ABSTRACT
We investigated the genetic backbones of 14 blaCTX-M-8-positive Escherichia coli isolates recovered from human stool samples and chicken meat. All isolates carried IncI1 plasmids with blaCTX-M-8 (blaCTX-M-8/IncI1), and most (9/14) belonged to a specific genetic lineage, namely, plasmid sequence type 113 (pST113). The genetic contexts of the nine blaCTX-M-8/IncI1 pST113 plasmids were similar, regardless of the source. These results suggest the probable local transfer of blaCTX-M-8/IncI1 between humans and chickens with genetically diverse E. coli.
KEYWORDS: blaCTX-M-8, IncI1 plasmid, Escherichia coli, retail chicken meat, human
TEXT
Escherichia coli isolates harboring CTX-M-type extended-spectrum β-lactamase (ESBL) genes have become a global concern because they are widely disseminated in clinical settings, livestock, healthy humans, companion animals, and wild animals (1). CTX-M-type ESBL-producing E. coli isolates in livestock/retail meat need special attention because food contamination could be a major cause of their transfer to humans (2, 3). To assess local transmission of the CTX-M-type ESBL gene between livestock/retail meat and humans in Japan, we analyzed the genetic backbones of CTX-M-8-producing E. coli, since its spread is still expected to be limited, at least in Japan (4–6). As described below, our findings suggest the possible horizontal transfer of plasmids of specific genetic lineages bearing the CTX-M-8 β-lactamase gene between humans and chicken meat.
We collected CTX-M-type ESBL-producing E. coli from several sources, such as ill patients, healthy people handling food (including employees of retail meat shops and meat producers), and retail foods (chicken meat, beef, and pork) (4, 7, 8). The isolates from ill patients were collected from hospitals spread across Japan, while those from healthy people handling food and retail foods were collected in Aichi Prefecture, Japan (4, 7, 8). Only 14 CTX-M-8-producing E. coli isolates were identified. Six were from stool samples from healthy food handlers, and eight were from imported chicken meat from Brazil (Table 1). All isolates were resistant to cefotaxime but susceptible to ceftazidime, imipenem, gentamicin, and fosfomycin (Table 1). We performed whole-genome sequencing (WGS) analysis of 14 CTX-M-8-producing E. coli isolates with the MiSeq platform and an A5-miseq assembler to investigate their genetic backbones (9). Multilocus sequence typing (MLST) was performed by transferring the WGS data to the MLST 1.8 server (10), and the presence of antibiotic resistance genes was confirmed by transferring the WGS data to the ResFinder 2.1 server (11). MLST showed highly diverse backbones; 14 isolates were classified into 12 different sequence types (STs), although strain ST131 was found in both human stool samples and chicken meat and ST1144 was found in stool samples from two different people (Table 1). The susceptibility-testing results and carriage of antibiotic resistance genes were quite consistent (Table 1). The replicon types of plasmids carried by 14 CTX-M-8 producers were confirmed by transferring the WGS data to the PlasmidFinder 1.3 server (12). The numbers of plasmids carried by CTX-M-8 producers were estimated by S1 nuclease pulsed-field gel electrophoresis (PFGE) analysis (13) and simple agarose gel electrophoresis of plasmids extracted with the Plasmid Miniprep System (Promega) (Table 1). All 14 CTX-M-8 producers had IncI1 plasmids, as well as several plasmids with different incompatibility groups (Table 1).
TABLE 1.
Characteristics of 14 E. coli isolates carrying blaCTX-M-8a
| Sample | Source | ST | CTX resistance transfer by conjugation | Plasmid replicon type | No. of plasmids | MIC (μg/ml) |
Antimicrobial resistance gene(s) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTX | CAZ | IPM | GM | TC | CP | FOM | CIP | |||||||
| HU23 | Human stool sample | 131 | Yes | IncI1, IncFIB, IncFIC (FII) | 3 | 8 | 0.5 | 0.25 | 0.5 | 0.5 | 4 | 0.5 | 0.25 | blaCTX-M-8 |
| HU447 | Human stool sample | 1144 | Yes | IncI1, Col156, IncFIC (FII), IncFIB, IncQ1 | 5 | 4 | 2 | 0.06 | 1 | 64 | 4 | 0.5 | ≤0.03 | blaCTX-M-8, blaTEM-1B, strA/B, sul2, tetA, dfrA8 |
| HU476 | Human stool sample | 1144 | Yes | IncI1, Col156, IncFIC (FII), IncFIB, IncQ1 | 5 | 8 | 0.5 | 0.12 | 0.5 | 64 | 8 | 0.5 | ≤0.03 | blaCTX-M-8, blaTEM-1B, strA/B, sul2, tetA, dfrA8 |
| HU485 | Human stool sample | 23 | Yes | IncI1, ColpVC, IncFIB | 3 | 8 | 0.5 | 0.25 | 0.5 | 1 | 4 | 1 | ≤0.03 | blaCTX-M-8 |
| HU493 | Human stool sample | 1170 | Yes | IncI1, IncFIB, IncFIA, IncFIC (FII) | 3 | 8 | 1 | 0.25 | 1 | 1 | 4 | 1 | 0.12 | blaCTX-M-8 |
| HU590 | Human stool sample | 2526 | Yes | IncI1, IncFIB (K) | 2 | 8 | 1 | 0.25 | 0.5 | 64 | 8 | 1 | 0.25 | blaCTX-M-8, aadA5, sul2, tetA, dfrA17 |
| CH11 | Chicken meat | 345 | Yes | IncI1, IncFII, IncFIB, IncQ1, IncN | 5 | 8 | 0.5 | 0.12 | 0.5 | 64 | 4 | 0.5 | 0.25 | blaCTX-M-8, blaTEM-1B, aadA1, strA/B, sul1, sul2, tetA, dfrA1 |
| CH41 | Chicken meat | 351 | Yes | IncI1, IncQ1, IncFII, IncFIB | 3 | 8 | 1 | 0.25 | 0.5 | 64 | 4 | 0.5 | 0.25 | blaCTX-M-8, aadA1, strA/B, sul1, sul2, tetA, dfrA1 |
| CH42 | Chicken meat | 88 | Yes | IncI1, IncX1, IncFIC (FII), IncFIB | 4 | 8 | 1 | 0.25 | 0.5 | 128 | 8 | 1 | 0.25 | blaCTX-M-8, blaTEM-1B, tetA |
| CH49b | Chicken meat | 224 | Yes | IncI1, IncX4, ColpVC, IncFII, IncY, IncX1, IncFIB | 7 | 8 | 1 | 0.25 | 1 | 128 | 64 | 1 | 32 | blaCTX-M-8, aadA1, aadA2, aph(3′)-Ia, cmlA1, sul3, tetA, dfrA12 |
| CH56 | Chicken meat | 131 | No | IncI1, IncFIB, IncFIC(FII), IncX1 | 4 | 8 | 0.5 | 0.25 | 0.5 | 0.5 | 4 | 0.5 | 0.25 | blaCTX-M-8, blaTEM-1B |
| CH110 | Chicken meat | 4576 | Yes | IncI1, IncFII, IncQ1, IncFIB | 4 | 8 | 0.5 | 0.25 | 0.5 | 32 | 4 | 1 | ≤0.03 | blaCTX-M-8, blaTEM-1B, aadA1, strA/B, sul1, sul2, tetA, dfrA1 |
| CH365 | Chicken meat | 602 | Yes | IncI1, Col156, IncFIB, IncFIC (FII) | 4 | 8 | 1 | 0.25 | 0.5 | 0.5 | 4 | 1 | ≤0.03 | blaCTX-M-8 |
| CH407 | Chicken meat | 101 | Yes | IncI1, IncFIB, IncFIC(FII) | 2 | 8 | 0.5 | 0.12 | 0.5 | 1 | 8 | 1 | 0.25 | blaCTX-M-8 |
Abbreviations: CTX, cefotaxime; CAZ, ceftazidime; IPM, imipenem; GM, gentamicin; TC, tetracycline; CP, chloramphenicol; FOM, fosfomycin; CIP, ciprofloxacin.
Strain CH49 has amino acid substitutions S83L and D87N in GyrA and S80I in ParC, which confer ciprofloxacin resistance.
A broth-mating conjugation experiment was performed to transfer the cefotaxime resistance phenotype of 14 CTX-M-8 producers to E. coli J53 (azide resistant), and 13 conjugants were selected on Luria-Bertani (LB) agar plates containing sodium azide (150 μg/ml) and cefotaxime (1 μg/ml) (Table 1). Further, plasmids were extracted from 14 CTX-M-8 producers and introduced into the E. coli DH10B strain by electroporation. Fourteen cefotaxime-resistant E. coli DH10B transformants were selected on LB agar plates containing cefotaxime (1 μg/ml) (Table 2). As expected, blaCTX-M-8 was detected in these cefotaxime-resistant conjugants and transformants. The plasmids were extracted from 14 E. coli DH10B transformants with the Qiagen Plasmid Midi kit and subjected to PFGE. DNA bands corresponding to the plasmids were extracted and used as a DNA template for WGS analysis as described above. The assembled contigs derived from the plasmids were transferred to the PlasmidFinder 1.3 and ResFinder 2.1 servers to investigate the replicon types and presence of antibiotic resistance genes, respectively (11), and plasmid MLST was performed through the pMLST 1.4 server (12). Although the sizes of the 14 plasmids, which were estimated by S1 nuclease PFGE analysis of 14 cefotaxime-resistant E. coli DH10B transformants, varied from 82 to 105 kbp, these plasmids were assigned to the IncI1 group and carried blaCTX-M-8 as the only antibiotic resistance gene (Table 2). The 14 IncI1 plasmids were assigned to five plasmid STs (pSTs); 9 were pST113, 1 was pST114, 2 were pST131, 1 was pST132, and 1 was pST235 (newly assigned) (Table 2). IncI1 pST113 plasmids were dominant in E. coli isolates from humans (3/6) and retail chicken meat (6/8) (Table 2). These results indicated the possibility that the blaCTX-M-8 spread in E. coli in Japan was mainly due to the horizontal transfer of IncI1 plasmids belonging to a specific genetic lineage, such as pST113, regardless of their sources, rather than due to the distribution of a clonal E. coli strain producing CTX-M-8. To date, CTX-M-8-producing E. coli isolates have been found in Germany (14), French Guiana (15), Tunisia (16), Kenya (17), Spain (18), and Brazil (19–21), and IncI1 pST113 plasmids harboring blaCTX-M-8 have been reported (22, 23). Preferential carriage of blaCTX-M-8/IncI1 pST113 plasmids has also been reported in CTX-M-8-producing Enterobacteriaceae isolates, including E. coli and Salmonella spp. in Germany, whose carriage may be related to contaminated food (14). The carriage of IncI1 plasmids pST114, pST131, and pST132 is lower than that of IncI1 pST113 in this study, and these plasmids were found in CTX-M-8 producers from both humans and poultry in Brazil (22, 23). Worldwide dissemination of the blaCTX-M-8 gene might also be mediated by specific IncI1 plasmids such as pST113 and less-well-known plasmids pST114, pST131, and pST132.
TABLE 2.
Characteristics of 14 E. coli transformants carrying IncI1 plasmids with blaCTX-M-8
| Transformant | Plasmid replicon type | Plasmid ST | Approximate plasmid size estimated by S1 PFGE | MIC (μg/ml) of cefotaxime | β-Lactamase gene |
|---|---|---|---|---|---|
| E. coli(pHU23) | IncI1 | pST113 | 91,831 bpa | 8 | blaCTX-M-8 |
| E. coli(pHU447) | IncI1 | pST131 | 90 kb | 4 | blaCTX-M-8 |
| E. coli(pHU476) | IncI1 | pST131 | 91 kb | 8 | blaCTX-M-8 |
| E. coli(pHU485) | IncI1 | pST114 | 82 kb | 8 | blaCTX-M-8 |
| E. coli(pHU493) | IncI1 | pST113 | 94 kb | 8 | blaCTX-M-8 |
| E. coli(pHU590) | IncI1 | pST113 | 88 kb | 8 | blaCTX-M-8 |
| E. coli(pCH11) | IncI1 | pST113 | 101,377 bpa | 8 | blaCTX-M-8 |
| E. coli(pCH41) | IncI1 | pST113 | 87 kb | 8 | blaCTX-M-8 |
| E. coli(pCH42) | IncI1 | pST113 | 86 kb | 8 | blaCTX-M-8 |
| E. coli(pCH49) | IncI1 | pST113 | 84 kb | 8 | blaCTX-M-8 |
| E. coli(pCH56) | IncI1 | pST113 | 87 kb | 8 | blaCTX-M-8 |
| E. coli(pCH110) | IncI1 | pST235 | 92 kb | 8 | blaCTX-M-8 |
| E. coli(pCH365) | IncI1 | pST132 | 105 kb | 8 | blaCTX-M-8 |
| E. coli(pCH407) | IncI1 | pST113 | 83 kb | 8 | blaCTX-M-8 |
| E. coli DH10B | 0.06 |
Plasmid size was determined by WGS analysis, gap-closing PCR, and subsequent Sanger sequencing.
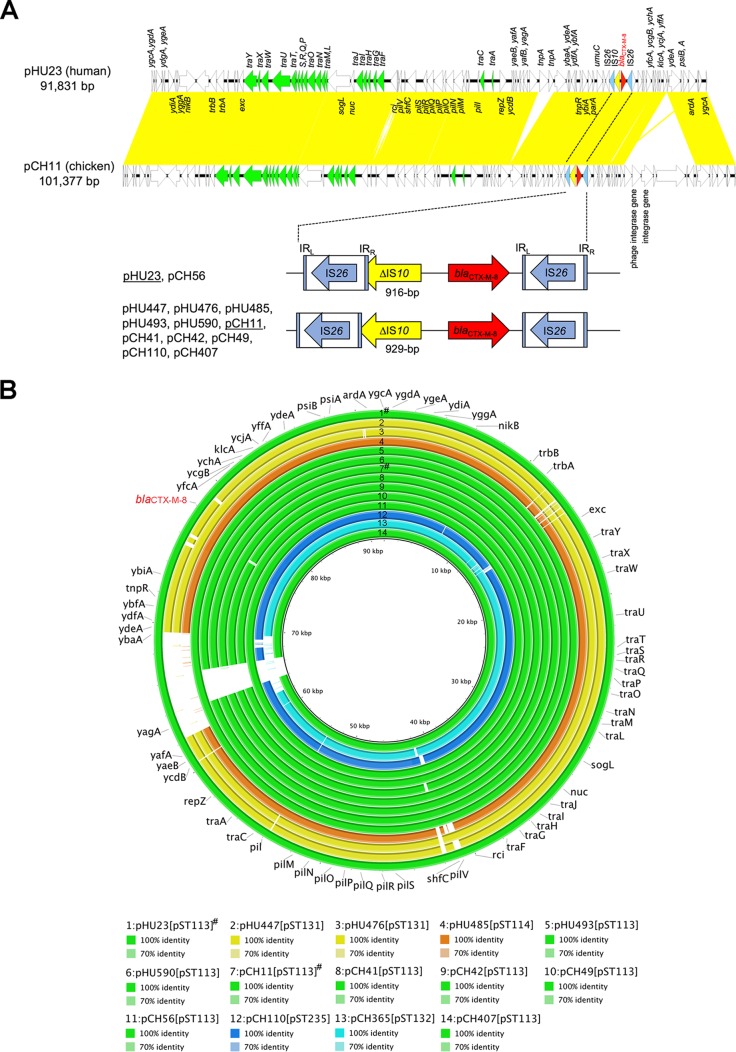
To further evaluate the genetic backbones of blaCTX-M-8/IncI1 plasmids from humans and chicken meat, we determined the complete nucleotide sequences of representative blaCTX-M-8/IncI1 pST113 plasmids, pHU23 from humans, and pCH11 from chicken meat by gap-closing PCR and Sanger sequencing based on the draft sequences of these plasmids. The plasmid sequences were submitted to the Microbial Genome Annotation Pipeline (http://www.migap.org) for annotations. Figure 1A was prepared on the basis of the complete sequences of pHU23 and pCH11 with Easyfig (24). The backbones of the plasmids, including the protein-coding genes traA to traY responsible for plasmid transfer and the protein-coding genes pilI to pilV responsible for pilus formation, were identical, and both had no antibiotic resistance gene, except for blaCTX-M-8 (Fig. 1A). The nucleotide sequence of the pHU23 plasmid showed 97% query coverage and >99% nucleotide identity to that of pCH11. Both plasmids were slightly different in terms of the presence or absence of several putative transposase and integrase genes and hypothetical protein genes (Fig. 1A). Comparison of 14 IncI1/blaCTX-M-8 plasmids (pHU23 and pCH11 with complete sequences and 12 plasmids with draft sequences) was performed on the basis of the complete sequence of the pHU23 plasmid with BRIG software (25), and nine blaCTX-M-8/IncI1 pST113 plasmids showed high similarity, regardless of the source (Fig. 1B).
FIG 1.
(A) Genetic comparison of the pHU23 plasmid (GenBank accession no. AP017892) with the pCH11 plasmid (GenBank accession no. AP017893). The open reading frames are represented by arrows and color coded according to their functions. blaCTX-M-8 is red. Insertion sequences ΔIS10 and IS26 are yellow and blue, respectively. The tra region is green. Yellow shading indicates regions with high genetic identity. IRL, left inverted repeat; IRR, right inverted repeat. (B) Comparison of 14 blaCTX-M-8/IncI1plasmids by using BRIG software. The comparison was performed on the basis of the pHU23 plasmid (91,831 bp), whose nucleotide sequences were completely determined. Color coding is based on pST types as follows: green, pST113; yellow, pST131; orange, pST114; blue, pST235; cyan, pST132. Plasmids marked with the symbol # were completely sequenced, while those with draft sequences are not marked.
In addition, a common IS10 element with a partially truncated 3′ end was upstream of the blaCTX-M-8 gene, although its location slightly differed between pHU23 and pCH11 (Fig. 1A). The blaCTX-M-8 gene was flanked by two IS26 elements. Although the WGS analyses of the remaining 12 IncI1 plasmids could not determine the extended genetic region around blaCTX-M-8, considering the corresponding regions of these plasmids, the genetic region around blaCTX-M-8 of pCH56 was identical to that of pHU23 with 916-bp ΔΙS10, while those around blaCTX-M-8 of pHU447, pHU476, pHU485, pHU493, pHU590, pCH41, pCH42, pCH49, pCH110, and pCH407 were identical to that of pCH11 with 929-bp ΔΙS10 (Fig. 1A). The genetic context around blaCTX-M-8 in the pCH365 plasmid could not be categorized because the terminal end of the contigs carrying blaCTX-M-8 neighbored the middle of the IS10 element. However, the assembled 2,158-bp sequence of the contigs was the same as that of pHU23 and pCH11. Therefore, the DNA sequence around blaCTX-M-8 showed low diversity among the 14 IncI1 plasmids analyzed, as well as low overall diversity (Fig. 1B), indicating that the E. coli isolates from healthy individuals and retail chicken meat had blaCTX-M-8/IncI1 plasmids with almost the same sequences. These results can potentially explain the possible horizontal transfer of blaCTX-M-8/IncI1 plasmids with specific genetic lineages between humans and retail chicken meat.
In conclusion, this study is the first to identify and evaluate the genetic relatedness of CTX-M-8-producing E. coli derived from different origins (i.e., humans and retail chicken meat), and we revealed the possible horizontal transfer of blaCTX-M-8/IncI1 plasmids with a specific genetic lineage, such as pST113. In Japan, CTX-M-8-producing E. coli has been mainly found in retail chicken meat imported from Brazil (4, 26) but has rarely been found in other sources such as patients in clinical settings and livestock (5, 6, 8). Our findings suggest that carriage of CTX-M-8-producing E. coli in humans might be attributed to the horizontal transfer of blaCTX-M-8/IncI1 harbored by genetically diverse E. coli lineages through imported chicken meat. The food handlers analyzed in this study might have acquired CTX-M-8-producing E. coli and/or its blaCTX-M-8/IncI1 plasmids by handling chicken meat. The carriage of antibiotic resistance genes by E. coli in retail meat should be regularly and carefully monitored to prevent their further dissemination to humans.
Accession number(s).
The complete nucleotide sequences of pHU23 from healthy humans and pCH11 from chicken meat were deposited in the DDBJ database under accession numbers AP017892 and AP017893, respectively.
ACKNOWLEDGMENT
This study was supported by grants from the Food Safety Commission, Cabinet Office, Government of Japan (Research Program for Risk Assessment Study on Food Safety, no. 1504).
REFERENCES
- 1.Woerther PL, Burdet C, Chachaty E, Andremont A. 2013. Trends in human fecal carriage of extended-spectrum β-lactamases in the community: toward the globalization of CTX-M. Clin Microbiol Rev 26:744–758. doi: 10.1128/CMR.00023-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dahms C, Hubner NO, Kossow A, Mellmann A, Dittmann K, Kramer A. 2015. Occurrence of ESBL-producing Escherichia coli in livestock and farm workers in Mecklenburg-Western Pomerania, Germany. PLoS One 10:e0143326. doi: 10.1371/journal.pone.0143326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Been M, Lanza VF, de Toro M, Scharringa J, Dohmen W, Du Y, Hu J, Lei Y, Li N, Tooming-Klunderud A, Heederik DJ, Fluit AC, Bonten MJ, Willems RJ, de la Cruz F, van Schaik W. 2014. Dissemination of cephalosporin resistance genes between Escherichia coli strains from farm animals and humans by specific plasmid lineages. PLoS Genet 10:e1004776. doi: 10.1371/journal.pgen.1004776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kawamura K, Goto K, Nakane K, Arakawa Y. 2014. Molecular epidemiology of extended-spectrum β-lactamases and Escherichia coli isolated from retail foods including chicken meat in Japan. Foodborne Pathog Dis 11:104–110. doi: 10.1089/fpd.2013.1608. [DOI] [PubMed] [Google Scholar]
- 5.Suzuki S, Shibata N, Yamane K, Wachino J, Ito K, Arakawa Y. 2009. Change in the prevalence of extended-spectrum-β-lactamase-producing Escherichia coli in Japan by clonal spread. J Antimicrob Chemother 63:72–79. doi: 10.1093/jac/dkn463. [DOI] [PubMed] [Google Scholar]
- 6.Hiroi M, Yamazaki F, Harada T, Takahashi N, Iida N, Noda Y, Yagi M, Nishio T, Kanda T, Kawamori F, Sugiyama K, Masuda T, Hara-Kudo Y, Ohashi N. 2012. Prevalence of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in food-producing animals. J Vet Med Sci 74:189–195. doi: 10.1292/jvms.11-0372. [DOI] [PubMed] [Google Scholar]
- 7.Nakane K, Kawamura K, Goto K, Arakawa Y. 2016. Long-term colonization by blaCTX-M-harboring Escherichia coli in healthy Japanese people engaged in food handling. Appl Environ Microbiol 82:1818–1827. doi: 10.1128/AEM.02929-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hattori T, Nakane K, Kawamura K. 2011. Multidrug-resistant phenotype and prevalence of CTX-M-27 in extended-spectrum β-lactamase-producing uropathogenic Escherichia coli. J Jpn Soc Clin Microbiol 21:25–34. (In Japanese.) [Google Scholar]
- 9.Coil D, Jospin G, Darling AE. 2015. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 31:587–589. doi: 10.1093/bioinformatics/btu661. [DOI] [PubMed] [Google Scholar]
- 10.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Ponten T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, Moller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barton BM, Harding GP, Zuccarelli AJ. 1995. A general method for detecting and sizing large plasmids. Anal Biochem 226:235–240. doi: 10.1006/abio.1995.1220. [DOI] [PubMed] [Google Scholar]
- 14.Eller C, Leistner R, Guerra B, Fischer J, Wendt C, Rabsch W, Werner G, Pfeifer Y. 2014. Emergence of extended-spectrum β-lactamase (ESBL) CTX-M-8 in Germany. J Antimicrob Chemother 69:562–564. doi: 10.1093/jac/dkt387. [DOI] [PubMed] [Google Scholar]
- 15.Woerther PL, Angebault C, Jacquier H, Clermont O, El Mniai A, Moreau B, Djossou F, Peroz G, Catzeflis F, Denamur E, Andremont A. 2013. Characterization of fecal extended-spectrum-β-lactamase-producing Escherichia coli in a remote community during a long time period. Antimicrob Agents Chemother 57:5060–5066. doi: 10.1128/AAC.00848-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jouini A, Slama KB, Klibi N, Sallem RB, Estepa V, Vinué L, Saenz Y, Ruiz-Larrea F, Boudabous A, Torres C. 2013. Lineages and virulence gene content among extended-spectrum β-lactamase-producing Escherichia coli strains of food origin in Tunisia. J Food Prot 76:323–327. doi: 10.4315/0362-028X.JFP-12-251. [DOI] [PubMed] [Google Scholar]
- 17.Kiiru J, Kariuki S, Goddeeris BM, Butaye P. 2012. Analysis of β-lactamase phenotypes and carriage of selected β-lactamase genes among Escherichia coli strains obtained from Kenyan patients during an 18-year period. BMC Microbiol 12:155. doi: 10.1186/1471-2180-12-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vinué L, Saenz Y, Martinez S, Somalo S, Moreno MA, Torres C, Zarazaga M. 2009. Prevalence and diversity of extended-spectrum β-lactamases in faecal Escherichia coli isolates from healthy humans in Spain. Clin Microbiol Infect 15:954–957. doi: 10.1111/j.1469-0691.2009.02803.x. [DOI] [PubMed] [Google Scholar]
- 19.Bonnet R, Sampaio JL, Labia R, De Champs C, Sirot D, Chanal C, Sirot J. 2000. A novel CTX-M β-lactamase (CTX-M-8) in cefotaxime-resistant Enterobacteriaceae isolated in Brazil. Antimicrob Agents Chemother 44:1936–1942. doi: 10.1128/AAC.44.7.1936-1942.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dropa M, Lincopan N, Balsalobre LC, Oliveira DE, Moura RA, Fernandes MR, da Silva QM, Matte GR, Sato MI, Matte MH. 2016. Genetic background of novel sequence types of CTX-M-8- and CTX-M-15-producing Escherichia coli and Klebsiella pneumoniae from public wastewater treatment plants in Sao Paulo, Brazil. Environ Sci Pollut Res Int 23:4953–4958. doi: 10.1007/s11356-016-6079-5. [DOI] [PubMed] [Google Scholar]
- 21.Aizawa J, Neuwirt N, Barbato L, Neves PR, Leigue L, Padilha J, Pestana de Castro AF, Gregory L, Lincopan N. 2014. Identification of fluoroquinolone-resistant extended-spectrum β-lactamase (CTX-M-8)-producing Escherichia coli ST224, ST2179 and ST2308 in buffalo (Bubalus bubalis). J Antimicrob Chemother 69:2866–2869. doi: 10.1093/jac/dku218. [DOI] [PubMed] [Google Scholar]
- 22.Ferreira JC, Penha Filho RA, Andrade LN, Berchieri A Jr, Darini AL. 2016. Evaluation and characterization of plasmids carrying CTX-M genes in a non-clonal population of multidrug-resistant Enterobacteriaceae isolated from poultry in Brazil. Diagn Microbiol Infect Dis 85:444–448. doi: 10.1016/j.diagmicrobio.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 23.Ferreira JC, Penha Filho RA, Andrade LN, Berchieri A Jr, Darini AL. 2014. IncI1/ST113 and IncI1/ST114 conjugative plasmids carrying blaCTX-M-8 in Escherichia coli isolated from poultry in Brazil. Diagn Microbiol Infect Dis 80:304–306. doi: 10.1016/j.diagmicrobio.2014.09.012. [DOI] [PubMed] [Google Scholar]
- 24.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimojima Y, Ida M, Inomata M, Higuchi Y, Takano C, Kawamura M, Konishi N, Hatakeyama K, Nakama A, Kai A. 2011. Extended-spectrum β-lactamase (ESBL)-producing Escherichia coli isolated from meat. Ann Rep Tokyo Metr Inst Pub Health 62:145–150. (In Japanese.) [Google Scholar]