Abstract
Aim: To investigate the causative genetic mutations in 12 Pakistani families with nonsyndromic or syndromic hearing loss.
Methods: Mutations in the most common causative gene for hearing loss, GJB2, were evaluated by Sanger sequencing. Targeted next-generation sequencing or whole-exome sequencing was used to analyze the genomic DNA samples from 11 probands with hearing loss. Sanger sequencing was performed to verify all identified variants.
Results: We found pathogenic, or likely to be pathogenic, mutations in all 12 families, including six known mutations in GJB2, SLC26A4, LHFPL5, and USH2A and eight novel mutations in ESPN, MYO7A, LRTOMT, PCDH15, USH2A, or EPS8L2. Notably, four compound heterozygous mutations in the MYO7A and USH2A genes were detected in two consanguineous families. In addition, the novel frameshift mutation in EPS8L2 was first documented in Pakistan.
Conclusions: Our study increases the spectrum of mutations associated with hearing loss in the Pakistani population. In addition, our study highlights the fact that compound heterozygous mutations, although rare, can occur in consanguineous families.
Keywords: : hearing loss, gene, mutation, targeted next-generation sequencing, whole-exome sequencing
Introduction
Hearing loss is the most common congenital sensorineural disorder and affects 1 in 500–1000 newborns (Morton and Nance, 2016). More than 50% of childhood prelingual hearing impairments are caused by genetic factors, while the remaining cases are mainly attributed to environmental factors such as antibiotic use, noise, and infection (Morton and Nance, 2016). Hearing loss can occur in syndromic or nonsyndromic (NSHL) forms. There are nearly 400 types of syndromic deafness, which are accompanied by other clinical abnormalities such as retinitis pigmentosa or goiter, and accounts for 30% of deafness cases. Meanwhile, NSHL or isolated deafness represents 70% of hearing loss cases. The prevalence of profound bilateral hearing loss is 1.6 per 1000 in the Pakistani population and 70% of deafness cases occur in consanguineous families (Elahi et al., 1998).
NSHL and syndromic hearing loss are genetically heterogeneous, wherein most cases show autosomal recessive inheritance, although autosomal dominant, X-linked, or mitochondrial transmission modes also exist (Morton, 1991). To date, more than 100 genetic loci and 80 genes underlying NSHL have been identified (Hereditary Hearing Loss Homepage). The mutation spectrum of deafness genes may vary among different ethnic groups. For the Pakistani population, mutations in GJB2, SLC26A4, MYO15A, TMC1, OTOF, and CDH23 were previously shown to be the major cause of hearing loss (Shafique et al., 2014).
In this study, we used Sanger sequencing and next-generation sequencing (NGS) to analyze 12 consanguineous Pakistani families with hearing loss. We found mutations in eight known genes to confirm the extreme heterogeneity of this disorder.
Materials and Methods
Study subjects
A group of 12 consanguineous families with hearing loss were recruited from Pakistan. A clinical questionnaire was used to rule out any history of other diseases and environmental factors such as antibiotic use, noise, and infection that could cause hearing loss. Clinical information and blood samples were obtained from the probands and their family members after informed consent was provided. Genomic DNA was extracted from peripheral blood leukocytes using a QIAamp DNA Blood Midi Kit (Qiagen) according to the manufacturer's protocols. Genetic testing was performed in accordance with the Helsinki Declaration and approved by the Peking Union Medical College Institutional Review Board.
Screening for GJB2 mutations
The two coding exons and the flanking intronic sequences of GJB2 were amplified by polymerase chain reaction (PCR) and then subjected to Sanger sequencing after purification. Primer sequences are listed in Supplementary Table S1 (Supplementary Data are available online at www.liebertpub.com/gtmb).
Targeted NGS
Genomic DNA (20 ng) was amplified to prepare libraries using a designed panel containing 62 genes that are associated with hearing loss (Life Technologies). The amplifiable libraries were diluted to 100 pM and emulsion PCR was performed to obtain template-positive ion sphere particles (ISPs) according to the manufacturer's instructions (Life Technologies). ISPs were loaded and sequenced on an Ion 318V2 Chip using an Ion Torrent Personal Genome Machine (Life Technologies).
Whole-exome sequencing
For whole-exome sequencing, genomic DNA (100 ng) was amplified to prepare an exome library using the Ion AmpliSeq™ Exome Panel (Life Technologies). The amplifiable libraries were diluted to 100 pM. Then, we performed emulsion PCR using a OneTouch 2 instrument (Life Technologies) with an Ion PI Template OT2 200 Kit V3 (Life Technologies). ISP enrichment was achieved using the Ion OneTouch ES enrichment system (Life Technologies). An Ion Proton I chip (Life Technologies) was prepared and loaded according to the manufacturer's instructions.
Data analysis
Sequence data were aligned to the GRCh37/hg19 reference sequence with the Torrent Mapping Alignment Program. Genotype calling of multiallelic substitutions and indels was performed using a Torrent Variant Caller (version 4.4.3). After variant detection, ANNOVAR was used for annotation (Wang et al., 2010). In addition, sequence data were visualized using the Integrative Genome Viewer (IGV) (Robinson et al., 2011; Thorvaldsdóttir et al., 2013). To assess missense mutation pathogenicity, three prediction programs (SIFT, Polyphen2, and Mutation Taster) (Kumar et al., 2009; Adzhubei et al., 2010; Schwarz et al., 2014) and two conservation programs (PhyloP and GERP++) were used (Davydov et al., 2010; Pollard et al., 2010). Effects on splicing were evaluated with Human Splicing Finder (HSF) (Desmet et al., 2009).
Mutation confirmation
To validate mutations detected by NGS, specific fragments were PCR amplified using site-specific primers (Supplementary Table S1) and analyzed by Sanger sequencing. Novel mutations were further analyzed in 200 ethnically matched control individuals.
Results
Clinical manifestations
This study enrolled 12 families that had 71 individuals, of whom 34 were affected by hearing loss. All families were derived from consanguineous mating, indicating a possible autosomal recessive inheritance, although other inheritance patterns, such as autosomal dominant, were also considered. The age of the patients ranged from 10 to 52 years. All of the probands had bilateral, prelingual, moderate to profound hearing impairment. The patients were otherwise healthy and developed normally, except those from F1 and F4. The proband F1 suffered from night blindness at the age of eight and Usher syndrome was diagnosed (Ahmed et al., 2008). The affected individuals in F4 had goiter and presented with Pendred syndrome (Coyle et al., 1998) (Table 1).
Table 1.
Clinical Features of Probands in Twelve Pakistani Families with Hearing Loss
| Family | No. of probands | Gender | Age (years) | Severity | Other symptoms |
|---|---|---|---|---|---|
| F1 | IV-7 | Male | 10 | Profound | Night blindness |
| F2 | IV-1 | Female | 18 | Profound | No |
| F3 | IV-1 | Male | 16 | Moderate | No |
| F4 | IV-4 | Male | 38 | Profound | Goiter |
| F5 | V-5 | Male | 12 | Profound | No |
| F6 | IV-2 | Female | 12 | Profound | No |
| F7 | III-4 | Male | 52 | Profound | No |
| F8 | V-1 | Female | 16 | Profound | No |
| F9 | IV-1 | Male | 21 | Profound | No |
| F10 | V-5 | Male | 13 | Severe | No |
| F11 | IV-2 | Male | 25 | Moderate | No |
| F12 | IV-2 | Male | 12 | Profound | No |
Mutation identification and verification
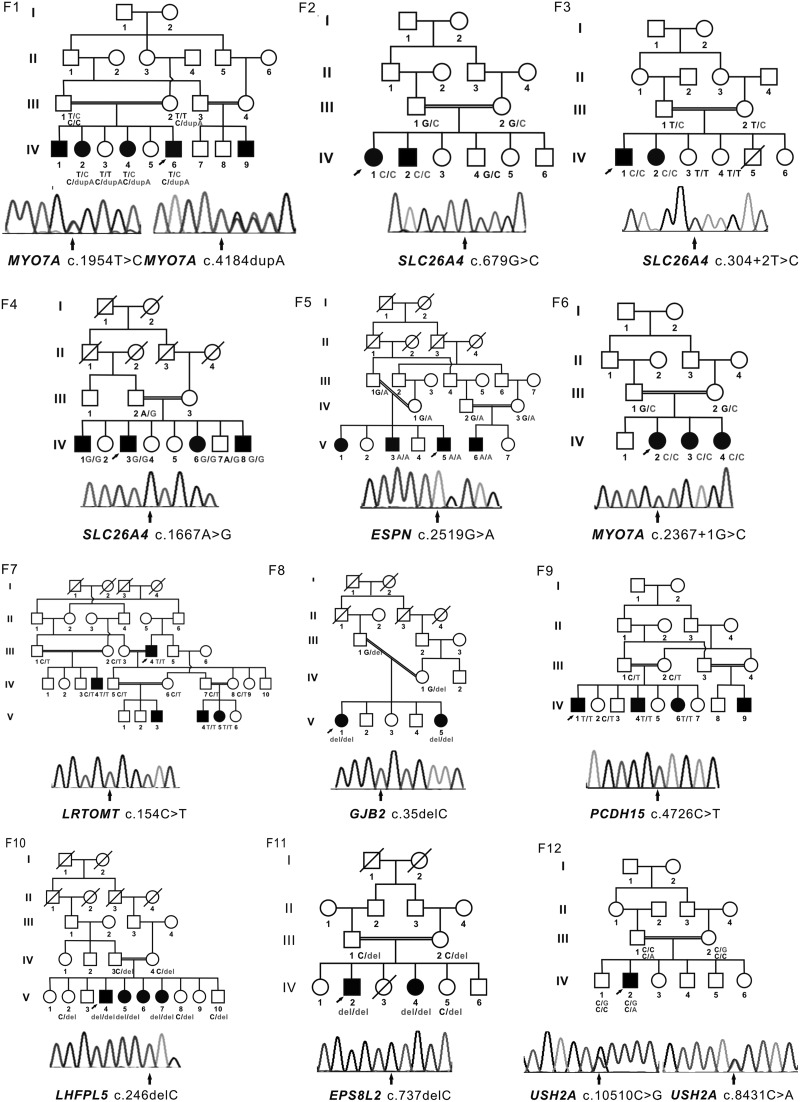
Details of the candidate variants are summarized in Figure 1 and Table 2. Screening for GJB2 mutations identified a homozygous c. 35del G mutation (p. Gly12Valfs*2) in F8. Subsequently, the other 11 probands were selected for targeted NGS to identify pathogenic mutations. For each sample, 70 MB of data were obtained, and the mean depth reached to 216 × coverage with 95.86% of the target region covered. These data showed the detection of ∼300 variants. To identify plausible pathogenic mutations, a series of filter criteria were applied. First, only homozygous or compound heterozygous variants were selected as candidates arising from an autosomal recessive inheritance pattern. Second, all variants with a minor allele frequency <1% (dbSNP142 and Exome Aggregation Consortium [ExAC]) were retained. Third, only nonsynonymous variants in the coding region or splicing variants were selected. Finally, that status of the variants was confirmed visually using IGV (Robinson et al., 2011; Thorvaldsdóttir et al., 2013). The remaining variants were further verified using Sanger sequencing. If we did not find candidate variants with these filters, we focused on heterozygous de novo variants following an autosomal dominant inheritance pattern.
FIG. 1.
Family pedigrees and the segregation of mutations in known genes. Squares and circles indicate males and females, respectively, whereas solid and open symbols represent affected and unaffected individuals, respectively. Slashes indicate deceased family members. Arrows indicate the probands or the mutant sites. Del, deletion; dup, duplication.
Table 2.
Mutations Identified in Twelve Pakistani Families with Hearing Loss
| Family | Gene | Transcript ID | cDNA | Predicted protein effect | ExAC(OAF) | PhyloP | GERP++ | SIFT/Polyphen2/mutation taster | References |
|---|---|---|---|---|---|---|---|---|---|
| F1 | MYO7A | NM_000260 | c. 1954 T > C | p. Cys652Arg | 0 | 1.972 | 5.21 | D/D/D | Novel |
| c. 4184dupA | p. Gln1395Thrfs*9 | 0 | NA | NA | NA/NA/NA | Novel | |||
| F2 | SLC26A4 | NM_000441 | c. 679 G > C | p. Ala227Pro | 0 | 2.536 | 5.41 | D/D/D | Jiang et al. (2010) |
| F3 | SLC26A4 | NM_000441 | c. 304 + 2 T > C | p. Met103Lysfs*4 | 0.000008237 | NA | NA | NA/NA/D | Anwar et al. (2009) |
| F4 | SLC26A4 | NM_000441 | c. 1667 A > G | p. Tyr556Cys | 0.00002487 | 2.117 | 5.51 | D/D/D | Coyle et al. (1998) |
| F5 | ESPN | NM_031475 | c. 2519G > A | p. Trp840* | 0 | 1.945 | 4.32 | NA/NA/D | Novel |
| F6 | MYO7A | NM_000260 | c. 2367 + 1G > C | NA | 0 | 2.272 | 4.87 | NA/NA/D | Novel |
| F7 | LRTOMT | NM_001145308 | c. 154C > T | p. Arg52Trp | 0 | 0.085 | 0.993 | D/D/N | Novel |
| F8 | GJB2 | NM_004004 | c. 35del G | p. Gly12Valfs*2 | 0.006040 | NA | NA | NA/NA/NA | Zelante et al. (1997) |
| F9 | PCDH15 | NM_001142769 | c. 4726 C > T | p. Gln1576* | 0 | 0.188 | 0.931 | NA/NA/D | Novel |
| F10 | LHFPL5 | NM_182548 | c. 246del C | p. Gly82Glyfs*3 | 0 | NA | NA | NA/NA/NA | Shabbir et al. (2006) |
| F11 | EPS8L2 | NM_022772 | c.737delC | p. Ala246Alafs*6 | 0 | NA | NA | NA/NA/NA | Novel |
| F12 | USH2A | NM_206933 | c.10510 C > G | p. Pro3504Ala | 0.001964 | 0.562 | 3.3 | T/B/D | van Huet et al. (2015) |
| c.8431 C > A | p. Pro2811Thr | 0.0003544 | 2.731 | 5.79 | T/D/D | Novel |
ExAC, exome aggregation consortium; OAF, overall allele frequency; NA, not available; D, deleterious; N, neutral; T, tolerated; B, benign.
In total, we found eight homozygous and four compound heterozygous candidate variants in 10 different consanguineous deafness families using targeted NGS (Fig. 1; Table 2). All variants cosegregated with the phenotype within each family and none of the novel mutations was present in chromosomes from 200 ethnically matched control individuals. Five mutations were reported previously, including c.679G > C [p. Ala227Pro] and c.304 + 2 T > C [p. Met103Lysfs*4] in SLC26A4, c.246delC [p. G82Gfs*3] in LHFPL5, c.1667A > G [p. Tyr556Cys] in SLC26A4, and c.10510 C > G [p. Pro3504Ala] variants in USH2A. Among the remaining seven novel variants, two nonsense variants (c. 2519G > A [p. Trp840*] in ESPN and c. 4726 C > T [p. Gln1576*] in PCDH15) and the frameshift variant (c. 4184dupA [p. Gln1395Thrfs*9] in MYO7A) were predicted to lead to either synthesis of truncated protein products or to degradation by nonsense-mediated mRNA decay (NMD). HSF predicted a splicing variant (c. 2367 + 1G > C) in MYO7A (Desmet et al., 2009) that would produce an aberrantly spliced transcript, whereas three missense variants (c. 1954 T > C [p. Cys652Arg] in MYO7A, c. 154C > T [p. Arg52Trp] in LRTOMT, and c.8431 C > A [p. Pro2811Thr] in USH2A) changed conserved amino acids. Two of the three prediction programs (SIFT, Polyphen2, or Mutation Taster) predicted these mutations to be possibly damaging (Kumar et al., 2009; Adzhubei et al., 2010; Schwarz et al., 2014).
In addition, we performed whole-exome sequencing on the proband from F11 in which we failed to identify a causative mutation by Sanger sequencing and targeted NGS. We identified a homozygous frameshift variant (c.737delC [p. Ala246Alafs*6]) in EPS8L2 in this individual (Fig. 1; Table 2). The homozygous variant cosegregated with deafness within the family, but was absent from public databases (dbSNP142 and ExAC) and the 200 control individuals. To our knowledge, this is the first report of mutations in EPS8L2 in a Pakistani family with NSHL.
Discussion
In this study, we found 14 candidate mutations that accounted for deafness in all 12 families studied. Of these, three affected families harbored mutations in SLC26A4 (3/12 = 25%) and two in MYO7A (2/12 = 16.67%). The other six families each harbored one or two pathogenic mutation in GJB2, ESPN, LRTOMT, PCDH15, LHFPL5, USH2A, or EPS8L2. This finding indicates that SLC26A4 and MYO7A mutations make a notable contribution to populations with hearing loss in Pakistan, which is in line with previously reported studies, although the mutation frequency was higher (Shahzad et al., 2013).
Among the 14 mutations, six were known mutations and eight were novel mutations. All of the mutations segregated with affected individuals in these families, but were not detectable in 200 control individuals, indicating that they were not merely common silent polymorphisms. Two mutations (c.679G > C [p. Ala227Pro] and c. 304 + 2 T > C [p. Met103Lysfs*4]) in SLC26A4 as well as the c.246delC (p. G82Gfs*3) LHFPL5 mutation were previously shown to be associated with NSHL (Shabbir et al., 2006; Anwar et al., 2009; Jiang et al., 2010). In addition, the c.1667A > G (p. Tyr556Cys) SLC26A4 mutation in F4 causes Pendred syndrome (Coyle et al., 1998). Meanwhile, the potential significance of the c.10510 C > G [p. Pro3504Ala] variant in USH2A was unclear (van Huet et al., 2015), yet the c.10510 C > A [p. Pro3504Thr] variant in the same position was reported to be pathogenic (Dreyer et al., 2008). Although mutations in GJB2 have previously been reported to be the most common cause of NSHL in the Pakistani population (Bukhari et al., 2013; Shafique et al., 2014), only one known mutation (c. 35del G [p. Gly12Valfs*2]) (Zelante et al., 1997) was detected in these 12 families. This result might be due to the small sample size in this study.
Because the frameshift mutations in MYO7A (c. 4184dupA [p. Gln1395Thrfs*9]) and EPS8 L2 (c.737delC [p. Ala246Alafs*6]) produce stop codons in the middle of the two genes, they may activate NMD pathways and could conceivably underlie the pathogenesis of hearing loss through a loss-of-function mechanism (Dahmani et al., 2015). With respect to the nonsense mutations in ESPN (c. 2519G > A [p. Trp840*]) and PCDH15 (c. 4726 C > T [p. Gln1576*]) that are located in the last exons of both genes, these mutant mRNAs might escape degradation by the NMD pathway and are predicted to result in truncation of the encoded proteins. However, direct RNA analysis must be performed to verify this possibility. ESPN encodes the calcium-insensitive actin-bundling protein espin (Naz et al., 2004). Amino acids 13 and 19 of the espin carboxy-terminus are known to be required for actin bundling and microvillar elongation activity, respectively (Bartles et al., 1998; Loomis et al., 2003). Hence, the nonsense c. 2519G > A mutation lacking the 15 amino acids of the espin C-terminus is presumed to be pathogenic. PCDH15 encodes three alternative, evolutionarily conserved unique cytoplasmic domains (CD1, CD2, and CD3) (Ahmed et al., 2006, 2008). Although ∼74 different PCDH15 mutations have been reported to cause NSHL, only the c.4542dupA ([p. P1515Tfs*4]) mutation and c.1103delT [p.Leu368Trpfs*58] mutation specifically affected the CD2 isoform (Pepermans et al., 2014; Bonnet et al., 2016). The c. 4726 C > T [p. Gln1576*] mutation found in this study adds a third example to illuminate the key function of the PCDH15 CD2 isoform in mature auditory hair cells. In general, missense variants in PCDH15 are associated with “Deafness, autosomal recessive 23” (DFNB23, OMIM 609533) (Ahmed et al., 2008), whereas more severe pathogenic variants (e.g., splicing, frameshift, nonsense, and large deletions) cause Usher syndrome type I (OMIM 276900). The p. Gln1576* mutation that produces a PCDH15 protein lacking the C-terminal 214 amino acids might underlie NSHL because of its relatively mild effect on protocadherin-15. Both missense mutations (c. 154C>T [p. Arg52Trp] in LRTOMT and c. 1954 T > C [p. Cys652Arg] in MYO7A) are believed to be pathogenic because of their location and conservation of the affected amino acids, and because neither change has been observed in a series of normal controls. The c. 154C > T mutation is present within a mutational hot spot in the LRTOMT2 gene (Ichinose et al., 2015), whereas the c. 1954 T > C mutation in MYO7A is located in the crucial region of the myosin head that is thought to be involved in transducing energy from the site of ATP hydrolysis to the regulatory domain (Mburu et al., 1997). Thus, the pathogenicity of both mutations is highly probable. USH2A encodes a protein that contains laminin EGF motifs, a pentaxin domain, and several fibronectin type III motifs. Mutations within this gene are associated with Usher syndrome type IIa and retinitis pigmentosa (Eudy et al., 1998; Rivolta et al., 2000). In this study, the c.8431 C > A and c.10510 C > G mutations identified in USH2A were predicted to affect the fibronectin type III 14 and 20 motifs, respectively. Based on the low frequency of these mutations in population databases and the cosegregation with the phenotype, these mutations are likely to be pathogenic.
Notably, we identified compound heterozygous MYO7A and USH2A mutations in consanguineous families F1 and F12, indicating that the cause of deafness in this family was not due to inheritance of the same mutation from both parents, as is often the case for autosomal recessive disorders in consanguineous marriages.
In summary, we found 14 pathogenic mutations that accounted for deafness in all 12 Pakistani families studied. Because the methods used for sequence analysis offer high efficiency and accuracy, as well as cost-effectiveness, this study provides support for the clinical adoption of NGS to screen rare pathogenic genes associated with hearing loss.
Supplementary Material
Acknowledgments
We thank all the individuals for their participation in this study. This work was supported by the National Natural Science Foundation of China (Grant number 81230015), the Beijing Municipal Science and Technology Commission (Grant number Z151100003915078), and the National Key Research and Development Program (Grant number 2016YFC1000504).
Author Disclosure Statement
No competing financial interests exist.
Web Resources
Hereditary Hearing Loss Homepage: http://hereditaryhearingloss.org
ANNOVAR: http://wannovar.usc.edu
IGV: http://www.broadinstitute.org/igv
SIFT: http://sift.jcvi.org
Polyphen2: http://genetics.bwh.harvard.edu/pph2
Mutation Taster: http://www.mutationtaster.org
References
- Adzhubei IA, Schmidt S, Peshkin L, et al. (2010) A method and server for predicting damaging missense mutations. Nat Methods 7:248–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed ZM, Goodyear R, Riazuddin S, et al. (2006) The tip-link antigen, a protein associated with the transduction complex of sensory hair cells, is protocadherin-15. J Neurosci 26:7022–7034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed ZM, Riazuddin S, Aye S, et al. (2008) Gene structure and mutant alleles of PCDH15: nonsyndromic deafness DFNB23 and type 1 Usher syndrome. Hum Genet 124:215–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anwar S, Riazuddin S, Ahmed ZM, et al. (2009) SLC26A4 mutation spectrum associated with DFNB4 deafness and Pendred's syndrome in Pakistanis. J Hum Genet 54:266–270 [DOI] [PubMed] [Google Scholar]
- Bartles JR, Zheng L, Li A, et al. (1998) Small espin: a third actin bundling protein and potential forked protein ortholog in brush border microvilli. J Cell Biol 143:107–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet C, Riahi Z, Chantot-Bastaraud S, et al. (2016) An innovative strategy for the molecular diagnosis of Usher syndrome identifies causal biallelic mutations in 93% of European patients. Eur J Hum Genet 24:1730–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukhari I, Mujtaba G, Naz S. (2013) Contribution of GJB2 mutations to hearing loss in the Hazara Division of Pakistan. Biochem Genet 51:524–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyle B, Reardon W, Herbrick JA, et al. (1998) Molecular analysis of the PDS gene in Pendred syndrome. Hum Mol Genet 7:1105–1112 [DOI] [PubMed] [Google Scholar]
- Dahmani M, Ammar-Khodja F, Bonnet C, et al. (2015) EPS8L2 is a new causal gene for childhood onset autosomal recessive progressive hearing loss. Orphanet J Rare Dis 10:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydov EV, Goode DL, Sirota M, et al. (2010) Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput Biol 6:e1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmet FO, Hamroun D, Lalande M, et al. (2009) Human splicing finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res 37:e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyer B, Brox V, Tranebjaerg L, et al. (2008) Spectrum of USH2A mutations in Scandinavian patients with Usher syndrome type II. Hum Mutat 29:451. [DOI] [PubMed] [Google Scholar]
- Elahi MM, Elahi F, Elahi A, et al. (1998). Paediatric hearing loss in rural Pakistan. J Otolaryngol 27:348–353 [PubMed] [Google Scholar]
- Eudy JD, Weston MD, Yao S, et al. (1998) Mutation of a gene encoding a protein with extracellular matrix motifs in Usher syndrome type IIa. Science 280:1753–1757 [DOI] [PubMed] [Google Scholar]
- Ichinose A, Moteki H, Hattori M, et al. (2015) Novel mutations in LRTOMT associated with moderate progressive hearing loss in autosomal recessive inheritance. Ann Otol Rhinol Laryngol 124 (Suppl 1):142S–147S [DOI] [PubMed] [Google Scholar]
- Jiang L, Feng Y, Chen H, et al. (2010) [An investigation of SLC26A4 gene mutation in nonsyndromic hearing impairment in Hunan province of China]. Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi 24:587–591(Article in Chinese) [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4:1073–1081 [DOI] [PubMed] [Google Scholar]
- Loomis PA, Zheng L, Sekerkova G, et al. (2003) Espin cross links cause the elongation of microvillus type parallel actin bundles in vivo. J Cell Biol 163:1045–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mburu P, Liu XZ, Walsh J, et al. (1997) Mutation analysis of the mouse myosin VIIA deafness gene. Genes Funct 1:191–203 [DOI] [PubMed] [Google Scholar]
- Morton CC, Nance WE. (2016) Newborn hearing screening—a silent revolution. N Engl J Med 354:2151–2164 [DOI] [PubMed] [Google Scholar]
- Morton NE. (1991) Genetic epidemiology of hearing impairment. Ann N Y Acad Sci 630:16–31 [DOI] [PubMed] [Google Scholar]
- Naz S, Griffith AJ, Riazuddin S, et al. (2004) Mutations of ESPN cause autosomal recessive deafness and vestibular dysfunction. J Med Genet 41:591–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepermans E, Michel V, Goodyear R, et al. (2014) The CD2 isoform of protocadherin-15 is an essential component of the tip-link complex in mature auditory hair cells. EMBO Mol Med 6:984–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Hubisz MJ, Rosenbloom KR, et al. (2010) Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res 20:110–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivolta C, Sweklo EA, Berson EL, et al. (2000) Missense mutation in the USH2A gene: association with recessive retinitis pigmentosa without hearing loss. Am J Hum Genet 66:1975–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, et al. (2011) Integrative genomics viewer. Nat Biotechnol 29:24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JM, Cooper DN, Schuelke M, et al. (2014) MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods 11:361–362 [DOI] [PubMed] [Google Scholar]
- Shabbir MI, Ahmed ZM, Khan SY, et al. (2006) Mutations of human TMHS cause recessively inherited non-syndromic hearing loss. J Med Genet 43:634–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafique S, Siddiqi S, Schraders M, et al. (2014) Genetic spectrum of autosomal recessive non-syndromic hearing loss in Pakistani families. PLoS One 9:e100146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahzad M, Sivakumaran TA, Qaiser TA, et al. (2013) Genetic analysis through otoseq of Pakistani families segregating prelingual hearing loss. Otolaryngol Head Neck Surg 149:478–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP. (2013) Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14:178–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Huet RA, Pierrache LH, Meester-Smoor MA, et al. (2015) The efficacy of microarray screening for autosomal recessive retinitis pigmentosa in routine clinical practice. Mol Vis 21:461–476 [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelante L, Gasparini P, Estivill X, et al. (1997) Connexin26 mutations associated with the most common form of non-syndromic neurosensory autosomal recessive deafness (DFNB1) in Mediterraneans. Hum Mol Genet 6:1605–1609 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.