H2O, 50 mM β-mercaptoethanol) was added along with 20 μl of freshly prepared 0.1% SDS and 40 μl of chloroform to100 μl of bacterial culture. Permeabilization was achieved by subjecting test tubes containing the cell-chloroform/SDS mixture to vortex mixing (one at a time) for 30 s followed by a 10-min settling period.
A 100-μl volume of supernatant from each tube was divided into aliquots in quadruplicate and placed into a flat-bottom 96-well plate. At time zero, the assay was initiated by using a multichannel pipette to add 20 μl of o-nitrophenyl-β-d-galactopyranoside (ONPG; 4 mg/ml). Once the yellow chromophore was visualized, the reaction was terminated by the addition of 50 μl of 1 M Na2CO3 and then absorbance at A420 and A550 was measured. β-Galactosidase activity was calculated in Miller units from the absorbance data as follows: Miller units = 1,000 × [A420 − (1.75 × A550)]/(T × V × A600). A420 and A550 are read from the reaction mixture, A600 reflects cell density in the mid-logarithmic phase, T represents the time of the reaction in minutes before addition of 1 M Na2CO3, and V represents the volume of the culture used in the assay in milliliters (38).
Statistical analysis.
A paired-sample Student t test was conducted to compare the levels of blaKPC expression of E. coli transformants containing parent plasmids Tn4401a, Tn4401b, and Tn4401h. A paired-sample Student t test was also used to compare the levels of β-galactosidase activity of lacZ fusion constructs containing Tn4401a, Tn4401b, Tn4401h, P1, P1+IVS, P2, or P2+IVS.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Clinical Microbiology laboratory at UVaHS for their outstanding contributions to this study.
This research was conducted with support from a NIH T32 training grant (NIH grant 3185T32AI007 046-37). E.W. was supported by the Summer Research Internship Program (SRIP) and the University of Virginia School of Medicine. This work was also supported by the National Institute for Health Research (NIHR grant G0800778) and by the NIHR Oxford Biomedical Research Centre (N.S., A.E.S., and D.W.C.).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00025-17.
REFERENCES
- 1.Centers for Disease Control and Prevention (CDC). 2013. Antibiotic resistance threats in the United States. CDC, Atlanta, GA, USA. [Google Scholar]
- 2.Mathers AJ, Peirano G, Pitout JD. 2015. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev 28:565–591. doi: 10.1128/CMR.00116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Naas T, Cuzon G, Villegas MV, Lartigue MF, Quinn JP, Nordmann P. 2008. Genetic structures at the origin of acquisition of the beta-lactamase bla KPC gene. Antimicrob Agents Chemother 52:1257–1263. doi: 10.1128/AAC.01451-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowers JR, Kitchel B, Driebe EM, MacCannell DR, Roe C, Lemmer D, de Man T, Rasheed JK, Engelthaler DM, Keim P, Limbago BM. 2015. Genomic analysis of the emergence and rapid global dissemination of the clonal group 258 Klebsiella pneumoniae pandemic. PLoS One 10:e0133727. doi: 10.1371/journal.pone.0133727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chmelnitsky I, Shklyar M, Leavitt A, Sadovsky E, Navon-Venezia S, Ben Dalak M, Edgar R, Carmeli Y. 2014. Mix and match of KPC-2 encoding plasmids in Enterobacteriaceae-comparative genomics. Diagn Microbiol Infect Dis 79:255–260. doi: 10.1016/j.diagmicrobio.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 6.Sheppard AE, Stoesser N, Wilson DJ, Sebra R, Kasarskis A, Anson LW, Giess A, Pankhurst LJ, Vaughan A, Grim CJ, Cox HL, Yeh AJ; Modernising Medical Microbiology (MMM) Informatics Group, Sifri CD, Walker AS, Peto TE, Crook DW, Mathers AJ. 2016. Nested Russian doll-like genetic mobility drives rapid dissemination of the carbapenem resistance gene blaKPC. Antimicrob Agents Chemother 60:3767–3778. doi: 10.1128/AAC.00464-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luo Y, Yang J, Ye L, Guo L, Zhao Q, Chen R, Chen Y, Han X, Zhao J, Tian S, Han L. 2014. Characterization of KPC-2-producing Escherichia coli, Citrobacter freundii, Enterobacter cloacae, Enterobacter aerogenes, and Klebsiella oxytoca isolates from a Chinese hospital. Microb Drug Resist 20:264–269. doi: 10.1089/mdr.2013.0150. [DOI] [PubMed] [Google Scholar]
- 8.Cuzon G, Naas T, Nordmann P. 2011. Functional characterization of Tn4401, a Tn3-based transposon involved in blaKPC gene mobilization. Antimicrob Agents Chemother 55:5370–5373. doi: 10.1128/AAC.05202-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen L, Chavda KD, Mediavilla JR, Jacobs MR, Levi MH, Bonomo RA, Kreiswirth BN. 2012. Partial excision of blaKPC from Tn4401 in carbapenem-resistant Klebsiella pneumoniae. Antimicrob Agents Chemother 56:1635–1638. doi: 10.1128/AAC.06182-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bryant KA, Van Schooneveld TC, Thapa I, Bastola D, Williams LO, Safranek TJ, Hinrichs SH, Rupp ME, Fey PD. 2013. KPC-4 is encoded within a truncated Tn4401 in an IncL/M plasmid, pNE1280, isolated from Enterobacter cloacae and Serratia marcescens. Antimicrob Agents Chemother 57:37–41. doi: 10.1128/AAC.01062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pecora ND, Li N, Allard M, Li C, Albano E, Delaney M, Dubois A, Onderdonk AB, Bry L. 2015. Genomically informed surveillance for carbapenem-resistant Enterobacteriaceae in a health care system. mBio 6:e01030. doi: 10.1128/mBio.01030-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baraniak A, Izdebski R, Fiett J, Herda M, Derde LP, Bonten MJ, Adler A, Carmeli Y, Goossens H, Hryniewicz W, Brun-Buisson C, Gniadkowski M; MOSAR WP2, WP3, and WP5 Study Groups. 2015. KPC-like carbapenemase-producing Enterobacteriaceae colonizing patients in Europe and Israel. Antimicrob Agents Chemother 60:1912–1917. doi: 10.1128/AAC.02756-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, Kumarasamy K, Livermore DM, Maya JJ, Nordmann P, Patel JB, Paterson DL, Pitout J, Villegas MV, Wang H, Woodford N, Quinn JP. 2013. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis 13:785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Curiao T, Morosini MI, Ruiz-Garbajosa P, Robustillo A, Baquero F, Coque TM, Canton R. 2010. Emergence of bla KPC-3-Tn4401a associated with a pKPN3/4-like plasmid within ST384 and ST388 Klebsiella pneumoniae clones in Spain. J Antimicrob Chemother 65:1608–1614. doi: 10.1093/jac/dkq174. [DOI] [PubMed] [Google Scholar]
- 15.Gootz TD, Lescoe MK, Dib-Hajj F, Dougherty BA, He W, Della-Latta P, Huard RC. 2009. Genetic organization of transposase regions surrounding blaKPC carbapenemase genes on plasmids from Klebsiella strains isolated in a New York City hospital. Antimicrob Agents Chemother 53:1998–2004. doi: 10.1128/AAC.01355-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cuzon G, Naas T, Truong H, Villegas MV, Wisell KT, Carmeli Y, Gales AC, Venezia SN, Quinn JP, Nordmann P. 2010. Worldwide diversity of Klebsiella pneumoniae that produce beta-lactamase blaKPC-2 gene. Emerg Infect Dis 16:1349–1356. doi: 10.3201/eid1609.091389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kitchel B, Rasheed JK, Patel JB, Srinivasan A, Navon-Venezia S, Carmeli Y, Brolund A, Giske CG. 2009. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob Agents Chemother 53:3365–3370. doi: 10.1128/AAC.00126-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen L, Chavda KD, Fraimow HS, Mediavilla JR, Melano RG, Jacobs MR, Bonomo RA, Kreiswirth BN. 2013. Complete nucleotide sequences of blaKPC-4- and blaKPC-5-harboring IncN and IncX plasmids from Klebsiella pneumoniae strains isolated in New Jersey. Antimicrob Agents Chemother 57:269–276. doi: 10.1128/AAC.01648-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez TMI, Vazquez GJ, Aquino EE, Robledo IE. 2016. Genetic environment of the KPC gene in Acinetobacter baumanii ST2 clone from Puerto Rico and genomic insights into its drug resistance. J Med Microbiol 65:784–792. doi: 10.1099/jmm.0.000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naas T, Cuzon G, Truong HV, Nordmann P. 2012. Role of ISKpn7 and deletions in blaKPC gene expression. Antimicrob Agents Chemother 56:4753–4759. doi: 10.1128/AAC.00334-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kos VN, Déraspe M, McLaughlin RE, Whiteaker JD, Roy PH, Alm RA, Corbeil J, Gardner H. 2015. The resistome of Pseudomonas aeruginosa in relationship to phenotypic susceptibility. Antimicrob Agents Chemother 59:427–436. doi: 10.1128/AAC.03954-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathers AJ, Stoesser N, Sheppard AE, Pankhurst L, Giess A, Yeh AJ, Didelot X, Turner SD, Sebra R, Kasarskis A, Peto T, Crook D, Sifri CD. 2015. Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae at a single institution: insights into endemicity from whole-genome sequencing. Antimicrob Agents Chemother 59:1656–1663. doi: 10.1128/AAC.04292-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.CLSI. 2015. Performance standards for antimicrobial susceptibility testing. M100-S19. Clinical and Laboratory Standards Institute, Wayne, PA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mathers AJ, Carroll J, Sifri CD, Hazen KC. 2013. Modified Hodge test versus indirect carbapenemase test: prospective evaluation of a phenotypic assay for detection of Klebsiella pneumoniae carbapenemase (KPC) in Enterobacteriaceae. J Clin Microbiol 51:1291–1293. doi: 10.1128/JCM.03240-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simons RW, Houman F, Kleckner N. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 26.Kitchel B, Rasheed JK, Endimiani A, Hujer AM, Anderson KF, Bonomo RA, Patel JB. 2010. Genetic factors associated with elevated carbapenem resistance in KPC-producing Klebsiella pneumoniae. Antimicrob Agents Chemother 54:4201–4207. doi: 10.1128/AAC.00008-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roth AL, Kurpiel PM, Lister PD, Hanson ND. 2011. bla(KPC) RNA expression correlates with two transcriptional start sites but not always with gene copy number in four genera of Gram-negative pathogens. Antimicrob Agents Chemother 55:3936–3938. doi: 10.1128/AAC.01509-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tavares CP, Pereira PS, Marques EDA, Faria C Jr, Araújo Herkenhoff de Souza MDP, de Almeida R, Alves CDF, Asensi MD, D'Alincourt Carvalho-Assef AP. 2015. Molecular epidemiology of KPC-2-producing Enterobacteriaceae (non-Klebsiella pneumoniae) isolated from Brazil. Diagn Microbiol Infect Dis 82:326–330. doi: 10.1016/j.diagmicrobio.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 29.Shen P, Wei Z, Jiang Y, Du X, Ji S, Yu Y, Li L. 2009. Novel genetic environment of the carbapenem-hydrolyzing beta-lactamase KPC-2 among Enterobacteriaceae in China. Antimicrob Agents Chemother 53:4333–4338. doi: 10.1128/AAC.00260-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Queenan AM, Bush K. 2007. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev 20:440–458, table of contents. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ellington MJ, Findlay J, Hopkins KL, Meunier D, Alvarez-Buylla A, Horner C, McEwan A, Guiver M, McCrae LX, Woodford N, Hawkey P. 2016. Multicentre evaluation of a real-time PCR assay to detect genes encoding clinically relevant carbapenemases in cultured bacteria. Int J Antimicrob Agents 47:151–154. doi: 10.1016/j.ijantimicag.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 32.Chen L, Mediavilla JR, Endimiani A, Rosenthal ME, Zhao Y, Bonomo RA, Kreiswirth BN. 2011. Multiplex real-time PCR assay for detection and classification of Klebsiella pneumoniae carbapenemase gene (bla KPC) variants. J Clin Microbiol 49:579–585. doi: 10.1128/JCM.01588-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poirel L, Walsh TR, Cuvillier V, Nordmann P. 2011. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis 70:119–123. doi: 10.1016/j.diagmicrobio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 34.Adler A, Khabra E, Chmelnitsky I, Giakkoupi P, Vatopoulos A, Mathers AJ, Yeh AJ, Sifri CD, De Angelis G, Tacconelli E, Villegas MV, Quinn J, Carmeli Y. 2014. Development and validation of a multiplex PCR assay for identification of the epidemic ST-258/512 KPC-producing Klebsiella pneumoniae clone. Diagn Microbiol Infect Dis 78:12–15. doi: 10.1016/j.diagmicrobio.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 35.Dallenne C, Da Costa A, Decre D, Favier C, Arlet G. 2010. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J Antimicrob Chemother 65:490–495. doi: 10.1093/jac/dkp498. [DOI] [PubMed] [Google Scholar]
- 36.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 37.Mathers AJ, Cox HL, Kitchel B, Bonatti H, Brassinga AK, Carroll J, Scheld WM, Hazen KC, Sifri CD. 2011. Molecular dissection of an outbreak of carbapenem-resistant enterobacteriaceae reveals intergenus KPC carbapenemase transmission through a promiscuous plasmid. mBio 2:e00204-11. doi: 10.1128/mBio.00204-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Griffith KL, Wolf RE Jr. 2002. Measuring beta-galactosidase activity in bacteria: cell growth, permeabilization, and enzyme assays in 96-well arrays. Biochem Biophys Res Commun 290:397–402. doi: 10.1006/bbrc.2001.6152. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.