ABSTRACT
Here, we report the description of a colistin-heteroresistant Klebsiella pneumoniae isolate fortuitously isolated from the stool sample of a patient with suspicion of tuberculosis in a public hospital of Marseille, France. In the colistin-resistant subpopulation, a mutation in the mgrB gene leading to a premature stop codon was found, and the hypermucoviscous phenotype was lost. Susceptibility to other antibiotics remained unchanged. To our knowledge, this is the first identification of such a colistin-heteroresistant Klebsiella pneumoniae isolate in France.
KEYWORDS: Klebsiella pneumoniae, heteroresistance, colistin, mgrB
TEXT
Klebsiella pneumoniae is a significant human pathogen belonging to the Enterobacteriaceae family (1). The increase of multidrug-resistant Enterobacteriaceae led to the renewed use of colistin as a treatment of last resort (2), conducive to the emergence of colistin resistance among K. pneumoniae strains worldwide (3). Colistin has bactericidal action against Gram-negative pathogens, targeting the lipid A moiety of lipopolysaccharide (LPS) and leading to cell membrane disruption (4). The main known mechanisms of colistin resistance induce lipid A modifications through the addition of 4-amino-4-deoxy-l-arabinose or phosphoethanolamine (5), mostly mediated by mutations in the two-component system PmrA/PmrB or PhoP/PhoQ (6) or its negative regulator, MgrB (7–9), or by acquisition of the mcr-1 plasmid-mediated gene (10), resulting in reduction of the polymyxin affinity to LPS.
Heteroresistance can be defined as the presence of subpopulations with various susceptibilities to an antibiotic within an isolate (11). Few reports of colistin heteroresistance exist, because it cannot be assessed by the recommended MIC testing methods (12). Here, we isolated a colistin-heteroresistant Klebsiella pneumoniae strain from the stool sample from a patient in the public hospital Hôpital Nord of Marseille, France. This isolation was fortuitously performed during research on Mycobacteriaceae, and the isolate was detected after 7 days by culture on an innovative selective medium containing the BBL MGIT PANTA antibiotic mixture (BD, USA), which included 2.38 μg/ml of polymyxin B. (PANTA includes polymyxin B, amphotericin B, nalidixic acid, trimethoprim, and azlocillin.)
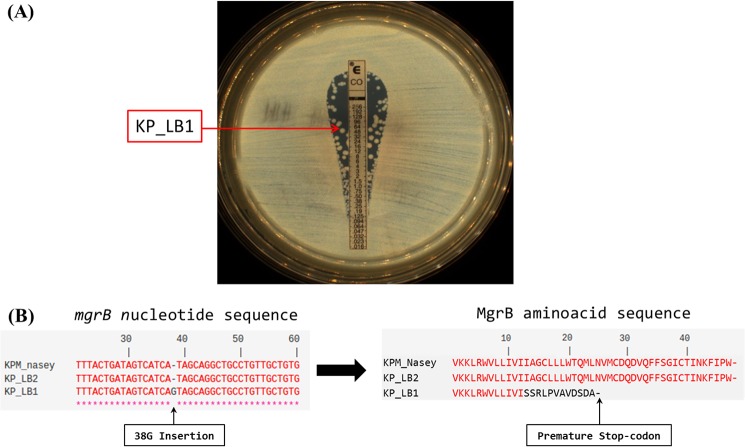
Colistin heteroresistance was detected by MIC testing using colistin Etest strips (bioMérieux, Marcy l'Étoile, France), where colonies were observed within the clear zone of inhibition (Fig. 1A). Two subpopulations were separated by subculture on Columbia agar plus 5% sheep blood (COS [bioMérieux, Marcy-l'Étoile, France]). Both were identified as Klebsiella pneumoniae by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (Microflex; Bruker Daltonics, Bremen, Germany) (13). They exhibited different colony morphotypes, and their colistin MIC was assessed by broth microdilution method and interpreted according to the EUCAST guidelines (http://www.eucast.org). The colistin-resistant subpopulation, named LB1, had a colistin MIC of 128 μg/ml and exhibited gray and plate colonies on COS medium, while the colistin-susceptible subpopulation, named LB2, had a colistin MIC of 1 μg/ml and exhibited white and hypermucoviscous colonies, with a positive string test (14). Their stability was assessed by 15 daily subcultures on COS medium, and both the morphologies and the colistin susceptibilities were maintained.
FIG 1.
(A) Colistin MIC determination using a colistin Etest strip. The arrow indicates the colistin-resistant subpopulation LB1. (B) Alignments of mgrB nucleotide sequences and amino acid MgrB protein sequences of the two subpopulations of colistin-heteroresistant Klebsiella pneumoniae with KPM_Nasey retrieved from GenBank (accession no. KF852765).
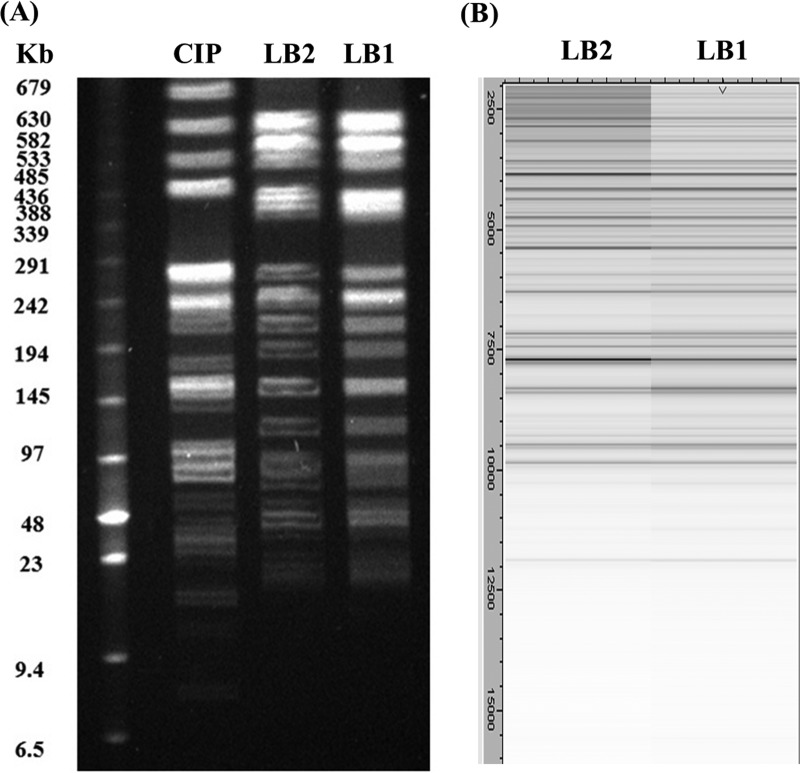
Susceptibility testing of the two subpopulations to 22 antibiotics was performed by the disk diffusion method, and their growth rates were compared with those of colistin-susceptible (15) and mcr-1-positive K. pneumoniae strains as controls (3, 16). The two subpopulations presented identical growth rates (see Fig. S1 in the supplemental material) and had the same susceptibility patterns, except for colistin, for which the isolates were not defined as multidrug-resistant pathogens (Table 1). Faced with this phenotype similarity, we explored the existence of heteroresistance by comparative methods, including comparison of MALDI-TOF MS mass spectra (17), pulsed-field gel electrophoresis (PFGE) with XbaI digestion (18), and multilocus sequence typing (MLST) analysis, using the Pasteur Institute database (http://bigsdb.pasteur.fr/klebsiella/klebsiella.html). Comparison of the MALDI-TOF MS mass spectra and PFGE gave identical protein profiles (Fig. 2), and the sequence type found by MLST was ST86, which had been reported as a hypermucoviscous (hypervirulent) strain, causing life-threatening infections (14, 19). These results confirmed that we had isolated a colistin-heteroresistant Klebsiella pneumoniae isolate, as LB1 and LB2 were isogenic strains.
TABLE 1.
Antibiotic susceptibility testing of LB1 and LB2 isogenic isolates by the disk diffusion methoda
| Antibiotic disk content (μg) | Breakpoint(s) from CA-SFM,b 2013 (mm) | Inhibition diam (mm) |
Susceptibility | |
|---|---|---|---|---|
| LB1 | LB2 | |||
| AMX (25) | 16–21 | 6 | 6 | R |
| AMC (30) | 16–21 | 14 | 14 | R |
| TIC (75) | 22–24 | 6 | 6 | R |
| TIM (85) | 22–24 | 20 | 20 | R |
| TZP (85) | 17–21 | 25 | 25 | S |
| FEP (30) | 21–24 | 33 | 33 | S |
| CRO (30) | 23–26 | 32 | 32 | S |
| CTX (30) | 23–26 | 36 | 36 | S |
| FOX (30) | 15–22 | 26 | 26 | S |
| IPM (10) | 17–24 | 32 | 32 | S |
| ETP (10) | 26–28 | 31 | 31 | S |
| ATM (30) | 21–27 | 35 | 35 | S |
| NAL (30) | 15–20 | 21 | 21 | S |
| CIP (5) | 22–25 | 28 | 28 | S |
| OFL (5) | 22–25 | 36 | 36 | S |
| AMK (30) | 15–17 | 20 | 20 | S |
| GEN (15) | 16–18 | 20 | 20 | S |
| TOB (10) | 16–18 | 23 | 23 | S |
| FOF (50) | 14 | 19 | 19 | S |
| SXT (25) | 13–16 | 6 | 6 | R |
| NIT (300) | 15 | 12 | 12 | R |
| CST (50) | 15 | 9 | 17 | R (LB1)/S (LB2) |
Both isolates were resistant (R) to amoxicillin (AMX), amoxicillin-clavulanate (AMC), ticarcillin (TIC), ticarcillin-clavulanate (TIM), nitrofurantoin (NIT), and trimethoprim-sulfamethoxazole (SXT) and susceptible (S) to piperacillin-tazobactam (TZP), cefepime (FEP), ceftriaxone (CRO), cefotaxime (CTX), cefoxitin (FOX), aztreonam (ATM), imipenem (IPM), ertapenem (ETP), nalidixic acid (NAL), ciprofloxacin (CIP), ofloxacin (OFX), amikacin (AMK), gentamicin (GEN), tobramycin (TOB), and fosfomycin (FOF). LB1 was resistant to colistin (CST), while LB2 was susceptible.
Comité de l'Antibiogramme—Société Française de Microbiologie (http://www.sfm-microbiologie.org/UserFiles/files/casfm/CASFM2013vjuin.pdf).
FIG 2.
(A) PFGE profiles of XbaI-digested genomic DNAs showing genomic relatedness among the CIP 82.91, LB2, and LB1 K. pneumoniae strains. DNA sizes of the low-range pulsed-field gel marker (Biolabs, New England) are indicated in kilobases on the left. (B) MALDI-TOF MS spectra from LB1 and LB2 strain comparison with GelView using Flex Analysis software.
Colistin resistance genes mgrB, pmrA, pmrB, phoP, and phoQ were amplified and sequenced from the two subpopulations (3), and the presence of the recently described mcr-1 gene was investigated by reverse transcription-PCR (RT-PCR) (20). Mutation was absent in pmrA, pmrB, phoP, and phoQ, and the amplification of mcr-1 remained negative. However, sequence analysis of mgrB showed a single nucleotide insertion (38G) in colistin-resistant subpopulation LB1, leading to a premature stop codon and, based on the predicted protein sequence, an inactive MgrB regulator (Fig. 1B). MgrB mutations following colistin exposure were reported (21, 22) as an in vitro mutant selection (23) and also without previous exposure (24). To our knowledge, the patient did not receive previous polymyxin treatment, but as the isolate was detected on a polymyxin-containing medium, the mutation of the resistant population could be induced (23) or the subpopulation selected, allowing the growth of the bacteria (25, 26).
As in a recent report highlighting the association between biofilm formation and heteroresistance (27), we described different stable morphotypes of the colonies exhibiting different susceptibilities to colistin, without previously described reversion (7, 28). The loss of hypervirulence and fitness of strains after acquisition of colistin resistance was previously demonstrated for hypermucoviscous K. pneumoniae ST23 strains (29).
There are only 2 reports of genomic analysis of colistin-heteroresistant K. pneumoniae isolates, which were multidrug-resistant bacteria: one OXA-48 carbapenemase-producing strain with a mutation in protein PhoP (7) and five extended-spectrum β-lactamase (ESBL)-expressing isolates with mutations in the phoQ, lpxM, and yciM genes, as well as two with mutations on the mgrB gene (the presence of an insertion sequence and deletion) (30). This is the first description of a colistin-heteroresistant Klebsiella pneumoniae isolate in France and the first description of a truncated MgrB protein after an insertion of a single nucleotide.
Colistin-heteroresistant K. pneumoniae isolates among isolates that had been classified as susceptible in clinical practice showed higher prevalence rates than isolates classified as colistin resistant in some studies (26, 28). These findings may raise concern about the choice of antimicrobial susceptibility testing methods to assess heteroresistance, as the prevalence of colistin-resistant isolates is widely underestimated (31). Indeed, in clinical practice, the MICs are often assessed with automated techniques such as Vitek2 (bioMérieux, Marcy l'Étoile, France), and the only reliable method to assess the colistin MIC is broth microdilution according to EUCAST (http://www.eucast.org/ast_of_bacteria/warnings/#c13111), but these techniques have been reported as unreliable in detecting heteroresistance (32, 33). Currently, only agar-based methods, especially Etest, can detect heterogeneous populations and could be used concomitantly as screening tests (7, 11, 34). These studies highlight that heteroresistance should be further studied, as the resistant population can be selected and lead to rapid development and therapeutic failure (12). Because this strain was isolated fortuitously, we believe that screening of colistin-resistant strains from fecal samples should be performed at least in intensive care units in order to isolate such patients.
Supplementary Material
ACKNOWLEDGMENTS
We thank Tradonline for English correction.
We have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00356-17.
REFERENCES
- 1.Podschun R, Ullmann U. 1998. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev 11:589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biswas S, Brunel J-M, Dubus J-C, Reynaud-Gaubert M, Rolain J-M. 2012. Colistin: an update on the antibiotic of the 21st century Expert Rev Anti Infect Ther 10:917–934. doi: 10.1586/eri.12.78. [DOI] [PubMed] [Google Scholar]
- 3.Olaitan AO, Diene SM, Kempf M, Berrazeg M, Bakour S, Gupta SK, Thongmalayvong B, Akkhavong K, Somphavong S, Paboriboune P, Chaisiri K, Komalamisra C, Adelowo OO, Fagade OE, Banjo OA, Oke AJ, Adler A, Assous MV, Morand S, Raoult D, Rolain JM. 2014. Worldwide emergence of colistin resistance in Klebsiella pneumoniae from healthy humans and patients in Lao PDR, Thailand, Israel, Nigeria and France owing to inactivation of the PhoP/PhoQ regulator mgrB: an epidemiological and molecular study. Int J Antimicrob Agents 44:500–507. doi: 10.1016/j.ijantimicag.2014.07.020. [DOI] [PubMed] [Google Scholar]
- 4.Hancock RE, Chapple DS. 1999. Peptide antibiotics. Antimicrob Agents Chemother 43:1317–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baron S, Hadjadj L, Rolain J-M, Olaitan AO. 2016. Molecular mechanisms of polymyxin resistance: knowns and unknowns. Int J Antimicrob Agents 48:583–591. doi: 10.1016/j.ijantimicag.2016.06.023. [DOI] [PubMed] [Google Scholar]
- 6.Olaitan AO, Morand S, Rolain J-M. 2014. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol 5:643. doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jayol A, Nordmann P, Brink A, Poirel L. 2015. Heteroresistance to colistin in Klebsiella pneumoniae associated with alterations in the PhoPQ regulatory system. Antimicrob Agents Chemother 59:2780–2784. doi: 10.1128/AAC.05055-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cannatelli A, Giani T, D'Andrea MM, Di Pilato V, Arena F, Conte V, Tryfinopoulou K, Vatopoulos A, Rossolini GM, COLGRIT Study Group. 2014. MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob Agents Chemother 58:5696–5703. doi: 10.1128/AAC.03110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cannatelli A, D'Andrea MM, Giani T, Di Pilato V, Arena F, Ambretti S, Gaibani P, Rossolini GM. 2013. In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob Agents Chemother 57:5521–5526. doi: 10.1128/AAC.01480-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 11.El-Halfawy OM, Valvano MA. 2015. Antimicrobial heteroresistance: an emerging field in need of clarity. Clin Microbiol Rev 28:191–207. doi: 10.1128/CMR.00058-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ah Y-M, Kim A-J, Lee J-Y. 2014. Colistin resistance in Klebsiella pneumoniae. Int J Antimicrob Agents 44:8–15. doi: 10.1016/j.ijantimicag.2014.02.016. [DOI] [PubMed] [Google Scholar]
- 13.Seng P, Rolain J-M, Fournier PE, La Scola B, Drancourt M, Raoult D. 2010. MALDI-TOF-mass spectrometry applications in clinical microbiology. Future Microbiol 5:1733–1754. doi: 10.2217/fmb.10.127. [DOI] [PubMed] [Google Scholar]
- 14.Shon AS, Bajwa RPS, Russo TA. 2013. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence 4:107–118. doi: 10.4161/viru.22718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bakour S, Sahli F, Touati A, Rolain J-M. 2015. Emergence of KPC-producing Klebsiella pneumoniae ST512 isolated from cerebrospinal fluid of a child in Algeria. New Microbes New Infect 3:34–36. doi: 10.1016/j.nmni.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rolain J-M, Kempf M, Leangapichart T, Chabou S, Olaitan AO, Le Page S, Morand S, Raoult D. 2016. Plasmid-mediated mcr-1 gene in colistin-resistant clinical isolates of Klebsiella pneumoniae in France and Laos. Antimicrob Agents Chemother 60:6994–6995. doi: 10.1128/AAC.00960-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egli A, Tschudin-Sutter S, Oberle M, Goldenberger D, Frei R, Widmer AF. 2015. Matrix-assisted laser desorption/ionization time of flight mass-spectrometry (MALDI-TOF MS) based typing of extended-spectrum B-lactamase producing E. coli—a novel tool for real-time outbreak investigation. PLoS One 10:e0120624. doi: 10.1371/journal.pone.0120624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raoult D, Audic S, Robert C, Abergel C, Renesto P, Ogata H, La Scola B, Suzan M, Claverie J-M. 2004. The 1.2-megabase genome sequence of Mimivirus. Science 306:1344–1350. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- 19.Cubero M, Grau I, Tubau F, Pallarés R, Dominguez MA, Liñares J, Ardanuy C. 2016. Hypervirulent Klebsiella pneumoniae clones causing bacteraemia in adults in a teaching hospital in Barcelona, Spain (2007–2013). Clin Microbiol Infect 22:154–160. doi: 10.1016/j.cmi.2015.09.025. [DOI] [PubMed] [Google Scholar]
- 20.Chabou S, Leangapichart T, Okdah L, Le Page S, Hadjadj L, Rolain J-M. 2016. Real-time quantitative PCR assay with Taqman* probe for rapid detection of MCR-1 plasmid-mediated colistin resistance. New Microbes New Infect 13:71–74. doi: 10.1016/j.nmni.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.López-Camacho E, Gómez-Gil R, Tobes R, Manrique M, Lorenzo M, Galván B, Salvarelli E, Moatassim Y, Salanueva IJ, Pareja E, Codoñer FM, Alvarez-Tejado M, Garcillán-Barcia MP, De la Cruz F, Mingorance J. 2014. Genomic analysis of the emergence and evolution of multidrug resistance during a Klebsiella pneumoniae outbreak including carbapenem and colistin resistance. J Antimicrob Chemother 69:632–636. doi: 10.1093/jac/dkt419. [DOI] [PubMed] [Google Scholar]
- 22.Falagas ME, Rafailidis PI, Matthaiou DK. 2010. Resistance to polymyxins: mechanisms, frequency and treatment options. Drug Resist Updat 13:132–138. doi: 10.1016/j.drup.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 23.Cannatelli A, Di Pilato V, Giani T, Arena F, Ambretti S, Gaibani P, D'Andrea MM, Rossolini GM. 2014. In vivo evolution to colistin resistance by PmrB sensor kinase mutation in KPC-producing Klebsiella pneumoniae is associated with low-dosage colistin treatment. Antimicrob Agents Chemother 58:4399–4403. doi: 10.1128/AAC.02555-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olaitan AO, Morand S, Rolain J-M. 2016. Emergence of colistin-resistant bacteria in humans without colistin usage: a new worry and cause for vigilance. Int J Antimicrob Agents 47:1–3. doi: 10.1016/j.ijantimicag.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 25.Li J, Rayner CR, Nation RL, Owen RJ, Spelman D, Tan KE, Liolios L. 2006. Heteroresistance to colistin in multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 50:2946–2950. doi: 10.1128/AAC.00103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poudyal A, Howden BP, Bell JM, Gao W, Owen RJ, Turnidge JD, Nation RL, Li J. 2008. In vitro pharmacodynamics of colistin against multidrug-resistant Klebsiella pneumoniae. J Antimicrob Chemother 62:1311–1318. doi: 10.1093/jac/dkn425. [DOI] [PubMed] [Google Scholar]
- 27.Silva A, Sousa AM, Alves D, Lourenço A, Pereira MO. 2016. Heteroresistance to colistin in Klebsiella pneumoniae is triggered by small colony variants sub-populations within biofilms. Pathog Dis 74:ftw036. doi: 10.1093/femspd/ftw036. [DOI] [PubMed] [Google Scholar]
- 28.Meletis G, Tzampaz E, Sianou E, Tzavaras I, Sofianou D. 2011. Colistin heteroresistance in carbapenemase-producing Klebsiella pneumoniae. J Antimicrob Chemother 66:946–947. doi: 10.1093/jac/dkr007. [DOI] [PubMed] [Google Scholar]
- 29.Choi M-J, Ko KS. 2015. Loss of hypermucoviscosity and increased fitness cost in colistin-resistant Klebsiella pneumoniae sequence type 23 strains. Antimicrob Agents Chemother 59:6763–6773. doi: 10.1128/AAC.00952-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Halaby T, Kucukkose E, Janssen AB, Rogers MRC, Doorduijn DJ, Van Der Zanden AGM, Al Naiemi N, Vandenbroucke-Grauls CMJE, Van Schaik W. 2016. Genomic characterization of colistin heteroresistance in Klebsiella pneumoniae during a nosocomial outbreak. Antimicrob Agents Chemother 60:6837–6843. doi: 10.1128/AAC.01344-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Falagas ME, Makris GC, Dimopoulos G, Matthaiou DK. 2008. Heteroresistance: a concern of increasing clinical significance? Clin Microbiol Infect 14:101–104. doi: 10.1111/j.1469-0691.2007.01912.x. [DOI] [PubMed] [Google Scholar]
- 32.Lo-Ten-Foe JR, de Smet AMGA, Diederen BMW, Kluytmans JAJW, van Keulen PHJ. 2007. Comparative evaluation of the VITEK 2, disk diffusion, Etest, broth microdilution, and agar dilution susceptibility testing methods for colistin in clinical isolates, including heteroresistant Enterobacter cloacae and Acinetobacter baumannii. Antimicrob Agents Chemother 51:3726–3730. doi: 10.1128/AAC.01406-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan TY, Ng SY. 2007. Comparison of Etest, Vitek and agar dilution for susceptibility testing of colistin. Clin Microbiol Infect 13:541–544. doi: 10.1111/j.1469-0691.2007.01708.x. [DOI] [PubMed] [Google Scholar]
- 34.Napier BA, Band V, Burd EM, Weiss DS. 2014. Colistin heteroresistance in Enterobacter cloacae is associated with cross-resistance to the host antimicrobial lysozyme. Antimicrob Agents Chemother 58:5594–5597. doi: 10.1128/AAC.02432-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.