Abstract
Intestinal cells are covered by mucus. In the small intestine, a single unattached mucus is present whereas the colon has both an inner attached mucus layer and an outer loose mucus. The attached mucus of the colon is impenetrable to bacteria while the loose mucus acts as a habitat for commensal bacteria. In germ-free (GF) mice, small intestinal mucus is attached to the epithelium and the inner colon mucus is penetrable. O-glycosylation plays an important role in the host–microbiota interactions as the commensal bacteria use glycans as nutrient sources and attachment sites. While mucus protein composition is relatively homogenous along the intestine, its main component the Muc2 mucin shows regiospecific O-glycan patterns. We have now analyzed the glycosyltransferase relative concentrations in the epithelial cells along the intestine in GF and conventionally raised mice and compared this with the O-glycans formed. As Muc2 is the main O-glycosylated product in mucus, we made the simplified assumption that most of the glycosyltransferases found in the epithelial cells are involved in Muc2 O-glycan biosynthesis. The O-glycosyltransferase abundances along the intestine correlated well with the Muc2 O-glycan patterns. Some of the glycosyltransferases involved in the O-glycan elongation were decreased in GF mice, something that is in concordance with the observed shorter Muc2 O-glycans.
Keywords: colon, mass spectrometry, mouse, mucus, small intestine
Introduction
The mucus layer forms a physical barrier and defense system along the intestine with varying local properties. The ileum has an easily removable and thin mucus layer that is penetrable to bacteria (Ermund et al. 2013). By contrast, the mucus layer in the colon consists of an inner mucus layer (~50 µm) that is firmly attached to the epithelial cells where mucus is devoid of commensal bacteria, and an outer mucus layer (~100 µm) that is easily removed by aspiration and is a habitat for commensal bacteria (Atuma et al. 2001; Johansson et al. 2008). In germ-free (GF) mice, small intestinal mucus is attached to the epithelium and the colon mucus is penetrable showing that the formation of normal mucus layer is dependent on the commensal microbiota (Johansson et al. 2015). The mucus layers have a stable core proteome (Johansson et al. 2008; Rodríguez-Piñeiro et al. 2013), suggesting that the diverse mucus properties are caused by different posttranslational processing within the cell or following secretion, especially of its main component, the heavily glycosylated gel-forming mucin Muc2.
While Muc2 holds 30 potential N-glycosylation sites and defects in N-glycosylation will result in accumulation of Muc2 in ER, O-glycosylation is far more abundant as it increases Muc2 mass five times (Asker et al. 1998). Muc2 glycosylation plays an important role in protecting the protein backbone from host digestive and bacterial proteases, but also helps to bind water for the formation of a mucus gel. Commensal bacteria can adhere to the glycans and use them as a nutritional source (Koropatkin et al. 2012). Mucus O-glycosylation has been shown to have a regiospecific distribution (Holmén Larsson et al. 2013) and it is different from GF compared with conventionally raised (ConvR) mice (Johansson et al. 2015).
Mucin-type O-glycosylation in mammals is initiated by the family of 20 UDP-GalNAc:polypeptide N-acetylgalactosaminyl-transferases (GalNAc-T) that catalyze the addition of GalNAc (via an O-glycosidic linkage) to the hydroxyl groups of serines or threonines in the protein substrate (Tran and Ten Hagen 2013). The GalNAc transferases have distinct but partly overlapping substrate specificities and are expressed differently in cells (Bennett et al. 2012). It has been suggested that the major initiators in human O-glycosylation are GalNAc-T1, -T2 and -T3 (Kong et al. 2015). GalNAc-T7 and GalNAc-T10 have been suggested to be a follow-up enzyme at the initiation step of O-glycosylation as they have an activity toward already glycosylated peptides (Bennett et al. 1999; Revoredo et al. 2016).
After the initial addition of GalNAc, extension of the sugar chain occurs in a stepwise manner, yielding higher order glycans. The most common extension in mouse intestine, but not in human, is the Core1 structure made by the Core1 β1,3-galactosyltransferase (C1galt1) adding Gal in a β1,3-linkage (Tran and Ten Hagen 2013). C1galt1 synthesis requires a specific chaperon cosmic C1galt1c1 (Ju and Cummings 2002). The Core3 structure is catalyzed by β1,3-N-acetylglucosaminyltransferase 6 (B3gnt6), which adds a GlcNAc in β1,3-linkage to the GalNAc (Tran and Ten Hagen 2013). Core1 and Core3 structures can be further modified to form Core2 and Core4 structures, respectively, by the β1,6-N-acetylglucosaminyltransferases that add a GlcNAc in β1,6-linkage to the existing GalNAc. There are three β1,6-N-acetylglucosaminyltransferases in mammals, two of which catalyze the formation of the Core2 structure (Gcnt1 and 2) and one that can catalyze the formation of either the Core2 or Core4 structure (Gcnt3) (Tran and Ten Hagen 2013). Further elongation and/or termination of O-glycans involves large array of glycosyltransferases, fucosyltransferases, sulfotransferases and sialyltransferases (Potapenko et al. 2010).
As Muc2 is the major O-glycosylated product in mucus of the small intestine and colon, we made the simplified assumption that most of the glycosylation transferases found in the epithelium are involved in Muc2 O-glycosylation. This allowed us to correlate the Muc2 O-glycosylation profile of the small intestine and colon with the relative abundancies of glycosyltransferases detected by proteomics.
Results
O-glycosylation initiation
Mucus from small and large intestine of ConvR and GF C57B/6 mice was scraped and the Muc2 mucin was partially purified from duodenum (ConvR), mid-jejunum (ConvR), ileum (ConvR, GF), proximal colon (ConvR), middle colon (ConvR, GF) and distal colon (ConvR). The mucus purification was based on mucus insolubility in guanidinium chloride. O-glycans were released by beta elimination and analyzed by graphitized carbon capillary-LC-MS/MS (Holmén Larsson et al. 2013). For quantifying glycosyltransferase protein expression, epithelial cells from the duodenum, jejunum, ileum, proximal colon and distal colon were isolated from ConvR and GF mice. Cells were lysed and proteins digested with endoproteinase LysC and resulting peptides were analyzed with nanoLC-MS/MS. The relative amounts of O-glycans and the relative concentrations of glycosyltransferases per cell were estimated as described in Materials and methods.
Peptidylgalactosaminyltransferases, GalNAc-Ts
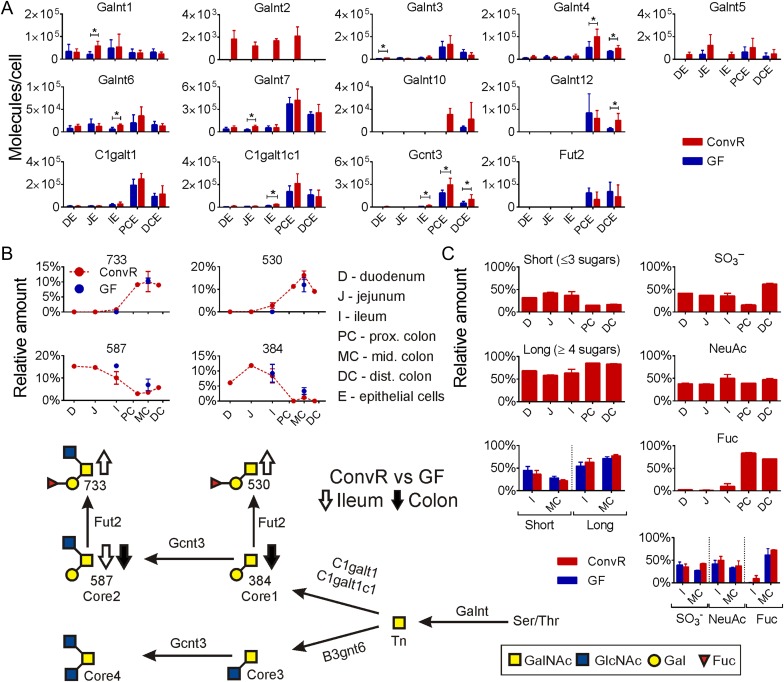
Out of 19 GalNAc-T found in mice, we could identify 9 (Galnt1, 2, 3, 4, 5, 6, 7, 10, 12) (Figure 1A). Galnt7 was the most abundant GalNAc-T and together with Galnt3, and 4 had the highest abundance in the colon compared with the small intestine (Figure 1A). Galnt1, 2, 5 and 6 had similar levels in small intestine and colon. Galnt10 and 12 were not detected in the small intestine. Most of the GalNAc-Ts showed increased levels in both small and large intestine of colonized mice (Figure 1A). One transferase, Galnt2 was only found in ConvR mice intestine (Figure 1A). Together, the results indicate a more active mucin biosynthesis and coupled O-glycosylation in the presence of microbiota.
Fig. 1.
The main mucin-type O-glycosylation pathways in mouse intestine. (A) Relative protein amounts for GalNAc transferases and for the enzymes responsible forming core structures along the intestine in ConvR and GF mice. Significance changes based on multiple t-test are shown with an asterisk. (B) Relative amounts of major core glycans are shown along the intestine in ConvR mice and compared with GF mice in ileum and middle colon, glycans are named by their molecular mass. (C) Relative amounts of short and long Muc2 glycans along ConvR mice intestine and comparison ConvR vs GF (left panel); relative amounts of sulfated, sialylated and fucosylated Muc2 glycans along ConvR mice intestine and comparison ConvR vs GF mice (right panel). Samples for Muc2 glycan analysis and proteomics from intestinal epithelial cells (E) are marked as follows: D, duodenum; J, jejunum; I, ileum; PC, proximal colon; MC, middle colon; DC, distal colon. Arrows (up/down) on biosynthesis pathway show the direction of the alteration in O-glycan amounts in the presence of microbiota. This figure is available in black and white in print and in color at Glycobiology online.
Core1 β1,3-Galactosyltransferase (C1galt1) and Its Chaperone Cosmc (C1galt1c1)
These two enzymes had similar relative protein abundance patterns along the intestine where both showed to be significantly more abundant in colon (Figure 1A). Colonization increased the expression of these proteins in all tissues, with C1galt1c1 being increased 2.5 times in ConvR mouse ileum compared with the GF mouse (Figure 1A). Interestingly, the product of C1galt1, the Core1 disaccharide, glycan 384 (glycan names are given according to their molecular mass) was more abundant in the small intestine than in the colon for both mice (Figure 1B, Table I) which is in keeping with the findings of Holmén Larsson et al. (2013). This glycan also had a somewhat higher abundance in the GF mice colon (3.3% of the glycans) compared with ConvR mice (1.2%) (Figure 1B, Table I) as was also previously observed by Johansson et al. (2015). As the C1galt1 enzyme abundance is much lower in the small intestine compared with the colon of both the ConvR and GF mice, the observed higher level of the Core1 glycan 384 in the small intestine is likely caused by reduced Core1 disaccharide elongation and perhaps by increased fucosylation in the colon. This is further supported by the overall finding that shorter glycans (≤3 sugars) were more abundant in the small intestine while longer glycans (≥4 sugars) were more abundant in the colon (Figure 1C).
Table I.
Structures of main O-linked oligosaccharides and their relative amounts
| Mass labelsa | Core | Proposed oligosaccharide sequence/compositionb,c,d | Duodenum (ConvR)e | Jejunum (ConvR)e | Ileum (ConvR)e | Ileum (GF)e | Proximal colon (ConvR)e | Middle colon (ConvR)e | Middel colon (GF)e | Distal colon (ConvR)e |
|---|---|---|---|---|---|---|---|---|---|---|
| Neutral | ||||||||||
| 384 | 1 | Gal-GalNAcol | 6.1% | 11.8% | 8.4% | 9.2% | – | 1.2% | 3.3% | – |
| 530 | 1 | Fuc-Gal-GalNAcol | – | – | 2.8% | – | 11.3% | 16.2% | 12.0% | 9.1% |
| 587 | 2 | Gal-(GalNAc-6)GalNAcol | 15.3% | 14.7% | 10.1% | 15.4% | 3.0% | 3.6% | 7.0% | 5.7% |
| 733 | 2 | Fuc-Gal-(GalNAc-6)GalNAcol | – | – | 0.8% | – | 9.1% | 10.1% | 10.4% | 8.9% |
| 749 | 2 | Gal-(Gal-4GlcNAc-6)GalNAcol | 17.9% | 13.1% | 6.6% | 7.9% | – | – | 2.9% | – |
| 895a | 2 | Gal-(Fuc-Gal-3/4GlcNAc-6)GalNAcol | – | – | – | – | 2.0% | 3.9% | 1.1% | 2.3% |
| 895b | 2 | Gal-(Fuc-Gal-4/3GlcNAc-6)GalNAcol | – | – | – | – | 5.6% | 1.2% | 2.1% | 2.6% |
| 1041a | 2 | Fuc-Gal-(Fuc-Gal-3/4GlcNAc-6)GalNAcol | – | – | 1.2% | – | 3.6% | 5.5% | 2.5% | 2.4% |
| 1041b | 2 | Fuc-Gal-(Fuc-Gal-4/3GlcNAc-6)GalNAcol | – | – | 0.3% | – | 13.5% | 4.1% | 7.1% | 4.3% |
| One acidic residue | ||||||||||
| 667 | 2 | Gal-(SO3 −-GlcNAc-6)GalNAcol | 10.0% | 15.7% | 15.2% | 20.4% | 0.6% | 1.4% | 5.8% | 1.8% |
| 813 | 2 | Fuc-Gal-(SO3 −-GlcNAc-6)GalNAcol | 0.3% | – | 1.9% | – | 3.0% | 6.7% | 4.8% | 8.5% |
| 829a | 2 | Gal-(Gal-(SO3 −)GlcNAc-6)GalNAcol | 4.2% | 3.4% | 2.0% | 2.3% | – | – | 1.0% | 0.4% |
| 829b | 2 | Gal-(SO3 −-Gal-GlcNAc-6)GalNAcol | 2.3% | 2.0% | 1.1% | 1.5% | – | 0.3% | 0.5% | 0.4% |
| 975a | 2 | Gal-(Fuc-Gal-(SO3 −-)GlcNAc-6)GalNAcol | – | – | – | – | 1.0% | 1.0% | 0.9% | 0.9% |
| 975b | 2 | Fuc-Gal-(SO3 −-Gal-GlcNAc-6)GalNAcol | – | – | – | – | – | 2.2% | 0.7% | 2.4% |
| 1032 | 2 | Gal-(HexNAc-Gal-(SO3 −-)GlcNAc-6)GalNAcol | 1.8% | 1.3% | – | 1.3% | – | – | 0.6% | – |
| 1040 | 2 | [NeuAc]1 Gal-(Gal-GlcNAc-6)GalNAcol | – | 0.9% | 1.5% | – | – | 0.7% | – | – |
| 1121 | 2 | Fuc-Gal-(Fuc-Gal-(SO3 −)GlcNAc-6)GalNAcol | – | – | – | – | 3.8% | 2.6% | 3.3% | 1.7% |
| 1243a | 2 | [NeuAc]1 Gal-(HexNAc-Gal-GlcNAc-6)GalNAcol | 10.5% | 12.9% | 13.1% | 15.2% | 0.6% | 0.5% | 3.1% | – |
| 1243b | 2 | [NeuAc]1 HexNAc-Gal-3(Gal-GlcNAc-6)GalNAcol | 0.3% | 0.9% | 6.5% | 7.4% | – | 0.5% | 3.2% | – |
| 1389 | 2 | [NeuAc]1 Fuc-Gal-(HexNAc-Gal-4GlcNAc-6)GalNAcol | 0.3% | – | 1.9% | – | 2.8% | 1.5% | 2.0% | 1.4% |
| Multiple acidic residues | ||||||||||
| 1266 | 2 | [NeuAc]1 [SO3 −]1 Fuc-Gal-(Gal-GlcNAc-6)GalNAcol | – | – | – | – | – | 3.1% | – | 2.3% |
| 1323 | 2 | [NeuAc]1 HexNAc-Gal-(Gal-(SO3 −-)GlcNAc-6)GalNAcol | 5.8% | 6.2% | 5.7% | 8.0% | – | 1.7% | 1.8% | 1.4% |
| 1331 | 2 | [NeuAc]1 Gal-(Gal-(NeuAc-)GlcNAc-6)GalNAcol | – | – | – | – | 0.8% | – | – | – |
| 1469 | 2 | [NeuAc]1 Fuc-Gal-(HexNAc-Gal-(SO3 −-)GlcNAc-6-)GalNAcol | 0.2% | – | 1.4% | – | 2.1% | 3.5% | 3.4% | 2.4% |
| 1477 | 2 | Gal-(GlcNAc-(SO3 −-)Gal-GlcNAc-Gal-(SO3 −-)GlcNAc-6)GalNAcol | 2.7% | – | – | – | 2.5% | 0.8% | – | |
| 1526a | 2 | [NeuAc]1 HexNAc-Gal-(HexNAc-Gal-(SO3 −)GlcNAc-6)GalNAcol | 1.0% | 1.0% | 1.2% | – | – | – | – | – |
| 1526b | 4 | [NeuAc]1 GlcNAc-(SO3 −-Gal-GlcNAc-Gal-GlcNAc-6)GalNAcol | – | – | – | – | – | 1.4% | 0.2% | 2.7% |
| 1535 | 2 | [NeuAc]2 Gal-(HexNAc-Gal-GlcNAc-6)GalNAcol | 0.8% | 1.3% | 1.5% | – | 6.1% | 3.9% | 2.8% | 0.9% |
| 1565 | 2 | [NeuAc]1 Gal-(SO3 −-Gal-GlcNAc-Gal-(SO3 −)GlcNAc-6)GalNAcol | – | – | – | – | – | 5.2% | – | 2.9% |
| 1681 | 2 | [NeuAc]2 Fuc-Gal-(HexNAc-Gal-GlcNAc-6)GalNAcol | – | – | – | – | 8.6% | 3.7% | 5.2% | 0.8% |
| 1688 | 2 | [NeuAc]1 HexNAc-Gal-(Gal-GlcNAc-Gal-(SO3 −)GlcNAc-6)GalNAcol | 4.6% | 3.5% | 2.4% | 3.3% | – | – | 0.8% | 0.6% |
| 1711 | 2 | [NeuAc]1 Fuc-Gal-(SO3 −-Gal-GlcNAc-Gal-(SO3 −)GlcNAc-6)GalNAcol | – | – | – | – | 0.9% | 2.2% | 0.2% | 3.6% |
| 1730 | 4 | [NeuAc]1 GlcNAc-(HexNAc-Gal-GlcNAc-Gal-(SO3 −)GlcNAc-6)GalNAcol | 0.2% | 0.4% | 0.7% | – | – | – | – | 0.8% |
| 1738 | 2 | [NeuAc]2 HexNAc-Gal-(HexNAc-Gal-GlcNAc-6)GalNAcol | 1.2% | 1.7% | 1.6% | 1.1% | 1.0% | 0.5% | 0.8% | – |
| 1769 | 2 | [NeuAc]1 Gal-(HexNAc-(SO3 −)Gal-GlcNAc-Gal-(SO3 −)GlcNAc-6)GalNAcol | – | – | – | – | – | 1.2% | – | 2.6% |
| 1835a | 2 | [NeuAc]1 Fuc-Gal-GlcNAc-Gal-(SO3 −)-GlcNAc-Gal-(HexNAc-6)GalNAcol | – | – | – | – | – | 1.6% | – | 5.3% |
| 1884a | [NeuAc]2 [Fuc]1 [HexNAc]3 [Gal]2 GalNAcol | – | – | – | – | 1.5% | – | 0.7% | – | |
| 1884b | 4 | [NeuAc]2 HexNAc-Gal-GlcNAc-(Fuc-Gal-4GlcNAc-6)GalNAcol | – | – | – | – | 2.9% | – | 1.6% | – |
| 1915 | 2 | [NeuAc]1 Fuc-Gal-(HexNAc-(SO3 -)-Gal-GlcNAc-Gal-(SO3 -)-GlcNAc-6)GalNAcol | – | – | – | – | – | – | – | 5.3% |
| 2103 | 2 | [NeuAc]2 HexNAc-Gal-GlcNAc-Gal-(HexNAc-Gal-GlcNAc-6)GalNAcol | 1.5% | 1.8% | 7.1% | 3.3% | – | – | 1.6% | – |
| 2126 | 2 | [NeuAc]2 Fuc-Gal-(HexNAc-Gal-GlcNAc-Gal-(SO3 -)GlcNAc-6)GalNAcol | – | – | – | – | 2.7% | – | 1.4% | – |
| 2249 | 4 | [NeuAc]2 Fuc-Gal-GlcNAc-(HexNAc-Gal-GlcNAc-Gal-GlcNAc-6)GalNAcol | – | – | – | – | 1.5% | – | 1.2% | – |
| 2425 | 2 | [NeuAc]2 SO3 --Gal-GlcNAc-Gal-(HexNAc-Gal-GlcNAc-Gal-(SO3 -)GlcNAc-6)GalNAcol | – | – | – | – | – | – | – | 3.9% |
| 2548 | 2 | [NeuAc]2 [SO3 -]1 HexNAc-Gal-GlcNAc-Gal-(HexNAc-Gal-GlcNAc-Gal-GlcNAc-6)GalNAcol | 1.5% | 1.4% | 1.8% | 1.5% | – | – | 0.4% | – |
| 2557 | 4 | [NeuAc]2 Fuc-Gal-GlcNAc-(Fuc-Gal-GlcNAc-Gal-GlcNAc-Gal-GlcNAc-6)GalNAcol | – | – | – | – | 1.9% | – | 0.6% | – |
| 2628 | 2 | [NeuAc]2 [SO3 -]1 HexNAc-Gal-GlcNAc-Gal-(HexNAc-Gal-GlcNAc-Gal-(SO3 -)GlcNAc-6)GalNAcol | – | – | – | – | – | – | – | 1.9% |
aMass labels are used as the name for individual glycans and based on the molecular mass of the glycan. More than one glycan with identical mass is marked by a or b.
bProposed structures were interpreted from LC-MSn analysis data collected in the negative and positive ion mode. Assumptions: Hex residues are Gal, deoxyHex are Fuc, HexNAc within chain are GlcNAc and HexNAcol are GalNAcol. The Fuc-Gal linkage is assumed to be α1-2.
cResidues attached to C6 of GalNAcol are highlighted in bold.
dSugar residues within [ ] have not been possible to localize and the numbers are given as subscript. A number of low abundant oligosaccharides are only given as monosaccharide composition as it has not been possible to elucidate their sequence.
eRelative percentage was calculated by normalizing to total identified peak areas. The peak areas for singly and doubly charged oligosaccharides, which has been compensated for difference in ionization efficiency with the factor of 0.4 and 0.3, respectively (for more detail, see Olson et al. 2005).
Core β1,6-N-acetylglucosaminyltransferases
B3gnt6 transferase that catalyzes the formation of the Core3 disaccharide core structure was not found in the current proteomic study, as reflected in low amounts of Core3 and Core4 glycans observed in mouse intestine (Table I) as has also been observed previously (Thomsson et al. 2012; Holmén Larsson et al. 2013). The only additional β1,6-N-acetylglucosaminyltransferase identified in the current analysis was Gcnt3, responsible for the formation of both the Core2 and Core4 trisaccharide core structures. Gcnt3 showed a much higher abundance in colon compared with the small intestine (Figure 1A). Similar to the Core1 transferase and its disaccharide product, the major Gcnt3 product the Core2 trisaccharide, glycan 587, showed an inverse relationship with expression and product levels, with higher levels of glycan 587 found in the small intestine compared with the colon (Figure 1A and B, Table I). This inverse relationship is also observed in the ileum and colon of the GF mice, which express lower level of Gcnt3 but have higher levels of the Core2 trisaccharide, glycan 587, compared with ConvR mice (Figure 1B, Table I). Again this inverse relationship suggests further elongation of the Core2 glycans in the colon compared with the small intestine (Figure 1C).
Fucosyltransferase, Fut2
Fut2 was the only identified enzyme able to add Fuc to Gal and thus all such linkages can be assumed to be α1,2. Both fucosylated Core1 glycan 530 and Core2 glycan 733 were highly abundant in colon (Figure 1B, Table I), as described previously (Holmén Larsson et al. 2013). The Fut2 transferase was thus detected in colon, but not in the small intestine (Figure 1A). Still small amounts of fucosylated Core1 and Core2 glycans were detected in ConvR mice ileum, but not in GF mice ileum (Figure 1B and C), The absence of Fut2 in the small intestine was accordingly accompanied by the accumulation of the Core1 glycan 384 and Core2 glycan 587 (Figure 1B, Table I). Further modifications and extensions of these core glycans resulted in a large pool of differently elongated glycans that differ along the intestine (Holmén Larsson et al. 2013). These subsequent elongation steps will be discussed separately for the small intestine and colon.
O-glycan elongation in the small intestine
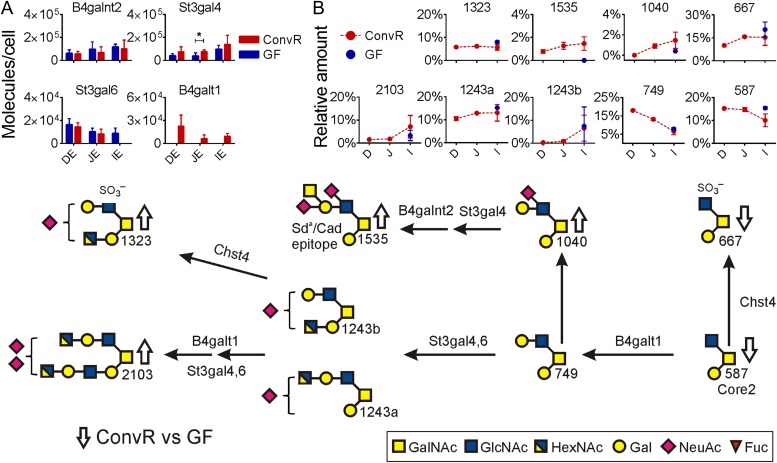
Fewer glycosyltransferases that elongate the core structures were detected in the small intestine compared with the colon, 4 vs. 10 as shown in Figures 2A and 3A. One β1,4-galactosyltransferase, two sialyltransferases and one β1,4-N-acetylgalactosaminyltransferase were detected in the small intestine.
Fig. 2.
Main mucin-type O-glycosylation pathways in mouse small intestine. (A) Relative concentrations for glycosyltransferases detected in small intestine of ConvR and GF mice. Significant protein changes based on multiple t-test are shown with an asterisk. (B) Relative amounts of most abundant glycans are shown in ConvR mice small intestine and compared with GF mice in ileum, identified glycans are named by their molecular mass. Arrows (up/down) on biosynthesis pathway show the alteration in O-glycan amounts in the presence of microbiota. This figure is available in black and white in print and in color at Glycobiology online.
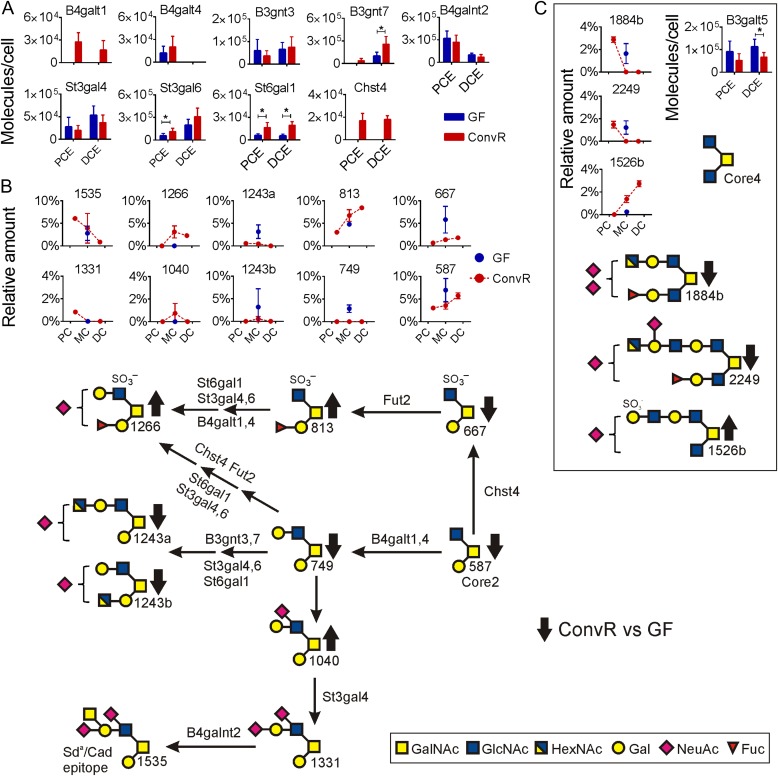
Fig. 3.
The main mucin-type O-glycosylation pathways in mouse colon. (A) Relative concentrations for glycosyltransferases detected in colon of ConvR and GF mice. Significant protein changes based on multiple t-test are shown with an asterisk. (B) Relative amounts of most abundant glycans are shown in ConvR mice colon and compared with GF mice. Identified glycans are named by their molecular mass. (C) Relative amounts of Core4 glycans and the B3galt5 transferase. Arrows (up/down) on biosynthesis pathway show the alteration in O-glycan amounts in the presence of microbiota. This figure is available in black and white in print and in color at Glycobiology online.
β1,4-Galactosyltransferase, B4galt1
β1,4-Galactosyltransferases are responsible for elongation of Core2 or Core4 structures by a Gal residue. The B4galt1 was the only transferase detected and this was found only in ConvR small intestine and not in GF mice (Figure 2A). The most abundant direct product of this enzyme was the elongated Core2 glycan749 that was of highest intensity in the ConvR proximal small intestine, the same location as the highest amount of the B4galt1 transferase (Figure 2B, Table I). Small amounts of the 749 glycan were also found in GF mice ileum, although the corresponding enzyme was not detected.
Sialyltransferases, St3gal4 and St3gal6
Two sialyltransferases were identified in the small intestine: β-galactoside α2,3-sialyltransferase 4 and 6 (St3gal4 and St3gal6). The abundance of St3gal4 increased in the distal direction, whereas St3Gal6 decreased (Figure 2A). Also the abundance of the sialylated glycans increased in the distal direction (glycans 1040, 1234a and b, 1323, 1535 and 2103, Figure 2B). In the presence of microbiota, the St3gal4 enzyme increased at the same time as the St3gal6 was unaffected in proximal and mid small intestine (Figure 2A). However, in ileum St3gal6 was undetectable in the ConvR mice, but well in GF, suggesting that it had decreased after bacterial colonization. The largest glycan difference in ConvR mice ileum compared with GF was the increased levels of the disialylated glycan 2103 (Figure 2B, Table I). Several other sialylated products were only detected in ConvR mice ileum (Table I, glycans 1469, 1526, 1535), probably reflecting the increased levels of St3gal4 transferase in ConvR mice small intestine.
β1,4-N-Acetylgalactosaminyltransferase 2, B4galnt2
The addition of GalNAc to sialylated acceptors creates the Sda/Cad epitope (Dall'Olio et al. 2014). The main Sda/Cad epitope carrying glycan in mice was the glycan 1535 found throughout the small intestine as was the necessary enzyme B4galnt2 (Figure 2B, Table I). Although there was no major change in the levels of the B4galnt2 transferase upon bacterial colonization, the Sda/Cad glycan 1535 was absent in GF mice small intestine (Figure 2A and B).
O-glycan elongation in colon
More glycosyltransferases were detected in colon than in small intestine (Figure 3A). The B4galnt2 transferase responsible for Sda/Cad epitope, increased in amounts from ileum to become one of the most abundant glycosyltransferases in proximal colon. This is reflected in the high concentration of the Sda/Cad glycan 1535 at this location. The sialyltranferase St3gal4 required for making the Sda/Cad precursor had on the other hand a higher abundance in distal colon (Figure 3A). This regional difference of these two enzymes has also been observed in human colon (van der Post and Hansson 2014).
β1,3-N-Acetylglucosaminyltransferases, β1,4-Galactosyltransferases
The B3gnt7 transferase had a high abundance in mouse distal colon, whereas the B3gnt3 had an even distribution (Figure 3A). These enzymes are responsible for elongation of glycans by GlcNAc and together with the B4galt enzymes (B4galt1 and B4galt4) synthesize lactosamine elongations. More of these enzymes were found in colon as compared with the small intestine thus these enzymes are responsible for the tendency of longer glycans in colon (Figure 1C). Bacterial colonization increased especially the B3gnt7 and B4galt1 transferase levels, for the later transferase from levels below detection. This suggests that the glycans in GF mice will be less elongated, something that was supported by the glycan analysis showing increased levels of short glycans in GF mice colon (Figure 1C). For example, the Core1 disaccharide 384 (Figure 1B), Core2 trisaccharide 587 and its sulfated product 667 showed higher amounts in GF mouse colon (Figure 3B).
β1,3-Galactosyltransferase 5, B3galt5
The B3galt5 enzyme catalyzing the transfer of Gal to GlcNAc-based acceptors with a preference for the Core3 structure (GlcNAcβ1,3-GalNAc) was found only in colon and had higher abundance in GF mice (Figure 3C). As stated before, no Core3 glycans were detected in mice small intestine or large intestine. However, some Core4 glycans were found with two Core4 glycans 1884b and 2249 that had higher abundance in GF mice colon (Figure 3C, Table I). The Core4 glycan 1526b, which had not been elongated on its C3 branch, had a higher abundance in ConvR mice colon (Figure 3C, Table I). This is in line with the lower level of the B3galt5 transferase after bacterial colonization.
Sialyltransferases, St3gal4, St3gal6 and St6gal1
In addition to the two St3gal transferases found in small intestine, also an enzyme adding sialic acid in the 6-position (St6gal1) was found in colon. All three sialyltransferases increased in amount toward distal colon and two of these enzymes, St3gal6 and St6gal1, increased significantly upon colonization (Figure 3A). This was reflected in slightly higher amounts of the longer sialylated glycans in ConvR mice colon, for example glycans 1040 and 1266 (Figures 1C and 3B).
Sulfotransferase 4, Chst4
The only sulfotransferase identified in mouse intestine was Chst4. This was only detected in ConvR colon (Figure 3A). However, relatively large amounts of sulfated glycans were found in the small and large intestine of both ConvR and GF mice (Figure 1C). This suggests that the sulfotransferases were expressed in too low protein concentrations to be detected in GF and ConvR small intestine or that additional enzymes not annotated in the available databases were responsible. The sulfated Core2 glycan 667 was abundant in GF mice colon and decreased upon colonization (Figure 3B). This glycan can be fucosylated to give glycan 813, a compound found in increased amounts in colonized mice (Figure 3B) although the responsible Fut2 transferase showed similar levels in GF and ConvR mice colon (Figure 1A).
Other glycans
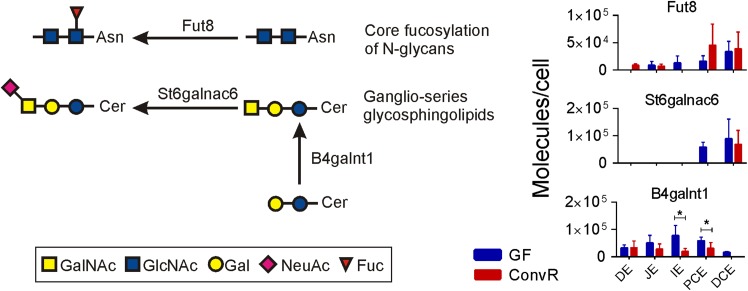
Additional glycosyltransferases not involved in O-glycan biosynthesis were also detected. Fut8, α1,6-fucosyltransferase responsible for the fucosylation of the N-glycan core structure, was found at low concentration in the small intestine and at higher levels especially in the ConvR mice proximal colon (Figure 4). B4galnt1 and St6galnac6, responsible for synthesizing ganglio-series glycosphingolipids (Potapenko et al. 2010), were more abundant in GF mice ileum and colon (Figure 4). This indicates that also glycosphingolipids biosynthesis is altered by microbiota.
Fig. 4.
Non-O-glycan transferases altered in mouse intestine by microbiota. Examples of transferases involved in N-glycan and glycosphingolipid biosynthesis. The relative amounts of these glycans were not analyzed. Significant protein changes based on multiple t-test are shown with an asterisk. This figure is available in black and white in print and in color at Glycobiology online.
Discussion
Protein O-glycosylation is initiated by GalNAc-Ts, among which GalNAc-T1, -T2, -T3 have been suggested to be most important in humans (Kong et al. 2015). In mouse intestine, Galnt1 is most abundant in small intestine while Galnt3 is more abundant in colon. Galnt7 and Galnt10 belong to the same enzyme family and are active on already glycosylated peptides (Bennett et al. 1999; Revoredo et al. 2016). Galnt7 is more abundant than Galnt10, but both are most abundant in colon and are adding GalNAc directly next to or with an intervening Pro to a GalNAc substituted Thr (van der Post et al. 2013; Revoredo et al. 2016). It can be suggested that these enzymes are important for the generation of a denser O-glycosylation that may be of functional importance for protecting the mucin protein core from bacterial proteases. These two enzymes are more abundant in ConvR mice compared with GF as are most of the other Galnt enzymes, also in agreement with an important role for GalNAc-Ts in generating the protection of the mucin protein core. Human GalNAc-T3 has been shown to glycosylate a specific Thr and by this block a bacterial protease cleavage in MUC2 that when cleaved will disrupt the MUC2 polymer and thus the mucus layer (van der Post et al. 2013). Accordingly, Galnt3 was found to be abundant in both mouse and human colon. However, there was no difference in Galnt3 expression between ConvR and GF mice, except in ConvR duodenum where twice as much Galnt3 was found. Galnt2, detected only in ConvR mice, has been shown to play a role in lipid metabolism (Kathiresan et al. 2008; Teslovich et al. 2010), a process known to be affected by bacteria (Rabot et al. 2010; Velagapudi et al. 2010).
A majority of the ileum and colon glycans in mouse are based on the Core2 substructures for which the Core1 and Core2 (C1galt1, C1galt1c1, Gcnt3) transferases are required. Mice have, in contrast to humans, less of Core3 and Core4 glycans as only a few glycans with a Core4 substructure were possible to identify. The Core4 1884 glycan was highly increased in the intestinal epithelial cell-specific Core1 knockout mice (IEC C1galt1−/−) (Thomsson et al. 2012). This animal was shown to have an altered microbiota as compared with ConvR mice (Sommer et al. 2014). The IEC C1galt1−/− mice were also more susceptible to dextran sodium sulfate-induced colitis. This supports a role for the mucin glycans as interaction partners for bacteria, and the importance of sufficiently long O-glycans on the mucins that requires longer time for the bacteria to degrade glycans and the mucus (Sommer et al. 2014; Bergstrom et al. 2016). However, bacterial effect on the host is even more complex than just extending glycans. The Core4 glycans (1884b and 2249) with extensions on both the C3 and C6 branches were in contrast to other longer glycans decreased in the presence of bacteria. The Core4 glycan 1526 with the extension on only the C6 branch was increased, showing that the presence of microbiota could also lower the transferase levels as for the B3galt5 that can extend C3 branch by adding Gal.
Most typical for intestinal colonization was increased glycosylation as more glycosyltransferases and longer glycans (Figure 1C) were detected in ConvR mice intestine. In the GF mice ileum and mid colon, several shorter glycans were highly abundant as the Core1 and Core2 glycans 384 and 587, and sulfated Core2 glycan 667. The increased relative amounts of short glycans were accompanied by lower levels of enzymes responsible for elongation of the core structures (for example B3gnt7, B4galt1). This nicely illustrates that glycan elongation is just as important as acceptor biosynthesis and exemplifies the complex relation between transferase levels and glycan amounts. The insoluble form of the Muc2 mucin was prepared from epithelial scrapings of washed tissue and thus reflects intracellular and inner mucus layer material where there is no or little bacteria. The Muc2 glycans can be expected to be intact, suggesting that the variation in length was not due to host or bacterial glycosidases.
Many glycans in the mouse intestine are mono- or disialylated (Holmén Larsson et al. 2013). The main sialylation enzymes identified in this study were St3gal6 and St6gal1, highest in colon, and St3gal4, highest in small intestine. Generally, the relative concentration of sialyltransferases increased upon colonization. The Muc2 glycan levels were in agreement with this as less sialylated glycans were found in GF mouse ileum and colon. The functional importance of these alterations is not understood, but sialic acid is implicated in both bacteria binding and masking bacterial binding sites (Miller-Podraza et al. 1999; Mahdavi et al. 2002).
Sulfated glycans are abundant in both the small intestine and colon, and are at higher relative amounts in the distal colon. The only enzyme responsible for sulfation that could be identified by proteomics was Chst4. This was found only in ConvR colon reflecting the higher levels of sulfated glycans in colonized mouse colon.
Mice without gut microbiota have far fewer epithelial cells expressing the α1,2-fucosylated epitope in the small intestine. The increase in this epitope upon colonization has been suggested to be caused by microbes interacting with host type 3 innate lymphoid cells that signal to the epithelial cells to produce the fucosylation enzyme (Goto et al. 2014). Fut2 gene upregulation and the correspondingly increased glycan fucosylation were observed in the small intestine of cystic fibrosis (CF) mice known to have bacterial overgrowth (Thomsson et al. 2002). GF mice small intestine lacks the fucosylated glycans 530 and 733 found in the ConvR mice. The only fucosyltransferase identified capable of adding fucose to galactose was Fut2, but not detected in the GF or ConvR small intestine. In colon on the other hand, Fut2 was detected with similar abundances in both GF and ConvR mice, something that correspond to the many fucosylated glycans found. Together, the results indicate that mouse Fut2 expression and fucosylation is highly dependent on the small intestinal luminal bacteria whereas colon fucosylation is independent of bacteria.
Glycosyltransferases along the length of the human colon have previously been identified by proteomics after enriching for membrane proteins (van der Post and Hansson 2014). We identified similar number of glycosyltransferases by analyzing whole epithelial cell fraction, suggesting that this approach was sensitive enough and membrane enrichment was not required for identifying most of the O-glycosylation transferases. However, as always for mass spectrometry-based proteomics, highly complex samples may mask low abundant proteins as for example glycosyltransferases. For example, Chst4 was detected at low amounts in ConvR colon (in seven out of nine biological replicates) and not detected in ConvR small intestine and GF mice where sulfated glycans were still observed. Fut2 was not found in small intestine despite trace amounts of fucosylated glycans, but present in colon (averagely in six out of nine biological replicates). A recent study comparing mRNA levels in ConvR and GF mice intestinal tissues showed expression of Chst4 and Fut2 mRNA along the entire intestine in both animals but with a higher abundance in colon (Larsson et al. 2012). In a deep and quantitative proteome analysis of 28 mouse tissues including duodenum, jejunum, ileum and colon, Chst4 was not observed at all and Fut2 only in colon (Geiger et al. 2013). This illustrates the current limitations of mass spectrometry-based proteomics when it comes to low abundant proteins in complex cellular contexts. Targeted proteomics using isotopically labeled standard peptides known to quantify attomole levels (Gallien et al. 2012) could increase the sensitivity in detecting transferases predicted to be involved in O-glycosylation.
Generally, the glycans of ConvR mice intestine were more sialylated, sulfated (colon only), fucosylated and longer. The fucosylation of mouse small intestinal Muc2 was low compared with colon and the glycans shorter, and this was also reflected by the less glycosyltranferases detected with proteomics. Studies have shown that there is a regiospecific bacterial colonization along the intestine, suggesting a role for the mucin O-glycans in the selection of bacterial flora (Donaldson et al. 2015). Already 3 weeks following the introduction of intestinal bacteria into GF mice, the glycosylation pattern was normalized (Johansson et al. 2015). This suggests that the mouse glycosylation machinery responds rapidly to bacterial products. Mucin O-glycosylation has been shown to be dynamic and glycan patterns are commonly altered as a secondary effect to inflammation, for example in CF and inflammatory bowel disease (IBD) (Karlsson et al. 2000; Thomsson et al. 2002; Holmén et al. 2003; Larsson et al. 2011). In patients with IBD, it was further shown that the MUC2 glycosylation pattern was reversed to normal when a patient with active disease went into remission (Larsson et al. 2011).
The O-glycosyltransferase abundances along the intestine correlate relatively well with the Muc2 O-glycan pattern. GF mice tend to have shorter glycans and fewer enzymes involved in the glycan elongation. It could be speculated that there is less need for long glycans in GF as compared with mice colonized with commensal bacteria. Glycans are required for protecting the mucins from degradation, but can also act as a nutritional source for bacteria. However, the basic machinery and signaling pathways used by the host to recognize and adapt to the intestinal bacteria by altering glycosyltransferase expression are currently unknown.
Materials and methods
Partial purification of the Muc2 mucin and its oligosaccharide analysis
Mucin oligosaccharide analysis was performed as described previously (Holmén Larsson et al. 2013). Briefly, mucus was scraped from the small and large intestine of ConvR and GF C57BL/6 mice. The insoluble Muc2 mucin was partially purified from duodenum (ConvR), mid-jejunum (ConvR), ileum (ConvR, GF), proximal colon (ConvR), middle colon (ConvR, GF) and distal colon (ConvR) by repeated 6 M guanidinium hydrochloride (GuHCl) extraction. The GuHCl insoluble fractions containing Muc2 were reduced, alkylated and analyzed by Ag-PAGE composite gel electrophoresis. Gel was stained by Alcian blue to detect the negatively charged glycans, and the protein bands were blotted onto a PVDF membrane. The stained Muc2 monomeric bands were excised and the O-glycans released by reductive beta elimination, desalted and analyzed by graphitized carbon capillary-LC-MS/MS in negative ion mode.
Glycan peak areas were processed with Skyline version 3.5 (MacLean et al. 2010). In order to compensate for the difference in ionization efficiency between the oligosaccharides, response factors were used (Olson et al. 2005). All identified O-linked oligosaccharides and their relative amounts are presented in Supplementary data, Table SI. Most abundant O-linked oligosaccharides and their relative amounts are presented in Table I.
Proteomics analysis of epithelial cells
Proteomic analysis of epithelial cells and calculation of protein abundances were performed as described previously (Johansson et al. 2015). Briefly, intestinal tissues from duodenum, mid-jejunum, ileum, proximal colon and distal colon were incubated in phosphate buffered saline (PBS) containing 3 mM EDTA and 1 mM DTT at 37°C for 60 min. Thereafter, epithelial cells were dissociated from the tissue by vigorous shaking in PBS and pelleting by centrifugation at 1000 × g for 5 min. Samples for proteomics analysis were prepared according to filter aided sample preparation (FASP) protocol (Wiśniewski et al. 2009). Briefly, cells were lysed by boiling in 4% sodium dodecyl sulfate, buffer was exchanged to 8 M urea on 30 kDa cutoff filters and proteins were digested overnight with endopeptidase LysC (Wako Chemicals, Richmond, VA). The resulting peptides were eluted and 30 μg of peptides was fractionated by SAX StageTips into two fractions (pH11 and pH3) (Rappsilber et al. 2007). Samples were analyzed with EASY-nLC system (Thermo Scientific, Odense, Denmark) connected to a Q-Exactive (Thermo Scientific, Bremen, Germany). MS raw files were processed with MaxQuant software version 1.3.0.5 (Cox et al. 2011). The summed peptide intensities for each protein were divided by the number of theoretically observable peptides (all fully LysC-digested peptides between 800 and 2500 Da) to compensate that the longer proteins with more peptides will produce higher intensity. The new corrected intensity values were normalized to the total number of protein molecules present in a mammalian cell—109 (Milo 2013). Statistical significance for glycosyltransferases was determined in Prism version 6 using multiple t-test Holm–Sidak method, with alpha = 5.000%. Each comparison was analyzed individually, without assuming a consistent standard deviation. Glycosyltransferase relative concentrations are presented in Supplementary data, Table SII.
Supplementary data
Supplementary Material
Funding
The Swedish Research Council; The Swedish Cancer Foundation; The Knut and Alice Wallenberg Foundation; IngaBritt and Arne Lundberg Foundation; Sahlgren's University Hospital (ALF); Wilhelm and Martina Lundgren's Foundation; National Institute of Allergy and Infectious Diseases (U01AI095473, the content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Health); and The Swedish Foundation for Strategic Research.
Conflict of interest statement
None declared.
Abbreviations
ConvR, conventionally raised; CF, cystic fibrosis; IBD, inflammatory bowel disease; GalNAc-T, peptidyl-GalNAc transferase; GF, germ-free; GuHCl, guanidinium hydrochloride; PBS, phosphate buffered saline.
References
- Asker N, Axelsson MAB, Olofsson SO, Hansson GC. 1998. Dimerization of the human MUC2 mucin in the endoplasmic reticulum is followed by a N-glycosylation-dependent transfer of the mono- and dimers to the golgi apparatus. J Biol Chem. 273:18857–18863. [DOI] [PubMed] [Google Scholar]
- Atuma C, Strugala V, Allen A, Holm L. 2001. The adherent gastrointestinal mucus gel layer: thickness and physical state in vivo. Am J Physiol Gastrointest Liver Physiol. 280:G922–G929. [DOI] [PubMed] [Google Scholar]
- Bennett EP, Hassan H, Hollingsworth MA, Clausen H. 1999. A novel human UDP-N-acetyl-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase, GalNAc-T7, with specificity for partial GalNAc-glycosylated acceptor substrates. FEBS Lett. 460:226–230. [DOI] [PubMed] [Google Scholar]
- Bennett EP, Mandel U, Clausen H, Gerken TA, Fritz TA, Tabak LA. 2012. Control of mucin-type O-glycosylation: A classification of the polypeptide GalNAc-transferase gene family. Glycobiology. 22:736–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom K, Fu J, Johansson MEV, Liu X, Gao N, Wu Q, Song J, McDaniel JM, McGee S, Chen W et al. 2016. Core 1- and 3-derived O-glycans collectively maintain the colonic mucus barrier and protect against spontaneous colitis in mice. Mucosal Immunol. 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. 2011. Andromeda: A peptide search engine integrated into the MaxQuant environment. J Proteome Res. 10:1794–1805. [DOI] [PubMed] [Google Scholar]
- Dall'Olio F, Malagolini N, Chiricolo M, Trinchera M, Harduin-Lepers A. 2014. The expanding roles of the Sd(a)/Cad carbohydrate antigen and its cognate glycosyltransferase B4GALNT2. Biochim Biophys Acta. 1840:443–453. [DOI] [PubMed] [Google Scholar]
- Donaldson GP, Lee SM, Mazmanian SK. 2015. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol. 14:20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermund A, Schütte A, Johansson MEV, Gustafsson JK, Hansson GC. 2013. Studies of mucus in mouse stomach, small intestine, and colon. I. Gastrointestinal mucus layers have different properties depending on location as well as over the Peyer's patches. Am J Physiol Gastrointest Liver Physiol. 305:G341–G347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallien S, Duriez E, Crone C, Kellmann M, Moehring T, Domon B. 2012. Targeted proteomic quantification on Quadrupole-Orbitrap Mass Spectrometer. Mol Cell Proteomics. 11:1709–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger T, Velic A, Macek B, Lundberg E, Kampf C, Nagaraj N, Uhlen M, Cox J, Mann M. 2013. Initial quantitative proteomic map of 28 mouse tissues using the SILAC mouse. Mol Cell Proteomics. 12:1709–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y, Obata T, Kunisawa J, Sato S, Ivanov II, Lamichhane A, Takeyama N, Kamioka M, Sakamoto M, Matsuki T et al. 2014. Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science. 345:1254009-1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmén JM, Olson FJ, Karlsson H, Hansson GC. 2003. Two glycosylation alterations of mouse intestinal mucins due to infection caused by the parasite Nippostrongylus brasiliensis. Glycoconj J. 19:67–75. [DOI] [PubMed] [Google Scholar]
- Holmén Larsson JM, Thomsson KA, Rodríguez-Piñeiro AM, Karlsson H, Hansson GC. 2013. Studies of mucus in mouse stomach, small intestine, and colon. III. Gastrointestinal Muc5ac and Muc2 mucin O-glycan patterns reveal a regiospecific distribution. Am J Physiol Gastrointest Liver Physiol. 305:G357–G363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MEV, Jakobsson HE, Holmén-Larsson J, Schütte A, Ermund A, Rodríguez-Piñeiro AM, Arike L, Wising C, Svensson F, Bäckhed F et al. 2015. Normalization of host intestinal mucus layers requires long-term microbial colonization. Cell Host Microbe. 18:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MEV, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. 2008. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci USA. 105:15064–15069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju T, Cummings RD. 2002. A unique molecular chaperone Cosmc required for activity of the mammalian core 1 beta 3-galactosyltransferase. Proc Natl Acad Sci USA. 99:16613–16618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson NG, Olson FJ, Jovall PA, Andersch Y, Enerback L, Hansson GC. 2000. Identification of transient glycosylation alterations of sialylated mucin oligosaccharides during infection by the rat intestinal parasite Nippostrongylus brasiliensis. Biochem J. 350:805–814. [PMC free article] [PubMed] [Google Scholar]
- Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, Cooper GM, Roos C, Voight BF, Havulinna AS et al. 2008. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 40:189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y, Joshi HJ, Schjoldager KT-BG, Madsen TD, Gerken TA, Vester-Christensen MB, Wandall HH, Bennett EP, Levery SB, Vakhrushev SY et al. 2015. Probing polypeptide GalNAc-transferase isoform substrate specificities by in vitro analysis. Glycobiology. 25:55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koropatkin NM, Cameron EA, Martens EC. 2012. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol. 10:323–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson E, Tremaroli V, Lee YS, Koren O, Nookaew I, Fricker A, Nielsen J, Ley RE, Bäckhed F. 2012. Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through MyD88. Gut. 61:1124–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson JMH, Karlsson H, Crespo JG, Johansson MEV, Eklund L, Sjövall H, Hansson GC. 2011. Altered O-glycosylation profile of MUC2 mucin occurs in active ulcerative colitis and is associated with increased inflammation. Inflamm Bowel Dis. 17:2299–2307. [DOI] [PubMed] [Google Scholar]
- MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. 2010. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments In: Bioinformatics. 26: 966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahdavi J, Sondén B, Hurtig M, Olfat FO, Forsberg L, Roche N, Angstrom J, Larsson T, Teneberg S, Karlsson K-A et al. 2002. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Science. 297:573–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller-Podraza H, Bergström J, Teneberg S, Milh MA, Longard M, Olsson BM, Uggla L, Karlsson KA. 1999. Helicobacter pylori and neutrophils: Sialic acid-dependent binding to various isolated glycoconjugates. Infect Immun. 67:6309–6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milo R. 2013. What is the total number of protein molecules per cell volume? A call to rethink some published values. Bioessays. 35:1050–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson FJ, Bäckström M, Karlsson H, Burchell J, Hansson GC. 2005. A MUC1 tandem repeat reporter protein produced in CHO-K1 cells has sialylated core 1 O-glycans and becomes more densely glycosylated if coexpressed with polypeptide-GalNAc-T4 transferase. Glycobiology. 15:177–191. [DOI] [PubMed] [Google Scholar]
- Post S, van der, Subramani DB, Bäckström M, Johansson MEV, Vester-Christensen MB, Mandel U, Bennett EP, Clausen H, Dahlén G, Sroka A et al. 2013. Site-specific O-glycosylation on the MUC2 mucin protein inhibits cleavage by the Porphyromonas gingivalis secreted cysteine protease (RgpB). J Biol Chem. 288:14636–14646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potapenko IO, Haakensen VD, Lüders T, Helland A, Bukholm I, Sørlie T, Kristensen VN, Lingjaerde OC, Børresen-Dale A-L. 2010. Glycan gene expression signatures in normal and malignant breast tissue; possible role in diagnosis and progression. Mol Oncol. 4:98–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabot S, Membrez M, Bruneau A, Gérard P, Harach T, Moser M, Raymond F, Mansourian R, Chou CJ. 2010. Germ-free C57BL/6J mice are resistant to high-fat-diet-induced insulin resistance and have altered cholesterol metabolism. FASEB J. 24:4948–4959. [DOI] [PubMed] [Google Scholar]
- Rappsilber J, Mann M, Ishihama Y. 2007. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2:1896–1906. [DOI] [PubMed] [Google Scholar]
- Revoredo L, Wang S, Bennett EP, Clausen H, Moremen KW, Jarvis DL, Ten Hagen KG, Tabak LA, Gerken TA. 2016. Mucin-type o-glycosylation is controlled by short- And long-range glycopeptide substrate recognition that varies among members of the polypeptide GalNAc transferase family. Glycobiology. 26:360–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Piñeiro AM, Bergström JH, Ermund A, Gustafsson JK, Johansson MEV, Hansson GC, Schuette A. 2013. Gastrointestinal mucus proteome reveals Muc2 and Muc5ac accompanied by a set of core proteins - 2. Studies of mucus in mouse stomach, small intestine, and colon. Am J Physiol Gastrointest Liver Physiol. 305:G348–G356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer F, Adam N, Johansson MEV, Xia L, Hansson GC, Bäckhed F. 2014. Altered mucus glycosylation in core 1 O-glycan-deficient mice affects microbiota composition and intestinal architecture. PLoS One. 9:85254–85254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ et al. 2010. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 466:707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsson KA, Hinojosa-Kurtzberg M, Axelsson KA, Domino SE, Lowe JB, Gendler SJ, Hansson GC. 2002. Intestinal mucins from cystic fibrosis mice show increased fucosylation due to an induced Fucα1-2 glycosyltransferase. Biochem J. 367:609–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsson KA, Holmén-Larsson JM, Angström J, Johansson MEV, Xia L, Hansson GC. 2012. Detailed O-glycomics of the Muc2 mucin from colon of wild-type, core 1- and core 3-transferase-deficient mice highlights differences compared with human MUC2. Glycobiology. 22:1128–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran DT, Ten Hagen KG. 2013. Mucin-type O-glycosylation during development. J Biol Chem. 288:6921–6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Post S, Hansson GC. 2014. Membrane protein profiling of human colon reveals distinct regional differences. Mol Cell Proteomics. 13:2277–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velagapudi VR, Hezaveh R, Reigstad CS, Gopalacharyulu P, Yetukuri L, Islam S, Felin J, Perkins R, Borén J, Oresic M et al. 2010. The gut microbiota modulates host energy and lipid metabolism in mice. J Lipid Res. 51:1101–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniewski JR, Zougman A, Mann M. 2009. Combination of FASP and StageTip-based fractionation allows in-depth analysis of the hippocampal membrane proteome. J Proteome Res. 8:5674–5678. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.