Highlight
A shift in C storage triggered by defoliation has a significant impact on nitrate uptake in an N-status-dependent manner. Cytokinins are possible co-ordinators of C and N metabolism and N acquisition at the whole-plant level.
Keywords: Carbohydrate, carbon, cytokinin, fructan, Lolium perenne, nitrate transporter (NRT), nitrate uptake, nitrogen, nitrogen use efficiency (NUE), perennial ryegrass.
Abstract
The mechanisms linking C/N balance to N uptake and assimilation are central to plant responses to changing soil nutrient levels. Defoliation and subsequent regrowth of grasses both impact C partitioning, thereby creating a significant point of interaction with soil N availability. Using defoliation as an experimental treatment, we investigated the dynamic relationships between plant carbohydrate status and NO3–-responsive uptake systems, transporter gene expression, and nitrate assimilation in Lolium perenne L. High- and low-affinity NO3– uptake was reduced in an N-dependent manner in response to a rapid and large shift in carbohydrate remobilization triggered by defoliation. This reduction in NO3– uptake was rescued by an exogenous glucose supplement, confirming the carbohydrate dependence of NO3– uptake. The regulation of NO3– uptake in response to the perturbation of the plant C/N ratio was associated with changes in expression of putative high- and low-affinity NO3– transporters. Furthermore, NO3– assimilation appears to be regulated by the C–N status of the plant, implying a mechanism that signals the availability of C metabolites for NO3– uptake and assimilation at the whole-plant level. We also show that cytokinins may be involved in the regulation of N acquisition and assimilation in response to the changing plant C/N ratio.
Introduction
Grasses are well adapted to tolerate and recover from the severe and frequent defoliation associated with grazing (Lestienne et al., 2006). The most obvious developmental aspect of this is the physical protection of the meristem at the crown. Less overt, but no less critical, are the metabolic aspects of this tolerance. For instance, leaf regrowth and the re-establishment of photosynthesis in the leaf prior to its transition from sink to source is dependent upon the remobilization of stored nutrient resources, particularly carbon (C) and nitrogen (N) (Ourry et al., 1989; Lestienne et al., 2006). In the period immediately following defoliation, photosynthetic capacity is temporarily compromised. This interrupts C assimilation and the capacity to assimilate newly acquired inorganic N into biomass, which is a process requiring reducing equivalents, ATP, and C skeletons generated from respiration of sucrose derived from stored C (Dawar et al., 2010; Nunes-Nesi et al., 2010). Indeed, it has been established that root growth, inorganic N uptake, respiration, and nitrate assimilation decline rapidly after defoliation (Boucaud and Bigot, 1989; Ourry et al., 1989; Richards, 1993; Louahlia et al., 2008).
Some evidence exists to suggest that C and N metabolism is co-ordinated via a sensing of plant C/N balance, and regulated by the expression of genes involved in photosynthesis, metabolic pathways, protein degradation, and N assimilation (Stitt and Krapp, 1999; Coruzzi and Zhou, 2001; Palenchar et al., 2004; Gutiérrez et al., 2007). Microarray data from Arabidopsis transiently exposed to a matrix of C and N treatments has indicated that a large proportion of genes respond to a C/N interaction, suggesting C and/or N, or a metabolic product of C and N assimilation (e.g. an amino acid), might act as a signal in the regulation of gene expression (Gutiérrez et al., 2007). Despite several studies proposing possible mechanisms underlying C and N signalling in Arabidopsis, a definitive mechanistic understanding remains elusive. The dramatic shift in C/N metabolic partitioning resulting from defoliation and regrowth in forage grasses, such as Lolium perenne L., has the potential to reveal insights into the integration of C/N assimilation in plants, something not possible to observe in dicotyledonous species.
Fructans, polymers of fructose that are based on sucrose, are the major storage carbohydrate in temperate forage grasses, such as L. perenne (Turner et al., 2006). We have previously confirmed that the fructan pool in L. perenne is degraded when NO3– is supplied to N-deficient plants (Roche et al., 2016), consistent with earlier work by Morvan-Bertrand et al. (1999) and Louahlia et al. (2008), and suggested that both root and shoot fructan pools support nitrate uptake and assimilation (Roche et al., 2016). In response to defoliation, early shoot regrowth is also sustained by remobilization of the C stored as fructans in elongating leaf bases and mature leaf sheathes (Morvan-Bertrand et al., 1999, 2001). Logically, the importance of root-derived C in supporting regrowth must also be considered in terms of the greater root biomass fraction that exists following defoliation. Thus, C and N remobilization under the new metabolic condition induced by defoliation must necessarily be co-ordinated at a whole-plant level.
The cytokinins, in addition to multiple roles in plant growth and development, are well known to play a role in regulating NO3– uptake systems in response to fluctuations in N availability (Krouk et al., 2011). The isoprenoid cytokinins are biosynthesized via adenosine phosphate-isopentenyltransferase (IPT). The first formed cytokinins are the nucleotides. The riboside forms are regarded as the main translocated forms, and the free bases the biologically active forms detected by receptors (Sakakibara, 2006; Lomin et al., 2015). The signalling pathway in Arabidopsis involves a multi-step signalling network linking between the receptors and the response regulators (RRs) (Hwang et al., 2012). The cis-forms of the cytokinins derive from the turnover of specific tRNA moities. Cytokinin homeostasis is determined by the rate of ctyokinin degradation which is catalysed by cytokinin oxidase/dehydrogenase (CKX) and by conjugation. For a general overview of the cytokinins, see Jameson (2017).
It has been shown that trans-zeatin (tZ)-type cytokinins (which carry a hydroxyl on the isoprenoid side chain) are the major forms in xylem sap, while isopentenyl adenine (iP-type) and cis-zeatin (cZ)-type cytokinins are the predominant forms in phloem sap (Takei et al., 2004; Hirose et al., 2008). Matsumoto-kitano et al. (2008) showed that iP-type cytokinins translocated from the shoot to the root in Arabidopsis were functional. It has been proposed that cytokinins could be involved in local and long-distance signalling to co-ordinate responses at the whole-plant level, and to communicate N status between multiple organs (Takei et al., 2001; Sakakibara et al., 2006; Matsumoto-Kitano et al., 2008; Kudo et al., 2010; Shtratnikova et al., 2015). A split-root experiment in Arabidopsis has demonstrated that NITRATE TRANSPORTERNRT2.1 and NAR2.1 regulation, driven by shoot–root systemic N signalling, is mediated by cytokinin biosynthesis (Ruffel et al., 2011). Further, genome-wide analyses for cytokinin-responsive genes in Arabidopsis have indicated that exogenous cytokinin can down-regulate root-localized transporters including AtNRT1.1, AtNRT2.1, AtNRT2.2, and AtNRT1.5 (Brenner et al., 2005; Li et al., 2007). These gene families have been functionally identified as major components of the NO3– uptake and xylem loading system (Liu et al., 1999; Lin et al., 2008). In contrast, cytokinin also up-regulates shoot-localized AtNRT genes under both high and low N conditions (Kiba et al., 2011).
Additionally, global gene profiling coupled with physiological analysis in Arabidopsis has indicated that 74% of cytokinin-regulated genes may be significantly affected by glucose, either agonistically or antagonistically (Kushwah and Laxmi, 2014). Additionally, up-regulation of AtIPT3, and some Type A ARABIDOPSIS RESPONSE REGULATOR (ARR) genes and CYTOKININ RESPONSE FACTOR (CRF) genes by glucose suggests a potential role for glucose in modulating cytokinin signalling (Kushwah and Laxmi, 2014). It is thus plausible that cytokinins may play an active role in balancing the C–N status of the plant (Wang and Ruan, 2016).
Cytokinin deficiency, resulting from overexpression of CKX, in Arabidopsis causes an increase in the root/shoot ratio, with inhibited shoot growth and a larger root system (Werner et al., 2003, 2008). This observation indicates that cytokinin deficiency may trigger a shift in C allocation from shoot to root, enhancing soil ‘foraging’ capacity for nitrate under limited N availability (Ruffel et al., 2011). Once taken up, the increased NO3– induces cytokinin biosynthesis in a nitrate-dependent manner (Krouk et al., 2011). Subsequently, the cytokinins translocated to the shoot could regulate C allocation to keep shoot and root development in balance (Wang and Ruan, 2016).
With an ultimate goal of finding ways to better manage N input in grassland-based agricultural systems such as dairy farming, we investigated the nitrate uptake response to two independent but coinciding metabolic demands on fructan: defoliation and nitrate addition. Of particular interest was the degree of metabolic constraint, if any, that the defoliation–regrowth cycle and associated fructan depletion had on inorganic N uptake efficiency. Given the C cost associated with inorganic N uptake and assimilation (Roche et al., 2016), we hypothesized that this metabolic demand is not adequately met in the days immediately following defoliation, due to regrowth competing with N uptake and assimilation for the stored C. To test this hypothesis, we carried out a stable isotope N uptake experiment, measured fructan content, and profiled cytokinin metabolites and relevant genes in L. perenne subjected to defoliation and glucose supplementation in a hydroponic system.
Materials and methods
Plant growth
Seeds of L.perenne L. cv. Grasslands Nui were germinated in Eppendorf tubes filled with perlite (tube tips removed) and the tubes placed in unfertilized soil for 11 weeks at 22°C. The basic N-free Hoagland medium (pH 6.0; Bioworld, USA) was supplemented with 0.05 mM or 5 mM KNO3 as a sole N source every week. Eight-week-old plants were defoliated at 4 cm above ground level. After 3 weeks regrowth, the plants were transferred to a hydroponic system which contained basic N-free Hoagland medium supplemented with either 0.05 mM or 5 mM KNO3, consistent with the previous growth conditions. The pH of treatment solutions was maintained at 6.0. Based on preliminary experiments, a 1 week adaptation phase in liquid culture medium was required for plants to retain pre-transfer competence (as assessed by leaf gas exchange and stomatal conductance measurements). After the 1 week adaptation, half the plants were again defoliated. The time zero of the experiments is defined by the second defoliation of plants.
K15NO3 uptake measurement and isotope analysis
Root uptake of NO3– was determined by 15N labelling. At 0 h and 48 h after defoliation, plants were gently blotted on tissue paper and then immediately rinsed with 0.1 mM CaSO4 for 1 min to remove any adsorbed compounds on the root surface, followed by 1 h exposure to basic N-free Hoagland medium supplemented with either 0.05 mM or 5 mM 15N-labelled KNO3 (atom % 15N: 10%). During the uptake experiments, the incubation solutions were aerated by an aquarium pump. At the end of the incubation period, roots were immediately rinsed with 0.1 mM CaSO4 for 1 min. Leaf sheaths and roots were separated, frozen in liquid nitrogen immediately, and stored at –80°C. The incubation timing was based on that used by Brackin et al. (2015). For this study, five plants were pooled as one biological replicate, and each treatment had five independent biological replicates. A 1 g aliquot of fresh samples was ground to a fine powder and freeze-dried for 3 d. Total N and 15N in the samples were determined with an isotope ratio mass spectrometer (Waikato University, New Zealand). A flow diagram of the experimental processes is shown in Supplementary Fig. S1 at JXB online.
Glucose treatment
After 42 h of regrowth as described above, 100 plants grown in solutions with 5 mM KNO3 were then supplemented with either 0.1% or 1% (w/v) glucose (25 plants each) or with 0.1% or 1% (w/v) mannitol (25 plants each) as an osmotic control. After 6 h, K15NO3 uptake measurements were carried out by exposing roots to 5 mM 15N-labelled KNO3 (atom % 15N: 10%) solutions as described above. A flow diagram of the experimental processes is shown in Supplementary Fig. S2.
Target gene sequence determination
Sequences of candidate target gene family members in perennial ryegrass were determined through BLAST searching the NCBI database and an RNA-Seq transcriptome database using prfectBLAST 2.0 software. An Illumina HiSeq2000 genome analyser (Macrogen Ltd, Seoul, Korea) and a pool of combined RNA samples extracted from multiple developmental stages of leaves, flower spikes, and seeds of perennial ryegrass cv. Nui were used to generate the transcriptome database containing 169 862 assembled sequence contigs of 595 bp in average length. All available orthologue sequences of the target gene families in the GenBank database for perennial ryegrass and closely related species were used as local BLAST search query sequences. The putative sequences of interest were verified via BLAST searching the GenBank database and via multiple sequence alignment with representative orthologue sequences in closely related species.
Neighbor–Joining (NJ) phylogenetic trees of the newly identified sequences of LpNRT1, LpNRT2, LpNR, LpNiR, LpCKX, LpRR, and LpFEH, and their orthologues were created using Clustal X software with 1000 bootstrap replicates. The phylogenetic trees were visualized with TreeView software (Supplementary Fig. S3). Each tree was rooted with an outgroup orthologue sequence from an unrelated species. The LpRR tree was shown in our previous study (Roche et al., 2016). The GenBank accession numbers for the nucleotide sequences determined in this work are listed in Supplementary Table S1.
RNA isolation and quantitative RT-PCR
Total RNA was extracted from up to 100 mg of frozen samples using an RNase Plant Mini Kit (Qiagen, Germany), following the manufacturer’s protocols. Three independent tissue samples (each comprising tissues from five plants) for each treatment were used as biological replicates. Up to 1 µg of extracted RNA was converted to cDNA using a QuantiTect Reverse Transcription Kit according to the manufacturer’s instructions (Qiagen).
The relative expression levels were determined using reverse transcription quantitative PCR (RT-qPCR). Specific PCR primers were designed for each family member of the target genes, using Primer Premier 6.20. Primer sequences for reference genes and target genes can be found in Supplementary Table S1. A volume of 15 µl was used for all qPCRs containing 1 µl of 10-fold diluted cDNA, the relevant primers, and home-made SYBR Green master mix (Song et al., 2012). PCR products were Sanger-sequenced to confirm homology to genes which were already identified in NCBI gene databases. For RT-qPCR, three technical replicates for each of three biological replicates were carried out for each sample set. The relative expression (fold change) of each target gene was corrected using the geometric means of the two reference genes, LpELONGATION FACTOR (eEF-1α) and LpGAPDH, and calculated using the 2–∆∆Ct method as described in previous studies (Schmittgen and Livak, 2008; Song et al., 2015).
Fructan analysis
Perennial ryegrass samples were harvested and immediately flash-frozen in liquid nitrogen. Ground and freeze-dried plant materials (25 mg) were weighed and placed into 2 ml Eppendorf tubes for fructan extraction. After addition of 750 µl of boiling milliQ water, samples were vortexed for 5 s, placed on a heating block at 90°C for 15 min, and then cooled down to room temperature and centrifuged at 20 000 g for 10 min. The supernatants were transferred to filter tubes, centrifuged at 20 000 g for 2 min, and transferred to LC vials. Extracts were analysed using an Agilent 1290 Infinity LC System (Agilent Technologies, Waldbronn, Germany) coupled to an Agilent 6550 Accurate-Mass QTOF LC-MS system with a dual Agilent Jet Stream source operating in negative mode. The QTOF was tuned for a mass range of 70–1700 m/z. A 2 µl aliquot of the sample was injected onto an Acquity UPLC HSS T3 C18 column (2.1 × 50 mm, 1.8 µm) combined with a 2.1 mm×5 mm, 1.8 µm VanGuard pre-column (Waters Corporation, Milford, MA, USA) held at 40°C. Compounds were eluted using the linear gradient reported in Roche et al. (2017). Data were collected in centroid mode with an acquisition rate of 4 scans s−1 and 1975 transients per spectrum.
Endogenous cytokinin quantification
Extraction and qualification of cytokinins from ~3 mg DW samples were performed as described previously (Dobrev and Kamınek, 2002; Antoniadi et al., 2015) using the LC-MS/MS system consisting of an ACQUITY UPLC® System (Waters) and a Xevo® TQ-S (Waters) triple quadrupole mass spectrometer (Svačinová et al., 2012).
Results
Regrowth 48 h after defoliation
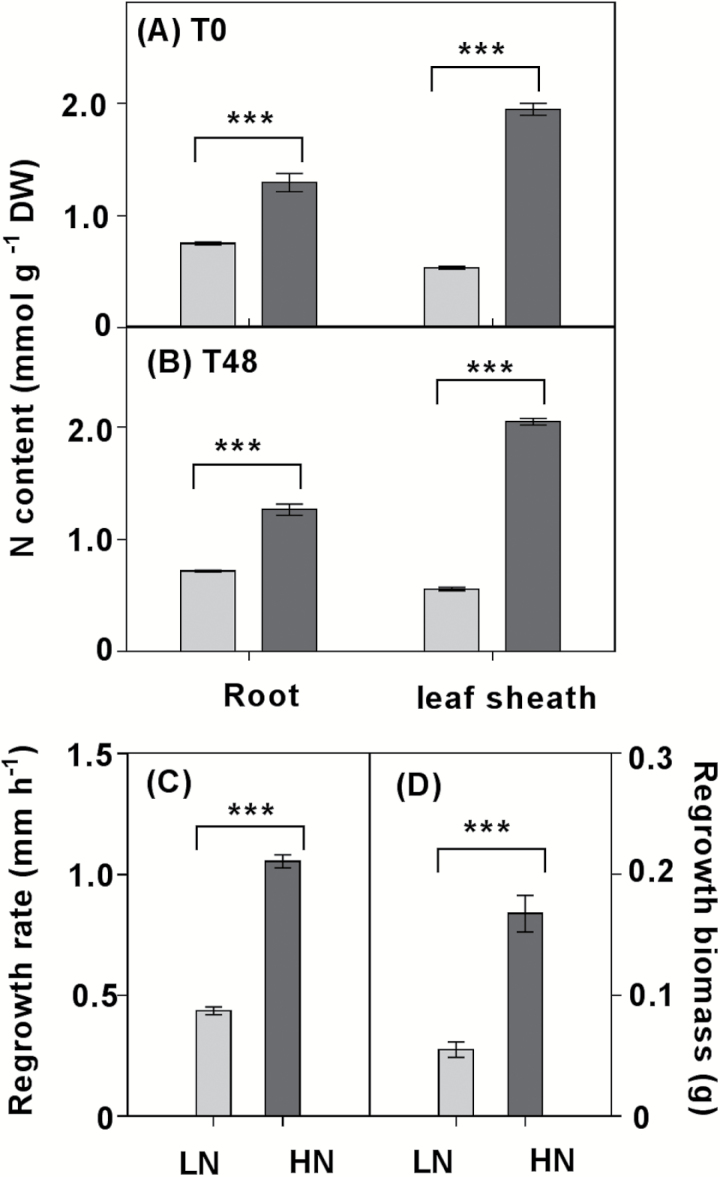
N content was measured in roots and leaf sheaths of 12-week-old L. perenne plants 8 d after they were transferred from soil to a hydroponic solution and then defoliated to 40 mm above the crown. For the purpose of this study, the leaf sheath is defined as tissue from crown to 40 mm above, including elongating leaf bases and mature leaf sheaths, and this same part was harvested for the analysis in the intact plants. As expected, leaf sheath and root N content, regrowth rate, and biomass were significantly greater in plants grown under high NO3– (HN, 5 mM) conditions than in plants grown under low NO3– (LN, 0.05 mM) (Fig. 1).
Fig. 1.
Growth parameters 48 h after defoliation of plants grown under low (0.05 mM) or high (5 mM) nitrate supply (LN versus HN). (A, B) Nitrogen (N) content before and 48 h after defoliation; (C) regrowth rate; (D) biomass 48 h after defoliation. Values in (A), (B), and (D) are means ±SE (n=5 pools of five plants each). Values in (C) are means ±SE (n=36 plants). Means were tested for significance using a two-tailed t-test. Asterisks denote significantly different means between LN plants (grey bars) and HN plants (black bars) at ***P<0.001.
Water-soluble carbohydrates and fructan exohydrolase gene transcript levels
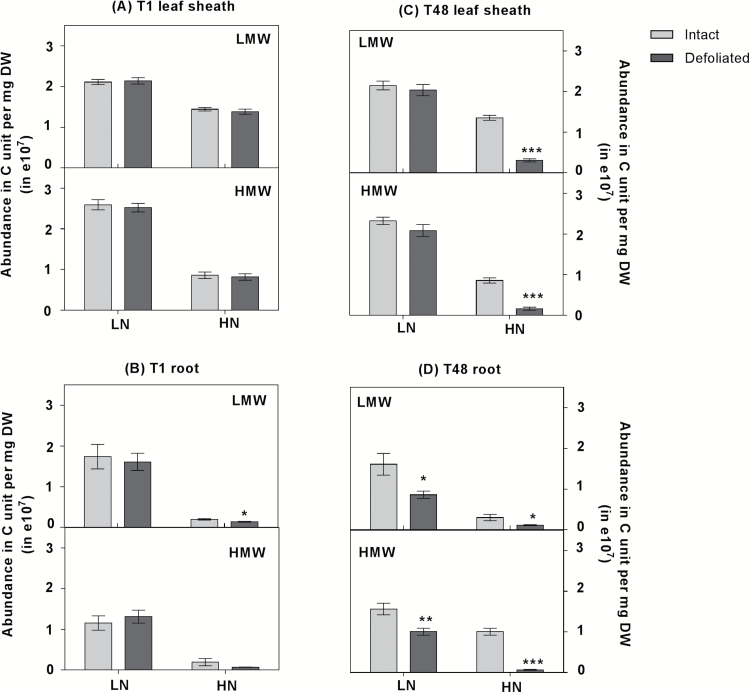
Water-soluble carbohydrates (WSCs) were profiled in order to assess the dynamics of C metabolism during the first 48 h of regrowth. WSCs with a degree of polymerization (DP) of 3–8 are referred to here as low molecular weight (LMW) WSCs, and those with DP9 to DP20 are referred to as high molecular weight (HMW) WSCs. With the exception of disaccharides, which showed similar relative abundance in LN and HN plants (Supplementary Fig. S4), LMW and HMW WSCs were found in greater abundance in LN roots and leaf sheaths compared with HN plants (Fig. 2). The detailed changes in the abundance of oligosaccharides with increasing DP are shown in Supplementary Fig. S4. During the first hour after defoliation, the remobilization of fructans occurred only in roots of HN plants, while no differences were observed in either roots or leaf sheaths of LN plants relative to intact plants (Fig. 2A, B; Supplementary Fig. S4). In comparison with the intact plants, the total concentration of LMW and HMW WSCs was significantly reduced 48 h after defoliation in both roots and leaf sheaths of HN plants, whereas in LN plants depletion of the fructan pool was only observed in roots, implying that the nature of defoliation-induced remobilization of the fructan pool after 48 h was tissue specific (Fig. 2C, D). In roots, the extent of exhaustion of the fructan pool of HN plants was more severe compared with that in LN plants (Fig. 2B, D; Supplementary Fig. S4).
Fig. 2.
Abundance in carbon (C) units of water-soluble carbohydrates (WSCs). WSCs were measured in plants grown under high (HN) and low nitrate (LN) supply, 1 h and 48 h after defoliation. Relative abundance in C units was calculated by multiplying peak intensity by degree of polymerization (DP). WSCs with a DP from three to eight are referred to here as low molecular weight (LMW) WSCs, and DP9 to DP20 are referred to as high molecular weight (HMW) WSCs. The total concentrations of LMW and HMW WSCs are shown in (A) leaf sheaths and (B) roots after 1 h; (C) leaf sheaths and (D) roots after 48 h. Values are means ±SE (n=5 pools of five plants each). Means were tested for significance using a two-tailed t-test. Asterisks denote significantly different means between intact plants (grey) and defoliated plants (black) (*P<0.05, **P<0.01, ***P<0.001).
Lp1-FEH and Lp6-FEH have been functionally characterized as fructan exohydrolases in L. perenne (Lothier et al., 2007, 2014). Within 48 h of defoliation, Lp6-FEH expression was induced in leaf sheath and root of HN plants, and also in LN roots (Supplementary Fig. S5). Similarly, Lp1-FEH transcription in response to defoliation increased significantly in LN roots and leaf sheaths after 48 h, where the fructan content was highest (Fig. 2D; Supplementary Fig. S5).
Nitrate uptake rate and N allocation
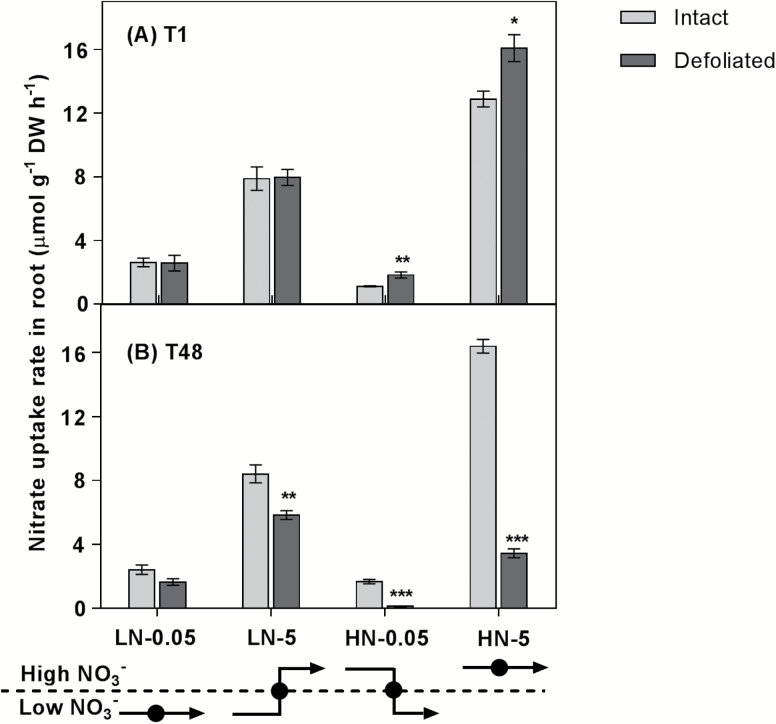
To better understand changes in NO3– uptake in response to defoliation, plants were exposed to 0.05 mM and 5 mM NO3– to stimulate either the nitrate high-affinity transport system (HATS) or the low-affinity transport system (LATS), respectively. The concentrations chosen were well below or above the point at which HATS or LATS would be saturated (Siddiqi et al., 1990; Garnett et al., 2013). During the first 1 h after defoliation, we observed no difference in NO3– uptake rate via HATS or LATS in LN plants compared with intact plants, whereas NO3– uptake via both HATS and LATS in defoliated HN plants was significantly greater relative to intact plants (Fig. 3A). In intact plants, HATS uptake in LN plants was greater than that in HN plants, while a much greater LATS uptake was induced in HN plants relative to LN plants (Fig. 3A). At 48 h after defoliation, the uptake rate of both HATS and LATS in HN plants dropped by 93% and 79% relative to intact plants, respectively (Fig. 3B). The HN plants, which had a relatively severe depletion of the fructan pool, showed markedly greater reduction in both HATS and LATS NO3– uptake than that of LN plants 48 h after defoliation, implying a dependence on C for the uptake of nitrate.
Fig. 3.
The impact of defoliation on NO3– uptake in L. perenne. In order to investigate HATS and LATS uptake capacity, plants were grown under low NO3– (LN: 0.05 mM) or high NO3– (HN: 5 mM), and then defoliated (or left intact). Plants were then exposed for 1 h to either 0.05 mM or 5 mM 15N-labelled NO3– either immediately after defoliation or 48 h after defoliation. During the period of 15N labelling, the NO3– uptake rate was measured. Values are means ±SE (n=5 pools of five plants each). Means were tested for significance using a two-tailed t-test. Asterisks denote significantly different means between intact plants (grey bars) and defoliated plants (black bars) (*P<0.05, **P<0.01, ***P<0.001).
To investigate the impact of defoliation on N distribution, 15N allocation to leaf sheaths following the incubations in 15N-labelled NO3– was measured at 1 h and 48 h after defoliation. Relative allocation was determined as the proportion of labelled 15N in leaf sheaths to that in the total plant (roots and leaf sheaths). During the first hour after defoliation, relatively less 15N was allocated to leaf sheaths in defoliated LN and HN plants than in the controls (Supplementary Fig. S8). In contrast, despite the decline in NO3– uptake rate 48 h after defoliation (Fig. 3B), the 15N allocation to leaf sheaths was relatively greater in both HN and LN defoliated plants than in intact plants (Supplementary Fig. S8).
NRT, NR, and NiR transcript levels
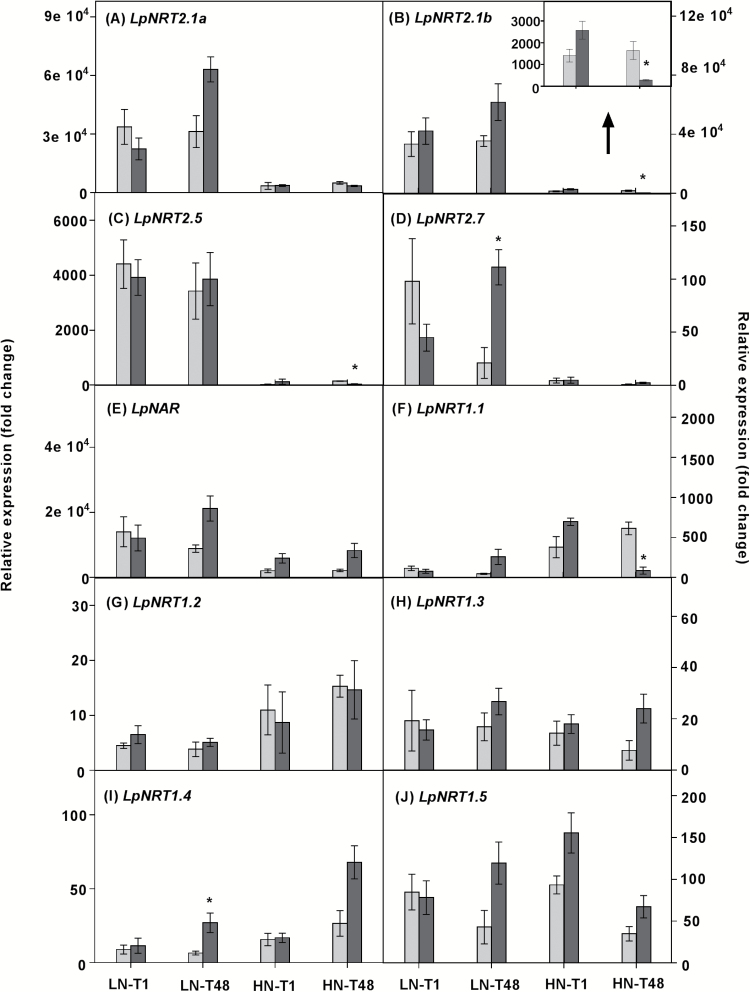
In our hydroponic system, we expected that HATS and LATS transcripts would be relatively abundant in LN and HN plants, respectively. Indeed, as expected, NRT2 and NAR transcript levels in LN roots were generally greater than those in HN roots (Fig. 4A–E). This was particularly evident for the putative HAT candidate genes LpNRT2.1a and LpNRT2.1b, where transcript expression in roots of LN plants was 20- to 50-fold greater relative to HN plants (Fig. 4A, B). No significant difference in transcript abundance of LpNRT2.1a and LpNRT2.1b was observed between defoliated and intact LN plants during the first hour or 48 h after defoliation. Although transcript levels were relatively low in HN roots, a reduction in LpNRT2.1b transcript abundance was apparent 48 h after defoliation (Fig. 4B). Notably, within 1 h of defoliation, LpNRT2.1b expression was slightly but not significantly induced in HN roots (P=0.07; Fig. 4B). However, there was no significant difference in LpNRT2.1a transcription in HN roots following 48 h of defoliation (Fig. 4A). Despite their relatively lower presence, LpNRT2.5 and LpNRT2.7 showed greater expression in LN roots than in HN roots, suggesting that these two putative transporters may play a role in the HATS (Fig. 4C, D). Similarly to LpNRT2.1a and LpNRT2.1b, LpNAR displayed greater expression in LN roots relative to HN roots, confirming an important role for LpNAR in the function of NRT2.1 in NO3– uptake (Fig. 4E).
Fig. 4.
Expression of putative low- and high-affinity (LpNRT1, LpNRT2, and LpNAR) NO3– transporter genes in roots of L.perenne. Plants were grown at either 0.05 mM (LN) or 5 mM (HN) NO3–, and then defoliated (or left intact). The expression level of LpNRT2.1b in HN roots is shown in the inserted bar graph in (B). Each data point is normalized against the reference genes eEF-1α and GAPDH. Values are means ±SE (n=3 pools of five plants each). Means were tested for significance using a two-tailed t-test. An asterisk denotes significantly different means between intact plants (grey bars) and defoliated plants (black bars) at P<0.05.
Although the transcript levels of LpNRT1.1 were relatively quite low, the expression pattern contrasts with those of the two putative HATS genes (LpNRT2.1a and LpNRT2.1b), with LpNRT1.1 being generally higher in HN conditions than in LN conditions, suggesting that the putative NRT1.1 may play an important role in the LATS (Fig. 4F). However, in HN roots, the transcript level of LpNRT1.1 was reduced 48 h after defoliation, showing a similar pattern to that of LpNRT2.1b (Fig. 4F). Unlike LpNRT1.1, other LpNRT1 genes, LpNRT1.2, 1.3, 1.4, and 1.5, showed extremely low, but constitutive expression patterns regardless of treatment, indicating that these putative NRT1 genes might be less sensitive to changing N conditions (Fig. 4G–J).
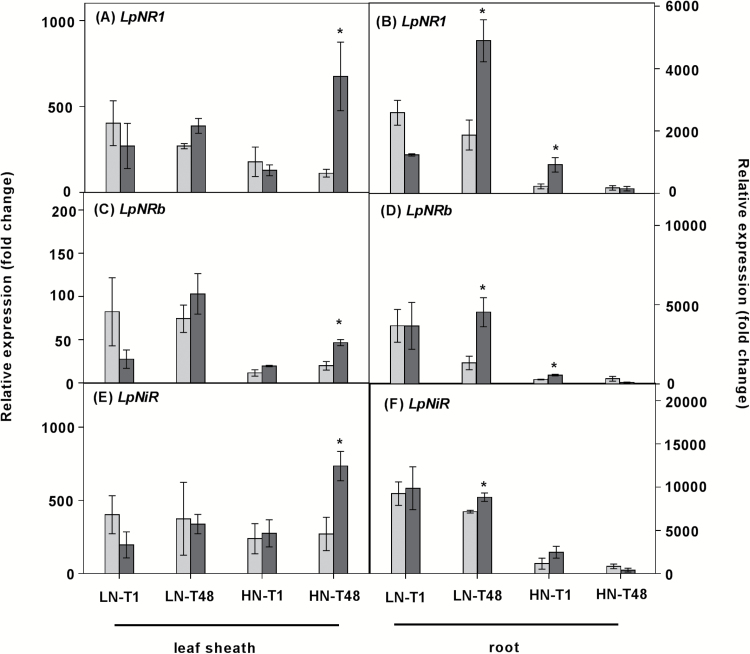
In order to determine whether N assimilation is tightly co-ordinated, we assessed the influence of the C/N balance on the expression of nitrate reductase (NR) and nitrite reductase (NiR) genes. Similar to nitrate transporter gene expression, transcript levels of genes involved in nitrate assimilation (LpNR1, LpNRb, and LpNiR) were more abundant in plants grown under LN conditions relative to those under HN conditions (Fig. 5). With exposure to HN conditions, expression of LpNR1 and LpNRb in sheaths was significantly induced 48 h after defoliation, whereas LpNR expression in roots showed no significant difference 48 h after defoliation (Fig. 5A–D). Notably, in HN roots, expression of LpNR1 and LpNRb was induced during the first hour after defoliation (Fig. 5B, D), which is consistent with the stimulation of NO3– uptake in HN plants following 1 h defoliation (Fig. 3A). The transcripts of LpNiR showed a similar pattern to that of LpNR1 and LpNRb. In LN conditions, expression of LpNR1, LpNRb, and LpNiR was induced in roots 48 h after defoliation, whereas there was no change in leaf sheaths (Fig. 5B, D, F).
Fig. 5.
Expression of putative nitrate reductase (LpNR) and nitrite reductase (LpNiR) genes in plants. Plants were grown at either 0.05 mM (LN) or 5 mM (HN) NO3–, and then defoliated (or left intact). Each data point is normalized against the reference genes eEF-1α and GAPDH. Values are means ±SE (n=3 pools of five plants each). Means were tested for significance using a two-tailed t-test. An asterisk denotes significantly different means between intact plants (grey bars) and defoliated plants (black bars) at P<0.05.
Glucose supplement
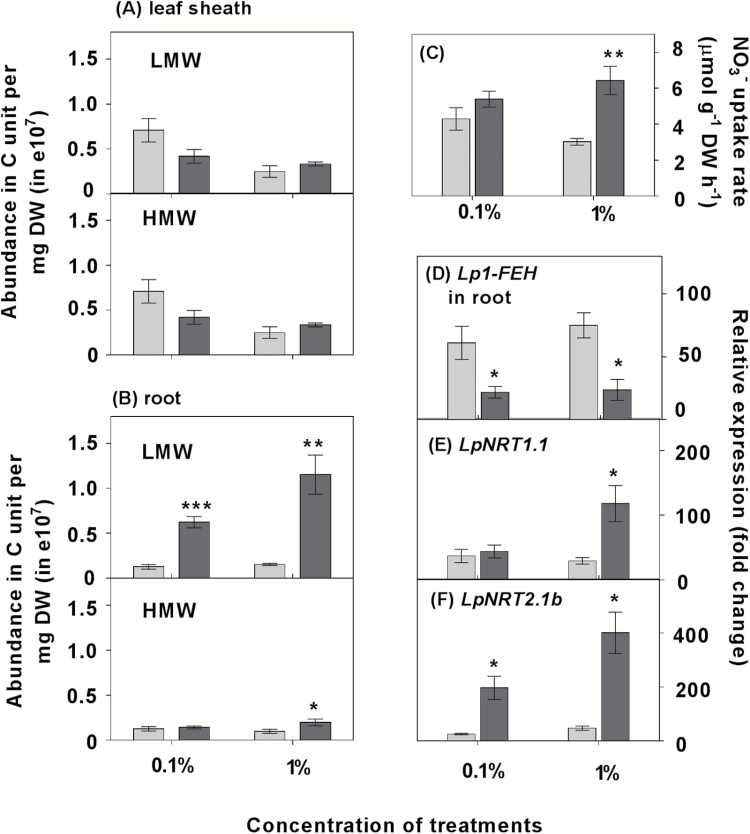
To distinguish further the relationship between carbon change dynamics following defoliation and N uptake responses, a subset of the HN plants were subjected to exogenous glucose treatment 42 h after defoliation. After 6 h of treatment with glucose, fructan abundance in roots increased compared with the mannitol control (Fig. 6B), whereas there was no significant difference in fructan abundance in the leaf sheaths (Fig. 6A). The detailed changes in the abundance of oligosaccharides with increasing DP are shown in Supplementary Fig. S6. Glucose supplementation led to repression of Lp1-FEH and Lp6-FEH expression in the roots but not in the leaf sheaths (Fig. 6D; Supplementary Fig. S7).
Fig. 6.
WSC content, NO3– uptake, and Lp1-FEH, LpNRT1.1, and LpNRT2.1b expression in roots with 6 h of supplemental glucose. Plants were grown in HN conditions and supplied with 0.1% or 1% glucose 42 h after defoliation. After 6 h, the impact of supplemental 0.1% or 1% glucose treatment on: (A, B) the relative abundance (in carbon units) of water-soluble carbohydrate (WSC); (C) NO3– uptake rate; and (D–F) Lp1-FEH, LpNRT1.1, and LpNRT2.1 expression in roots. Values are means ±SE (n=5 pools of five plants each). Means were tested for significance using a two-tailed t-test. Asterisks denote significantly different means between mannitol (grey) and glucose (black) treatment (*P<0.05, **P<0.01, ***P<0.001).
After 6 h of glucose treatment, a subset of HN defoliated plants was moved to 5 mM 15N isotopically labelled NO3– for NO3– uptake measurement. NO3– uptake increased significantly in the plants exposed to 1% glucose compared with those in the mannitol treatment (Fig. 6C). The 1% glucose supplement induced a significant decline in the 15N allocation to leaf sheaths (Supplementary Fig. S8).
Expression of both LpNRT2.1b and LpNRT1.1 was significantly enhanced by exposure to glucose (Fig. 6E, F). There was no significant change in the transcript abundance of LpNRT2.1a following glucose treatment (Supplementary Fig. S9).
Changes in endogenous cytokinin content following defoliation
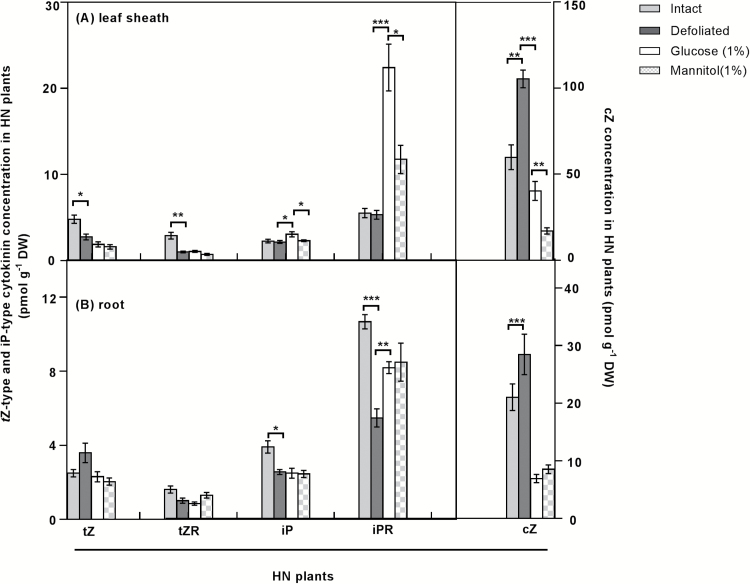
To better understand the role of cytokinin in communicating the root–shoot C/N balance at the whole-plant level, cytokinins were quantified in L. perenne plants. Overall, the total cytokinin complement was greater in the leaf sheaths and roots of HN plants compared with LN plants, irrespective of defoliation. Under HN, both tZ and tZ riboside (tZR) had decreased in the leaf sheaths 48 h after defoliation, whereas both iP and iP riboside (iPR) decreased in the roots 48 h after defoliation, relative to intact controls (Fig. 7). In the LN plants, with the exception of an accumulation of iPR in leaf sheaths 48 h after defoliation, there was little change in other cytokinins (Supplementary Fig. S10). In addition, iP and iPR in leaf sheaths was greater in HN defoliated plants following the 6 h 1% glucose treatment relative to HN defoliated plants without glucose treatment. Although there was no significant difference in the iPR content between the glucose and mannitol treatments, the iPR content was greater in HN roots with 1% glucose treatment relative to HN defoliated roots without glucose or mannitol addition (Fig. 7). Substantial amounts of cZ-type cytokinins were detected in HN plants. Among them, cZ accumulated in leaf sheaths in the 48 h following defoliation of HN plants, compared with the leaf sheaths of intact plants (Fig. 7). Further, cZ content decreased in leaf sheaths and roots of plants treated with glucose relative to HN defoliated plants without glucose or mannitol treatment. However, this decrease was also observed in the 1% mannitol treatment (Fig. 7).
Fig. 7.
Cytokinin concentrations in high N (HN) roots and leaf sheaths 48 h after defoliation. Values are means ±SE (n=5 pools of five plants each). Asterisks denote significantly different means between intact plants (grey bars) and defoliated plants (black bars), or between defoliated plants (black bars) and defoliated plants supplemented with 1% glucose (white bars) or 1% mannitol (hatched bars) (*P<0.05, **P<0.01, ***P<0.001). Means were tested for significance using a two-tailed t-test.
Expression of cytokinin oxidase/dehydrogenase and cytokinin response regulator gene family members.
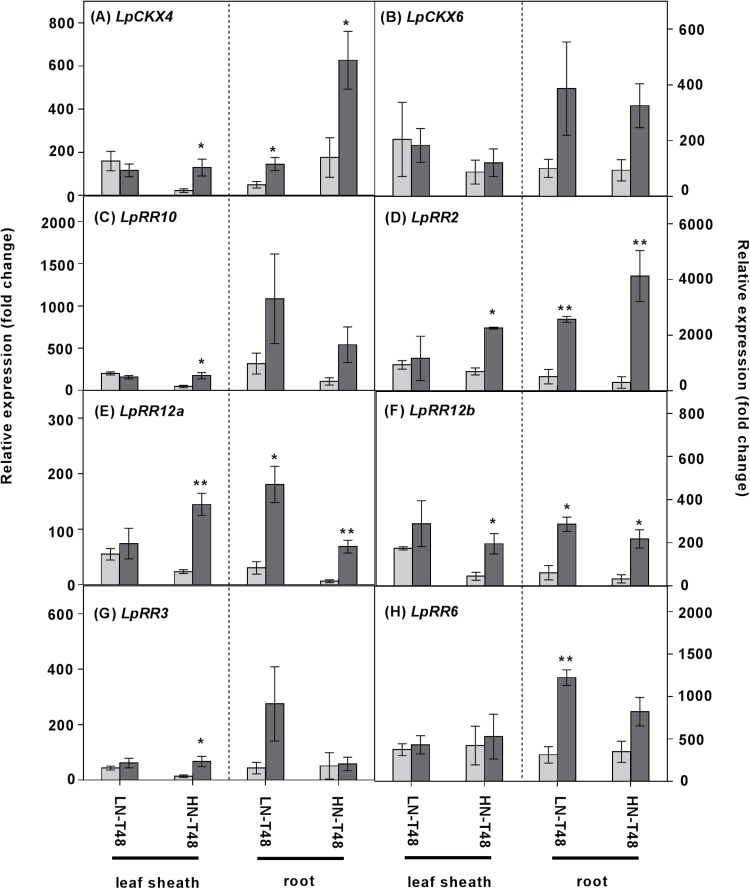
In roots, significant LpCKX4 induction was observed in LN and HN plants 48 h after defoliation (Fig. 8A), and in HN leaf sheaths 48 h after defoliation, relative to intact plants (Fig. 8A). No significant difference was observed during the first hour after defoliation, when compared with intact plants (Supplementary Fig. S11).
Fig. 8.
The impact of 48 h defoliation on the putative LpCKX and LpRR gene expression. Plants were grown at either 0.05 mM (LN) or 5 mM (HN) NO3-, and then defoliated (or left intact). Each data point is normalized against the reference genes eEF-1α and GAPDH. Values are means ±SE (n=3 pools of five plants each). Means were tested for significance using a two-tailed t-test. Asterisks denote significantly different means between intact plants (grey bars) and defoliated plants (black bars) (*P<0.05, **P<0.01).
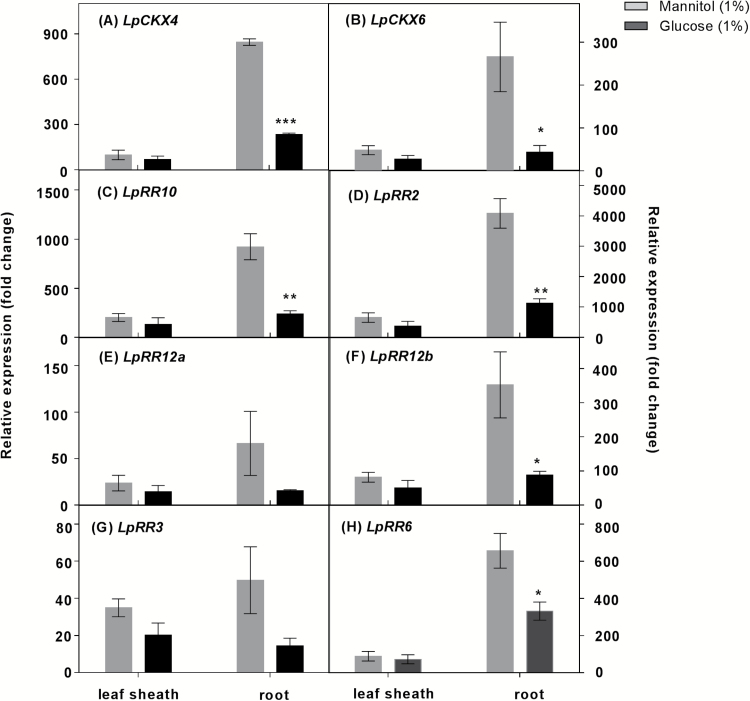
In comparison with intact plants, increases in expression of the four Type-B RR genes (LpRR2, LpRR10, LpRR12a, and LpRR12b) and two Type-A RR genes (LpRR3 and LpRR6) occurred in leaf sheaths of HN plants 48 h after defoliation, whereas no significant differences were displayed in leaf sheaths of LN plants (Fig. 8C–H). In roots, transcript levels of LpRR genes were greater in both LN and HN conditions 48 h after defoliation (Fig. 8C–H). Expression of LpCKX4, LpCKX6, and LpRR genes was strongly reduced in roots following the 1% glucose treatment of HN defoliated plants (Fig. 9A–F, H). Under 0.1% glucose treatment, there was no significant difference in expression of these gene families (Supplementary Fig. S12).
Fig. 9.
Effects of addition of 1% glucose on putative LpCKX and LpRR gene expression in plants grown in 5 mM NO3–, 48 h after defoliation. Each data point is normalized against the reference genes eEF-1α and GAPDH. Values are means ±SE (n=3 pools of five plants each). Means were tested for significance using a two-tailed t-test. Asterisks denote significantly different means between mannitol (grey bars) and glucose (black bars) treatment (*P<0.05, **P<0.01, ***P<0.001).
Discussion
An understanding of the mechanisms linking C/N balance to N uptake and assimilation is vital for optimizing plant growth in pasture-based production systems where defoliation by grazing is followed by N fertilizer addition. As shown previously, the fructan carbohydrate pool in elongating leaf bases and sheaths may be severely depleted in order to sustain regrowth following the removal of photosynthetically active tissues in grasses such as L. perenne (Morvan-Bertrand et al., 1999, 2001). Here, by experimentally defoliating plants to interfere with the C/N balance and by mitigating this with exogenous glucose supplementation, we have uncovered the dynamic relationships between the carbohydrate status of the plant, the nitrate-responsive uptake system, transporter gene expression, and assimilation. We confirm and extend previous studies (Louahlia et al., 1999) by showing that NO3– HATS and LATS uptake changes in response to the rapid and large shift in C status induced by defoliation. We also explored the involvement of cytokinins in the regulation of the C/N balance in response to defoliation. The results are discussed in relation to the internal C/N balance under defoliation.
Nitrate uptake and assimilation rely on labile carbohydrate availability
Consistent with previous studies (Louahlia et al., 2008; Roche et al., 2016), long-term N limitation led to significant accumulation of fructans in both roots and shoots relative to N-sufficient plants, which is important in the context of this study (Fig. 2). Two days after defoliation, severe fructan depletion occurred in both roots and leaf sheaths of HN plants. However, in LN plants, this depletion was only significant in roots, and was incomplete. We suggest that sufficient N in the HN plants was a stimulating factor in remobilization of the available fructan pool in leaf sheaths, which provided the required energy and C skeletons for regrowth. However, efficient regrowth following defoliation requires balanced pools of stored C for energy, cell biosynthesis, and C skeletons for nitrate reduction, together with stored N. LN plants clearly had insufficient N to utilize the stored C efficiently. These observations indicate that carbohydrate remobilization to support regrowth occurs in an N-dependent manner.
The regulation of root nitrate uptake by C metabolites from photosynthesis has previously been demonstrated through studying the diurnal stimulation of NO3– uptake. Sugars transported from shoots to roots played an important role in this diurnal stimulatory effect (Rideout and Raper, 1994; Delhon et al., 1996; Lejay et al., 1999). Using defoliation as a treatment in the present study allowed us to reveal the response of root NO3– uptake to changes in endogenous C. During the first hour following defoliation, we observed the immediate depletion of the fructan pool in roots of HN plants (Fig. 2B). This immediate remobilization of the fructan pool in HN roots was associated with a rapid and significant stimulation of root NO3– uptake rate (Figs 2B, 3A). Given that there is a substantial amount of sucrose in phloem immediately after defoliation (Morvan-Bertrand et al., 2001), this stimulation of NO3– uptake by the root may be attributed to the initial increase in sucrose followed by fructan remobilization in roots, suggesting an instantaneous response of root NO3– uptake to C availability. In contrast, 48 h after defoliation, low- and high-affinity NO3– uptake decreased following depletion of the fructan pool in the roots, irrespective of the N status of the plants (Figs 2, 3). Interestingly, a relatively greater fructan depletion in HN plants than in LN plants was associated with a much stronger repression of nitrate uptake rate in HN plants than in LN plants, indicating that NO3– uptake is a C-dependent process. These observations strongly suggest that root NO3– uptake is modulated by C metabolites at the whole-plant level, which is consistent with the suggestion that photosynthate supply is closely balanced with mineral uptake and assimilation (Lejay et al., 1999; Coruzzi and Zhou, 2001; Nunes-Nesi et al., 2010).
NO3– assimilation is regulated by the C–N status of the plant
According to Scheurwater et al. (2002), the intact shoot of the eight grass species that they tested was the predominant site for nitrate reduction. This can be explained by the fact that nitrate assimilation is more efficient in the shoot when leaves are photosynthesizing in fully saturating light conditions. Under these conditions, nitrate assimilation in roots is inefficient because sucrose has to be synthesized, transported to roots, and respired in order to generate ATP and NAD(P)H for N assimilation, rather than being directly utilized in the shoot (Nunes-Nesi et al., 2010). Our results also support tissue-specific N assimilation being regulated by local C availability. With relative high C availability in HN roots 1 h after defoliation, NO3– uptake was induced instantaneously compared with intact plants (Fig. 3A). This induction of nitrate uptake in HN roots was concomitant with stimulation of LpNR1 and LpNRb expression in HN roots (Fig. 5B, D). Given that light was non-saturating for expanding leaf tissue immediately following defoliation, because the leaf tissue was shaded by the sheath, we suggest that a relatively greater proportion of NO3– was assimilated locally in HN roots during the first hour after defoliation. However, in the days (48 h) after defoliation, when root C was reduced in HN plants and the young leaves were emerging, relatively more N was allocated to shoots compared with intact plants, thereby stimulating shoot growth (Fig. 2D; Supplementary Fig. S8). This was concomitant with changes in the transcript levels of nitrate reductase genes. In HN plants, expression of LpNR and LpNiR in leaf sheaths was induced following the strong C remobilization in roots, whereas expression of LpNR and LpNiR was repressed in roots (Figs 2C, D, 5). These results suggested that a shift in nitrate assimilation from roots to shoots occurred when root carbon availability was reduced, supporting the suggestion that local C availability regulates the tissue-specific assimilation of N.
With low N status associated with a high C/N ratio, the transcripts of LpNR and LpNiR genes were more abundant in roots compared with leaf sheaths (Figs 2C, D, 5). We suggest that under low NO3– availablility, roots with high C/N had sufficient carbohydrate to assimilate most of the acquired NO3– into amino acids locally, and thus little N was allocated to shoots in LN plants, thereby potentiating root growth and root foraging. Interestingly, 48 h after defoliation, the significant C remobilization in LN roots was accompanied by a strong induction of LpNR and LpNiR expression in roots, whereas there was no change in the fructan pool and the expression of LpNR and LpNiR in leaf sheaths (Figs 2C, D, 5). These observations imply a mechanism that signals the availability of C metabolites for NO3– uptake and assimilation at the whole-plant level. Within tissues, where stored C is available, it is apparent that NO3– is assimilated locally.
HATS and LATS regulation at the transcriptional level in response to a changing C/N ratio
Four NRT families have been characterized as being involved in nitrate uptake in Arabidopsis (Tsay et al., 2007). Among them, NRT1 and NRT2 are regarded as the main components controlling the HATS and LATS that function at low (<250 µM) or high (>1 mM) nitrate levels, respectively (Siddiqi et al., 1990; Tsay et al., 1993; Huang et al., 1999; Y.Y. Wang et al., 2012). In our study, as expected, the expression of LpNRT1.1 and LpNRT2.1b was significantly repressed in HN plants 48 h after defoliation, coinciding with reduced NO3– uptake activity (Figs 3B, 4B, F). Interestingly, within 1 h of defoliation, the increased expression of LpNRT2.1b in HN roots (P=0.07) also coincided with the rapid induction of NO3– uptake activity (Figs 3A, 4B). This is consistent with observations that regulation of root NO3– uptake is correlated with changes in transcript levels of NRT2.1 in response to N and C treatment (Zhuo et al., 1999; Lejay et al., 2003).
Conversely, but as expected, this was not the case in LN status plants which showed lower reductions in NO3– uptake rate following defoliation relative to HN plants, and only small increases in both LpNRT1.1 and LpNRT2.1 expression (Figs 3B, 4B, F). Indeed, the regulation of high-affinity nitrate uptake is complicated in low nitrate conditions. This result is consistent with previous studies showing that large and rapid increases in HATS uptake in low NO3– conditions are not accompanied by increased AtNRT2.1 expression (De Jong et al., 2014). One possible interpretation for the discrepancy between NRT2.1 transcript and nitrate uptake involves the potential for post-transcriptional regulation of existing transporter levels and activity, for example by phosphorylation (Laugier et al., 2012; X. Wang et al., 2012).
Exogenous glucose mitigates reduced nitrate uptake rate in defoliated plants
Exogenous glucose supplied for 6 h to defoliated HN plants largely restored LMW WSC levels in roots, which appears to have relieved the C limitation on NO3– uptake and assimilation (Fig. 6B). The NO3– uptake rate in defoliated plants supplemented with glucose attained a level greater than that in plants maintained in 5 mM NO3– but without glucose supply. In contrast, application of 1% mannitol had no impact on the NO3– uptake rate compared with plants kept in 5 mM NO3–, confirming the carbohydrate dependence of nitrate uptake as described above (Figs 3B, 6C). Interestingly, increasing C availability in roots was correlated with a decline in N allocation to leaf sheaths, which supports our observations of C and N interactions in nitrate uptake and metabolism (Fig. 6B; Supplementary Fig. S8).
The NO3– uptake rescue by glucose was positively correlated with the transcript levels of LpNRT1.1 and LpNRT2.1b (Fig. 6C, E, F), genes that are regarded as the major players in the LATS and HATS for nitrate, respectively (Y.Y. Wang et al., 2012). Glucose and sucrose have been found to regulate the expression of genes involved in metabolism and nutrient uptake (Li et al., 2006; Lejay et al., 2008). However, up-regulation of AtNRT2.1 by sugars is not attributed to well-established specific sucrose or glucose sensing, although, as shown by De Jong et al. (2014), glucose-regulated expression of AtNRT2.1, independent of nitrate-mediated mechanisms, can operate through HEXOKINASE1-mediated oxidative pentose phosphate pathway (OPPP) metabolism.
Cytokinin is involved in the regulation of N acquisition and assimilation in response to changes in C/N
Cytokinins have been proposed as candidate signalling molecules relaying N status between the root and shoot (Kudo et al., 2010; Ruffel et al., 2014). We show here that the cytokinin content of intact HN status plants is greater than that in plants of LN status (Fig. 7; Supplementary Fig. 10). This is consistent with the observations that N supply leads to cytokinin accumulation in roots, xylem sap, and shoots in maize (Takei et al., 2001), suggesting that cytokinin serves to signal N availability from the root to shoot (Sakakibara et al., 2006). After defoliation under HN, a decrease was observed in the tZ-type cytokinin in the leaf sheaths and the iP-type cytokinin in the roots which was associated with increased CKX expression (Figs 7, 8A, B). Interestingly, 1% glucose addition to defoliated HN plants induced iP and iPR accumulation in leaf sheaths and roots, and was associated with a significant decline in expression of LpCKX4 and LpCKX6 in roots, implying that iP-type cytokinins might be an integrating signal of the relative availability of C and N metabolites in plants (Figs 7, 9A, B).
In contrast, cZ-type cytokinins increased in the HN leaf sheaths, as did expression of the LpRR genes, 48 h after defoliation (Figs 7, 8). cZ-type cytokinins are recognized by receptors and activate response regulators in rice and maize (Veach et al., 2003; Yonekura-Sakakibara et al., 2004; Lomin et al., 2011; Roche et al., 2016). In this context, it is possible that the LpRR genes could be responding to the cis-cytokinins with the downstream activation of N assimilation in the leaf sheaths. Consistently, while cZ-type cytokinin content declined, LpRR expression was reduced in roots following 6 h of 1% glucose addition (Figs 7, 9). However, this decline of cZ also occurred in plants with 1% mannitol treatment (Fig. 7). The decreased cZ content under 1% mannitol treatment was not consistent with salt or osmotic stress which is suggested to induce cZ (Havlová et al., 2008; Vyroubalová et al., 2009; Macková et al., 2013). The cZ response to mannitol reflects the less well understood biology of the cis-cytokinin forms. As the LpRR expression decreased under glucose treatment but not under mannitol treatment, a cytokinin response to mannitol is not supported. Taken together with the increased NO3– uptake rate, fructan levels, and RR gene profiles under glucose treatment, the glucose supplement may have caused a cytokinin response via its impact on C metabolism.
Conclusions
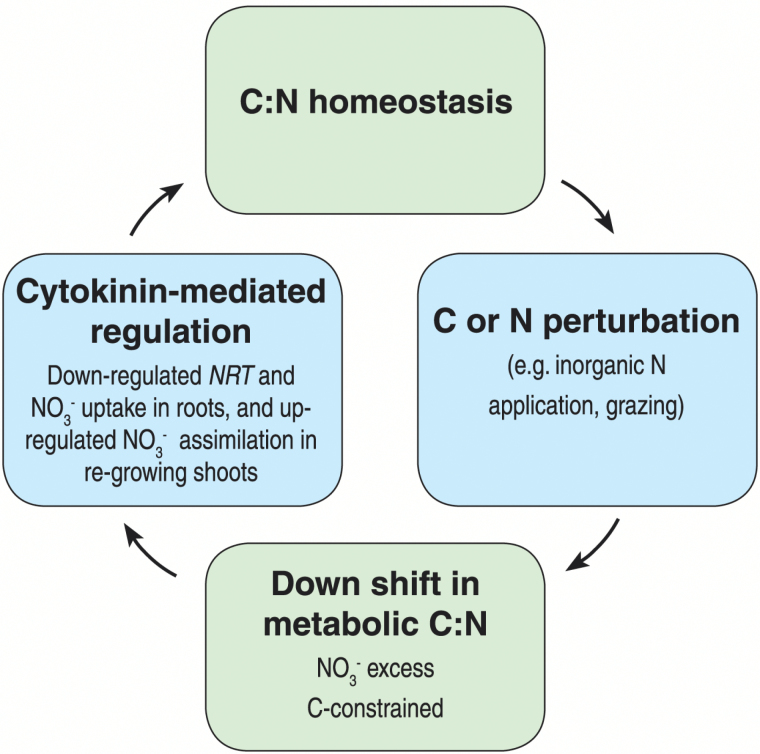
This study provides clear evidence that the rapid and large shifts in C storage triggered by defoliation have significant impact on nitrate uptake in an N-status-dependent manner. Our results for the perennial grass, L. perenne, are consistent with those in Arabidopsis, where cytokinins act as a systemic N signal regulating N uptake and assimilation (Kiba et al., 2011; Ruffel et al., 2011; Shtratnikova et al., 2015). By perturbing C assimilation and metabolism with a defoliation–regrowth treatment, we propose that cytokinin-mediated NO3– uptake and assimilation may also act to balance C and N resources at the whole-plant level. Building on our results and those of previous workers, we present a model (Fig. 10) whereby cytokinins serve as a signal to integrate C and N metabolism via tissue-specific function: cytokinin-mediated reduction in NO3– uptake in roots and an increase in NO3– assimilation in re-growing shoots under low C/high N status (low N demand); and a concomitant shift of C allocation from shoots to roots which serves to stimulate NO3– assimilation in roots under high C/low N status (high N demand) (Fig. 10). As we develop our understanding of the interaction between defoliation and N uptake, this knowledge may be applied to pasture production for grazing in order to increase N uptake and utilization efficiency.
Fig. 10.
A proposed model depicting the role of cytokinin in C:N homeostasis. Cytokinin restores the C:N balance by down-regulating NO3– uptake in a C-status-dependent manner: when grazing or inorganic N application leads to a decrease in the C:N balance, cytokinin serves as a signal to mediate the down-regulation of NO3– uptake in roots and the up-regulation of NO3– assimilation in the re-growing shoots, thereby re-balancing the C and N resources at the whole-plant level.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Sequences of qPCR primers used in this work.
Figs S1 and S2. Flow diagrams of the experimental processes
Fig. S3. Phylogenetic trees of LpNRT1, LpNRT2, LpNR, and LpNiR gene families.
Fig. S4. Relative abundance in carbon (C) units of water-soluble carbohydrates (WSCs) in HN and LN plants after defoliation
Fig. S5. Expression of putative Lp6-FEH and Lp1-FEH in plants grown under either 0.05 mM (LN) or 5 mM NO3– (HN) supply, 1 h and 48 h after defoliation.
Fig. S6. The impact of glucose on relative abundance in carbon (C) units of water-soluble carbohydrates (WSCs).
Fig. S7. Lp6-FEH and Lp1-FEH expression in leaf sheaths with 6 h of supplemental glucose.
Fig. S8. The impact of defoliation or glucose addition on 15N translocation to shoots.
Fig. S9. Effects of glucose addition on putative LpNRT2.1a gene expression in roots grown in 5 mM NO3-, 48 h after defoliation.
Fig. S10. Cytokinin concentrations in LN roots and leaf sheaths 48 h after defoliation.
Fig. S11. The impact of 1 h defoliation on putative LpCKX and LpRR gene expression.
Fig. S12. Effects of 0.1% glucose addition on putative LpCKX and LpRR gene expression in plants grown in 5 mM NO3–, 48 h after defoliation.
Supplementary Material
Acknowledgements
We thank Ballance AgriNutrients for the principle financial support for this research and for a scholarship for QG. ON was funded by the Czech Science Foundation (no. 17-06613S). We thank Dave Conder and Eva Hirnerová for technical assistance.
References
- Antoniadi I, Plačková L, Simonovik B, Doležal K, Turnbull C, Ljung K, Novák O. 2015. Cell-type-specific cytokinin distribution within the Arabidopsis primary root apex. The Plant Cell 27, 1955–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucaud J, Bigot J. 1989. Changes in the activities of nitrogen assimilation enzymes of Lolium perenne L. during regrowth after cutting. Plant and Soil 114, 121–125. [Google Scholar]
- Brackin R, Näsholm T, Robinson N, Guillou S, Vinall K, Lakshmanan P, Schmidt S, Inselsbacher E. 2015. Nitrogen fluxes at the root–soil interface show a mismatch of nitrogen fertilizer supply and sugarcane root uptake capacity. Scientific Reports 5, 15727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner WG, Romanov GA, Köllmer I, Bürkle L, Schmülling T. 2005. Immediate-early and delayed cytokinin response genes of Arabidopsis thaliana identified by genome-wide expression profiling reveal novel cytokinin-sensitive processes and suggest cytokinin action through transcriptional cascades. The Plant Journal 44, 314–333. [DOI] [PubMed] [Google Scholar]
- Coruzzi GM, Zhou L. 2001. Carbon and nitrogen sensing and signaling in plants: emerging ‘matrix effects’. Current Opinion in Plant Biology 4, 247–253. [DOI] [PubMed] [Google Scholar]
- Dawar K, Zaman M, Rowarth JS, Blennerhassett J, Turnbull MH. 2010. The impact of urease inhibitor on the bioavailability of nitrogen in urea and in comparison with other nitrogen sources in ryegrass (Lolium perenne L.). Crop and Pasture Science 61, 214–221. [Google Scholar]
- de Jong F, Thodey K, Lejay LV, Bevan MW. 2014. Glucose elevates NITRATE TRANSPORTER2.1 protein levels and nitrate transport activity independently of its HEXOKINASE1-mediated stimulation of NITRATE TRANSPORTER2.1 expression. Plant Physiology 164, 308–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhon P, Gojon A, Tillard P, Passama L. 1996. Diurnal regulation of NO3– uptake in soybean plants IV. Dependence on current photosynthesis and sugar availability to the roots. Journal of Experimental Botany 47, 893–900. [Google Scholar]
- Dobrev PI, Kamínek M. 2002. Fast and efficient separation of cytokinins from auxin and abscisic acid and their purification using mixed-mode solid-phase extraction. Journal of Chromatography A 950, 21–29. [DOI] [PubMed] [Google Scholar]
- Garnett T, Conn V, Plett D, et al. 2013. The response of the maize nitrate transport system to nitrogen demand and supply across the lifecycle. New Phytologist 198, 82–94. [DOI] [PubMed] [Google Scholar]
- Gutiérrez RA, Lejay LV, Dean A, Chiaromonte F, Shasha DE, Coruzzi GM. 2007. Qualitative network models and genome-wide expression data define carbon/nitrogen-responsive molecular machines in Arabidopsis. Genome Biology 8, R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havlová M, Dobrev PI, Motyka V, Storchová H, Libus J, Dobrá J, Malbeck J, Gaudinová A, Vanková R. 2008. The role of cytokinins in responses to water deficit in tobacco plants over-expressing trans-zeatin O-glucosyltransferase gene under 35S or SAG12 promoters. Plant, Cell and Environment 31, 341–353. [DOI] [PubMed] [Google Scholar]
- Hirose N, Takei K, Kuroha T, Kamada-Nobusada T, Hayashi H, Sakakibara H. 2008. Regulation of cytokinin biosynthesis, compartmentalization and translocation. Journal of Experimental Botany 59, 75–83. [DOI] [PubMed] [Google Scholar]
- Huang NC, Liu KH, Lo HJ, Tsay YF. 1999. Cloning and functional characterization of an Arabidopsis nitrate transporter gene that encodes a constitutive component of low-affinity uptake. The Plant Cell 11, 1381–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sheen J, Müller B. 2012. Cytokinin signaling networks. Annual Review of Plant Biology 63, 353–380. [DOI] [PubMed] [Google Scholar]
- Jameson PE. 2017. Cytokinins. In: Thomas B, Murray BG, Murphy DJ, eds. Encyclopedia of applied plant sciences, Vol. 1 Waltham, MA: Academic Press; 391–402. [Google Scholar]
- Kiba T, Kudo T, Kojima M, Sakakibara H. 2011. Hormonal control of nitrogen acquisition: roles of auxin, abscisic acid, and cytokinin. Journal of Experimental Botany 62, 1399–1409. [DOI] [PubMed] [Google Scholar]
- Krouk G, Ruffel S, Gutiérrez RA, Gojon A, Crawford NM, Coruzzi GM, Lacombe B. 2011. A framework integrating plant growth with hormones and nutrients. Trends in Plant Science 16, 178–182. [DOI] [PubMed] [Google Scholar]
- Kudo T, Kiba T, Sakakibara H. 2010. Metabolism and long-distance translocation of cytokinins. Journal of Integrative Plant Biology 52, 53–60. [DOI] [PubMed] [Google Scholar]
- Kushwah S, Laxmi A. 2014. The interaction between glucose and cytokinin signal transduction pathway in Arabidopsis thaliana. Plant, Cell and Environment 37, 235–253. [DOI] [PubMed] [Google Scholar]
- Laugier E, Bouguyon E, Mauriès A, Tillard P, Gojon A, Lejay L. 2012. Regulation of high-affinity nitrate uptake in roots of Arabidopsis depends predominantly on posttranscriptional control of the NRT2.1/NAR2.1 transport system. Plant Physiology 158, 1067–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejay L, Gansel X, Cerezo M, et al. 2003. Regulation of root ion transporters by photosynthesis: functional importance and relation with hexokinase. The Plant Cell 15, 2218–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejay L, Tillard P, Lepetit M, Olive Fd, Filleur S, Daniel-Vedele F, Gojon A. 1999. Molecular and functional regulation of two NO3– uptake systems by N- and C-status of Arabidopsis plants. The Plant Journal 18, 509–519. [DOI] [PubMed] [Google Scholar]
- Lejay L, Wirth J, Pervent M, Cross JM, Tillard P, Gojon A. 2008. Oxidative pentose phosphate pathway-dependent sugar sensing as a mechanism for regulation of root ion transporters by photosynthesis. Plant Physiology 146, 2036–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lestienne F, Thornton B, Gastal F. 2006. Impact of defoliation intensity and frequency on N uptake and mobilization in Lolium perenne. Journal of Experimental Botany 57, 997–1006. [DOI] [PubMed] [Google Scholar]
- Li W, Wang Y, Okamoto M, Crawford NM, Siddiqi MY, Glass AD. 2007. Dissection of the AtNRT2.1:AtNRT2.2 inducible high-affinity nitrate transporter gene cluster. Plant Physiology 143, 425–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Lee KK, Walsh S, Smith C, Hadingham S, Sorefan K, Cawley G, Bevan MW. 2006. Establishing glucose- and ABA-regulated transcription networks in Arabidopsis by microarray analysis and promoter classification using a Relevance Vector Machine. Genome Research 16, 414–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SH, Kuo HF, Canivenc G, et al. 2008. Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. The Plant Cell 20, 2514–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KH, Huang CY, Tsay YF. 1999. CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. The Plant Cell 11, 865–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomin SN, Yonekura-Sakakibara K, Romanov GA, Sakakibara H. 2011. Ligand-binding properties and subcellular localization of maize cytokinin receptors. Journal of Experimental Botany 62, 5149–5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomin SN, Krivosheev DM, Steklov MY, Arkhipov DV, Osolodkin DI, Schmülling T, Romanov GA. 2015. Plant membrane assays with cytokinin receptors underpin the unique role of free cytokinin bases as biologically active ligands. Journal of Experimental Botany 66, 1851–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lothier J, Lasseur B, Le Roy K, Van Laere A, Prud’homme MP, Barre P, Van den Ende W, Morvan-Bertrand A. 2007. Cloning, gene mapping, and functional analysis of a fructan 1-exohydrolase (1-FEH) from Lolium perenne implicated in fructan synthesis rather than in fructan mobilization. Journal of Experimental Botany 58, 1969–1983. [DOI] [PubMed] [Google Scholar]
- Lothier J, Van Laere A, Prud’homme MP, Van den Ende W, Morvan-Bertrand A. 2014. Cloning and characterization of a novel fructan 6-exohydrolase strongly inhibited by sucrose in Lolium perenne. Planta 240, 629–643. [DOI] [PubMed] [Google Scholar]
- Louahlia S, Laine P, MacDuff J, Ourry A, Humphreys M, Boucaud J. 2008. Interactions between reserve mobilization and regulation of nitrate uptake during regrowth of Lolium perenne L.: putative roles of amino acids and carbohydrates. Botany 86, 1101–1110. [Google Scholar]
- Louahlia S, Macduff J, Ourry A, Humphreys M, Boucaud J. 1999. Nitrogen reserve status affects the dynamics of nitrogen remobilization and mineral nitrogen uptake during recovery of contrasting cultivars of Lolium perenne from defoliation. New Phytologist 142, 451–462. [Google Scholar]
- Macková H, Hronková M, Dobrá J, et al. 2013. Enhanced drought and heat stress tolerance of tobacco plants with ectopically enhanced cytokinin oxidase/dehydrogenase gene expression. Journal of Experimental Botany 64, 2805–2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto-Kitano M, Kusumoto T, Tarkowski P, Kinoshita-Tsujimura K, Václavíková K, Miyawaki K, Kakimoto T. 2008. Cytokinins are central regulators of cambial activity. Proceedings of the National Academy of Sciences, USA 105, 20027–20031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morvan-Bertrand A, Boucaud J, Le Saos J, Prud’homme MP. 2001. Roles of the fructans from leaf sheaths and from the elongating leaf bases in the regrowth following defoliation of Lolium perenne L. Planta 213, 109–120. [DOI] [PubMed] [Google Scholar]
- Morvan-Bertrand A, Boucaud J, Prud’homme M-P. 1999. Influence of initial levels of carbohydrates, fructans, nitrogen, and soluble proteins on regrowth of Lolium perenne L. cv. Bravo following defoliation. Journal of Experimental Botany 50, 1817–1826. [Google Scholar]
- Nunes-Nesi A, Fernie AR, Stitt M. 2010. Metabolic and signaling aspects underpinning the regulation of plant carbon nitrogen interactions. Molecular Plant 3, 973–996. [DOI] [PubMed] [Google Scholar]
- Ourry A, Bigot J, Boucaud J. 1989. Protein mobilization from stubble and roots, and proteolytic activities during post-clipping re-growth of perennial ryegrass. Journal of Plant Physiology 134, 298–303. [Google Scholar]
- Palenchar PM, Kouranov A, Lejay LV, Coruzzi GM. 2004. Genome-wide patterns of carbon and nitrogen regulation of gene expression validate the combined carbon and nitrogen (CN)-signaling hypothesis in plants. Genome Biology 5, R91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards J. 1993. Physiology of plants recovering from defoliation. In: Bakes MJ, ed. Grassland for our world. Wellington, New Zealand: SIR Publishing, 85–94. [Google Scholar]
- Rideout JW, Raper CD., Jr 1994. Diurnal changes in net uptake rate of nitrate are associated with changes in estimated export of carbohydrates to roots. International Journal of Plant Sciences 155, 173–179. [DOI] [PubMed] [Google Scholar]
- Roche J, Love J, Guo Q, et al. 2016. Metabolic changes and associated cytokinin signals in response to nitrate assimilation in roots and shoots of Lolium perenne. Physiologia Plantarum 156, 497–511. [DOI] [PubMed] [Google Scholar]
- Roche J, Turnbull MH, Guo Q, Novák O, Späth J, Gieseg S, Jameson PE, Love J. 2017. Coordinated nitrogen and carbon remobilization for nitrate assimilation in leaf, sheath, and root and associated cytokinin signals during early regrowth of Lolium perenne. Annals of Botany. mcx014. [DOI] [PMC free article] [PubMed]
- Ruffel S, Gojon A, Lejay L. 2014. Signal interactions in the regulation of root nitrate uptake. Journal of Experimental Botany 65, 5509–5517. [DOI] [PubMed] [Google Scholar]
- Ruffel S, Krouk G, Ristova D, Shasha D, Birnbaum KD, Coruzzi GM. 2011. Nitrogen economics of root foraging: transitive closure of the nitrate–cytokinin relay and distinct systemic signaling for N supply vs. demand. Proceedings of the National Academy of Sciences, USA 108, 18524–18529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara H. 2006. Cytokinins: activity, biosynthesis, and translocation. Annual Review of Plant Biology 57, 431–449. [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Takei K, Hirose N. 2006. Interactions between nitrogen and cytokinin in the regulation of metabolism and development. Trends in Plant Science 11, 440–448. [DOI] [PubMed] [Google Scholar]
- Scheurwater I, Koren M, Lambers H, Atkin OK. 2002. The contribution of roots and shoots to whole plant nitrate reduction in fast- and slow-growing grass species. Journal of Experimental Botany 53, 1635–1642. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nature Protocols 3, 1101–1108. [DOI] [PubMed] [Google Scholar]
- Shtratnikova VY, Kudryakova N, Kudoyarova G, Korobova A, Akhiyarova G, Danilova M, Kusnetsov V, Kulaeva O. 2015. Effects of nitrate and ammonium on growth of Arabidopsis thaliana plants transformed with the ARR5::GUS construct and a role for cytokinins in suppression of disturbances induced by the presence of ammonium. Russian Journal of Plant Physiology 62, 741–752. [Google Scholar]
- Siddiqi MY, Glass AD, Ruth TJ, Rufty TW. 1990. Studies of the uptake of nitrate in barley: I. Kinetics of NO(3) influx. Plant Physiology 93, 1426–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Jiang L, Jameson PE. 2012. Co-ordinate regulation of cytokinin gene family members during flag leaf and reproductive development in wheat. BMC Plant Biology 12, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Jiang L, Jameson PE. 2015. Expression patterns of Brassica napus genes implicate IPT, CKX, sucrose transporter, cell wall invertase, and amino acid permease gene family members in leaf, flower, silique, and seed development. Journal of Experimental Botany 66, 5067–5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M, Krapp A. 1999. The interaction between elevated carbon dioxide and nitrogen nutrition: the physiological and molecular background. Plant, Cell and Environment 22, 583–621. [Google Scholar]
- Svačinová J, Novák O, Plačková L, Lenobel R, Holík J, Strnad M, Doležal K. 2012. A new approach for cytokinin isolation from Arabidopsis tissues using miniaturized purification: pipette tip solid-phase extraction. Plant Methods 8, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei K, Sakakibara H, Taniguchi M, Sugiyama T. 2001. Nitrogen-dependent accumulation of cytokinins in root and the translocation to leaf: implication of cytokinin species that induces gene expression of maize response regulator. Plant and Cell Physiology 42, 85–93. [DOI] [PubMed] [Google Scholar]
- Takei K, Yamaya T, Sakakibara H. 2004. Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyze the biosynthesis of trans-zeatin. Journal of Biological Chemistry 279, 41866–41872. [DOI] [PubMed] [Google Scholar]
- Tsay YF, Chiu CC, Tsai CB, Ho CH, Hsu PK. 2007. Nitrate transporters and peptide transporters. FEBS Letters 581, 2290–2300. [DOI] [PubMed] [Google Scholar]
- Tsay YF, Schroeder JI, Feldmann KA, Crawford NM. 1993. The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72, 705–713. [DOI] [PubMed] [Google Scholar]
- Turner LB, Cairns AJ, Armstead IP, Ashton J, Skøt K, Whittaker D, Humphreys MO. 2006. Dissecting the regulation of fructan metabolism in perennial ryegrass (Lolium perenne) with quantitative trait locus mapping. New Phytologist 169, 45–57. [DOI] [PubMed] [Google Scholar]
- Veach YK, Martin RC, Mok DW, Malbeck J, Vankova R, Mok MC. 2003. O-Glucosylation of cis-zeatin in maize. Characterization of genes, enzymes, and endogenous cytokinins. Plant Physiology 131, 1374–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyroubalová S, Václavíková K, Turecková V, Novák O, Smehilová M, Hluska T, Ohnoutková L, Frébort I, Galuszka P. 2009. Characterization of new maize genes putatively involved in cytokinin metabolism and their expression during osmotic stress in relation to cytokinin levels. Plant Physiology 151, 433–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Ruan Y-L. 2016. Shoot–root carbon allocation, sugar signalling and their coupling with nitrogen uptake and assimilation. Functional Plant Biology 43, 105–113. [DOI] [PubMed] [Google Scholar]
- Wang X, Bian Y, Cheng K, Zou H, Sun SS, He JX. 2012. A comprehensive differential proteomic study of nitrate deprivation in Arabidopsis reveals complex regulatory networks of plant nitrogen responses. Journal of Proteome Research 11, 2301–2315. [DOI] [PubMed] [Google Scholar]
- Wang YY, Hsu PK, Tsay YF. 2012. Uptake, allocation and signaling of nitrate. Trends in Plant Science 17, 458–467. [DOI] [PubMed] [Google Scholar]
- Werner T, Holst K, Pörs Y, Guivarc’h A, Mustroph A, Chriqui D, Grimm B, Schmülling T. 2008. Cytokinin deficiency causes distinct changes of sink and source parameters in tobacco shoots and roots. Journal of Experimental Botany 59, 2659–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmülling T. 2003. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. The Plant Cell 15, 2532–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Kojima M, Yamaya T, Sakakibara H. 2004. Molecular characterization of cytokinin-responsive histidine kinases in maize. Differential ligand preferences and response to cis-zeatin. Plant Physiology 134, 1654–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo D, Okamoto M, Vidmar JJ, Glass AD. 1999. Regulation of a putative high-affinity nitrate transporter (AtNrt2.1) in roots of Arabidopsis thaliana. The Plant Journal 17, 563–568. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.