ABSTRACT
Bacteria elicit an adaptive response against hostile conditions such as starvation and other kinds of stresses. Their ability to survive such conditions depends, in part, on stringent response pathways. (p)ppGpp, considered to be the master regulator of the stringent response, is a novel target for inhibiting the survival of bacteria. In mycobacteria, the (p)ppGpp synthetase activity of bifunctional Rel is critical for stress response and persistence inside a host. Our aim was to design an inhibitor of (p)ppGpp synthesis, monitor its efficiency using enzyme kinetics, and assess its phenotypic effects in mycobacteria. As such, new sets of inhibitors targeting (p)ppGpp synthesis were synthesized and characterized by mass spectrometry and nuclear magnetic resonance spectroscopy. We observed significant inhibition of (p)ppGpp synthesis by RelMsm in the presence of designed inhibitors in a dose-dependent manner, which we further confirmed by monitoring the enzyme kinetics. The Rel enzyme inhibitor binding kinetics were investigated by isothermal titration calorimetry. Subsequently, the effects of the compounds on long-term persistence, biofilm formation, and biofilm disruption were assayed in Mycobacterium smegmatis, where inhibition in each case was observed. In vivo, (p)ppGpp levels were found to be downregulated in M. smegmatis treated with the synthetic inhibitors. The compounds reported here also inhibited biofilm formation by the pathogen Mycobacterium tuberculosis. The compounds were tested for toxicity by using an MTT assay with H460 cells and a hemolysis assay with human red blood cells, for which they were found to be nontoxic. The permeability of compounds across the cell membrane of human lung epithelial cells was also confirmed by mass spectrometry.
KEYWORDS: (p)ppGpp, mycobacteria, stringent response
INTRODUCTION
Under stress, bacteria generate a response known as the “stringent response” to enhance their resilience under stressful and nutrient-limited conditions. This involves the production and accumulation of an altered nucleic acid base, (p)ppGpp (guanosine pentaphosphate or guanosine tetraphosphate). It is known that (p)ppGpp has a substantial role in the regulation of many physiological processes, including transcription, translation, replication, GTP homeostasis, viability, and virulence (1–4). In Bacillus subtilis, a (p)ppGpp-null mutant failed to survive under nonstressed conditions due to dysregulated GTP homeostasis (5).
Mycobacterium tuberculosis, the causative agent of tuberculosis (TB), is the major pathogen responsible for human mortality in the world. Successful treatment of TB requires the administration of multiple drugs for at least 6 months. The major reason for this prolonged treatment of TB is the bacterium's ability to survive in a dormant state, which makes it tolerant to various inhibitors used in the treatment regimen.
In mycobacteria, (p)ppGpp is synthesized and degraded by the bifunctional Rel protein, encoded by the rel gene. When this gene was deleted in M. smegmatis and M. tuberculosis, the knockout (KO) strains (Δrel) lost their long-term survival ability during nutrition starvation (6, 7). In addition, the Δrel strain of M. tuberculosis was unable to persist in mice (8) and unable to form tubercle lesions in guinea pigs (6), demonstrating the importance of (p)ppGpp in virulence. Specifically, the synthetase activity of the bifunctional Rel has been shown to be critical for the persistence of M. tuberculosis in mice (9). Many studies indicate that mycobacteria lacking (p)ppGpp are defective in biofilm formation and antibiotic tolerance (9–11). Most antibiotics target cellular components such as ribosomes and the cell wall, mainly affecting the bacterial metabolism. However, bacteria can adjust their metabolism to survive under different conditions, including entering a state of dormancy. Thus, there is a need to develop new inhibitor compounds to target the “alternate adaption strategies” of dormant bacteria (10). The stringent response is an attractive target with this goal in mind.
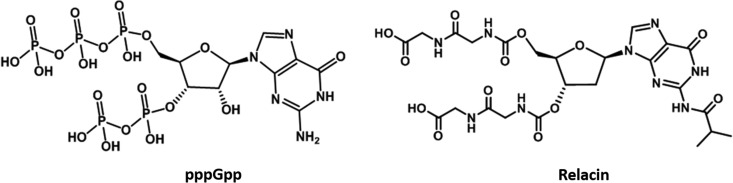
Active research is searching for compounds that can inhibit the synthesis of (p)ppGpp by Rel enzyme. Recently, a molecule named Relacin has been synthesized for inhibiting (p)ppGpp formation. It was designed based on the Rel/Spo (from Streptococcus equisimilis) crystal structure, where a 2′-deoxyguanosine-based analogue of alarmone molecule ppGpp with the original pyrophosphate moieties at positions 5′ and 3′ replaced by glycyl-glycine dipeptides (12, 13). Docking studies were carried out in which Relacin was modeled onto the Rel/SpoT synthetase site. A range of hydrogen bonds/hydrophobic interactions and occupation of the site was taken into consideration, thereby providing a structural basis for the inhibitory effect of Relacin (12, 13) (Fig. 1). However, the efficiency of these analogues is not appreciable. It has been reported that the isobutyryl group at C-2 of the guanine base in Relacin molecule is critical for inhibition. Therefore, we decided to synthesize a more potent compound by functionalization of the amine group at C-2 position of the guanine base in Relacin.
FIG 1.
Structure of pppGpp and Relacin.
RESULTS
Synthesis.
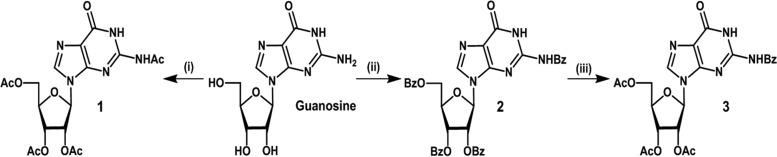
The amino group at the C-2 substituent of the guanine moiety in guanosine was modified with benzoyl and acetyl groups. The 2′, 3′, and 5′ positions were protected through acetylation.
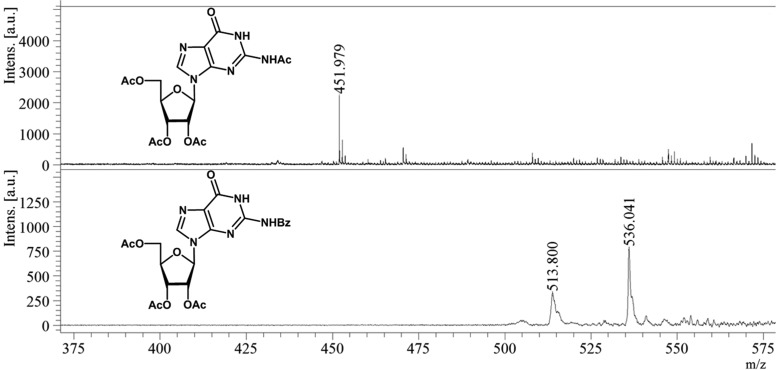
Synthesis of protected guanosine derivatives 1 and 3 were conducted as shown in scheme 1 (Fig. 2). Peracetylation of guanosine using acetic anhydride in pyridine afforded acetyl protected guanosine derivative 1, in a 56% yield. Incorporation of acetyl moiety at the available hydroxyl and amine sites was verified through physical techniques. The –NH proton in compound 1 resonated at 10.47 ppm as a singlet in a 1H nuclear magnetic resonance (NMR) spectrum, whereas the anomeric carbon in compound 1 resonated at 87.2 ppm in a 13C NMR spectrum. Peaks at 169.4 to 172.9 ppm corresponded to carbonyl moieties of ester and amide functionalities in compound 1. The molecular ion peak in compound 1 was observed at 451.97 [M] in a matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrum, as the base peak (Fig. 3).
FIG 2.
Reagents and conditions: scheme 1. (i) Acetic anhydride, 4-dimethylaminopyridine, pyridine, 0°C to room temperature, 12 h, 56%. (ii) Benzoyl chloride, 4-dimethylaminopyridine, pyridine, 0°C to room temperature, 12 h, 62%. (iii) a. Sodium hydroxide (0.1 M), methanol, room temperature, 12 h; b. acetic anhydride, 4-dimethylaminopyridine, pyridine, 0°C to room temperature, 12 h, 41% (after two steps). In the text, compound 1 is referred to as the acetylated compound (AC compound), and compound 3 is referred to as the acetylated benzoylated compound (AB compound).
FIG 3.
MALDI-TOF analysis of AC and AB compounds. (Top) MALDI-TOF analysis of acetylated compound (AC compound). The 451.9 m/z value corresponds to the acetylated derivative of guanosine (AC compound). (Bottom) MALDI-TOF analysis of acetylated benzoylated guanosine (AB compound). The 513.8 m/z value corresponds to the mass of AB compound, and the 536.0 m/z value is its sodiated adduct. Intens., intensity; a.u., arbitrary units.
Similarly, benzoylation of guanosine using benzoyl chloride in pyridine led to the formation of benzoyl protected guanosine intermediate compound 2, in 62% yield. O-debenzoylation of compound 2, by treatment with NaOH (0.1 M) in methanol (MeOH), followed by O-acetylation using acetic anhydride in pyridine, afforded protected guanosine compound 3, in a 41% yield (scheme 1, Fig. 2). The formation of compound 3 was confirmed by NMR spectroscopy and mass spectrometry. In the MALDI-TOF mass spectrum, the appearance of peak at 513.8 [M] and 536.04 [M + Na]+, as the base peak, corresponded to the molecular ion peak of compound 3 (Fig. 3). The −NH proton appeared as a broad singlet at 9.46 ppm in the 1H NMR spectrum, attributed to benzoyl amide functionality of compound 3. In the 13C NMR spectrum of compound 3, anomeric carbon appeared at 87.3 ppm, whereas carbonyl groups of ester functionalities resonated in the region of 169.9 to 171.0 ppm.
AB and AC compounds inhibited the in vitro and in vivo Rel activity.
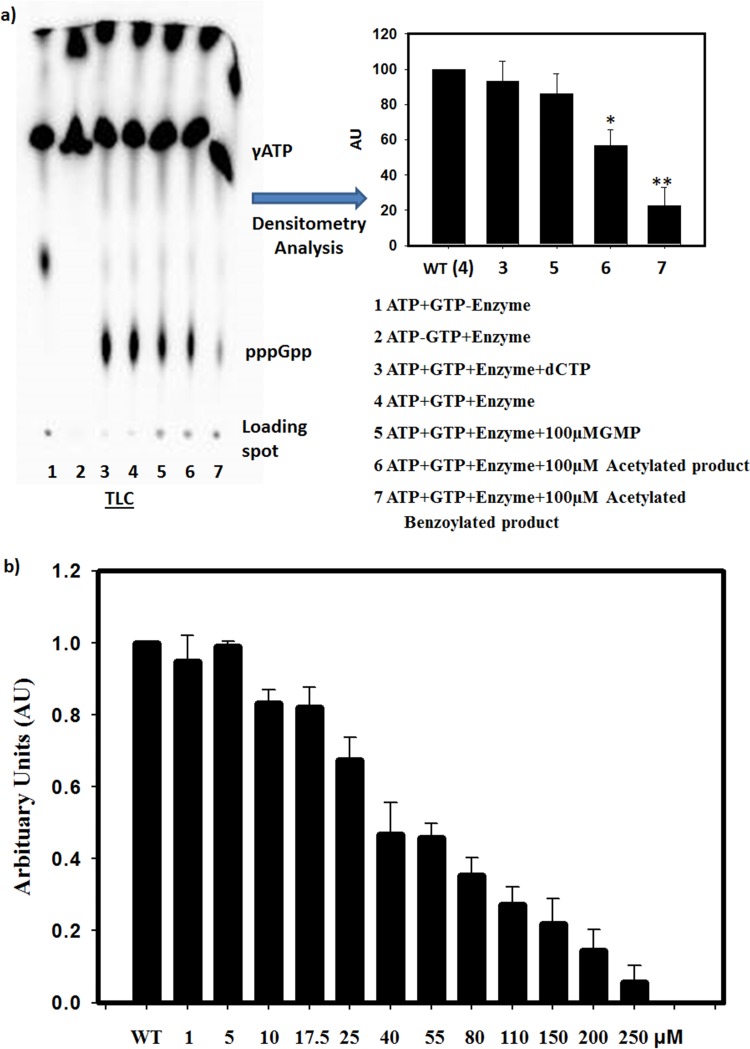
In vitro activity assays were performed in the presence of 100 μM acetylated compound (AC compound) and acetylated benzoylated compound (AB compound). At a 100 μM concentration, AC and AB compounds inhibited pppGpp synthesis by ∼30 and ∼75%, respectively (Fig. 4). The 50% inhibitory concentration was calculated to be nearly 40 μM by testing the inhibition in the presence of different doses of acetylated benzoylated compounds (Fig. 4b).
FIG 4.
(a) Inhibitory effects of AC and AB compounds on in vitro pppGpp synthesis at a 100 μM concentration. Each experiment was performed in three biological replicates. Densitometric analysis was performed, and the values obtained were normalized with respect to the wild type (WT or 4). A Student t test was carried out to confirm the significance. The P value was <0.05 in the case of 6 and 7. (b) Dose-dependent inhibition of in vitro pppGpp synthesis ranging from 1 to 250 μM.
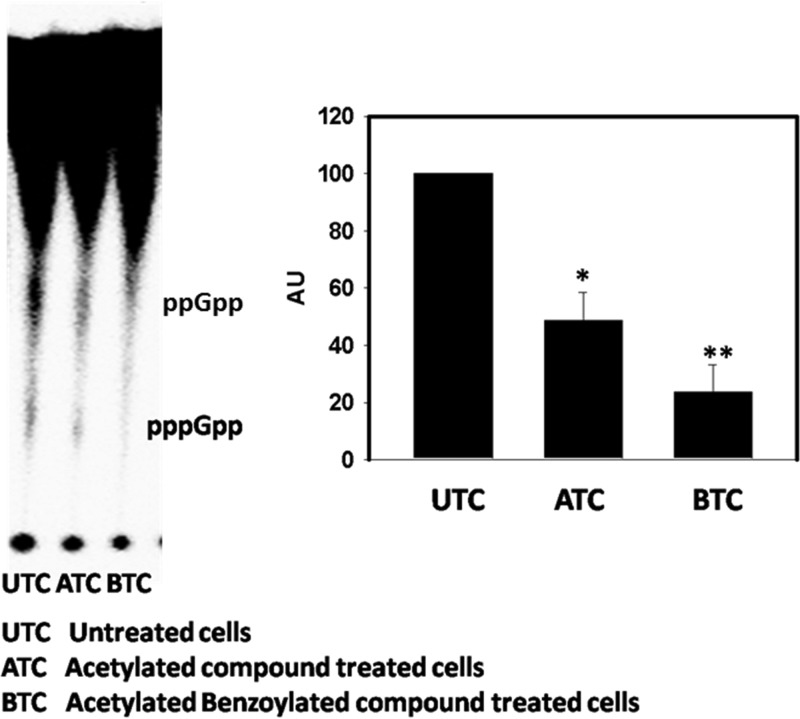
Next, we studied the effects of compounds on bacterial cells in minimal medium conditions and quantitated the in vivo (p)ppGpp levels in treated M. smegmatis cells in comparison to the untreated cells. The synthetic compounds inhibited (p)ppGpp synthesis in M. smegmatis. Densitometric analysis was done to check the decrease in (p)ppGpp levels by the application of ImageJ software. A significant decrease in (p)ppGpp levels was observed as determined by Student t test (Fig. 5). The AB compound was found to be more potent in inhibiting the in vivo (p)ppGpp synthesis.
FIG 5.
In vivo estimation of (p)ppGpp levels in M. smegmatis. The experiment was conducted in three biological replicates. A Student t test was performed to confirm the significance. The P value was found to be <0.05 for both the AC compound and the AB compound.
Previously, it has been reported that the mycobacterial cells devoid of the rel gene are morphologically different and elongated (4, 14). We analyzed the average lengths of the M. smegmatis cells treated with AB compound and found them to be elongated (see Fig. S6 in the supplemental material), which supports alterations in (p)ppGpp levels, indirectly.
Binding kinetics by isothermal titration calorimetry.
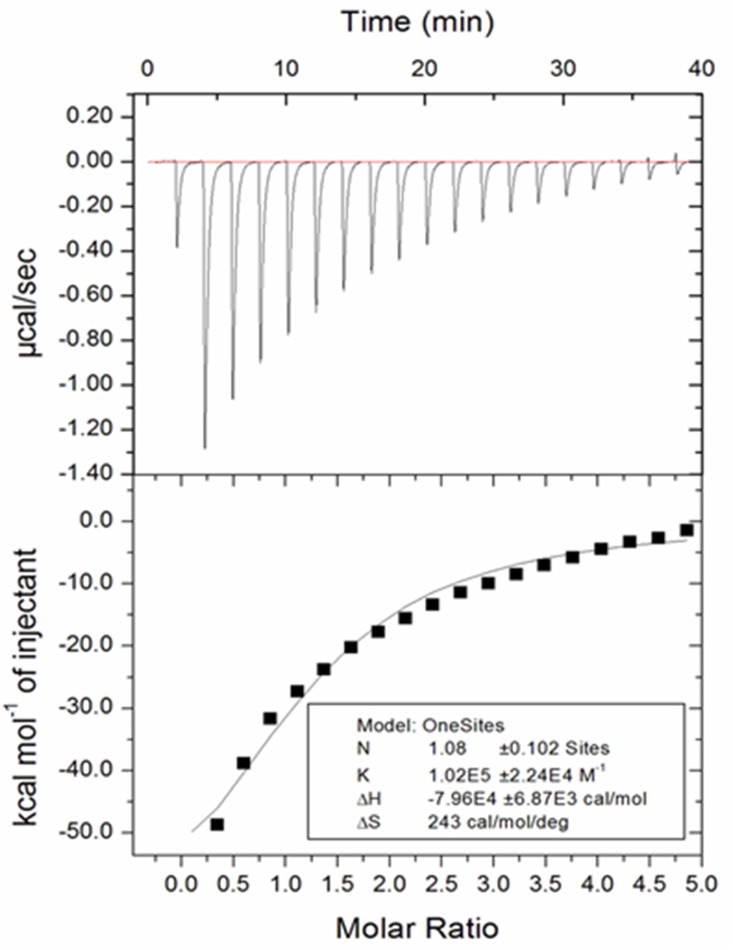
We performed a isothermal titration calorimetry based experiment to confirm the specific binding of the AB compound to the Rel molecule. From the isothermal titration calorimetry (ITC) curve of the Rel enzyme from M. smegmatis with the AB compound in a range of concentrations, a dissociation constant as ∼10 μM (Fig. 6) was obtained. The binding was predominantly driven by enthalpy. The n value obtained was 1, signifying one binding site of AB compound on the Rel enzyme (Fig. 6).
FIG 6.
Binding of the acetylated benzoylated compound to the Rel enzyme from M. smegmatis. An ITC curve corresponding to the binding of the synthetic compound to the Rel enzyme at 25°C is presented here. The upper panel shows the raw data for the titration of Rel enzyme with the AB compound, and the lower panel indicates the integrated heat of binding obtained from the raw data. The model for one-site binding was implicated to fit the curve. The “N” represents the number of binding site that is 1, and the “K” represents the association constant (Ka). The dissociation constant, Kd, can be calculated by reciprocating the Ka and was determined to be nearly 10 μM.
Enzyme kinetics of Rel.
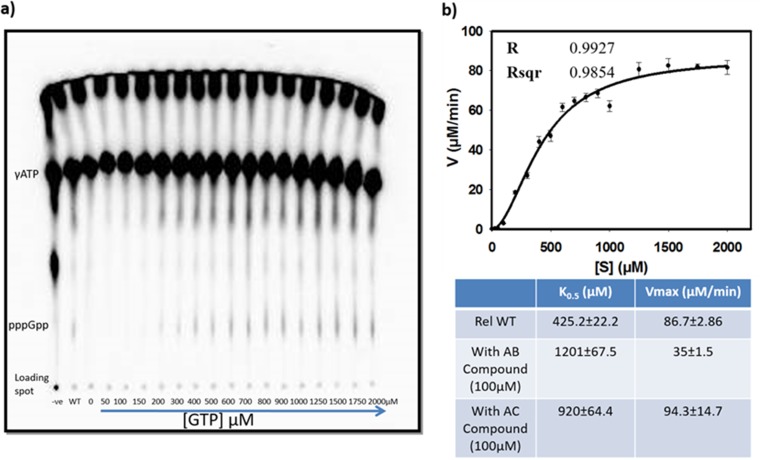
The enzyme kinetics for (p)ppGpp synthesis by RelMsm (i.e., the full-length Rel enzyme from M. smegmatis) in the presence of 100 μM AC and AB compounds were monitored to understand the level of inhibition in comparison to that of the Rel control. Substrate concentrations were varied from 0 to 2,000 μM for the kinetics study of Rel in the presence of AC compound. For the AB compound, a 0 to 3,000 μM range of substrate concentrations was used since a higher concentration of substrate was required to achieve saturation. The enzyme kinetics curve was observed to fit with the Hill equation.
The K0.5 was found to increase with the concomitant reduction in the Vmax value in the presence of AB compound. Such changes in the K0.5 and Vmax values indicate mixed inhibition (Fig. 7). We observed that the AC compound led to an increase in the K0.5 value, without a significant change in the Vmax value, suggesting competitive inhibition. The Hill coefficient value was observed to be more than 1 (>1), indicating positive cooperative binding where binding of one ligand molecule to the enzyme induces the binding of other ligand molecules.
FIG 7.
Enzyme kinetics of RelMsm. The rate of formation of ppGpp as a function of substrate GTP is shown here. (a) Formation of product with various substrate concentrations; (b) phosphorimage data of the same. The table below quantifies the K0.5 and Vmax of the reaction in the absence or presence of the synthesized inhibitors. Lanes: -ve, negative control (assay buffer); WT, control (where inhibitor was not added); other lanes, assay mixture with increasing concentrations of GTP. The concentration of the protein (RelMsm) was kept at 200 μg/ml. A Student t test was performed to analyze the significant change in Vmax and K0.5 values. The graph was fitted using the Hill equation. The Hill coefficient value was determined to be 1.89 ± 0.17.
Synthetic compounds affect cell survival.
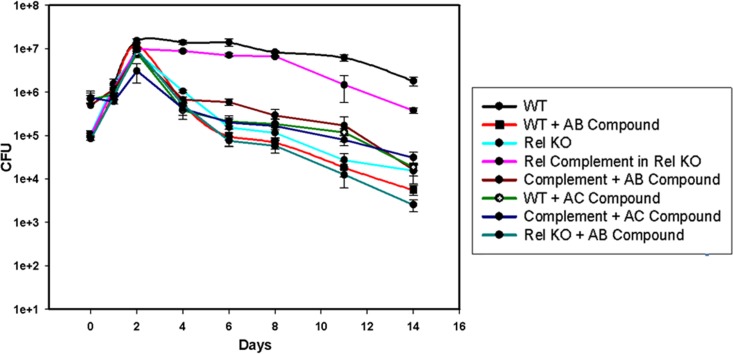
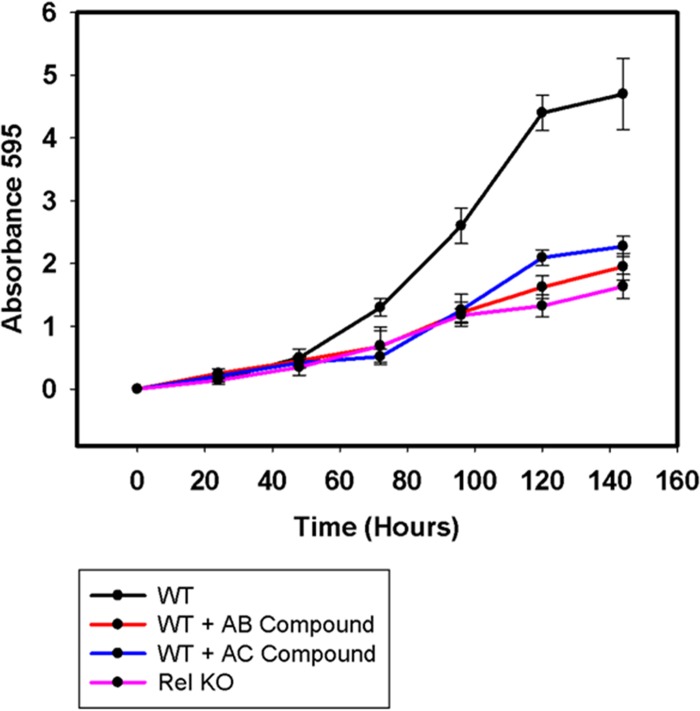
(p)ppGpp synthesis is important for the long-term survival of mycobacteria (15).Therefore, we were intrigued to look for the effects of the synthetic compounds on M. smegmatis survival. We found significant inhibition of long-term survival in the presence of the compounds (100 μM) in comparison to the wild-type untreated controls (Fig. 8). Both AC and AB compounds showed considerable inhibition. A Rel KO strain was used as a control which also showed the decreased long-term survival as reported by others (4). We used the Rel complemented knockout strain and found that long-term survival was restored only in the absence of compounds.
FIG 8.
Long-term survival in the presence or absence of synthetic compounds (100 μM) with Rel KO and Rel complement controls in triplicates. This experiment was performed in three biological replicates. Inhibition was found to be significant for both AB and AC compounds.
The Rel KO did not show further inhibition of long-term survival in the presence of the compounds (Fig. 8), supporting that Rel was the target of the compounds. Our compounds target Rel and inhibit (p)ppGpp synthesis, thereby affecting long-term survival in M. smegmatis.
Biofilm formation and quantification.
Bacterial adaptation to hostile conditions involves the activation of cascades and transitions to resilient phenotypes such as from planktonic to biofilm forms. Biofilms protect the bacteria from stress and induce tolerance to antibiotics. Biofilms are made up of microbial populations enclosed in a matrix. Biofilms are a thousand times more tolerant to antibiotics compared to the planktonic cells (10). It has been shown that tuberculosis bacteria incapable of forming biofilms cannot survive inside the host (15). Recent evidences indicate that M. tuberculosis display a biofilm-like phenotype during infection that could help it survive inside the host (16). Alarmone molecule (p)ppGpp has been directly linked to biofilm formation. It has been shown that M. tuberculosis and M. smegmatis rel KO strains are not effective in forming biofilms (9, 10). Therefore, (p)ppGpp formation and its associated pathways are seen as an important drug target for biofilm inhibition (17, 18).
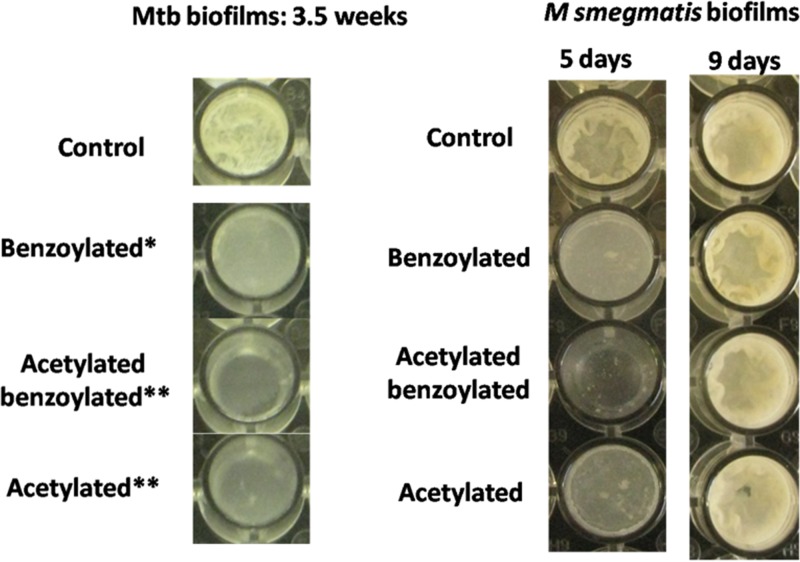
We analyzed biofilm formation in M. smegmatis in the presence of synthetic compounds at 100 μg/ml. A representative picture of biofilm formation is shown in Fig. 9.
FIG 9.
Biofilm formation in the presence of AC and AB compounds (100 μg/ml) compared to a control experiment in which compound was not added to M. tuberculosis and M. smegmatis.
We also quantified the biofilm in M. smegmatis in the presence or absence of the synthetic compounds in comparison to the appropriate controls at different time points (up to 144 h, Fig. 10). The biofilm was quantified by using a crystal violet assay (19). In order to determine whether compounds can disrupt already formed biofilms, biofilm disruption assays were performed where compounds were added just below the biofilm with the application of a 1-ml syringe after 72 h of growth. We assessed the change in biofilm morphology after 54 h of addition of the compound in comparison to the control (see Fig. S7 in the supplemental material). The AB and AC compounds were found to inhibit the formation of biofilms, as well as disrupt the preformed biofilms in M. smegmatis. It should be mentioned here that both compounds were not bactericidal in M. smegmatis, as determined by a CFU assay. In this assay, bacteria were cultured in the presence of the synthesized compounds. The bacterial cells from the early log phase were plated over the Luria-Bertani (LB), agar. We could not find a significant difference as determined by the Student t test in the CFU of treated cells in comparison to the untreated cells. Subsequently, the effect of the compounds on biofilm formation by M. tuberculosis was examined, and inhibition was observed, suggesting the reproducibility of compound effects in the different mycobacterial species and possible clinical relevance (Fig. 9).
FIG 10.
Biofilm quantification assay in the presence of compounds (100 μg/ml) in Sauton's media in three biological replicates. Biofilm formation was found to be decreased in the treated bacterial cells. Compounds were added at the zero time point.
Toxicity: hemolysis assay and microscopic studies.
The effects of synthetic compounds on healthy red blood cells (RBCs) and their toxicities were evaluated. The compounds (at 220 μg/ml) were found to be nontoxic, and the results were comparable to the negative controls as presented in Fig. S8 in the supplemental material. Healthy RBCs are biconcave in shape. The treated RBCs were analyzed under a microscope to visualize morphological changes, if any. RBCs treated with the synthetic compounds were observed to be biconcave (Fig. S8).
Compounds were permeable to the human lung epithelial cell line.
The permeability of compounds was tested using mass spectrometry to study drug permeability across the cell membrane. Cells were treated with compounds, and the cell lysates were analyzed using MALDI. The peaks corresponding to the compounds were absent in the control and present in the treated sample. This experiment was performed in three biological replicates to confirm the observation. Here, the cells were washed three times with phosphate-buffered saline (PBS) before lysis in order to prevent any carry forward of the inhibitor present outside the cell. Compounds were not detected in the final PBS wash (20).
MTT toxicity assay.
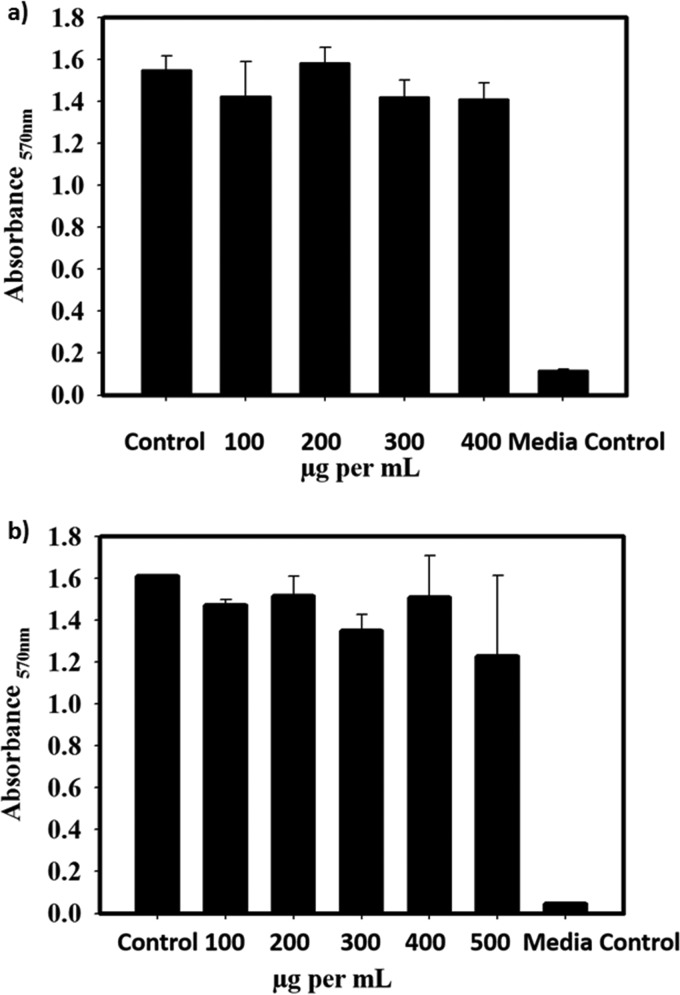
We performed an MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] assay to check the cell cytotoxicity in the presence of the inhibitors. Here, cells with media were taken as the control and compared to the sample (cell + media + compound) incubated for 36 h. Two concentration ranges, 100 to 500 μg/ml and 100 to 400 μg/ml, were tested for acetylated and acetylated benzoylated compounds, respectively. No cytotoxicity was observed for the treated samples, and they were comparable to the control (Fig. 11). The percent survival was calculated to be >95% up to a concentration of 400 μg/ml for both tested compounds.
FIG 11.
MTT assay results in the presence of a range of concentrations of synthetic compounds using the H460 cell line. The x axis indicates the concentration of the compound; the y axis indicates the absorbance at 570 nm. (a) Acetylated benzoylated (AB) compound; (b) acetylated (AC) compound.
DISCUSSION
Unlike the exponential phase, the stationary phase of bacteria is characterized by a low rate of translation, transcription, and replication (21). Therefore, many antibiotics that target these pathways are virtually ineffective in the stationary phase. Further, bacteria exposed to hostile conditions such as nutritional starvation and other kinds of stresses induce the stringent response, which is mediated by (p)ppGpp and helps the bacteria survive under such conditions (10). Antimicrobials that target stress induced stringent response pathways are very few; thus, such an approach offers a unique possibility (22).
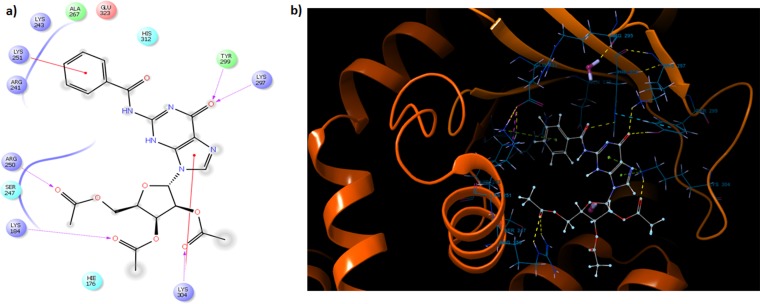
(p)ppGpp analogues such as Relacin have been shown to be effective in inhibiting ppGpp synthesis, stress responses, and key survival processes such as sporulation (12). In the latter study, it has been indicated that the isobutyryl group at the second position of guanine is critical for inhibition. We substituted it for the bulkier benzoyl group, as well as for the smaller acetyl group. We found the benzoyl group to be better a substituent for inhibition. Based on docking studies, we found the benzoyl ring at the C-2 position of the guanine base to be involved in a stacking interaction with the lysine residue at position 251 of the Rel enzyme from S. equisimilis (Fig. 12). The latter lysine residue was found to be conserved in the Rel enzyme from mycobacteria (see Fig. S9 in the supplemental material).
FIG 12.
The crystal structure of Relseq385, Rel/Spo from S. equisimilis, was utilized for in silico docking. The GDP molecule was removed from the active site of the protein structure. The acetylated benzoylated compound structure (in a low energy state) was positioned at the active site in place of GDP, and all of the rotatable bonds were kept flexible. (A) Two-dimensional representation of the interactions at the active site. The red line indicates the stacking interaction, whereas the purple dotted line shows hydrogen bonding. (B) Three-dimensional depiction of the active site with bound acetylated benzoylated compound.
We have shown here that (p)ppGpp analogs can be used to inhibit (p)ppGpp synthesis in acid-fast bacteria such as mycobacteria. Earlier, biofilm formation has been shown to be defective in a Rel mutant strain of M. smegmatis, and bacterial cells were found to be elongated (14). As expected, cells treated with the compounds used here were not able to form biofilm and were elongated, which is consistent with studies in the Rel mutant (14). The long treatment regime, antibiotic tolerance, and the emergence of multiple drug resistance in M. tuberculosis are attributed to its stress response (15). It is now well known that the bacterial strains with defective (p)ppGpp synthesis are metabolically compromised (1). Recent studies have suggested that the inhibition of (p)ppGpp production would have a detrimental effect on bacterial survival and virulence (9). Inhibition of the stringent response appears to be a promising approach to controlling pathogens, such as M. tuberculosis, the causative agent of tuberculosis, which is also known to persist inside the host. We observed that compounds were nontoxic, as demonstrated by the MTT assay and the RBC hemolysis assay. Also, compounds were found to be permeable to the cell membranes, as evidenced by the detection of synthetic compounds in cell lysates by mass spectrometry.
We monitored the enzyme kinetics for Rel from M. smegmatis and, interestingly, observed them to fit according to the Hill equation. Although the inhibition exhibited by these compounds is in the micromolar range, this suggests a novel strategy, in which the stress response of one of the most persistent pathogens, M. tuberculosis, can be potentially targeted. In the future, these compounds will be further improved by modifications in order to achieve the inhibition in nanomolar range and consequently evaluated for their use in humans.
MATERIALS AND METHODS
Synthesis.
All chemicals were purchased from Sigma-Aldrich and used without further purification. Solvents were dried and distilled before use. Silica gel (100 to 200 mesh and 230 to 400 mesh) was used for column chromatography, and thin-layer chromatography (TLC) analysis was performed on commercial plates coated with silica gel 60 F254. Visualization of the spots on TLC plates was achieved by UV radiation or spraying 5% sulfuric acid in ethanol. High-resolution mass spectra were obtained from a Q-TOF instrument by electrospray ionization (ESI) and MALDI-TOF. 1H and 13C NMR spectral analyses were performed on a spectrometer operating at 400 and 100 MHz, respectively. Chemical shifts are reported with respect to tetramethylsilane for 1H NMR and the central line (77.0 ppm) of CDCl3 for 13C NMR spectroscopy. Coupling constants (J) are reported in Hz. Standard abbreviations s, d, t, dd, br s, and m refer to singlet, doublet, triplet, doublet of doublet, broad singlet, and multiplet, respectively.
(i) N2,2′,3′,5′-O-Tetraacetylguanosine (compound 1).
Acetic anhydride (0.4 ml, 4.24 mmol) and 4-dimethylamino pyridine (0.009 g, 0.071 mmol) were added to a suspension of guanosine (0.2 g, 0.71 mmol) in pyridine (2 ml) at 0°C, stirred for 12 h at room temperature. The reaction mixture was diluted with CH2Cl2 (40 ml), washed with dilute aqueous HCl (2 × 10 ml), saturated aqueous NaHCO3 (1 × 10 ml), and brine (10 ml), dried (Na2SO4), filtered, and concentrated in vacuo, and purified (SiO2) (ethyl acetate [EtOAc]) to afford compound 1 (0.18 g, 56%); 1H NMR (CDCl3, 400 MHz) δ 12.15 (br s, 1 H), 10.47 (s, 1 H), 7.77 (s, 1 H), 5.90 to 5.86 (m, 2H), 5.54 (t, J = 4.5Hz, 1 H), 4.42 (dd, J = 4.5 Hz, 11.6 Hz, 1 H), 4.33 (dd, J = 4.8 Hz, 10 Hz, 1 H), 4.23 (dd, J = 5.8 Hz, 11.6 Hz, 1 H), 2.25 (s, 3 H), 2.04 (s, 3 H), 2.00 (s, 3 H), 1.99 (s, 3 H); 13C NMR (CDCl3, 100 MHz) δ 172.9, 170.9, 169.6, 169.4, 155.6, 147.9, 138.4, 121.8, 87.2, 79.8, 72.6, 70.6, 63.0, 24.1, 20.6, 20.4, 20.2; HRMS (ESI/TOF-Q) m/z: calculated for C18H21N5O9Na = 474.1237, found 474.1236 [M + Na]+, 925.2655 [2M + Na]+.
(ii) N2,2′-O,3′-O,5′-O-Tetrabenzoylguanosine (compound 2).
Benzoyl chloride (0.98 ml, 8.5 mmol) and DMAP (0.017 g, 0.14 mmol) was added to a suspended solution of guanosine (0.4 g, 1.41 mmol) in pyridine (3 ml) at 0°C and stirred for 12 h at room temperature. The reaction mixture was diluted with CH2Cl2 (60 ml), washed with dilute aqueous HCl (2 × 20 ml), saturated aqueous NaHCO3 (1 × 20 ml), and brine (20 ml), dried (Na2SO4), filtered and concentrated in vacuo and purified (SiO2) (petroleum ether:EtOAc = 1:1) to afford compound 2 (0.62 g, 62%); 1H NMR (CDCl3, 400 MHz) δ 11.94 (br s, 1 H), 9.53 (s, 1H), 8.15 (d, J = 7.6 Hz, 2 H), 7.97 (d, J = 6.8 Hz, 4 H), 7.84 (s, 1 H), 7.73 (d, J = 7.2 Hz, 2 H), 7.69 (d, J = 7.2 Hz, 1 H), 7.59 (t, J = 7.8 Hz, 4 H), 7.52 to 7.47 (m, 1 H), 7.44 (d, J = 8 Hz, 2 H), 7.40 (d, J = 8 Hz, 2 H), 7.23 (d, J = 7.6 Hz, 2 H), 6.92 (dd, J = 5.2 Hz, 7.6 Hz, 1 H), 6.45 (dd, J = 2 Hz, 5.2 Hz, 1 H), 6.18 (d, J = 2 Hz, 1 H), 4.89 to 4.83 (m, 2 H), 4.78 to 4.74 (m, 1 H); 13C NMR (CDCl3, 100 MHz) δ 167.3, 166.3, 166.0, 165.1, 155.2, 147.5, 139.0, 133.9, 133.8, 133.6, 131.6, 129.8, 129.7, 129.2, 129.0, 128.8, 128.7, 128.6, 128.5, 128.4, 128.3, 128.0, 122.3, 88.0, 79.3, 74.2, 70.7, 61.5; ESI-MS m/z: calculated for C38H29N5O9Na = 722.1863 [M + Na]+, found 722.1865.
(iii) N2-Benzoyl-2′,3′,5′-O-triacetylguanosine (compound 3).
Aqueous NaOH (2 M) (0.1 ml) was added to a solution of compound 2 (0.3 g, 0.43 mmol) in MeOH (2 ml), stirred for 12 h at room temperature, neutralized with Amberlite ion (H+) exchange resin, filtered, and concentrated in vacuo. Acetic anhydride (0.19 ml, 2.05 mmol) and 4-dimethylamino pyridine (0.005 g, 0.041 mmol) were added to the resulting product (0.16 g, 0.41 mmol) in pyridine (2 ml) at 0°C, stirred for 12 h at room temperature. The reaction mixture was diluted with CH2Cl2 (40 ml), washed with dilute aqueous HCl (2 × 10 ml), saturated aqueous NaHCO3 (1 × 10 ml) and brine (10 ml), dried (Na2SO4), filtered and concentrated in vacuo and purified (SiO2) (petroleum ether:EtOAc = 2:3) to afford compound 3 (0.09 g, 41%, after two steps); 1H NMR (CDCl3, 400 MHz) δ 12.2 (br s, 1 H), 9.46 (br s, 1 H), 8.02 (d, J = 8 Hz, 2 H), 7.72 (br s, 1 H), 7.59 (t, J = 7.3 Hz, 1 H), 7.48 (t, J = 7.3 Hz, 2 H), 5.99 to 5.87 (m, 3 H), 4.47 (dd, J = 3.4 Hz, 11.5 Hz, 1 H), 4.43 to 4.39 (m, 1 H), 4.35 (dd, J = 5.6 Hz, 11.5 Hz, 1 H), 2.10 (s, 3 H), 2.05 (s, 3 H), 1.97 (s, 3 H); 13C NMR (CDCl3, 100 MHz) δ 171.0, 169.9, 167.8, 155.4, 147.9, 133.8, 131.3, 129.0, 127.9, 87.2, 79.5, 72.9, 70.7, 62.8, 20.7, 20.6, 20.4; HRMS (ESI/TOF-Q) m/z: calculated for C23H23N5O9Na = 536.1393, found 536.1392 [M + Na]+, 1049.3358 [2M + Na]+.
The acetylated compound and the acetylated benzoylated compound were soluble in water up to 1 and 0.4 mg/ml, respectively. Both compounds were obtained as white powder.
Isothermal titration calorimetry.
ITC analysis was conducted on a ITC200 Micro-Calorimeter (GE Healthcare). The Rel enzyme (protein) was added into the cell component (200 μl), and the syringe (40 μl) was filled by the acetylated benzoylated (AB) compound (ligand). Protein was quantified by using a Bradford assay. Protein and ligand were prepared in the same buffer (20 mM Tris-HCl [pH 7.9] and 50 mM NaCl). Protein concentrations from 5 to 40 μM and ligand concentrations from 125 to 700 μM were used in all experiments. The data obtained were fitted using a single-binding-site model by Origin software (v7.0; MicroCal). The first data point was deleted, and the ligand buffer control accounting for the heat of dilution (of ligand) was subtracted. Peak integration and the calculation of stoichiometry, Ka, and other parameters were also accomplished using Origin software. The experiments were reproduced three times.
Activity assay.
pET 21b plasmid containing Rel cassette was used for Rel protein preparation. Escherichia coli DH5α and E. coli BL21(DE3) were utilized for plasmid purification and protein overexpression and purification, respectively. The identity of protein and its purity was confirmed by SDS-PAGE analysis, followed by mass spectrometry (23). To trace pppGpp synthesis, [γ-32P]ATP (125 μCi, 3,500 Ci mmol−1) was added to an assay mixture containing 200 μg of purified Rel full-length protein, 1 mM nonradioactive ATP, 50 mM HEPES (pH 7.4), 5 mM MgCl2, 50 mM NaCl, and 1 mM GTP in a reduced condition, as described previously (24–26). The compounds were added to the reaction mixture at the zero time point. The protein was precipitated by heating at 95°C for 5 min, and the reaction mixture was then subjected to centrifugation at 14,000 rpm. The supernatant was loaded on a polyethyleneimine cellulose sheet for TLC analysis, followed by phosphorimaging (Bio-Rad-PharosFX Plus molecular imager). The pppGpp spot was analyzed by densitometric analysis using ImageJ software (National Institutes of Health), and the levels of pppGpp were determined.
Enzyme kinetics.
Rel from M. smegmatis is a bifunctional protein with both (p)ppGpp synthesis and hydrolysis activities. We quantified the synthesis activity of the enzyme in the presence of synthetic compounds. The Vmax and K0.5 values were derived from the steady-state kinetic experiments for the full-length Rel protein. The radioactive pppGpp spots were quantified as described previously, and the standard curve was plotted (11, 27). Kinetic parameters, such as K0.5 and Vmax, were calculated using the Hill equation. A pppGpp synthesis assay was carried out using a range of substrate (GTP) concentrations in order to monitor the enzyme kinetics. A substrate (GTP) concentration range of 0 to 3,000 μM was used for the acetylated benzoylated (AB) compound, whereas in the case of the control and acetylated (AC) compound, the substrate concentration varied from 0 to 2,000 μM. The range was adjusted to achieve saturation. K0.5 and Vmax values derived from the enzyme kinetics in the presence of compounds were compared to the values obtained from enzyme kinetics in the absence of compounds. An assay mixture without enzyme was taken as the negative control.
In vivo quantification assay for (p)ppGpp synthesis.
M. smegmatis mc2155 strain was cultured in the presence of AB and AC compounds to an optical density at 600 nm (OD600) of 0.2 in 5 ml of 1× morpholinepropanesulfonic acid (MOPS) defined medium supplemented with 80 μg/ml Casamino Acids, 0.05% Tween 80, and 2% glucose at 37°C with agitation. Cultures were radiolabeled by adding [o-32P]phosphoric acid (specific activity, >3,000 mCi/mmol; Board of Radiation and Isotope Technology, India) directly to the growth medium to a final concentration of 100 μCi/ml. The cells were harvested at 72 h of growth, followed by lyophilization. Equal amounts of cells were suspended in 20 μl of 1× MOPS solution. The cells were lysed by the addition of 12 N formic acid and stored on ice for 20 min. The sample was subjected to centrifugation at 13,000 rpm and 4°C for 10 min. Then, 2 μl of supernatant, normalized to an OD260 of 2.0, was spotted onto a polyethylenimine-cellulose sheet (Merck) for TLC analysis (1.5 M KH2PO4 [pH 3.4]) (27). The TLC sheets were air dried and phosphorimaged, and the (p)ppGpp spots were examined by densitometry as described previously. The identity of the spots was further confirmed by MALDI-TOF mass spectrometry.
Long-term survival assay.
Strains were cultured in MB7H9 medium containing 0.02% (wt/vol) glucose and 0.05% (vol/vol) Tween 80 in the presence or absence of the compounds (100 μM) of interest. The antibiotics were not used in the culture in order to rule out their effects on long-term survival. The CFU were estimated at regular time intervals for 14 days. Bacterial cultures were vortexed before plating on an MB7H9 agar plate to prevent the aggregation of cells.
Biofilm formation assay.
For biofilm assay, M. smegmatis mc2155 was grown in Sauton's medium supplemented with 2% glycerol and 0.05% Tween 80 and utilized as the primary culture. The procedure was followed as illustrated elsewhere (28–31). Briefly, fully grown primary culture was washed with Sauton's media and then used as a secondary inoculum. Biofilm was cultured in a six-well cell culture plate (Laxbro) in which the primary inoculum was 100-fold diluted with Sauton's medium. Inhibitors (100 μg/ml) were added at the zero time point. Culture plates were incubated in a humidified incubator set at 37 °C and evaluated at different time points.
M. tuberculosis Erdman biofilms were inoculated with stationary-phase planktonic cultures into Sauton's medium at a 1:100 dilution in 96-well plates to a final volume of 200 μl per well. Biofilm cultures were incubated in airtight plastic bags to restrict oxygen for 3 weeks and then vented. The compounds were added to the biofilm cultures at the time of inoculation at the indicated concentrations. Photographs of biofilms were taken at the times indicated.
Biofilm quantification.
The biofilm formation was quantified in 96-well plates, and the protocol followed was as described previously (31). Briefly, primary culture was washed and resuspended in Sauton's medium to an OD590 of 0.0025. The inhibitors were added to the well containing 200 μl of inoculum at the zero time point, and biofilm formation was monitored for up to 144 h. The medium was removed from the wells, followed by washing with water. Subsequently, staining solution (1% crystal violet, 200 μl) was added to each well, and each plate was incubated for 20 min. The wells were washed with water and dried. The dye was quantitated after solubilization in dimethyl sulfoxide (DMSO; 200 μl), and the absorption at 590 nm was then recorded using a microplate reader. This experiment was performed in three biological replicates for each inhibitor.
Biofilm disruption assay.
In this assay, M smegmatis mc2155 was allowed to form a biofilm in Sauton's medium in a 6-well plate at 37°C humidified incubator as described earlier. Inhibitors were administered below the biofilm at 72 h by using 1-ml syringe. Biofilm growth was monitored until the 126-h time point, and the plates were monitored for biofilm disruption. This experiment was conducted in three biological replicates
Hemolysis assay and microscopy.
Hemolysis of human RBCs was monitored in the presence of synthetic compounds. Left over blood sample obtained from Health Center, Indian Institute of Science, Bangalore, India, was used for this study. Hemolysis assays were performed as described elsewhere (32). Briefly, each blood sample was pelleted by centrifugation (500 × g, 10 min), and supernatant (plasma) was gently removed (treated with bleach and discarded into biohazardous wastes). The RBC pellet was washed and resuspended in PBS (pH 7.4). An assay was performed at various concentrations of inhibitors (10 μl, 20 to 220 μg/ml) mixed with 190 μl of diluted RBCs (1:20 dilution) in a 96-well plate. Then, 10 μl of 20% sodium dodecyl sulfate was used in a positive-control well, and 10 μl of phosphate buffer was used as the negative control. The plate was incubated at 37°C for 1 h, and then 100 μl of the supernatant was transferred into Eppendorf tubes, followed by centrifugation for 10 min at 500 × g. The supernatant was transferred into a fresh Eppendorf tubes, and the absorbance of the supernatant was measured at 540 nm in a UV/visible spectrophotometer (Eppendorf). This experiment was performed in triplicates. RBCs treated with synthetic compounds were observed under a light microscope at ×40 magnification. These RBCs, suspended in PBS, were poured onto a slide, covered with a coverslip, cleaned, and placed under a microscope.
Cell culture.
The human lung epithelial cell line was cultured in Roswell Park Memorial Institute 1640 (RPMI 1640) medium supplemented with 10% fetal bovine serum. The cultures were grown in a humidified atmosphere with 5% CO2 in an incubator at 37°C for 2 to 3 days. At nearly 80% confluence, the medium was removed, and the cells were washed with PBS and detached using 0.25% trypsin-EDTA at 37°C for 2 to 3 min. Subsequently, 1 ml of fresh medium was added. The cells were pelleted down at 1,000 rpm for 5 min and resuspended in fresh medium (10 ml) in a tissue culture flask. The cells were mixed with trypan blue in a 1:1 ratio and observed using hemocytometer under a microscope in order to check the cell count.
Cell permeability assay.
Cell suspensions (nearly 2,500 cells, 0.2 ml) were added to each well of a 96-well plate, followed by incubation for 24 h for adherence. The culture medium was replaced with fresh medium with compounds at a concentration of 200 μg/ml. The cells were incubated for 6 h, the medium was removed, and the cells were washed with PBS. Adhered cells were removed using 0.25% trypsin-EDTA, and the cells were washed three times with PBS. The cell pellet was resuspended in 50 μl of PBS. Cells were lysed by heating them at 95°C for 10 min, followed by centrifugation at 15,000 rpm for 10 min. This soup was transferred to a labeled tube and analyzed by mass spectrometry.
Mass spectrometry.
MALDI was used for the peak identification of drugs. A 1-μl portion of cell lysate mixed with 1 μl of CCA (α-cyano-4-hydroxycinnamic acid) was spotted onto a plate and allowed to dry. Mass spectra were recorded on Ultraflex II MALDI-TOF/TOF mass spectrometer equipped with a SmartBeam (Bruker Daltonik, Germany) operated in positive-ion, reflectron mode. Three to five mass spectra were recorded and averaged for each individual sample using 1,000 to 1,500 laser shots over the entire spot on the MALDI target plate. A PBS wash was taken as the negative control.
MTT assay.
The effect of inhibitors on cell viability was studied using an MTT assay. This is a colorimetric assay, which measures the metabolic activity of the cell as a function of reduction of tetrazolium dye to insoluble formazan. H460 cell lines were used to determine the cytotoxicity. Cell suspensions (nearly 2,500 cells, 0.2 ml) were seeded into 96-well plates and incubated for 24 h for the cells to adhere. After this culture, the medium was replaced with fresh medium with the inhibitors in a concentration range of 100 to 500 μg/ml. This experiment was conducted in three biological replicates for each concentration of the inhibitor. After 36 h of incubation, 20 μl of MTT at a concentration of 0.5 mg/ml was added to each well, followed by incubation for 4 h. Later, the medium was removed, and the cells were washed with PBS. Then, 100 μl of DMSO was used to dissolve the formazan crystal, followed by measuring the absorbance at 570 nm using an enzyme-linked immunosorbent assay plate reader. Medium alone and medium with cells (without inhibitor) were used as controls. The percentage of cell survival was calculated using the following formula: cell survival (%) = (AbsorbanceTreatment/AbsorbanceControl) × 100.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the Proteomics Facility, Molecular Biophysics Unit, Indian Institute of Science, Bangalore, and the NMR facility, Department of Organic Chemistry, Indian Institute of Science, Bangalore, for the help with the characterization of the compounds.
K.S. carried out the experiments. K.F. tested the effects of compounds on M. tuberculosis biofilms and reproduced M. smegmatis biofilm work. N.B. did permeability and MTT toxicity assay. K.F. and N.B. contributed equally. K.M. and N.J. helped in the synthesis and in NMR based characterization of compounds. C.L.S. designed the work on M. tuberculosis and partly wrote the paper. K.S. and D.C. designed the experiments, analyzed the data, and wrote the paper.
K.S. acknowledges a research associateship from the Indian Institute of Science, Bangalore, India. K.F. is supported by a pilot award from the Center for Women's Infectious Disease Research at the Washington University School of Medicine. N.B. acknowledges Department of Biotechnology, Government of India, for the research fellowship. K.M. acknowledges CSIR, New Delhi, India, for a fellowship. N.J. acknowledges the Department of Science and Technology, Government of India, for funding the laboratory. C.L.S. is supported by a Beckman Young Investigator Award from the Arnold and Mabel Beckman Foundation, an Interdisciplinary Research Initiative grant from the Children's Discovery Institute of Washington University and St. Louis Children's Hospital, and National Institutes of Health grant 4R33AI111696. D.C. acknowledges a Centre of Excellence grant, Department of Biotechnology, Government of India, for funding the laboratory.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00443-17.
REFERENCES
- 1.Liu K, Bittner AN, Wang JD. 2015. Diversity in (p)ppGpp metabolism and effectors. Curr Opin Microbiol 24:72–79. doi: 10.1016/j.mib.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu K, Myers AR, Pisithkul T, Claas KR, Satyshur KA, Amador-Noguez D, Keck JL, Wang JD. 2015. Molecular mechanism and evolution of guanylate kinase regulation by (p)ppGpp. Mol Cell 57:735–749. doi: 10.1016/j.molcel.2014.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K. 2015. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol 13:298–309. doi: 10.1038/nrmicro3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dahl JL, Arora K, Boshoff HI, Whiteford DC, Pacheco SA, Walsh OJ, Lau-Bonilla D, Davis WB, Garza AG. 2005. The relA homolog of Mycobacterium smegmatis affects cell appearance, viability, and gene expression. J Bacteriol 187:2439–2447. doi: 10.1128/JB.187.7.2439-2447.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kriel A, Bittner AN, Kim SH, Liu K, Tehranchi AK, Zou WY, Rendon S, Chen R, Tu BP, Wang JD. 2012. Direct regulation of GTP homeostasis by (p)ppGpp: a critical component of viability and stress resistance. Mol Cell 48:231–241. doi: 10.1016/j.molcel.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klinkenberg LG, Lee JH, Bishai WR, Karakousis PC. 2010. The stringent response is required for full virulence of Mycobacterium tuberculosis in guinea pigs. J Infect Dis 202:1397–1404. doi: 10.1086/656524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatterji D, Ojha AK. 2001. Revisiting the stringent response, ppGpp and starvation signaling. Curr Opin Microbiol 4:160–165. doi: 10.1016/S1369-5274(00)00182-X. [DOI] [PubMed] [Google Scholar]
- 8.Dahl JL, Kraus CN, Boshoff HI, Doan B, Foley K, Avarbock D, Kaplan G, Mizrahi V, Rubin H, Barry CE III. 2003. The role of RelMtb-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. Proc Natl Acad Sci U S A 100:10026–10031. doi: 10.1073/pnas.1631248100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weiss LA, Stallings CL. 2013. Essential roles for Mycobacterium tuberculosis Rel beyond the production of (p)ppGpp. J Bacteriol 195:5629–5638. doi: 10.1128/JB.00759-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Syal K, Maiti K, Naresh K, Chatterji D, Jayaraman N. 2015. Synthetic glycolipids and (p)ppGpp analogs: development of inhibitors for mycobacterial growth, biofilm, and stringent response. Adv Exp Med Biol 842:309–327. doi: 10.1007/978-3-319-11280-0_20. [DOI] [PubMed] [Google Scholar]
- 11.Syal K, Bhardwaj N, Chatterji D. 2017. Vitamin C targets (p)ppGpp synthesis leading to stalling of long-term survival and biofilm formation in Mycobacterium smegmatis. FEMS Microbiol Lett 364:fnw282–fnw282. doi: 10.1093/femsle/fnw282. [DOI] [PubMed] [Google Scholar]
- 12.Wexselblatt E, Oppenheimer-Shaanan Y, Kaspy I, London N, Schueler-Furman O, Yavin E, Glaser G, Katzhendler J, Ben-Yehuda S. 2012. Relacin, a novel antibacterial agent targeting the stringent response. PLoS Pathog 8:e1002925. doi: 10.1371/journal.ppat.1002925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wexselblatt E, Kaspy I, Glaser G, Katzhendler J, Yavin E. 2013. Design, synthesis and structure-activity relationship of novel Relacin analogs as inhibitors of Rel proteins. Eur J Med Chem 70:497–504. doi: 10.1016/j.ejmech.2013.10.036. [DOI] [PubMed] [Google Scholar]
- 14.Gupta KR, Baloni P, Indi SS, Chatterji D. 2016. Regulation of growth, cell shape, cell division, and gene expression by second messengers (p)ppGpp and cyclic Di-GMP in Mycobacterium smegmatis. J Bacteriol 198:1414–1422. doi: 10.1128/JB.00126-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Primm TP, Andersen SJ, Mizrahi V, Avarbock D, Rubin H, Barry CE III. 2000. The stringent response of Mycobacterium tuberculosis is required for long-term survival. J Bacteriol 182:4889–4898. doi: 10.1128/JB.182.17.4889-4898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Islam MS, Richards JP, Ojha AK. 2012. Targeting drug tolerance in mycobacteria: a perspective from mycobacterial biofilms. Expert Rev Anti Infect Ther 10:1055–1066. doi: 10.1586/eri.12.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de la Fuente-Nunez C, Reffuveille F, Haney EF, Straus SK, Hancock RE. 2014. Broad-spectrum anti-biofilm peptide that targets a cellular stress response. PLoS Pathog 10:e1004152. doi: 10.1371/journal.ppat.1004152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nunes-Alves C. 2014. Biofilms: targeting (p)ppGpp disrupts biofilms. Nat Rev Micro 12:461–461. doi: 10.1038/nrmicro3302. [DOI] [Google Scholar]
- 19.O'Toole GA, Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signaling pathways: a genetic analysis. Mol Microbiol 28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 20.Elmquist A, Langel U. 2003. In vitro uptake and stability study of pVEC and its all-D analog. Biol Chem 384:387–393. doi: 10.1515/BC.2003.044. [DOI] [PubMed] [Google Scholar]
- 21.Nazir A, Harinarayanan R. 2016. (p)ppGpp and the bacterial cell cycle. J Biosci 41:277–282. doi: 10.1007/s12038-016-9611-3. [DOI] [PubMed] [Google Scholar]
- 22.Andresen L, Varik V, Tozawa Y, Jimmy S, Lindberg S, Tenson T, Hauryliuk V. 2016. Auxotrophy-based high-throughput screening assay for the identification of Bacillus subtilis stringent response inhibitors. Sci Rep 6:35824. doi: 10.1038/srep35824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Syal K, Tadala R. 2015. Modifications in trypsin digestion protocol for increasing the efficiency and coverage. Protein Pept Lett 22:372–378. doi: 10.2174/0929866522666150327132415. [DOI] [PubMed] [Google Scholar]
- 24.Jain V, Saleem-Batcha R, Chatterji D. 2007. Synthesis and hydrolysis of pppGpp in mycobacteria: a ligand mediated conformational switch in Rel. Biophys Chem 127:41–50. doi: 10.1016/j.bpc.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 25.Syal K, Chatterji D. 2015. Differential binding of ppGpp and pppGpp to Escherichia coli RNA polymerase: photo-labeling and mass spectral studies. Genes Cells 20:1006–1016. doi: 10.1111/gtc.12304. [DOI] [PubMed] [Google Scholar]
- 26.Syal K, Joshi H, Chatterji D, Jain V. 2015. Novel pppGpp binding site at the C-terminal region of the Rel enzyme from Mycobacterium smegmatis. FEBS J 282:3773–3785. doi: 10.1111/febs.13373. [DOI] [PubMed] [Google Scholar]
- 27.Murdeshwar MS, Chatterji D. 2012. MS_RHII-RSD, a dual-function RNase HII-(p)ppGpp synthetase from Mycobacterium smegmatis. J Bacteriol 194:4003–4014. doi: 10.1128/JB.00258-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naresh K, Bharati BK, Avaji PG, Chatterji D, Jayaraman N. 2011. Synthesis, biological studies of linear and branched arabinofuranoside-containing glycolipids and their interaction with surfactant protein A. Glycobiology 21:1237–1254. doi: 10.1093/glycob/cwr068. [DOI] [PubMed] [Google Scholar]
- 29.Naresh K, Bharati BK, Avaji PG, Jayaraman N, Chatterji D. 2010. Synthetic arabinomannan glycolipids and their effects on growth and motility of the Mycobacterium smegmatis. Org Biomol Chem 8:592–599. doi: 10.1039/B917070G. [DOI] [PubMed] [Google Scholar]
- 30.Naresh K, Bharati BK, Jayaraman N, Chatterji D. 2008. Synthesis and mycobacterial growth inhibition activities of bivalent and monovalent arabinofuranoside containing alkyl glycosides. Org Biomol Chem 6:2388–2393. doi: 10.1039/b803409e. [DOI] [PubMed] [Google Scholar]
- 31.Mathew R, Mukherjee R, Balachandar R, Chatterji D. 2006. Deletion of the rpoZ gene, encoding the omega subunit of RNA polymerase, results in pleiotropic surface-related phenotypes in Mycobacterium smegmatis. Microbiology 152:1741–1750. doi: 10.1099/mic.0.28879-0. [DOI] [PubMed] [Google Scholar]
- 32.Syal K, Maiti K, Naresh K, Avaji PG, Chatterji D, Jayaraman N. 2016. Synthetic arabinomannan glycolipids impede mycobacterial growth, sliding motility and biofilm structure. Glycoconj J 33:763–777. doi: 10.1007/s10719-016-9670-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.