Summary
Actin cytoskeleton dynamics plays vital roles in most forms of intracellular trafficking by promoting the biogenesis and transport of vesicular cargoes. Mounting evidence indicates that actin dynamics and membrane-cytoskeleton scaffolds also play essential roles in macroautophagy, the process by which cellular waste is isolated inside specialized vesicles called autophagosomes for recycling and degradation. Thus, branched-actin polymerization is necessary for the biogenesis of autophagosomes from the endoplasmic reticulum (ER) membrane. Actomyosin-based transport is then used to feed the growing phagophore with pre-selected cargoes and debris derived from different membranous organelles inside the cell. Mature autophagosomes then detach from the ER membrane by an unknown mechanism, and are transported and fused with lysosomes, endosomes and multi-vesicular bodies through mechanisms that involve actin- and microtubule-based motility, cytoskeleton-membrane scaffolds and signaling proteins. In this minireview, we highlight the considerable progress made recently towards understanding the diverse roles of the cytoskeleton in autophagy.
Introduction
Actin filament assembly and disassembly provides the mechanical forces for a wide range of cellular activities that involve membrane deformation, such as cell motility, phagocytosis, endocytosis and cytokinesis [1]. A large number of cytoskeletal proteins become involved in these processes, but while specific membrane structures (tubules, vesicles and filopodia) use specific subsets of proteins, some essential components and mechanisms are generally conserved. These include the Arp2/3 complex, activated by one of several nucleation promoting factors (NPFs), as well as actin filament elongation, severing and capping proteins [2]. Thus, it is beginning to emerge that cells use similar, and sometimes identical proteins to control the generation of endosomes from the plasma membrane [3], vesicles budding from the Golgi apparatus [4] and, more recently, the biogenesis of autophagosomes from the endoplasmic reticulum (ER) [5–8].
Autophagy is a highly regulated catabolic process, involving the formation of a double membrane cisterna in which cellular debris, protein aggregates, damaged organelles and pathogens are sequestered for degradation and/or recycling [9]. Numerous stimuli can trigger autophagy, but the most studied form, macroautophagy, is induced either by nutrient starvation (non-selective form) or by specific degradation targets (selective form). Examples of selective macroautopagy include mitophagy, aggrephagy, and xenophagy, referring respectively to the degradation of mitochondria, cytoplasmic aggregates, and pathogens [10]. Macroautophagy constitutes an evolutionarily conserved component of cellular homeostasis, and compromised autophagy results in the accumulation of damaged materials, which can lead to several human diseases, including cancer [11], neurodegenerative disorders [12], muscle degeneration [13] and cardiovascular diseases [14]. This minireview focuses on the role of the cytoskeleton in macroautophagy.
Autophagosome biogenesis and maturation
While our understanding of the molecular mechanisms of mammalian macroautophagy is still incipient, at least six discrete steps can be distinguished: initiation, formation of the omegasome, formation and expansion of the phagophore membrane, closure and detachment of the mature autophagosome, transport, and fusion of the autophagosome with lysosomes for degradation (Figure 1). In non-selective macroautophagy, autophagosome initiation is triggered by nutrient starvation, which activates the cellular sensor of energy homeostasis, AMPK (AMP-activated protein kinase). AMPK inhibits mTORC1 (mammalian target of rapamycin complex 1), which acts as a general repressor of autophagy. AMPK also activates the autophagosome initiating complex, consisting of ULK1 (Unc-51-like kinase 1, also known as Atg1 for autophagy-related protein 1) and two regulatory proteins, Atg13 and FIP200 (FAK-family kinase-interacting protein of 200 kDa) [15]. The ULK1 complex phosphorylates beclin-1, a component of the class III PI3K phosphatidylinositol 3-kinase complex that consists of a core catalytic subunit, Vps34 (vacuolar protein sorting 34), and three adaptor/regulatory subunits: beclin-1, Vps15 and Atg14 (also known as Barkor for Beclin 1-associated autophagy-related key regulator) [16]. The class III PI3K complex phosphorylates phosphatidylinositol at specific locations of the ER membrane, leading to an increase in the local concentration of phosphatidylinositol (3)-phosphate (PI(3)P) [17] (Figure 1A). These PI(3)P-rich sites act as precursors of autophagosome formation, by triggering the recruitment of a number of proteins, such as DFCP1 (double FYVE-containing protein 1) and WIPI (WD-repeat domain phosphoinositide-interacting) proteins, which are involved in the generation of a ring-like extension on the ER membrane known as the omegasome [9].
Figure 1.
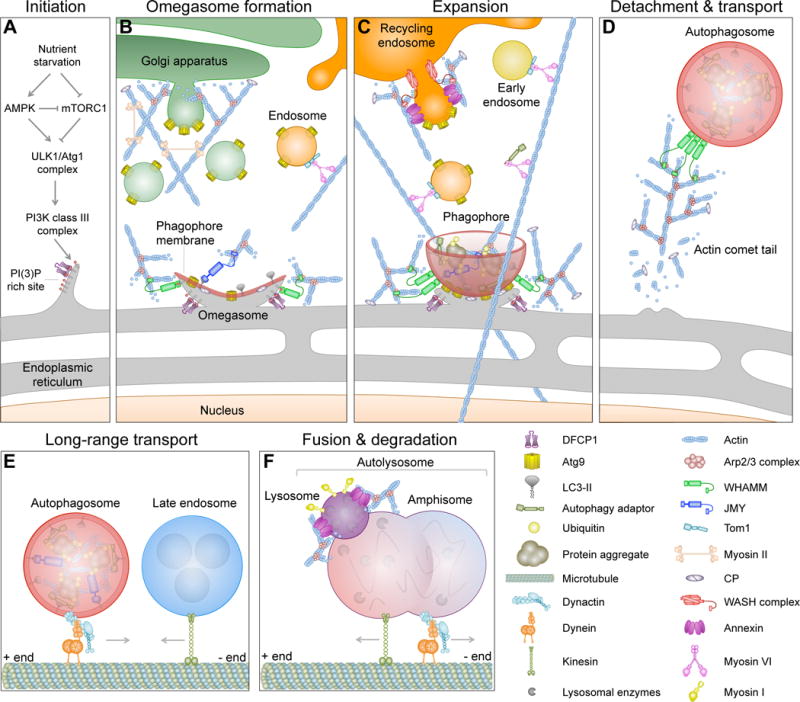
Stages of autophagy and the link to the cytoskeleton. (A) During nutrient starvation, the class III PI3K complex is recruited to the endoplasmic reticulum (ER), where it creates a PI(3)P-rich site that seeds the formation of the omegasome. Key autophagy proteins, such as WIPI1, WIPI2 and DFCP1, and the Arp2/3 complex NPF WHAMM are recruited to these sites. WHAMM activates the Arp2/3 complex to form an actin-branched network that provides mechanical forces for the formation of the omegasome. (B) A membrane cisterna called the phagophore (or isolation membrane) begins to form and expand on the omegasome. The expansion of the phagophore requires Atg9-rich membranes derived from different sources, including endosomes and the Golgi apparatus. LC3-II inserts into the phagophore membrane, where it serves as an anchor for autophagy adaptor proteins, and may also recruit JMY, another NPF of the Arp2/3 complex. (C) Together, JMY and WHAMM induce the formation of a branched actin network needed for the expansion of the phagophore membrane. Ubiquitinated cargo from different sources, including mitochondria and protein aggregates, is delivered to the growing phagophore using actomyosin-based transport. BAR domain proteins and annexins contribute to membrane remodeling and fusion events during expansion. (D) The mature autophagosome detaches from the omegasome and is initially transported using an actin comet tail mechanism. (E) The autophagosome is then transported over longer distances by dynein-dynactin along microtubules. (F) The autophagosome fuses with a late endosome to form an amphisome, which then fuses with an acidic lysosome to form an autolysosome. These fusion events depend on membrane-cytoskeleton adaptors, such as annexins, SNARE proteins, actin and myosin I. The acidic pH of the autolysosome activates hydrolytic enzymes that degrade the contents of the autophagosome.
The omegasome serves as a cradle for the formation of the preautophagosome membrane, commonly referred to as the phagophore or isolation membrane (Figure 1B). In higher eukaryotes, several membrane sources contribute to the expansion of the phagophore membrane, including specialized ER domains [18], the plasma membrane [19], endosomes [20–22], mitochondria [23], the Golgi apparatus [24, 25], and the ER-Golgi intermediate compartment (ERGIC) [26]. These sources contribute a key regulator of autophagy induction, the multi-spanning membrane protein Atg9. Atg9 acts as an essential precursor for the recruitment of the ubiquitin-like conjugation machinery, comprising a conserved Atg5-Atg12-Atg16 complex, that covalently lipidates cytosolic LC3 (known as LC3-I) to produce an LC3-phosphatidylethanolamine conjugate (LC3-II) [27, 28]. An increase in the ratio of LC3-II to LC3-I typically correlates with an increase in the number of autophagosomes [29]. LC3-II inserts into the phagophore membrane [30], where it functions as a universal anchor for a number of proteins involved in macroautophagy [31]. In selective forms of macroautophagy, several autophagy-specific adaptors, such as sequestosome-1/p62 [32], NDP52 (nuclear dot protein of 52 kDa) [33], optineurin [34], TAX1BP1 (Tax1-binding protein 1) [35] and NBR1 (next to BRCA1 gene 1 protein) [36], are able to directly tether ubiquitinated targets to LC3-II through their LC3-interacting region (LIR) (Figure 1C). LC3-II also plays an essential role during closure of the phagophore membrane [37], which generates the double-membrane autophagosome. Following their detachment, mature autophagosomes are transported using both actin- and microtubule-based mechanisms (Figure 1D–E). Autophagosomes are then fused either directly with lysosomes, to form autolysosomes, or with components of the endocytic pathway, such as endosomes or multivesicular bodies, to form amphisomes [38, 39] (Figure 1F). Ultimately, it is the SNARE-dependent fusion of autophagosomes (or in some cases amphisomes) with acidic lysosomes that leads to the degradation of the autolysosome content and inner membrane by lysosomal degradation enzymes [40].
Actin dynamics and autophagy
A role for actin in autophagy was first recognized ~25 years ago, when it was observed that starved cells treated with actin depolymerizing agents, such as cytochalasin D and latrunculin B, failed to produce autophagosomes [41]. Yet, following this initial report, actin assembly and autophagy were rarely considered together, until a flurry of recent studies reproduced and expanded upon these initial observations [42–44]. Other than the effect of actin depolymerizing agents on autophagy, these new studies demonstrate the colocalization of actin filaments with important autophagy markers [42, 43] (Figure 2). What is more, there appears to exist a reciprocal interconnection between autophagy and actin assembly, since a mouse knockout of Atg7, which prevents autophagosome formation, also displays severe defects in actin assembly due to reduced expression of proteins involved in the control of actin dynamics [44].
Figure 2.
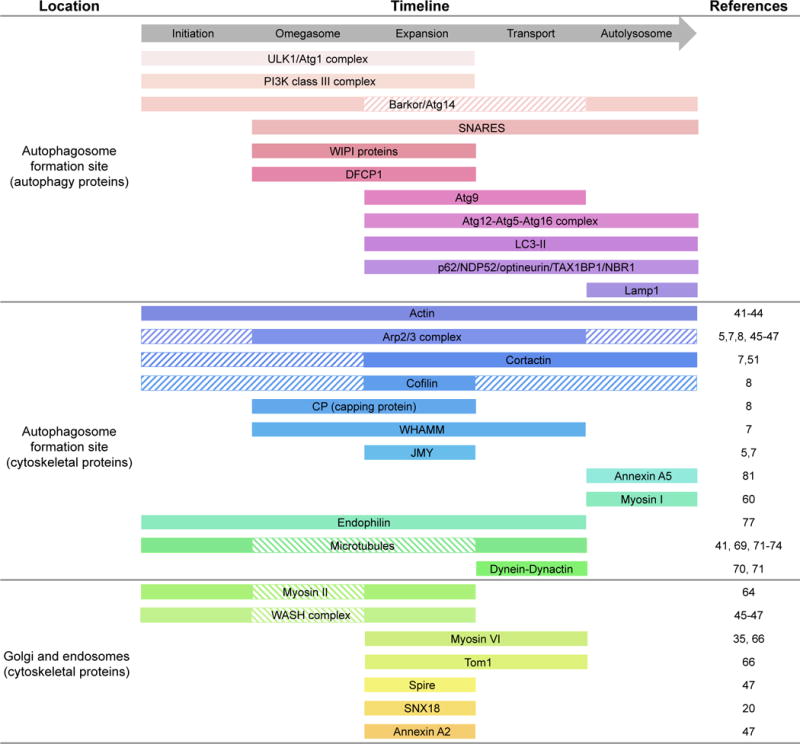
Autophagy timeline showing the arrival and departure of mammalian cytoskeleton assembly factors. Solid bars indicate the timing of appearance of autophagy and cytoskeletal proteins according to published evidence (references listed on the right), whereas striped bars indicate the time of arrival based on the known functions of proteins for which direct evidence is still lacking.
Arp2/3 complex and autophagy
A series of recent studies have specifically link the Arp2/3 complex to actin polymerization during the various steps of autophagy [5, 7, 8, 45–47] (Figure 2). The first step in actin polymerization, nucleation, is kinetically unfavorable, such that cells use filament nucleation machineries to promote actin polymerization in a spatiotemporally controlled manner [48]. Among these machineries, the Arp2/3 complex is unique in that it is the only nucleator capable of producing branched actin networks, which are essential for nearly all forms of membrane remodeling activities in cells [49], including not surprisingly autophagy. Thus, pharmacological inhibition of the Arp2/3 complex with CK-666 [5, 7, 47] or siRNA knockdown of Arp2 [47], a key subunit of the Arp2/3 complex, lead to a decrease in LC3-II levels and the number of autophagosomes. Inhibition of the Arp2/3 complex also leads to the formation of elongated tubular structures that are positive for the omegasome markers DFCP1 [8] and LC3-II [7], suggesting that proper omegasome formation is somehow halted by the inhibition of the Arp2/3 complex, i.e. branched actin network assembly (Figure 1B). In mature autophagosomes, the Arp2/3 complex appears to localize to highly dynamic puncta on the sides of autophagosomes [8], where it triggers the formation of actin comet tails [7]. Actin comet tails are characteristically linked to actin-driven motility of membranous organelles and pathogens [50], suggesting that the Arp2/3 complex plays a role in shaping the autophagosome membrane [7] (Figure 1C) and transporting autophagosomes after detachment (Figure 1D). In selective autophagy, the fusion of autophagosomes with lysosomes depends on the recruitment of cortactin to fusion sites by the ubiquitin-binding deacetylase HDAC6 [51] (Figure 1F). Cortactin is a well-known stabilizer of actin filament branches formed by the Arp2/3 complex [49], suggesting that the Arp2/3 complex is implicated in autophagosome-lysosome fusion. Conceivably, the Arp2/3 complex could play a similar role in non-selective macroautophagy.
Nucleation promoting factors and autophagy
A group of proteins known as nucleation-promoting factors (NPFs) are responsible for the spatiotemporal activation of the Arp2/3 complex in cells [2, 49]. Recent work on NPFs has reinforced the link between the Arp2/3 complex and authophagy (Figure 2). Different NPFs activate actin nucleation and branching at different subcellular locations and in connection with different activities and signaling inputs. NPFs share little in common, other than their C-terminal proline-rich, WH2 domain, central, acidic (PWCA) region, which mediates their interactions with profilin-actin, actin, and the Arp2/3 complex, respectively [48]. The PWCA region of NPFs is sufficient to activate the Arp2/3 complex in vitro, whereas the other domains are responsible for the regulation and localization of NPFs. Three NPFs, WHAMM (WASP homolog associated with actin, membranes and microtubules), JMY (junction-mediating and regulatory protein), and the WASH (WAS protein family homolog) complex, have been implicated in mammalian autophagy [5, 7, 45, 46, 52, 53], whereas another NPF, the WAVE (WASP family verprolin homologous) complex, has been implicated in plant autophagy [53].
First, the pentameric WASH complex was shown to colocalize with autophagosomes upon starvation [45, 46, 52]. The WASH complex colocalizes with markers of expanding phagosomes (Atg5, LC3-II and p62), but does not appear to colocalize with autolysosome markers (Lamp1), suggesting that its role in autophagy is limited to the early stages of autophagosome biogenesis [45]. WASH-complex deficiency in mouse embryos causes a marked increase in autophagy, which was initially interpreted as evidence that the complex inhibits autophagy through suppression of beclin-1 ubiquitination [45]. However, a more recent study showed that siRNA depletion of the WASH subunit of the complex leads to Atg9 accumulation at the Golgi and a loss of LC3-II-positive vesicles in starved cells [46], indicating a defect in the transport of Atg9-containing membranes for the expansion of the phagophore membrane. Thus, the role of the WASH complex in autophagy appears to involve retromer-mediated sorting of protein cargos to the growing phagorphore (Figure 1C). Future studies should clarify whether the WASH complex also plays an inhibitory [45] role during autophagy.
Recently, two related NPFs, WHAMM and JMY, were also implicated in autophagosome formation [5, 7]. Thus, depletion of both WHAMM and JMY by siRNA or mutations that impair their ability to interact with the Arp2/3 complex lead to decreased LC3-II levels and the number of autophagosomes [5, 7]. Removal of the first 169 amino acids of WHAMM or the first 119 residues of JMY results in loss of autophagosome localization, suggesting that both NPFs interact with the phagophore membrane through their N-terminal region (Figure 1B). JMY contains an LC3-interacting LIR motif within this region [5]. It is unclear whether WHAMM also contains a LIR motif, since its N-terminal so-called ER-binding domain displays notable sequence differences with JMY [7]. Moreover, WHAMM appears to bind directly and specifically to PI(3)P-containing membranes [54] (Figure 1B). Consistently, WHAMM precedes both LC3-II and JMY at autophagy sites (Figure 2), co-localizing with puncta of the early autophagy marker DFCP1 on the ER membrane, and propelling the movement of these puncta through an Arp2/3 complex-dependent actin comet tail mechanism [7] (Figure 1D). On the contrary, JMY does not appear to colocalize with actin comet tails [7]. JMY also contains three actin-binding WH2 domains, whereas WHAMM contains only two, such that JMY displays Arp2/3 complex-independent nucleation activity [55]. It is not surprising, but somewhat confirmatory, that these two related NPFs both function in autophagy, and yet they display clear differences in sequence, actin nucleation activity, and the time of localization to autophagy sites, suggesting that they play non-overlapping roles in this process. It is tempting to speculate that WHAMM is more directly involved in the formation of the omegasome and/or the early stages of phagophore expansion as well as later on during autophagosome comet-tail-driven motility, whereas JMY is recruited after LC3-II, playing a role during the intermediate stages of autophagosome maturation.
One study proposed that an Arp2/3 complex-dependent network of actin filaments might help shape and expand the phagophore membrane from the inside [8]. While this work did not identify the NPF that activates the Arp2/3 complex within this context, other actin-assembly factors were identified, including capping protein (CP), which binds filament barbed ends and stops monomer addition/dissociation, and cofilin, a filament severing protein. Knocking down CP, in particular, resulted in irregularly shaped phagophores that failed to mature into autophagosomes. These authors proposed that CP is trapped at the omegasome membrane through interaction with PI(3)P, thus freeing barbed end polymerization at sites of autophagosome formation [8].
Finally, the WAVE complex was found to be involved in autophagy in plants [53]. NAP1, a subunit of the pentameric WAVE complex, was shown to relocate from the cytoplasm to the ER membrane in response to pressure-induced stress, and this triggers Arp2/3 complex-dependent polymerization needed for phagophore formation and expansion [53]. Consistently, knocking out NAP1, or other subunits of the WAVE complex, reduces the number of autophagosomes during starvation and leads to autophagy defects that render these plats less tolerant to salt and nitrogen starvation [53]. The contribution of the WAVE complex to autophagy in plants, which lack genes for WHAMM and JMY, underscores the conserved role of the Arp2/3 complex in eukaryotic autophagy.
Actin-based motility and autophagy
Another in which actin participates in autophagy is by providing the tracks for myosin-based motility. Several actin-based motors, including myosins I, II and VI have been implicated in autophagy [56]. Myosin IC is widely expressed and plays diverse roles in eukaryotic cells [57], including in the transport of sphingolipid- and cholesterol-rich lipid rafts from the TGN to the plasma membrane [58]. Cholesterol is a critical component for autophagosome-lysosome fusion [59], which probably explains why myosin IC deficiency leads to autophagosome accumulation [60] (Figure 1F). While myosin IB is also found on lysosomes [61], it remains to be demonstrated whether this isoform also functions in autophagy.
Non-muscle myosin II (NMM2) is abundantly expressed and accomplishes countless functions [62], including the transport of autophagy-specific proteins from the TGN [63]. Thus, ULK1/Atg1 promotes the phosphorylation-dependent activation of myosin II to help drive autophagosome formation by regulating Atg9 trafficking from the TGN (or cis-Golgi) during phagophore expansion [64] (Figures 1B). In this way, ULK1 regulates both autophagosome initiation and trafficking of membranes for its maturation.
Myosin VI is the only pointed-end directed actin-based motor, and is involved in many cellular processes [65], including cargo delivery for phagophore expansion and lysosomal fusion [66] (Figure 1C). Myosin VI recruits cargo through adaptor proteins, including optineurin, NDP52, and TAX1BP1, that associate with damaged, ubiquitinated organelles or pathogens [35, 66]. Neuronal and fibroblast cells derived from myosin VI knockout mice display an accumulation of autophagosomes, consistent with inhibition of autophagosome clearance [66]. A similar phenotype is observed in HeLa cells in which the myosin VI-endosome adaptor protein Tom1 is knocked down [66]. Tom1 is a constituent of the ESCRT-0 complex essential for autophagosome maturation during autophagy [67]. Thus, the inhibition of autophagosome clearance in myosin VI-deficient cells is possibly due to compromised transport of endosomal components during phagophore expansion and/or autophagosome maturation, which are afterwards required for lysosomal fusion [66].
Microtubule-based motility and autophagy
Microtubule dynamics and microtubule-based motors have also been implicated in autophagy [68]. Thus, starvation-induced autophagosome formation appears to require the most dynamic, so-called labile microtubule subset [69], whereas the centripetal movement of mature autophagosomes prior to fusion with lysosomes may require stable microtubules [41, 69–73] (Figures 1E and 1F). Drugs that either stabilize of destabilize microtubules interfere with authophagy in different ways. Thus, limited treatment of HeLa cells with nocodazole, a microtubule-depolymerizing drug, depolymerizes primarily the labile microtubule subset and prevents the formation of starvation-induced autophagosomes [69]. More extensive nocodazole treatment of HeLa [69], HEK293 [73], and rat kidney [41] cells leads to complete depolymerization of both labile and stable microtubules, and fully inhibits autophagic flux. Autophagosome formation is also inhibited by treatment with Taxol, a drug that stabilizes microtubules and interferes with the dynamic turnover of the microtubule network [69, 73]. A key step in autophagosome clearance is the transport of autophagosomes dispersed throughout the cell toward lysosomes that concentrate near centrosomes [70]. In HeLa cells, mature autophagosomes accumulate around the centrosome, and the dynein-dynactin complex that drives microtubule minus-end directed movement promotes this accumulation and is essential for lysosomal fusion [70, 71]. Microtubule-dependent transport is particularly critical in neuronal cells, where autophagosomes formed in the axonal tip need to be transported over long distances toward the cell body for degradation [74]. The role of microtubule plus-end directed kinesin motors in macroautophagy is less well understood, although there is strong evidence that they participate in autophagosome maturation in neurons [74], and may also be important for maintaining lysosome homeostasis during autophagy [75].
Coordinated membrane and cytoskeleton dynamics during autophagy
A tight spatiotemporal coordination of actin cytoskeleton and membrane dynamics is a distinctive feature of many cellular processes, including cell migration, morphogenesis and endocytosis [76]. In these processes, Bin/Amphiphysin/Rvs (BAR) domain proteins have emerged as essential regulators, linking signaling pathways to actin cytoskeleton and membrane remodeling. BAR domain proteins feature a curved membrane-binding surface as well as other protein-protein, protein-membrane and signaling domains that contribute to the membrane binding activity and the recruitment of signaling and cytoskeletal proteins. Two BAR domain proteins, endophilin and SNX18 (sorting nexin 18), have been found in association with autophagosomes [20] (Figure 2). Endophilin was shown to stabilize mature autophagosomes by binding to the outer leaflet of the autophagosomal membrane [77]. The endophilin orthologue in plants, SH3P2, was also linked to autophagy and shown to interact with LC3-II and beclin-1, suggesting that endophilin arrives at autophagy sites during the phagophore expansion phase [78]. SNX18 was shown to bind to LC3 in vitro and to colocalize with LC3-II in nutrient-starved cells [20]. In these cells, SNX18 also colocalizes with makers of recycling endosomes, Rab11 and the transferrin receptor, implying that SNX18 is not involved in autophagosome maturation per se, but rather in the tubulation and delivery of endosomes to autophagosomes.
The annexin family of Ca2+-regulated phospholipid-binding proteins constitutes another group of adaptors implicated in membrane budding and fusion and the recruitment of binding partners to specific membranes [79]. Of the 12 annexins expressed in humans, several interact directly with actin, including A1, A2, A5 and A6, and may thus physically connect membranes to actin filaments [80]. Of these, annexins A2 and A5 have been implicated in autophagy [47, 81] (Figure 2). Annexin A2 regulates autophagosome formation by enabling trafficking of Atg9-containing vesicles, likely acting as a tether between recycling endosomes and actin networks assembled by the Arp2/3 complex or the WH2 domain-based nucleator Spire [47, 48] (Figure 1C). Annexin A2 expression increases with starvation; its knockdown abrogates starvation-induced autophagy whereas its overexpression upregulates autophagy [47]. Annexin A5 accumulates on lysosomal membranes during starvation, and overexpression and silencing experiments indicate that it induces autophagosome-lysosome fusion (Figure 1F), thus increasing lysosomal protein degradation [81]. An annexin role in autophagosome-lysosome fusion is further consistent with the observation that this step is both actin- [51] and Ca2+-dependent [38].
While the evidence is still limited, these two families of membrane adaptors likely play distinct roles in autophagy, with BAR domain proteins mediating the establishment and maintenance of membrane curvature and annexins participating in membrane fusion during the expansion of the phagophore membrane and amphisome formation. Both families share the ability to recruit binding partners, including signaling proteins and actin, which is a common factor in these events.
Additional actin-independent, membrane-binding scaffolds are implicated in autophagy [82]. For instance, Atg14 contains a Barkor/Atg14 autophagosome-targeting sequence (BATS) that targets it to PI(3)P-rich curved membranes, where it regulates the PI3K complex and stabilizes the curvature of the phagophore membrane [83]. Recent work further indicates that oligomeric Atg14 triggers the formation of a trans-SNARE complex to promote autophagosome-lysosome fusion [84]. Finally, the Atg5-Atg12-Atg16 complex may also be involved in establishing and maintaining membrane curvature during phagophore expansion [85]. Atg5-Atg12 oligomers are thought to bind to LC3-II directly, which then recruits a dimer of Atg16. Side-by-side association of Atg5-Atg12-Atg16 complexes might allow the formation of a continuous meshwork that could potentially scaffold the growing phagophore membrane [85].
GTPase signaling and autophagy
Most aspects of actin dynamics are controlled by Rho-family GTPases [86], and RhoA and Rac1 appear to be specifically implicated in autophagy, whereas Cdc42 is not [42]. Overexpression of constitutively active RhoA leads to an increase in the number of autophagosomes, whereas the expression of a dominant negative mutant of Rac1 increases the number of autophagosomes, even without starvation [42]. The effect of RhoA on autophagy appears to be mediated by rho-associated protein kinase (ROCK), although its function is disputed. In one study, pharmacological inhibition of ROCK in starved cells blocks the stimulation of autophagosome formation by RhoA expression [42]. In contrast, another study shows that ROCK inhibition during starvation leads to stimulation of the autophagic response and abnormal size and protein composition and size of autophagosomes [42, 87]. Future work should help clarify the diverging views on the role of RhoA in autophagy.
In a parallel way, Rab-family GTPases control membrane-remodeling events, and several Rab-family GTPases function as regulators of membrane fusion events or as cargo adaptors during autophagy [88]. Rab1, in particular, has been implicated in regulating actin assembly through interaction with WHAMM [89]. In mammalian cells, the number of autophagosomes increases with overexpression of Rab1, and a constitutively active form of Rab1 strongly colocalizes with LC3-II after induction of macroautophagy [90]. Rab1 expression also promotes the accumulation of WHAMM on the ER, which leads to increased membrane tubulation [89]. This effect could be due to inhibition of the Arp2/3 complex, since Rab1 inhibits the WHAMM-dependent activation of the Arp2/3 complex in vitro [89] and Arp2/3 complex inhibition by CK-666 also leads to an increase in WHAMM-coated tubules [7].
Concluding remarks
Growing evidence supports the notion that actin dynamics plays important roles throughout the various steps of autophagy (Figures 1 and 2). The Arp2/3 complex is at least in part responsible for actin polymerization in this process, but the specific function of actin branched networks in autophagy is still unknown. Future work should clarify why at least three different NPFs are required for this process, and how their activities are coordinated.
While the role of actin dynamics in non-selective macroautophagy in mammalian cells is undisputable, less is known about its role in other forms of autophagy, and specifically in selective forms of macroautophagy. In these processes, the degradation targets themselves appear to act as templates for autophagosomes biogenesis, bypassing the need for the formation of a ‘waste basket’ [91], and thus the requirement for actin polymerization-dependent forces to reshape the ER membrane. How the phagophore membrane is initiated on these targets is only beginning to emerge [92]. However, actin dynamics has been implicated in the formation of autophagosomes during mitophagy [93], as well as in lysosomal fusion during the clearance of protein aggregates [51] or bacteria [94], suggesting that while actin’s role may vary, its presence is likely ubiquitous in any form of macroautophagy.
Predictably, assembly factors that control actin dynamics in other membrane trafficking events, including formins and actin bundling proteins, are likely to also participate in autophagy. Indeed, a recent study implicates the actin nucleator Spire, which typically functions is combination with formins [48], in Atg9 trafficking to recycling endosomes [47]. These and other aspects of the cytoskeleton-autophagy connection will likely occupy scientists in the coming years.
Acknowledgments
This work was supported by NIH grants P01 GM087253.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Anitei M, Hoflack B. Bridging membrane and cytoskeleton dynamics in the secretory and endocytic pathways. Nat Cell Biol. 2011;14:11–19. doi: 10.1038/ncb2409. [DOI] [PubMed] [Google Scholar]
- 2.Suetsugu S, Gautreau A. Synergistic BAR-NPF interactions in actin-driven membrane remodeling. Trends Cell Biol. 2012;22:141–150. doi: 10.1016/j.tcb.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Kaksonen M, Toret CP, Drubin DG. Harnessing actin dynamics for clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2006;7:404–414. doi: 10.1038/nrm1940. [DOI] [PubMed] [Google Scholar]
- 4.Egea G, Lazaro-Dieguez F, Vilella M. Actin dynamics at the Golgi complex in mammalian cells. Curr Opin Cell Biol. 2006;18:168–178. doi: 10.1016/j.ceb.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 5.Coutts AS, La Thangue NB. Actin nucleation by WH2 domains at the autophagosome. Nat Commun. 2015;6:7888. doi: 10.1038/ncomms8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kast DJ, Dominguez R. WHAMM links actin assembly via the Arp2/3 complex to autophagy. Autophagy. 2015;11:1702–1704. doi: 10.1080/15548627.2015.1073434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kast DJ, Zajac AL, Holzbaur EL, Ostap EM, Dominguez R. WHAMM Directs the Arp2/3 Complex to the ER for Autophagosome Biogenesis through an Actin Comet Tail Mechanism. Curr Biol. 2015;25:1791–1797. doi: 10.1016/j.cub.2015.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mi N, Chen Y, Wang S, Chen M, Zhao M, Yang G, Ma M, Su Q, Luo S, Shi J, et al. CapZ regulates autophagosomal membrane shaping by promoting actin assembly inside the isolation membrane. Nat Cell Biol. 2015;17:1112–1123. doi: 10.1038/ncb3215. [DOI] [PubMed] [Google Scholar]
- 9.Ariosa AR, Klionsky DJ. Autophagy core machinery: overcoming spatial barriers in neurons. J Mol Med (Berl) 2016;94:1217–1227. doi: 10.1007/s00109-016-1461-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reggiori F, Komatsu M, Finley K, Simonsen A. Autophagy: more than a nonselective pathway. Int J Cell Biol. 2012;2012:219625. doi: 10.1155/2012/219625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.White E. The role for autophagy in cancer. J Clin Invest. 2015;125:42–46. doi: 10.1172/JCI73941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Menzies FM, Fleming A, Rubinsztein DC. Compromised autophagy and neurodegenerative diseases. Nat Rev Neurosci. 2015;16:345–357. doi: 10.1038/nrn3961. [DOI] [PubMed] [Google Scholar]
- 13.Sandri M. Autophagy in skeletal muscle. FEBS Lett. 2010;584:1411–1416. doi: 10.1016/j.febslet.2010.01.056. [DOI] [PubMed] [Google Scholar]
- 14.Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368:651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- 15.Wong PM, Puente C, Ganley IG, Jiang X. The ULK1 complex: sensing nutrient signals for autophagy activation. Autophagy. 2013;9:124–137. doi: 10.4161/auto.23323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jean S, Kiger AA. Classes of phosphoinositide 3-kinases at a glance. J Cell Sci. 2014;127:923–928. doi: 10.1242/jcs.093773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yla-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5:1180–1185. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
- 19.Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol. 2010;12:747–757. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knaevelsrud H, Soreng K, Raiborg C, Haberg K, Rasmuson F, Brech A, Liestol K, Rusten TE, Stenmark H, Neufeld TP, et al. Membrane remodeling by the PX-BAR protein SNX18 promotes autophagosome formation. J Cell Biol. 2013;202:331–349. doi: 10.1083/jcb.201205129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Puri C, Renna M, Bento CF, Moreau K, Rubinsztein DC. Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell. 2013;154:1285–1299. doi: 10.1016/j.cell.2013.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Longatti A, Tooze SA. Recycling endosomes contribute to autophagosome formation. Autophagy. 2012;8:1682–1683. doi: 10.4161/auto.21486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, Sekito T, Kondo-Kakuta C, Ichikawa R, Kinjo M, Ohsumi Y. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol. 2012;198:219–233. doi: 10.1083/jcb.201202061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mari M, Griffith J, Rieter E, Krishnappa L, Klionsky DJ, Reggiori F. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol. 2010;190:1005–1022. doi: 10.1083/jcb.200912089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ge L, Melville D, Zhang M, Schekman R. The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. Elife. 2013;2:e00947. doi: 10.7554/eLife.00947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamasaki M, Shibutani ST, Yoshimori T. Up-to-date membrane biogenesis in the autophagosome formation. Curr Opin Cell Biol. 2013;25:455–460. doi: 10.1016/j.ceb.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 28.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 29.Rubinsztein DC, Cuervo AM, Ravikumar B, Sarkar S, Korolchuk V, Kaushik S, Klionsky DJ. In search of an “autophagomometer”. Autophagy. 2009;5:585–589. doi: 10.4161/auto.5.5.8823. [DOI] [PubMed] [Google Scholar]
- 30.Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19:2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wild P, McEwan DG, Dikic I. The LC3 interactome at a glance. J Cell Sci. 2014;127:3–9. doi: 10.1242/jcs.140426. [DOI] [PubMed] [Google Scholar]
- 32.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 33.Verlhac P, Gregoire IP, Azocar O, Petkova DS, Baguet J, Viret C, Faure M. Autophagy receptor NDP52 regulates pathogen-containing autophagosome maturation. Cell Host Microbe. 2015;17:515–525. doi: 10.1016/j.chom.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 34.Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tumbarello DA, Manna PT, Allen M, Bycroft M, Arden SD, Kendrick-Jones J, Buss F. The Autophagy Receptor TAX1BP1 and the Molecular Motor Myosin VI Are Required for Clearance of Salmonella Typhimurium by Autophagy. PLoS Pathog. 2015;11:e1005174. doi: 10.1371/journal.ppat.1005174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 37.Fujita N, Hayashi-Nishino M, Fukumoto H, Omori H, Yamamoto A, Noda T, Yoshimori T. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol Biol Cell. 2008;19:4651–4659. doi: 10.1091/mbc.E08-03-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mauvezin C, Nagy P, Juhasz G, Neufeld TP. Autophagosome-lysosome fusion is independent of V-ATPase-mediated acidification. Nat Commun. 2015;6:7007. doi: 10.1038/ncomms8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fader CM, Colombo MI. Autophagy and multivesicular bodies: two closely related partners. Cell Death Differ. 2009;16:70–78. doi: 10.1038/cdd.2008.168. [DOI] [PubMed] [Google Scholar]
- 40.Saftig P, Klumperman J. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat Rev Mol Cell Biol. 2009;10:623–635. doi: 10.1038/nrm2745. [DOI] [PubMed] [Google Scholar]
- 41.Aplin A, Jasionowski T, Tuttle DL, Lenk SE, Dunn WA., Jr Cytoskeletal elements are required for the formation and maturation of autophagic vacuoles. J Cell Physiol. 1992;152:458–466. doi: 10.1002/jcp.1041520304. [DOI] [PubMed] [Google Scholar]
- 42.Aguilera MO, Beron W, Colombo MI. The actin cytoskeleton participates in the early events of autophagosome formation upon starvation induced autophagy. Autophagy. 2012;8:1590–1603. doi: 10.4161/auto.21459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reggiori F, Monastyrska I, Shintani T, Klionsky DJ. The actin cytoskeleton is required for selective types of autophagy, but not nonspecific autophagy, in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2005;16:5843–5856. doi: 10.1091/mbc.E05-07-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhuo C, Ji Y, Chen Z, Kitazato K, Xiang Y, Zhong M, Wang Q, Pei Y, Ju H, Wang Y. Proteomics analysis of autophagy-deficient Atg7−/−MEFs reveals a close relationship between F-actin and autophagy. Biochem Biophys Res Commun. 2013;437:482–488. doi: 10.1016/j.bbrc.2013.06.111. [DOI] [PubMed] [Google Scholar]
- 45.Xia P, Wang S, Du Y, Zhao Z, Shi L, Sun L, Huang G, Ye B, Li C, Dai Z, et al. WASH inhibits autophagy through suppression of Beclin 1 ubiquitination. EMBO J. 2013;32:2685–2696. doi: 10.1038/emboj.2013.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zavodszky E, Seaman MN, Moreau K, Jimenez-Sanchez M, Breusegem SY, Harbour ME, Rubinsztein DC. Mutation in VPS35 associated with Parkinson’s disease impairs WASH complex association and inhibits autophagy. Nat Commun. 2014;5:3828. doi: 10.1038/ncomms4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moreau K, Ghislat G, Hochfeld W, Renna M, Zavodszky E, Runwal G, Puri C, Lee S, Siddiqi F, Menzies FM, et al. Transcriptional regulation of Annexin A2 promotes starvation-induced autophagy. Nat Commun. 2015;6:8045. doi: 10.1038/ncomms9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dominguez R. The WH2 Domain and Actin Nucleation: Necessary but Insufficient. Trends Biochem Sci. 2016;41:478–490. doi: 10.1016/j.tibs.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rotty JD, Wu C, Bear JE. New insights into the regulation and cellular functions of the ARP2/3 complex. Nat Rev Mol Cell Biol. 2013;14:7–12. doi: 10.1038/nrm3492. [DOI] [PubMed] [Google Scholar]
- 50.Lamason RL, Welch MD. Actin-based motility and cell-to-cell spread of bacterial pathogens. Curr Opin Microbiol. 2016;35:48–57. doi: 10.1016/j.mib.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee JY, Koga H, Kawaguchi Y, Tang W, Wong E, Gao YS, Pandey UB, Kaushik S, Tresse E, Lu J, et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 2010;29:969–980. doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.King JS, Gueho A, Hagedorn M, Gopaldass N, Leuba F, Soldati T, Insall RH. WASH is required for lysosomal recycling and efficient autophagic and phagocytic digestion. Mol Biol Cell. 2013;24:2714–2726. doi: 10.1091/mbc.E13-02-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang P, Richardson C, Hawes C, Hussey PJ. Arabidopsis NAP1 Regulates the Formation of Autophagosomes. Curr Biol. 2016;26:2060–2069. doi: 10.1016/j.cub.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 54.Campellone KG, Webb NJ, Znameroski EA, Welch MD. WHAMM is an Arp2/3 complex activator that binds microtubules and functions in ER to Golgi transport. Cell. 2008;134:148–161. doi: 10.1016/j.cell.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zuchero JB, Coutts AS, Quinlan ME, Thangue NB, Mullins RD. p53-cofactor JMY is a multifunctional actin nucleation factor. Nat Cell Biol. 2009;11:451–459. doi: 10.1038/ncb1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kruppa AJ, Kendrick-Jones J, Buss F. Myosins, Actin and Autophagy. Traffic. 2016;17:878–890. doi: 10.1111/tra.12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Greenberg MJ, Ostap EM. Regulation and control of myosin-I by the motor and light chain-binding domains. Trends Cell Biol. 2013;23:81–89. doi: 10.1016/j.tcb.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brandstaetter H, Kendrick-Jones J, Buss F. Myo1c regulates lipid raft recycling to control cell spreading, migration and Salmonella invasion. J Cell Sci. 2012;125:1991–2003. doi: 10.1242/jcs.097212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fraldi A, Annunziata F, Lombardi A, Kaiser HJ, Medina DL, Spampanato C, Fedele AO, Polishchuk R, Sorrentino NC, Simons K, et al. Lysosomal fusion and SNARE function are impaired by cholesterol accumulation in lysosomal storage disorders. EMBO J. 2010;29:3607–3620. doi: 10.1038/emboj.2010.237. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 60.Brandstaetter H, Kishi-Itakura C, Tumbarello DA, Manstein DJ, Buss F. Loss of functional MYO1C/myosin 1c, a motor protein involved in lipid raft trafficking, disrupts autophagosome-lysosome fusion. Autophagy. 2014;10:2310–2323. doi: 10.4161/15548627.2014.984272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cordonnier MN, Dauzonne D, Louvard D, Coudrier E. Actin filaments and myosin I alpha cooperate with microtubules for the movement of lysosomes. Mol Biol Cell. 2001;12:4013–4029. doi: 10.1091/mbc.12.12.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ma X, Adelstein RS. The role of vertebrate nonmuscle Myosin II in development and human disease. Bioarchitecture. 2014;4:88–102. doi: 10.4161/bioa.29766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miserey-Lenkei S, Chalancon G, Bardin S, Formstecher E, Goud B, Echard A. Rab and actomyosin-dependent fission of transport vesicles at the Golgi complex. Nat Cell Biol. 2010;12:645–654. doi: 10.1038/ncb2067. [DOI] [PubMed] [Google Scholar]
- 64.Tang HW, Wang YB, Wang SL, Wu MH, Lin SY, Chen GC. Atg1-mediated myosin II activation regulates autophagosome formation during starvation-induced autophagy. EMBO J. 2011;30:636–651. doi: 10.1038/emboj.2010.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Buss F, Spudich G, Kendrick-Jones J. Myosin VI: cellular functions and motor properties. Annu Rev Cell Dev Biol. 2004;20:649–676. doi: 10.1146/annurev.cellbio.20.012103.094243. [DOI] [PubMed] [Google Scholar]
- 66.Tumbarello DA, Waxse BJ, Arden SD, Bright NA, Kendrick-Jones J, Buss F. Autophagy receptors link myosin VI to autophagosomes to mediate Tom1-dependent autophagosome maturation and fusion with the lysosome. Nat Cell Biol. 2012;14:1024–1035. doi: 10.1038/ncb2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rusten TE, Stenmark H. How do ESCRT proteins control autophagy? J Cell Sci. 2009;122:2179–2183. doi: 10.1242/jcs.050021. [DOI] [PubMed] [Google Scholar]
- 68.Mackeh R, Perdiz D, Lorin S, Codogno P, Pous C. Autophagy and microtubules - new story, old players. J Cell Sci. 2013;126:1071–1080. doi: 10.1242/jcs.115626. [DOI] [PubMed] [Google Scholar]
- 69.Geeraert C, Ratier A, Pfisterer SG, Perdiz D, Cantaloube I, Rouault A, Pattingre S, Proikas-Cezanne T, Codogno P, Pous C. Starvation-induced hyperacetylation of tubulin is required for the stimulation of autophagy by nutrient deprivation. J Biol Chem. 2010;285:24184–24194. doi: 10.1074/jbc.M109.091553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jahreiss L, Menzies FM, Rubinsztein DC. The itinerary of autophagosomes: from peripheral formation to kiss-and-run fusion with lysosomes. Traffic. 2008;9:574–587. doi: 10.1111/j.1600-0854.2008.00701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kimura S, Noda T, Yoshimori T. Dynein-dependent movement of autophagosomes mediates efficient encounters with lysosomes. Cell Struct Funct. 2008;33:109–122. doi: 10.1247/csf.08005. [DOI] [PubMed] [Google Scholar]
- 72.Fass E, Shvets E, Degani I, Hirschberg K, Elazar Z. Microtubules support production of starvation-induced autophagosomes but not their targeting and fusion with lysosomes. J Biol Chem. 2006;281:36303–36316. doi: 10.1074/jbc.M607031200. [DOI] [PubMed] [Google Scholar]
- 73.Kochl R, Hu XW, Chan EY, Tooze SA. Microtubules facilitate autophagosome formation and fusion of autophagosomes with endosomes. Traffic. 2006;7:129–145. doi: 10.1111/j.1600-0854.2005.00368.x. [DOI] [PubMed] [Google Scholar]
- 74.Maday S, Holzbaur EL. Autophagosome biogenesis in primary neurons follows an ordered and spatially regulated pathway. Dev Cell. 2014;30:71–85. doi: 10.1016/j.devcel.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Du W, Su QP, Chen Y, Zhu Y, Jiang D, Rong Y, Zhang S, Zhang Y, Ren H, Zhang C, et al. Kinesin 1 Drives Autolysosome Tubulation. Dev Cell. 2016;37:326–336. doi: 10.1016/j.devcel.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 76.McMahon HT, Boucrot E. Membrane curvature at a glance. J Cell Sci. 2015;128:1065–1070. doi: 10.1242/jcs.114454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Khan MM, Strack S, Wild F, Hanashima A, Gasch A, Brohm K, Reischl M, Carnio S, Labeit D, Sandri M, et al. Role of autophagy, SQSTM1, SH3GLB1, and TRIM63 in the turnover of nicotinic acetylcholine receptors. Autophagy. 2014;10:123–136. doi: 10.4161/auto.26841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhuang X, Wang H, Lam SK, Gao C, Wang X, Cai Y, Jiang L. A BAR-domain protein SH3P2, which binds to phosphatidylinositol 3-phosphate and ATG8, regulates autophagosome formation in Arabidopsis. Plant Cell. 2013;25:4596–4615. doi: 10.1105/tpc.113.118307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gerke V, Creutz CE, Moss SE. Annexins: linking Ca2+ signalling to membrane dynamics. Nat Rev Mol Cell Biol. 2005;6:449–461. doi: 10.1038/nrm1661. [DOI] [PubMed] [Google Scholar]
- 80.Hayes MJ, Rescher U, Gerke V, Moss SE. Annexin-actin interactions. Traffic. 2004;5:571–576. doi: 10.1111/j.1600-0854.2004.00210.x. [DOI] [PubMed] [Google Scholar]
- 81.Ghislat G, Aguado C, Knecht E. Annexin A5 stimulates autophagy and inhibits endocytosis. J Cell Sci. 2012;125:92–107. doi: 10.1242/jcs.086728. [DOI] [PubMed] [Google Scholar]
- 82.Carlsson SR, Simonsen A. Membrane dynamics in autophagosome biogenesis. J Cell Sci. 2015;128:193–205. doi: 10.1242/jcs.141036. [DOI] [PubMed] [Google Scholar]
- 83.Fan W, Nassiri A, Zhong Q. Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L) Proc Natl Acad Sci U S A. 2011;108:7769–7774. doi: 10.1073/pnas.1016472108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Diao J, Liu R, Rong Y, Zhao M, Zhang J, Lai Y, Zhou Q, Wilz LM, Li J, Vivona S, et al. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature. 2015;520:563–566. doi: 10.1038/nature14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kaufmann A, Beier V, Franquelim HG, Wollert T. Molecular mechanism of autophagic membrane-scaffold assembly and disassembly. Cell. 2014;156:469–481. doi: 10.1016/j.cell.2013.12.022. [DOI] [PubMed] [Google Scholar]
- 86.Ridley AJ. Rho GTPase signalling in cell migration. Curr Opin Cell Biol. 2015;36:103–112. doi: 10.1016/j.ceb.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mleczak A, Millar S, Tooze SA, Olson MF, Chan EY. Regulation of autophagosome formation by Rho kinase. Cell Signal. 2013;25:1–11. doi: 10.1016/j.cellsig.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 88.Ao X, Zou L, Wu Y. Regulation of autophagy by the Rab GTPase network. Cell Death Differ. 2014;21:348–358. doi: 10.1038/cdd.2013.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Russo AJ, Mathiowetz AJ, Hong S, Welch MD, Campellone KG. Rab1 recruits WHAMM during membrane remodeling but limits actin nucleation. Mol Biol Cell. 2016;27:967–978. doi: 10.1091/mbc.E15-07-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zoppino FC, Militello RD, Slavin I, Alvarez C, Colombo MI. Autophagosome formation depends on the small GTPase Rab1 and functional ER exit sites. Traffic. 2010;11:1246–1261. doi: 10.1111/j.1600-0854.2010.01086.x. [DOI] [PubMed] [Google Scholar]
- 91.Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009;11:1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- 92.Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, Sideris DP, Fogel AI, Youle RJ. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tang D, Kang R, Zeh HJ, 3rd, Lotze MT. High-mobility group box 1, oxidative stress, and disease. Antioxid Redox Signal. 2011;14:1315–1335. doi: 10.1089/ars.2010.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mostowy S. Multiple roles of the cytoskeleton in bacterial autophagy. PLoS Pathog. 2014;10:e1004409. doi: 10.1371/journal.ppat.1004409. [DOI] [PMC free article] [PubMed] [Google Scholar]


