ABSTRACT
Meiotic failure in oocytes is the major determinant of human zygote-originated reproductive diseases, the successful accomplishment of meiosis largely relay on the normal functions of many female fertility factors. Elmod2 is a member of the Elmod family with the strongest GAP (GTPase-activating protein) activity; although it was identified as a possible maternal protein, its actual physiologic role in mammalian oocytes has not been elucidated. Herein we reported that among Elmod family proteins, Elmod2 is the most abundant in mouse oocytes, and that inhibition of Elmod2 by specific siRNA caused severe meiotic delay and abnormal chromosomal segregation during anaphase. Elmod2 knockdown also significantly decreased the rate of oocyte maturation (to MII, with first polar body extrusion), and significantly greater numbers of Elmod2-knockdown MII oocytes were aneuploid. Correspondingly, Elmod2 knockdown dramatically decreased fertilization rate. To investigate the mechanism(s) involved, we found that Elmod2 knockdown caused significantly more abnormal mitochondrial aggregation and diminished cellular ATP levels; and we also found that Elmod2 co-localized and interacted with Arl2, a GTPase that is known to maintain mitochondrial dynamics and ATP levels in oocytes. In summary, we found that Elmod2 is the GAP essential to meiosis progression of mouse oocytes, most likely by regulating mitochondrial dynamics.
KEYWORDS: Arl2, ATP, Elmod2, GTPase-activating protein, meiosis, mitochondria, oocyte
Introduction
The GTPase superfamily is a large family of hydrolases that bind and hydrolyze GTP; and these activities depend upon the G domain, which is highly conserved in all GTPases. These molecules function in many distinct and important cellular processes, including signal transduction in conjugation with the intracellular domain of transmembrane receptors, protein biosynthesis, cell cycle kinetics and differentiation, macroautophagy, vesicle transport, and protein translocation through membranes.1-5 Abnormal GTPase activities are significant in the etiology of many human diseases.6-8 In mammalian oocytes, many members of GTPase superfamily have been identified to be involved in meiosis progression. Ran GTPase has been reported to regulate the polarized activation of Cdc42, which will than promote polar body protrusion and asymmetric division.9,10 Rab5a and Rab6a, 2 members of Rab subfamily, have been found to participate in chromosome alignment, kinetochore-microtubule attachment and spindle organization of mouse oocyte.11,12 During porcine oocyte maturation and early embryo development, small GTPase RhoA regulates cytoskeleton dynamics.13 Although GTPases can hydrolyze GTP on their own, their efficiency is greatly elevated by GTPase-activating proteins (GAPs); while guanine exchange factors (GEFs) help convert GDP into GTP to regenerate GTPases. All members of the GTPase family have their specific GEFs and GAPs.14
The Elmod (ELMO/CED-12 domain containing) family of GAPs includes Elmod1-Elmod3. The family members have only one conserved Elmo (engulfment and cell motility) domain of about 180 amino acids, and a conserved catalytic arginine residue that inserts between the β-γ phosphate bond helps to neutralize the negative charge produced in the transition state of GTP hydrolysis, thereby accelerating GTP hydrolysis as much as 5-fold.15 Another family of proteins closely related to the Elmods is Elmo (including Elmo1, Elmo2 and Elmo3), but these molecules do not contain the key arginine residue, and therefore they do not belong to the GAPs. ADP-ribosylation factors (Arfs), a subfamily of GTPases, are the primary targets of the Elmods.16 A study of in-vitro enzyme activity showed that the GTPase activation activity of Elmod2 was 40 times higher than Elmod1 and 1000 times higher than Elmod3; thus, Elmod2 was confirmed to be the most potent GAP of the Elmod family.17 Elmod2 was also reported to be a candidate gene for familial idiopathic pulmonary fibrosis (IPF).18 In another study where investigators attempted to ascertain the underlying mechanism(s) of action involved, Elmod2 knockdown inhibited TLR3 (Toll-like receptor 3)-dependent expression of Type-I and -III interferon (IFN), while Elmod2 overexpression enhanced IFN expression, indicating that Elmod2 was important for an appropriate immune response.19 Elmod2 was identified to function as an Arf-GAP so as to regulate arldipocyte triglyceride lipase (ATGL) transport and cellular lipid metabolism by modulating Arf1-COPI activity in lipid droplets.20 Elmod2 has been shown to be the main GAP of the Arfs, among which ADP-ribosylation factor-like 2 (Arl2) is the primary target, as it is a GTPase required for mitochondrial morphology, motility, and maintenance of ATP levels.21, 22 In the previous study, we have identified the importance of Arl2 in meiotic progression through inhibition of Arl2 by specific antibody transfection.23
Elmod2 and its known target GTPase Arl2 were both identified as maternal proteins in a proteomics study of mature mouse oocytes and Arl2 was predicted to be important for embryonic development.24 Elmod2 was also ranked among “proteins of particular interest“ due to its high mRNA level in MII oocytes.24 But there are no extant functional studies pertaining to the female reproductive system. In our preliminary study we found that Elmod2 mRNA was predominant among Elmod family members in mouse oocytes, and that Elmod2 protein was much more abundant in oocytes compared with theca cells (TCs) and cumulus cells (CCs). We therefore hypothesized that Elmod2 plays important roles in mouse oocyte meiosis.
Results
Elmod2 is a predominant GAP in mouse oocytes
We first examined the expression and localization patterns of Elmod2 in mouse ovaries as well as oocytes. RT-PCR showed that Elmod2 mRNA was the most abundant among all Elmo and Elmod family members in oocytes Fig. 1A, indicating that Elmod2 might be the principal GAP in oocytes. Immunoblotting showed that the protein level of Elmod2 significantly increased at 3 d post-natal (post-natal day, PND) (when assembly of primordial follicles occurs), and peaked at PND21 (when the first wave of follicular maturation occurs) Fig. 1B) in ovaries, indicating that Elmod2 was important for follicle assembly and development. Additionally, Elmod2 was more pronounced in oocytes than in theca cells (TCs) or cumulus cells (CCs) Fig. 1C, and the protein level in oocytes was constant during meiosis progression Fig. 1D, indicating that the role of Elmod2 might be primarily in oocytes. We also assessed the localization of Elmod2 in oocytes by immunofluorescence. During in-vitro maturation (IVM), Elmod2 was high in nuclei of oocytes at the GV stage, rich within spindles after resumption of meiosis. Elmod2 also concentrated on spindle poles of MI oocytes and co-localized with γ-tubule Fig. 1E and F.
Figure 1.
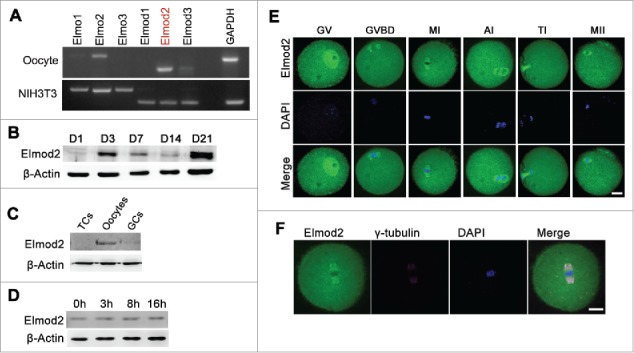
Elmod2 is an oocyte-prominent protein and primary GAP in mouse ovaries. (A) RT-PCR showed Elmod2 was the most abundant among all Elmo and Elmod family members in mouse oocytes. (B) Immunoblotting in ovaries at different post-natal day (PND) showed that protein level of Elmod2 significantly increased at PND3 and peaked at PND21. (C) Elmod2 was much more predominant in oocytes than in theca cells (TCs) and cumulus cells (CCs). (D) Immunoblotting showed that Elmod2 was constant from GV to MII stage. (E) Immunofluorescence in mouse oocytes showed that Elmod2 (green) was rich in nucleus of GV oocytes and concentrated within spindles regions during meiosis. DAPI in blue. (F) Immunofluorescence of oocyte at MI stage showed the co-localization of Elmod2 (green) and γ-tubulin (purple) on spindle poles. Scale bars, 20 μm.
Elmod2 is important for meiotic progression of mouse oocytes
The abundance of Elmod2 in mouse oocytes suggests that Elmod2 participates in meiotic progression. To further study Elmod2 function, we microinjected a specific siRNA into fully grown oocytes at GV stage, cultured them, and examined their meiotic progression at different stages. RT-PCR and western immunoblotting results showed that both Elmod2 mRNA and protein levels in oocytes were significantly reduced Fig. 2A and B. At 8 hours of culture, a significantly greater number of oocytes remained in GVBD, and fewer had progressed to MI compared with controls (GVBD oocytes, controls vs. knockdowns, 27.46 % vs. 55.46 %, respectively; MI oocytes, controls vs. knockdowns, 51.63 % vs. 28.17 %) Fig. 2C. In addition, Elmod2 knockdown caused severe abnormalities in spindle organization as well as chromosomal alignment Fig. 2D. Similarly, at 16 hours of culture, loss of Elmod2 significantly reduced the proportion of MII oocytes (controls vs. knockdowns, 65.03 % vs. 38.52 %) Fig. 2E. Moreover, the percentage of MII oocytes with uncongressed chromatids, defined as “abnormal MII” oocytes, dramatically increased (controls vs knockdowns, 52.37 % vs. 78.31 %) Fig. 2F and G.
Figure 2.
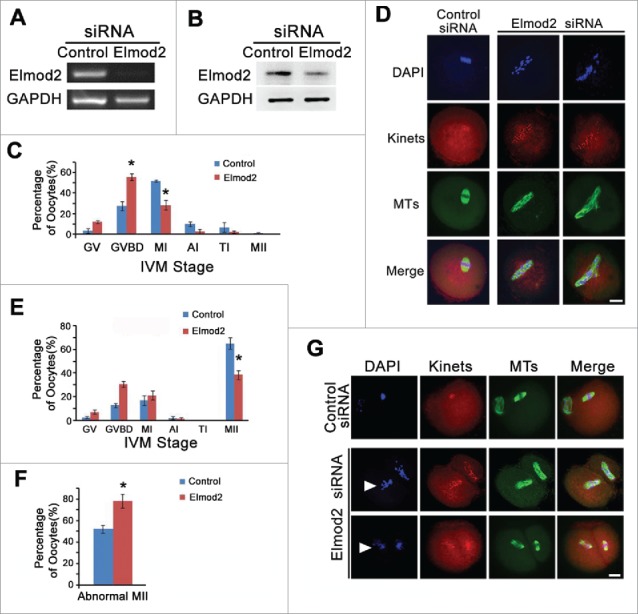
Elmod2 is important for meiosis progression of mouse oocytes. (A) RT-PCR showed mRNA level of Elmod2 was significantly reduced by specific siRNA. (B) Immunoblotting showed Elmod2 protein level was significantly reduced by specific siRNA. (C) Percentage of oocytes at each stage at 8 hours of in-vitro maturation (IVM), and there were significantly more GVBD and less MI oocytes in Elmod2-knockdown oocytes than in control. (D) Immunofluorescence showed severe abnormality in spindle organization and chromosome alignment after Elmod2 knockdown. (E and F) At 16 hours of IVM, there were significantly less MII oocytes and more MII oocytes with less-congressed chromosomes (abnormal MII oocytes) in Elmod2-knockdown oocytes than in control. (G) Representative confocal images showed oocytes after 16 hours of culture, and less-congressed chromosomes were arrow-headed. Microtubules (MTs) in green, kinetochores (Kinets) in red, chromosomes (DAPI) in blue. Scale bars, 20 μm. Significant difference was asterisk (*) labeled.
Elmod2 knockdown perturbs chromosomal segregation and increases oocyte aneuploidy
Since Elmod2 knockdown significantly disturbed meiotic progression, we wished to further examine whether chromosomal segregation in anaphase (which is the key for euploidy in matured oocytes), was also perturbed. Immunofluorescence showed that significantly more oocytes underwent asymmetric chromosomal separation after Elmod2 knockdown (controls vs. knockdowns, 1 of 17, 5.88 % vs. 7 of 10, 70 %) Fig. 3A and B. Accordingly, Elmod2 knockdown significantly increased the percentage of oocytes with aneuploidy (controls vs. knockdowns, 0 of 40 vs. 13 of 38) Fig. 3C and D.
Figure 3.
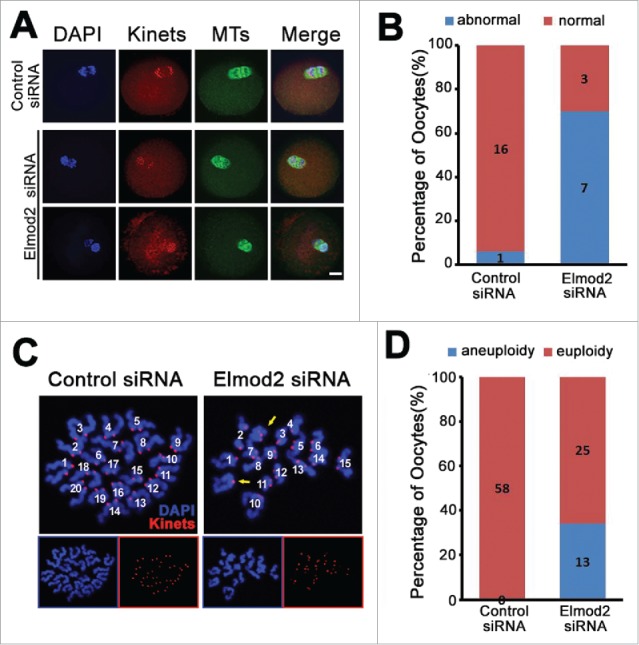
Elmod2 knockdown perturbs chromosome segregation and increases oocytes with aneuploidy. (A) Immunofluorescence staining of anaphase I (AI) oocytes showed that asymmetric chromosome separation occurred after Elmod2 knockdown. Chromosomes (DAPI) in blue, microtubules (MTs) in green, kinetochores (Kinets) in red. (B) Quantification showed that percentage of oocytes with asymmetric chromosomes separation (abnormal) significantly increased in Elmod2-knockdown group than in control. (C) Representative confocal images indicate euploid control oocytes, and aneuploid Elmod2-knockdown oocytes. Chromosomes (DAPI) in blue and kinetochores (Kinets) in red. (D) Quantification showed that percentage of oocytes with aneuploidy significantly increased in Elmod2-knockdown group than in control.
Elmod2 knockdown decreases normal fertilization
Since Elmod2 knockdown significantly increased the percentage of MII oocytes with less-congressed chromosomes and aneuploidy, we wished to further examine whether Elmod2 knockdown adversely affected fertilization. Results showed that at 9 hours after in-vitro fertilization (IVF), both the fertilization and 2-PN (pronuclei) rates were dramatically reduced (fertilization rate, controls vs. knockdowns, 64.87 % vs. 22.02 %; 2-PN rate, controls vs. knockdown, 70.41 % vs. 42.50 %) Fig. 4A and B.
Figure 4.
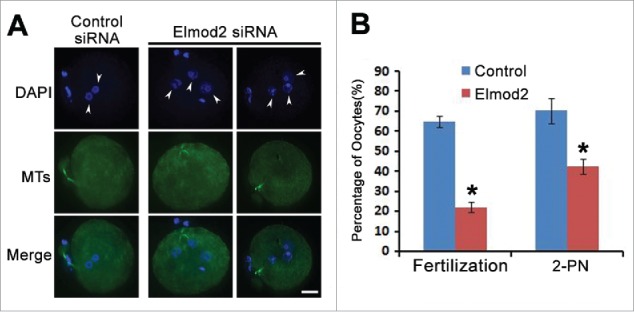
Elmod2 knockdown decreases normal fertilization. (A) Representative confocal images showed that multiple pronuclei (arrow-headed) occurred after Elmod2 knockdown. Chromosomes (DAPI) in blue, microtubules (MTs) in green. (B) Quantification showed that there were significantly less fertilized oocytes and less oocytes with 2-pronuclei (PN) in Elmod2-knockdown group than in control. Significant difference was asterisk (*) labeled. Scale bars, 20 μm.
Elmod2 is required for the normal distribution of mitochondria and maintenance of ATP content during oocyte meiosis
Here we tried to uncover the potential mechanism(s) by which Elmod2 functions in oocyte meiosis. Since in somatic cells Elmod2 was found to activate Arl221 (which is required for mitochondrial morphology and motility22), we postulated that Elmod2 in meiotic oocytes might function similarly. To verify this hypothesis, the morphologic features of mitochondria were evaluated by staining with fluorescently labeled mitochondria tracker. Confocal imaging showed that in control oocytes the distribution of mitochondria was very dynamic at various culture stages; i.e., before meiotic resumption (GV stage), mitochondria were more highly concentrated on the nuclear membrane but were largely distributed evenly within the cytoplasm; after meiotic resumption (GVBD, MI, MII), mitochondria were more concentrated around spindle microtubules, but also prevalent elsewhere within the cytoplasm. Notably, at MI and MII, mitochondria existed within the cytoplasm and also in small granules Fig. 5A. In contrast, in Elmod2-knockdown oocytes mitochondria always formed large aggregates around the chromosomes and remained in very low abundance within the cytoplasm, without any apparent dynamic localization from the GV to MII stages Fig. 5A. Accordingly, ATP levels in Elmod2-knockdown oocytes were significantly decreased compared with controls Fig. 5B. These observations confirmed that Elmod2 regulates meiosis by maintaining normal mitochondrial dynamics and ATP levels.
Figure 5.
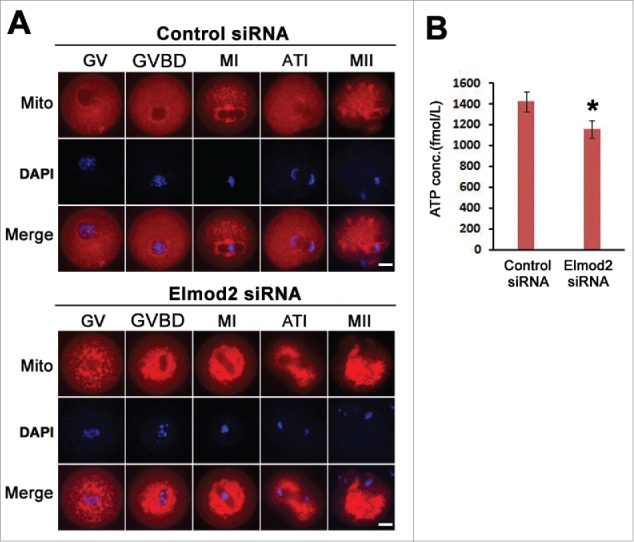
Elmod2 is required for normal distribution of mitochondria and maintenance of ATP content during oocyte meiosis. (A) Representative confocal images showed that abnormal mitochondria aggregation occurred after Elmod2 knockdown. Mitochondria (red) are labeled with fluorescent mitochondria tracker, chromosomes in blue. (B) ATP level was significantly reduced in Elmod2-knockdown oocytes than in control. Significant difference is asterisk (*) labeled. Scale bars, 20 μm.
Elmod2 interacts with Arl2 in mouse oocytes
Next, we tried to further investigate how Elmod2 maintains normal mitochondrial dynamics and ATP levels, and again focused on the known Elmod2 effector Arl2 in somatic cells.21 Immunofluorescence showed that Elmod2 was largely co-localized with Arl2 within spindles Fig. 6A, and furthermore, co-immunoprecipitation showed that they also interacted with one another Fig. 6B. These 2 experiments indicated that Elmod2 might function in meiosis mediated by Arl2.
Figure 6.
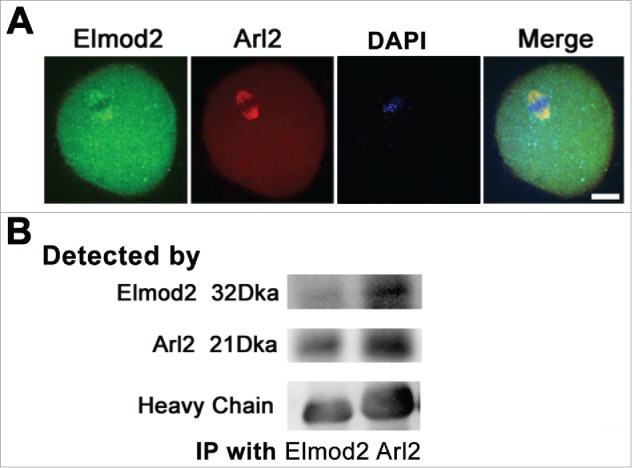
Elmod2 interacts with Arl2 in mouse oocytes. (A) Immunofluorescence staining showed that Elmod2 (green) and Arl2 (red) co-localized within spindle region of MI oocytes. (B) Co-immunoprecipitation (Co-IP) and western blot showed that Elmod2 interacted well with Arl2. Scale bars, 20 μm.
Discussion
Fully grown oocyte is a type of special cell rich in various maternal factors that are important for the resumption of meiosis (GVBD), completion of meiosis I&II, fertilization, and early embryonic development.25, 26 Certainly, both synthesis and function of these highly abundant proteins require an amount of mitochondrially generated ATP,27, 28 as normal mitochondrial dynamics and distribution are important for maintaining ATP levels.29-31 As was reported previously, translocation to the perinuclear region and aggregation into clusters of mitochondria lead to bursts in ATP production during spontaneous oocyte maturation in mice, and disruption of the mitochondrial clusters with cytochalasin B (by breaking down microfilaments) reduces the ATP production,31 suggesting that the status and redistribution of mitochondria in mammalian oocytes constitute a determining factor of oocyte energy supply. In the current study, Elmod2 knockdown caused abnormal mitochondrial distribution as well as significantly decreased ATP levels, indicating that Elmod2 maintained ATP levels by regulating mitochondrial dynamics.
The Arf GTPase family member Arl2 has been shown to be important for mitochondrial morphology, motility, and maintenance of ATP levels in mouse oocytes.22 In our previous study, Arl2 was found to distribute evenly within the cytoplasm at the GV stage, while upon meiotic resumption it was strongly concentrated within spindles23; this is very similar to the dynamic localization of Elmod2 during oocyte meiosis. In the present study, we showed that Elmod2 was co-localized and co-immunoprecipitated with Arl2, and that Elmod2 appeared to be important for normal mitochondrial dynamics and ATP levels. Since Elmod2 is known to be the GAP for Arl222, it is reasonable to conjecture that Elmod2 functions in oocyte meiosis by activating Arl2.
Many studies have shown that decreased ATP levels will cause universally adverse effects on normally diverse cellular processes, including cell cycle kinetics.32-35 For example, ATP is required for the release of the anaphase-promoting complex / cyclosome from inhibition by the mitotic checkpoint.33 Cdc123, another cell cycle regulator, is identified as an ATP-Grasp protein that functions in cell cycle regulation by catalyzing protein modifications in an ATP-dependent manner.34 In mouse oocytes, the decrease in mitochondrially derived ATP may induce disassembly of MII oocyte spindles.35 Per our cytologic analysis, Elmod2 knockdown caused a series of meiotic problems, including a delay in progression to meiosis I and II. Furthermore, inaccurate chromosomal segregation occurred during anaphase, and the number of oocytes with aneuploidy increased significantly. In parallel fashion, problematically mature oocytes (“abnormal MII oocytes”) also increased significantly, and eventually precipitated decreased fertilization and increased abnormal fertilization. Based on these results, the reduction in ATP levels after Elmod2 knockdown appeared to cause universal defects in oocyte meiosis and subsequent fertilization.
In conclusion, we have characterized Elmod2 as being a predominant GAP in mouse oocyte maturation, and thereby a likely master regulator of intracellular ATP levels and of important cellular activities such as meiosis and fertilization. Through these activities, Elmod2 might regulate ATP levels by modulating Arl2 activity. However, further studies are required to investigate whether Elmod2 also affects other Arfs and how Elmod2 regulates Arfs in oocyte meiosis.
Materials & methods
General chemicals and reagents and animals
Chemicals and reagents were obtained from Sigma unless otherwise stated. ICR mice used in this study were supplied by Vitalriver Experimental Animal Technical Co. (Beijing, China). All animal experiments were approved by the Animal Care and Use Committee of Nanjing Medical University and were performed in accordance with institutional guidelines.
Antibodies
Mouse monoclonal anti-β-actin (Cat #: A5316–100) was purchased from Santa Cruz (Dallas, Texas, USA). Mouse monoclonal anti-β-tubulin antibody (Cat #: sc-5274) antibody was purchased from Santa Cruz (Dallas, Texas, USA). Human anti-centromere CREST antibody (Cat #: 15–234) was purchased from Antibodies Inc. (Davis, CA, USA). Cy2-conjugated donkey anti-mouse IgG (Code: 715–225–150), Rhodamine (TRITC)-conjugated donkey anti-human IgG (Code: 709–025–149), Cy2-conjugated donkey anti-human IgG (Code: 709–225–149) and Cy2-conjugated donkey anti-Rabbit IgG (Code: 711–225–152) were purchased from Jackson ImmunoResearch Laboratory (West Grove, PA, USA). Horseradish Peroxidase (HRP)-conjugated goat anti rabbit IgG and HRP-conjugated goat anti mouse IgG were purchased from Vazyme (Nanjing, Jiangsu, China). Rabbit polyclonal anti-Elmod2 antibody was supplied by Shanghai Yingji Biotechnology Company (Shanghai, China). Rabbit polyclonal anti-Arl2 (Cat #: 10232–1-AP) was purchased from Proteintech (Chicago, IL, USA).
Oocytes collection and culture
Immature oocytes arrested in prophase I (GV stage) were obtained from the ovaries of 3–4 week-old ICR female mice. The mice were killed by cervical dislocation and the ovaries were isolated and placed in operation medium (Hepes) containing 10% fetal bovine serum (FBS) (Gibco). Oocytes were released from the ovary by puncturing the follicles with a hypodermic needle. Cumulus cells were washed off the cumulus-oocyte complexes by repeatedly mouth-pipetting and every 50 isolated denuded oocytes were placed in 100-ul droplets of culture medium under mineral oil in plastic dishes (BD). The culture medium was MEM+ (MEM with 0.01 mM EDTA, 0.23 mM Na-pyruvate, 0.2 mM pen / strep, 3 mg/ml BSA ) containing 20 % FBS. Oocytes were cultured at 37.0 °C, 5 % O2, 5 % CO2 in humidified atmosphere. Prior to IVM, all operation and culture medium include 2.5 nM milrinone to prevent resumption of meiosis.
siRNA knockdown
Sequences of all DNA templates for siRNA production are listed in Table 1. The sequence of control templates is a mock sequence that does not specifically bind to any mRNA from the mouse genome. DNA templates against 4 different coding for DNA sequence (CDS) regions of Elmod2 siRNA were designed online through BLOCK-iT™ RNAi Designer (http://rnaidesigner.invitrogen.com/rnaiexpress/) with some modification. Sequence specificity was verified through a blast homology search.
Table 1.
DNA oligos for siRNA production.
| Target Site | DNA templates |
|---|---|
| Elmod2 211–2351 | Oligo1: GGATCCTAATACGACTCACTATAGACAGGTGTATAGCGAACATCATGA2 |
| Oligo2:AATCATGATGTTCGCTATACACCTGTCTATAGTGAGTCGTATTAGGATCC2 | |
| Oligo3: GGATCCTAATACGACTCACTATATCATGATGTTCGCTATACACCTGTC[2] | |
| Oligo4:AAGACAGGTGTATAGCGAACATCATGATATAGTGAGTCGTATTAGGATCC2 | |
| Elmod2 522–5461 | Oligo1: GGATCCTAATACGACTCACTATAGATCAATCTCGTGTATTTCAGTGAA2 |
| Oligo2:AATTCACTGAAATACACGAGATTGATCTATAGTGAGTCGTATTAGGATCC2 | |
| Oligo3: GGATCCTAATACGACTCACTATATTCACTGAAATACACGAGATTGATC2 | |
| Oligo4:AAGATCAATCTCGTGTATTTCAGTGAATATAGTGAGTCGTATTAGGATCC2 | |
| Elmod2 772–7961 | Oligo1: GGATCCTAATACGACTCACTATAGAGGAAGAGCCAGAAAGCATTATGT2 |
| Oligo2:AAACATAATGCTTTCTGGCTCTTCCTCTATAGTGAGTCGTATTAGGATCC2 | |
| Oligo3: GGATCCTAATACGACTCACTATAACATAATGCTTTCTGGCTCTTCCTC2 | |
| Oligo4:AAGAGGAAGAGCCAGAAAGCATTATGTTATAGTGAGTCGTATTAGGATCC2 | |
| Elmod2 850–8741 | Oligo1: GGATCCTAATACGACTCACTATAGATTGCAATGCTGTGCTCACTTTGA22 |
| Oligo2:AATCAAAGTGAGCACAGCATTGCAATCTATAGTGAGTCGTATTAGGATCC2 | |
| Oligo3: GGATCCTAATACGACTCACTATATCAAAGTGAGCACAGCATTGCAATC2 | |
| Oligo4:AAGATTGCAATGCTGTGCTCACTTTGATATAGTGAGTCGTATTAGGATCC2 | |
| Control3 | Oligo1: GGATCCTAATACGACTCACTATACCTACGCCACCAATTTCGTTT2 |
| Oligo2:AAAAACGAAATTGGTGGCGTAGGTATAGTGAGTCGTATTAGGATCC2 | |
| Oligo3: GGATCCTAATACGACTCACTATAAAACGAAATTGGTGGCGTAGG2 | |
| Oligo4:AACCTACGCCACCAATTTCGTTTTATAGTGAGTCGTATTAGGATCC2 |
The numbers are the starting and ending position of the target sites in Elmod2 CDS (NM_001170691.1 in NCBI).
Two pairs of DNA oligos are needed for each double-stand siRNA. Oligo 2 is complementary with oligo 1 except an “AA” overhang at 5′; Oligo 3 is complementary with oligo 4 except an “AA” overhang at 5′. In each oligo, gene-specific sequences are underlined; other sequences are for recognition and binding by T7 RNA polymerase.
Control siRNA does not target to any mRNA sequence in mouse.
SiRNAs were produced using the T7 RiboMAXTM Express RNAi System (Promega) according to the manufacturer's instructions. Briefly, for each double-stranded siRNA against one of the 4 Elmod2 CDS regions, 2 pairs of synthesized complementary single-stranded DNA oligonucleotides were first annealed to form 2 double-stranded DNA templates. Subsequently, 2 complementary single-stranded siRNAs were separately synthesized in accordance with these 2 templates and then annealed to form a final double-stranded siRNA. Next, the siRNA was purified by conventional phenol / chloroform / isoproponal precipitation. Followed with a quality check on the agarose gel, the siRNA product was then aliquoted and stored at −80 °C. A ready-to-use siRNA mixture was prepared by mixing siRNAs against 4 target regions together.
Fully-grown immature oocytes were microinjected with Elmod2-targeting siRNA to knock down Elmod2 proteins. SiRNA was diluted with water to give a stock concentration of 1 mM, and 2.5 pico-liter solution was injected. The control siRNA was injected as control. To facilitate the siRNA-mediated mRNA degradation, oocytes were arrested at GV stage in MEM+ medium containing 2.5 μM milrinone for 24 hours, and then cultured in milrinone-free medium for further experiments.
In-vitro fertilization (IVF)
Oocytes were injected with siRNA and cultured to MII stage as described. Shortly before fertilization, oocytes were washed rapidly for 3 times with MEM+ medium to remove FBS. Spermatozoa were obtained from the epididymis of 10–18 weeks old B6D2F1 male mice and were then capacitated in 1 ml MEM+ for 1 hour. Subsequently, 10 µl of a sperm suspension containing 5–10×106/ml spermatozoa was added to 490 ul MEM+ medium, and oocytes washed off of FBS were added. 9 hours later, the oocytes were processed for immunoassaying and examined to determine the frequency of successful fertilization, by the identification of the formation of pronuclei. Fertilization rate is the proportion of eggs with 2 or more pronuclei in all the oocytes used for IVF, while 2-PN rate is the percentage of eggs with 2 pronuclei among all the fertilized eggs.
Immunofluorescence
Oocytes were briefly washed in PBS with 0.05 % polyvinylpyrrolidone (PVP), permeated in 0.5 % Triton X-100 / PHEM (60 mM PIPES, 25 mM Hepes pH 6.9, 10 mM EGTA, 8 mM MgSO4) for 5 minutes and washed 3 times rapidly in PBS / PVP. Next the oocytes were fixed in 3.7 % paraformaldehyde (PFA) / PHEM for 20 minutes, washed 3 times (10 minutes each) in PBS / PVP and blocked with blocking buffer (1 % BSA / PHEM with 100 mM glycine) at room temperature for 1 hour. Then the oocytes were incubated at 4 °C overnight with primary antibody diluted in blocking buffer. After being washed 3 times (10 minutes each) in PBS with 0.05 % tween-20 (PBST), the oocytes were incubated at room temperature for 45 minutes with secondary antibody diluted in blocking buffer (1:750 in all cases). Finally chromosomes were stained by 10 µg/ml Hochest 33342 (Sigma) and the oocytes were mounted onto a slide with mounting medium (0.5 % propgal gallate, 0.1 M Tris-Hcl, pH 7.4, 88 % Glycerol) and covered with a cover glass (0.13–0.17 µm thick). To maintain the dimension of the oocytes, 2 strips of double-stick tap (90 µm thick) were sticked between the slide and cover glass. Dilution of primary antibodies was as follows: anti-Elmod2, 1:200; anti-tubulin, 1:500; anti-human centromere, 1:500. The oocytes were examined with an Andor Revolution spining disk confocal workstation (Oxford instruments, Belfast, Northern Ireland).
Chromosome spread
Oocytes were exposed to Tyrode's buffer (pH 2.5) for 40–50 seconds to remove zona pellucidae, and then fixed in a drop of 1 % paraformal -dehyde with 0.15 % Triton X-100 on a glass slide. Kinetochores and chromosomes were then stained as that of immunofluorescence. Andor Revolution spining disk confocal workstation was used to examine chromosome and kinetochore numbers in oocytes.
Co-immunoprecipitaion
For immunoprecipitation experiments, 5 μg Rabbit anti Elmod2 or Rabbit anti Arl2 antibody was first coupled to 30 μl protein-A/G beads (Macgene, Beijing, China) for 4 hours at 4°C on a rotating wheel in 250 μl IP buffer (20 mM Tris-HCl, pH 8.0, 10 mM EDTA, 1 mM EGTA, 150 mM NaCl, 0.05 % Triton X-100, 0.05 % Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride) with 1:100 protease inhibitor (Sigma) and 1:500 phosphatase inhibitor (Sigma). Meanwhile, 3000 oocytes were lysed and ultra-sonicated in 250 µl IP buffer and then pre-cleaned with 30 ul protein A/G beads for 4 hours at 4 °C. After that, protein A/G-coupled Elmod2 or Arl2 antibody was incubated overnight at 4 °C with pre-cleaned oocyte lysate supernatant. Finally, after being washed for 3 times (10 minutes each with 250 ul IP buffer), the resulting beads with bound immuno complexes were subjected for western immnoblotting analysis.
Mitochondrial staining and ATP measurements
Oocytes were injected with Elmod2-spedific or control siRNA and cultured to MII stage as described. For mitochondrial staining, the oocytes were stained in Hepes containing 100 nM Mito Tracker (Invitrogen, m7521) and 10 ug/ml Hochest 33342 (Sigma) for 30 minutes. Subsequently the oocytes were examined with an Andor Revolution spin disk confocal workstation. For measurement of ATP, the oocytes were first lysed with 100 ul RIPA lysis solution on ice. The samples were then detected by enzyme-labeled instrument Synergy2 (BioTek, USA) to evaluate ATP level.
Data analysis and statistics
All experiments were repeated at least 3 times. Measurement on confocal images was done with Image J. Data were presented as average ± sem. Statistical comparisons were made with Student's test of EXCEL. Data were presented average ± sem. P < 0.05 was considered to be statistically significant.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Funding
This work was supported by the National Basic Research Program of China (973 Program; Grant No: 2013CB945504); General Program of the National Natural Science Foundation of China (Grant No: 31471406); General Program of the National Natural Science Foundation of China (Grant No: 31671561).
Author's contributions
D.Z. and X.-Y.Y. designed the research; C.-X.Z., L.-Y.S. and R.-C.L. performed most of the experiments and data analysis; J.-W.L. and B.Y. are 2 visiting fellows and assisted in the revision; B.-Q.X. and Z.-X.Y. assisted in experiments and data analysis; D.Z. and C.-X.Z. wrote the manuscript with the assistance of X.-Y.Y., L.-Y. S., R.-C.L. and Y.-H.L.. All authors read and approved the final manuscript.
References
- [1].Cromm PM, Spiegel J, Grossmann TN, Waldmann H. Direct modulation of small GTPase activity and function. Angew Chem Int Ed Engl 2015; 54:13516-37; PMID:26470842; http://dx.doi.org/ 10.1002/anie.201504357 [DOI] [PubMed] [Google Scholar]
- [2].Chwartz SL, Cao C, Pylypenko O, Rak A, Wandinger-Ness A. Rab GTPases at a glance. J Cell Sci 2007; 15:3905-10; http://dx.doi.org/ 10.1242/jcs.015909 [DOI] [PubMed] [Google Scholar]
- [3].Yang XZ, Li XX, Zhang YJ, Rodriguez-Rodriguez L, Xiang MQ, Wang HY, Zheng XF. Rab1 in cell signaling, cancer and other diseases. Oncogene 2016; 35:5699-5704; PMID:27041585; http://dx.doi.org/ 10.1038/onc.2016.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Yang S, Rosenwald A. Small GTPase proteins in macroautophagy. Small GTPases 2016; 20:1-6; http://dx.doi.org/ 10.1080/21541248.2016.1246280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ng EL, Gan BQ, Ng F, Tang BL. Rab GTPases regulating receptor trafficking at the late endosome-lysosome membranes. Cell Biochem Funct 2012; 30:515-23; PMID:22473705; http://dx.doi.org/ 10.1002/cbf.2827 [DOI] [PubMed] [Google Scholar]
- [6].Haga RB, Ridley AJ. Rho GTPases: Regulation and roles in cancer cell biology. Small GTPases 2016; 7:207-21; PMID:27628050; http://dx.doi.org/ 10.1080/21541248.2016.1232583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yamauchi Y, Miura Y, Kanaho Y. Machineries regulating the activity of the small GTPase Arf6 in cancer cells are potential targets for developing innovative anti-cancer drugs. Adv Biol Regul 2016; 18:30060-4. [DOI] [PubMed] [Google Scholar]
- [8].Hongu T, Yamauchi Y, Funakoshi Y, Katagiri N, Ohbayashi N, Kanaho Y. Pathological functions of the small GTPase Arf6 in cancer progression: Tumor angiogenesis and metastasis. Small GTPases 2016; 7:47-53; PMID:26909552; http://dx.doi.org/ 10.1080/21541248.2016.1154640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dehapiot B, Halet G. Ran GTPase promotes oocyte polarization by regulating ERM (Ezrin/Radixin/Moesin) inactivation. Cell Cycle 2013; 12(11):1672-8; PMID:23656777; http://dx.doi.org/ 10.4161/cc.24901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Dehapiot B, Carrière V, Carroll J, Halet G. Polarized Cdc42 activation promotes polar body protrusion and asymmetric division in mouse oocytes. Dev Biol 2013; 377(1):202-12; PMID:23384564; http://dx.doi.org/ 10.1016/j.ydbio.2013.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ma R, Hou X, Zhang L, Sun SC, Schedl T, Moley K, Wang Q. Rab5a is required for spindle length control and kinetochore-microtubule attachment during meiosis inoocytes. FASEB J 2014; 28(9):4026-35; PMID:24876181; http://dx.doi.org/ 10.1096/fj.14-250886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hou X, Zhang J, Li L, Ma R, Ge J, Han L, Wang Q. Rab6a is a novel regulator of meiotic apparatus and maturational progression in mouse oocytes. Sci Rep 2016; 6:22209; PMID:26915694; http://dx.doi.org/ 10.1038/srep22209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhang Y, Duan X, Cao R, Liu HL, Cui XS, Kim NH, Rui R, Sun SC. Small GTPase RhoA regulates cytoskeleton dynamics during porcine oocyte maturation and early embryo development. Cell Cycle 2014; 13(21):3390-403; PMID:25485583; http://dx.doi.org/ 10.4161/15384101.2014.952967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Cherfils J, Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev 2016; 93:269-309; http://dx.doi.org/ 10.1152/physrev.00003.2012 [DOI] [PubMed] [Google Scholar]
- [15].Scheffzek K, Ahmadian MR, Wittinghofer A. GTPase-activating proteins: helping hands to complement an active site. Trends Biochem Sci 1998; 23:257-62; PMID:9697416; http://dx.doi.org/ 10.1016/S0968-0004(98)01224-9 [DOI] [PubMed] [Google Scholar]
- [16].East MP, Bowzard JB, Dacks JB, Kahn RA. ELMO domains, evolutionary and functional characterization of a novel GTPase-activating protein (GAP) domain for Arf protein family GTPases. J Biol Chem 2012; 287:39538-53; PMID:23014990; http://dx.doi.org/ 10.1074/jbc.M112.417477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ivanova AA, East MP, Yi SL, Kahn RA. Characterization of recombinant ELMOD (cell engulfment and motility domain) proteins as GTPase-activating proteins (GAPs) for ARF family GTPases. J Biol Chem 2014; 289:11111-21; PMID:24616099; http://dx.doi.org/ 10.1074/jbc.M114.548529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hodgson U, Pulkkinen V, Dixon M, Peyrard-Janvid M, Rehn M, Lahermo P, Ollikainen V, Salmenkivi K, Kinnula V, Kere J, Tukiainen P, Laitinen T. ELMOD2 is a candidate gene for familial idiopathic pulmonary fibrosis. Am J Hum Genet 2006; 79:149-54; PMID:16773575; http://dx.doi.org/ 10.1086/504639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Pulkkinen V, Bruce S, Rintahaka J, Hodgson U, Laitinen T, Alenius H, Kinnula VL, Myllärniemi M, Matikainen S, Kere J. ELMOD2, a candidate gene for idiopathic pulmonary fibrosis, regulates antiviral responses. FASEB J 2010; 24:1167-77; PMID:19966137; http://dx.doi.org/ 10.1096/fj.09-138545 [DOI] [PubMed] [Google Scholar]
- [20].Suzuki M, Murakami T, Cheng J, Kano H, Fukata M, Fujimoto T. ELMOD2 is anchored to lipid droplets by palmitoylation and regulates adipocyte triglyceride lipase recruitment. Mol Biol Cell 2015; 26:2333-42; PMID:25904333; http://dx.doi.org/ 10.1091/mbc.E14-11-1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bowzard JB, Cheng D, Peng J, Kahn RA. ELMOD2 is an Arl2 GTPase-activating protein that also acts on Arfs. J Biol Chem 2007; 15:17568-80; http://dx.doi.org/ 10.1074/jbc.M701347200 [DOI] [PubMed] [Google Scholar]
- [22].Newman LE, Zhou CJ, Mudigonda S, Mattheyses AL, Paradies E, Marobbio CM, Kahn RA. The ARL2 GTPase Is Required for Mitochondrial Morphology, Motility, and Maintenance of ATP Levels. PLoS One 2014; 9:e99270; PMID:24911211; http://dx.doi.org/ 10.1371/journal.pone.0099270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Li R, Jin Z, Gao L, Liu P, Yang Z, Zhang D. Effective protein inhibition in intact mouse oocytes through peptide nanoparticle-mediated antibodytransfection. PeerJ 2016; 4:e1849; PMID:27114861; http://dx.doi.org/ 10.7717/peerj.1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhang P, Ni X, Guo Y, Guo X, Wang Y, Zhou Z, Huo R, Sha J. Proteomic-based identification of maternal proteins in mature mouse oocytes. BMC Genomics 2009; 10:348; PMID:19646285; http://dx.doi.org/ 10.1186/1471-2164-10-348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gianaroli L, Magli MC, Cavallini G, Crippa A, Capoti A, Resta S, Robles F, Ferraretti AP. Predicting aneuploidy in human oocytes: key factors which affect the meiotic process. Hum Reprod 2010; 9:2374-86; http://dx.doi.org/ 10.1093/humrep/deq123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Brunet S, Maro B. Cytoskeleton and cell cycle control during meiotic maturation of the mouse oocyte: integrating time and space. Reproduction 2005; 6:801-11; http://dx.doi.org/ 10.1530/rep.1.00364 [DOI] [PubMed] [Google Scholar]
- [27].Van Blerkom J. Mitochondrial function in the human oocyte and embryo and their role in developmental competence. Mitochondrion 2011; 11:797-813; PMID:20933103; http://dx.doi.org/ 10.1016/j.mito.2010.09.012 [DOI] [PubMed] [Google Scholar]
- [28].Dalton CM, Szabadkai G, Carroll J. Measurement of ATP in single oocytes: impact of maturation and cumulus cells on levels and consumption. J Cell Physiol 2014; 229:353-61; PMID:24002908; http://dx.doi.org/ 10.1002/jcp.24457 [DOI] [PubMed] [Google Scholar]
- [29].Wilding M, Dale B, Marino M, Matteo L, Alviggi C, Pisaturo M Lombardi L, De Placido G. Mitochondrial aggregation patterns and activity in human oocytes and preimplantation embryos. Hum Reprod 2001; 16:909-17; PMID:11331637; http://dx.doi.org/ 10.1093/humrep/16.5.909 [DOI] [PubMed] [Google Scholar]
- [30].Blerkom J, Davis P, Lee J. ATP content of human oocytes and developmental potential and outcome after in-vitro fertilization and embryo transfer. Hum Reprod 1995; 10:415-24; PMID:7769073; http://dx.doi.org/ 10.1093/oxfordjournals.humrep.a135954 [DOI] [PubMed] [Google Scholar]
- [31].Yu Y, Dumollard R, Rossbach A, Anthony Lai F, Swann K. Redistribution of Mitochondria Leads to Bursts of ATP Production during Spontaneous Mouse Oocyte Maturation. J Cell Physiol 2010; 224:672-80; PMID:20578238; http://dx.doi.org/ 10.1002/jcp.22171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Fu XF, Yao K, Du X, Li Y, Yang XY, Yu M, Li MZ, Cui QH. PGC-1α regulates the cell cycle through ATP and ROS in CH1 cells. J Zhejiang Univ Sci B 2016; 17:136-46; PMID:26834014; http://dx.doi.org/ 10.1631/jzus.B1500158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Miniowitz-Shemtov S, Teichner A, Sitry-Shevah D, Hershko A. ATP is required for the release of the anaphase-promoting complex/cyclosome from inhibition by the mitotic checkpoint. Proc Natl Acad Sci U S A 2010; 107:5351-6; PMID:20212161; http://dx.doi.org/ 10.1073/pnas.1001875107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Panvert M, Dubiez E, Arnold L, Perez J, Mechulam Y, Seufert W, Schmitt E. Cdc123, a Cell Cycle Regulator Needed for eIF2 Assembly, Is an ATP-Grasp Protein with Unique Features. Structure 2015; 23:1596-608; PMID:26211610; http://dx.doi.org/ 10.1016/j.str.2015.06.014 [DOI] [PubMed] [Google Scholar]
- [35].Zhang X, Wu XQ, Lu S, Guo YL, Ma X. Deficit of mitochondria-derived ATP during oxidative stress impairs mouse MII oocyte spindles. Cell Res 2006; 16:841-50; PMID:16983401; http://dx.doi.org/ 10.1038/sj.cr.7310095 [DOI] [PubMed] [Google Scholar]


