Highlight
We show subcellular localization of all six Arabidopsis thaliana arogenate dehydratases to chloroplasts and stromules. Furthermore, ADT2 and ADT5 have additional localization patterns suggesting non-enzymatic functions and roles as moonlighting proteins.
Keywords: Arogenate dehydratase, chloroplast division, moonlighting proteins, nuclear localization, phenylalanine biosynthesis, stromules.
Abstract
Arogenate dehydratases (ADTs) catalyze the final step in phenylalanine biosynthesis in plants. The Arabidopsis thaliana genome encodes a family of six ADTs capable of decarboxylating/dehydrating arogenate into phenylalanine. Using cyan fluorescent protein (CFP)-tagged proteins, the subcellular localization patterns of all six A. thaliana ADTs were investigated in intact Nicotiana benthamiana and A. thaliana leaf cells. We show that A. thaliana ADTs localize to stroma and stromules (stroma-filled tubules) of chloroplasts. This localization pattern is consistent with the enzymatic function of ADTs as many enzymes required for amino acid biosynthesis are primarily localized to chloroplasts, and stromules are thought to increase metabolite transport from chloroplasts to other cellular compartments. Furthermore, we provide evidence that ADTs have additional, non-enzymatic roles. ADT2 localizes in a ring around the equatorial plane of chloroplasts or to a chloroplast pole, which suggests that ADT2 is a component of the chloroplast division machinery. In addition to chloroplasts, ADT5 was also found in nuclei, again suggesting a non-enzymatic role for ADT5. We also show evidence that ADT5 is transported to the nucleus via stromules. We propose that ADT2 and ADT5 are moonlighting proteins that play an enzymatic role in phenylalanine biosynthesis and a second role in chloroplast division or transcriptional regulation, respectively.
Introduction
Arogenate dehydratases (ADTs; EC 4.2.1.911) catalyze the final step of phenylalanine biosynthesis through decarboxylation/dehydration of arogenate to form the aromatic amino acid phenylalanine (Fig. 1A; Laskar et al., 2010; Tzin and Galili, 2010). In plants phenylalanine serves as a precursor not only of proteins but also of many secondary metabolites, including phenylpropanoids (Herrmann and Weaver, 1999; Knaggs, 2003; Vogt, 2010). Phenylpropanoids have diverse functions, including structural support, pigmentation, and scent formation (Vogt, 2010), indicating the great importance of phenylalanine biosynthesis in plants.
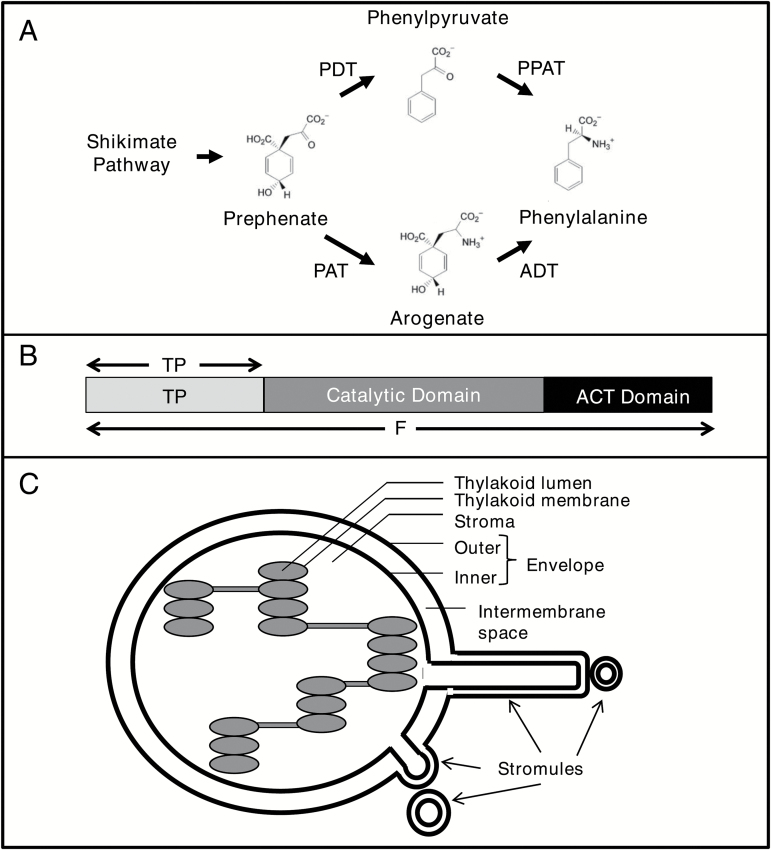
Fig. 1.
Phenylalanine synthesis, arogenate dehydratases, and stromules. (A) Phenylalanine can be synthesized in plants using either the prephenate (top) or the arogenate (bottom) pathway (Cho et al., 2007; Maeda and Dudareva, 2012). Prephenate is either decarboxylated/dehydrated to phenylpyruvate (PP) by a prephenate dehydratase (PDT) and PP is then transaminated by a phenylpyruvate aminotransferase (PPAT) to phenylalanine. Alternatively the two enzymatic steps are reversed, whereby prephenate is transaminated to arogenate by a prephenate aminotransferase (PAT) and arogenate is then decarboxylated/dehydrated to phenylalanine by an arogenate dehydratase (ADT). (B) A. thaliana ADT constructs were cloned in different lengths. The full-length (F) sequence represents the entire ADT ORF while an N-terminal construct only includes the transit peptide (TP). (C) Schematic diagram of a chloroplast showing the formation of stromules. Stromules are stroma-filled protrusions of the outer and inner membrane from chloroplasts. They can differ in length, forming long thread-like extensions or globular structures.
The Arabidopsis thaliana genome encodes a small gene family of six ADT genes, all sharing similar sequences and domain structures (Ehlting et al., 2005; Cho et al., 2007). Families of ADTs are common in plant genomes and have been identified in both monocot and dicot species (Tuskan et al., 2006; Yamada et al., 2008; Maeda et al., 2010, 2011). The presence of multiple isoforms suggests that ADTs might have evolved different properties, and/or that each ADT is either transcriptionally or post-translationally regulated to allow for distinct functional roles. For example, all six A. thaliana ADTs accept arogenate as a substrate, as their name suggests, but two of the six ADTs also accept prephenate [meaning they can act as ADTs and prephenate dehydratases (PDTs; Fig. 1A; Cho et al., 2007; Bross et al., 2011)]. Furthermore, ADTs differentially contribute to lignin content and bolting/flowering transition. Analysis of different A. thaliana ADT knockout mutants has indicated that specific ADTs preferentially contribute to the synthesis of different downstream products of the phenylpropanoid pathway (Corea et al., 2012b). In lines harboring an adt5 knockout, the amount of phenylalanine, and its proportions relative to tyrosine and tryptophan, were lower in stem tissues compared with the wild-type control (Corea et al., 2012a). Together, these data suggest that ADT5 plays a predominant role in phenylalanine biosynthesis for lignin deposition in stems. Also, the adt1, but not the adt4, knockout line exhibited a resistant late bolting/flowering phenotype compared with wild-type A. thaliana under different environmental conditions, which is consistent with a decreased level of ADT expression after cold treatment in resistant late bolting/flowering Beta vulgaris altissima (Hébrard et al., 2013). This suggests that ADTs may be functional targets of DNA methylation in the shoot apical meristem during vernalization, and that the accumulation of phenolic compounds may play a role in floral transition.
Plant ADTs, including all six A. thaliana isoforms, have three domains (Fig. 1B), a putative N-terminal transit peptide (TP), an internal catalytic domain, and a C-terminal ACT (aspartokinase–chorismate mutase–TyrA) domain (Cho et al., 2007). Both the catalytic and ACT domains are conserved across plant, bacterial, and fungal ADTs and PDTs, with the catalytic domain decarboxylating/dehydrating prephenate and/or arogenate (Cho et al., 2007; Bross et al., 2011) while the ACT domain is involved in allosteric regulation induced by ligand binding (Tan et al., 2008; Vivan et al., 2008). The N-terminal domain is unique to plant ADTs and is not found in the bacterial or fungal proteins. In A. thaliana ADTs, this domain is ~100–130 amino acids in length, and are likely to be chloroplast TPs according to sequence prediction programs, which is consistent with phenylalanine biosynthesis occurring in chloroplasts (Jung et al., 1986; Cho et al., 2007; Li and Chiu, 2010) and a chloroplastic localization identified for ADTs in protoplasts (Rippert et al., 2009).
Identifying the subcellular localization of proteins can help to define their functional role and has subsequently led to the identification of new unexpected roles (Sparkes and Brandizzi, 2012). This approach can be particularly helpful when dissecting and differentiating the biological roles of members within protein families (Karve et al., 2008). In this study, we provide evidence that most A. thaliana ADTs are targeted to the stroma and stromules (stroma-filled tubules; Köhler and Hanson, 2000) of chloroplasts and we show that this targeting is dependent on the presence of the TP. This subcellular localization is consistent with the enzymatic role of ADTs in phenylalanine biosynthesis and the proposed role of stromules in increasing metabolite transport (Natesan et al., 2005). In addition, we demonstrate that two of the ADTs, ADT2 and ADT5, have additional subcellular localization patterns that suggest novel, non-enzymatic functions.
Materials and methods
Growth conditions for bacteria and plants
Escherichia coli DH5α and DH10β strains (Invitrogen catalog nos 11319019 and 18290015, respectively) were used for the maintenance and amplification of plasmid DNA. Agrobacterium tumefaciens strain LBA4404 containing Ti helper plasmid pAL4404 (NCCB accession no. PC2760; Hoekema et al., 1983; Hellens et al., 2000) was used for transformation of Nicotiana benthamiana and A. thaliana. Agrobacterium tumefaciens strain GV3101 (Koncz and Schell, 1986; Hellens et al., 2000) was used for the transformation of the dominant negative myosin XI-2 (dnMyoXI-2) and dnMyoXI-K/GTD constructs (Avisar et al., 2008). Escherichia coli and A. tumefaciens were grown at 37°C in LB medium, and at 28°C in YEB medium, respectively (Vervliet et al., 1975; Bertani, 2004), with media supplemented with appropriate antibiotics.
Three- to five-week-old N. benthamiana were used for localization studies of A. thaliana ADTs and grown in incubators (Conviron) under 16 h light (80–100 µmol m−2 s−1) and 8 h dark, with light and dark temperatures set to 24°C and 22°C, respectively. Arabidopsis thaliana accession Columbia-0 (Col-0) was grown for 3–4 weeks with the same photoperiod and a light intensity of 150 µmol m−2 s−1. Arabidopsis thaliana plants grown for transient transformations were watered with a 20 mM l-ascorbic acid solution.
The adt2-1D mutation is an ethyl methanesulfonate-induced point mutation that causes a serine to be replaced by an alanine in the ACT regulatory domain, leading to an enzyme that is unable to respond to phenylalanine-mediated allosteric inhibition (Huang et al., 2010).
Cloning of ADT–CFP fusion constructs
Primers were designed (Table 1) to amplify full-length A. thaliana ADT genes (ADT1, At1g11790; ADT2, At3g07630; ADT3, At2g27820; ADT4, At3g44720; ADT5, At5g22630; and ADT6, At1g08250) (Ehlting et al., 2005; Cho et al., 2007) and tested (Lynnon BioSoft, Version 6). Amplified full-length ADT sequences contain the entire ORF, including the coding sequence for TPs, ADT/PDT catalytic and ACT regulatory domains. In addition, ADT2 was cloned as the TP sequence only (Fig. 1B). Primers were designed to include restriction enzyme cleavage and docking sites, to allow for directional integration of PCR fragments into the target vector (Table 1). ADT sequences were amplified with Platinum Taq Polymerase High Fidelity (Invitrogen catalog no. 11304011) with previously cloned ADT sequences as templates (Cho et al., 2007).
Table 1.
List of primer sequences
| Name a | Sequence (5'–3')b | Restriction enzyme recognition sequence |
|---|---|---|
| CFP-For | AT CGGACCGGTCGCCACC ATGGTGAGCAAGG | CpoI |
| CFP-Rev | TCA TCTAGATTACTTGTA CAGCTCGTCC | XbaI |
| CFP-Seq | GATCTGAGCTACACATGC | N/A |
| ADT1-F | AAGCTTATGGCTCTGAGGTGTTTTC | HindIII |
| ADT1-R | GGATCCTGTCTGACTAGATCCATTGG | BamHI |
| ADT2-F | AAGCTTATGGCAATGCACACTGTTCG | HindIII |
| ADT2-S | AAGCTTATGCGTGTTGCGT ATCAGGGAGTACG | HindIII |
| ADT2-R | GGATCCAAGAGCATTGTA GTGTCCACTGG | BamHI |
| ADT2-RTP | GGATCCTTAACGCGGGAGCCATTAG | BamHI |
| ADT3-F | GAATTCATGAGAACTCTCTTACCTTC | EcoRI |
| ADT3-R | GGATCCATCAATGAAAATGTTGATGACG | BamHI |
| ADT4-F | CTCGAGATGCAAGCCGCAACGTCG | XhoI |
| ADT4-R | GGATCCAATGCTTCTTCT GTGGATGTCATGG | BamHI |
| ADT5-F | CTCGAGATGCAAACCATTTCGCC | XhoI |
| ADT5-R | CCCGGGTTACGTCTTCGCTAG | SmaI |
| ADT6-F | GAATTCATGAAAGCTCTATCATC | EcoRI |
| ADT6-R | GGATCCATCGATGAAGTTGATG | BamHI |
a CFP-For/Rev, amplify cerulean cyan fluorescent protein sequence; CFP-Seq, pCB sequencing primer; F, complementary to the 5' end of the full-length ADT coding sequence; S, complementary to the 5' end of the catalytic domain; R, reverse primer complementary to the 3' end of the ADT coding sequence; RTP, reverse primer complementary to the 3' end of the transit peptide.
b Italics, restriction enzyme docking sites; bold, restriction enzyme recognition sequence, underline, nucleotides to maintain frame; dotted underline, pEZT-NL vector sequence; double underline, introduced start or stop codon; unformatted, original, unmodified template sequence.
The T-DNA binary vector pEZT-NL (Carnegie Cell Imaging Project, http://deepgreen.stanford.edu, last accessed 13 February 2017) was used in conjunction with the pAL4404 Ti-helper plasmid for expression in planta. ADT genes expressed from pEZT-NL are under the control of the Cauliflower mosaic virus (CaMV) 35S promoter and are translated as C-terminally tagged enhanced green fluorescent protein (EGFP) fusions. For co-localization studies, pEZT-NL was modified by replacing EGFP, flanked by CpoI and XbaI sites, with the cerulean cyan fluorescent protein (CFP)-coding sequence (Conley et al., 2009b). Both PCR-amplified CFP sequences and the empty pEZT-NL were double-digested with CpoI and XbaI; the resulting fragments were ligated and then transformed into E. coli. Positive transformants were selected on LB medium containing gentamicin. The resulting vector was renamed pCB. Using the appropriate restriction enzymes, ADT genes were cloned into pCB, and all resulting pCB-ADT vectors were sequenced to ensure proper fusion and sequence integrity of the ADT–CFP sequences.
To clone the native ADT5 promoter (proNat5), 1 kb upstream of the ADT5 start codon was PCR amplified with primers that added a 5' MauBI and a 3' XhoI restriction site. The amplified MauBI–XhoI fragment was used to replace the CaMV 35S promoter in pCB-ADT5, generating the vector proNat5::ADT5:CFP.
Cloning filamentous temperature sensitive Z2 (FtsZ2)–yellow fluorescent protein (YFP)
The Gateway-compatible vector pLIC6 encoding FtsZ2-1 cDNA was obtained from the ABRC (stock number DKLAT2G36250; Popescu et al., 2007). Restriction digest of pLIC6 with HindIII yielded a 2329 bp restriction fragment containing FtsZ2-1 flanked by attachmentB (attB) sites. This gel-purified fragment was then recombined into pDONR221 (Hartley et al., 2000). The entry clone created was digested with AseI and the expected 2608 bp fragment encoding FtsZ2-1 flanked by attL sites was isolated and recombined into the destination vector pEarleygate101 (Earley et al., 2006), generating an FtsZ2–YFP fusion construct with expression regulated by the CaMV 35S promoter.
Bacterial transformations
Plasmid DNA was isolated from overnight E. coli cultures using an alkaline lysis method (modified from Ish-Horowicz and Burke, 1981; Sambrook and Russell, 2001) and transformed into electrocompetent E. coli (ElectroMax DH5α, Invitrogen), or electrocompetent A. tumefaciens (Wise et al., 2006), using the Gene Pulser II System (Bio-Rad) set to 2.0 kV, 25 µF capacitance, and 200 Ω or 400 Ω resistance, respectively. Immediately following electroporation, E. coli and A. tumefaciens cells were incubated for 1 h in non-selective LB medium, before plating on selective medium. Correct insertion of amplicons into plasmid DNA of positive E. coli transformants was confirmed by restriction enzyme digestion and sequencing of isolated plasmid DNA. pCB-ADTs were transformed into A. tumefaciens LBA4404 containing Ti-helper plasmid pAL4404 (Hoekema et al., 1983; Hellens et al., 2000).
Organelle markers
To identify stromules, the TP of the small subunit of tobacco RuBisCO fused to YFP (TP-ssRuBisCO–YFP; Nelson et al., 2007) was used in co-localization experiments. This fusion construct is under the control of a CaMV 35S promoter and, after translation, the TP guides the fusion protein to the chloroplast stroma where it can be used to identify stromules (Nelson et al., 2007).
The T-DNA-containing binary vector pEarleygate301-YFP, encoding A. thaliana NUCLEOPORIN1 fused to YFP (NUP1–YFP) is under the expression of its native promoter (Lu et al., 2010) and was used as a nuclear marker.
Agrobacterium tumefaciens GV3101 containing pCB302 encoding the dominant negative form of either N. benthamiana myosin XI-2 or myosin XI-K/GTD was used to inhibit stromule formation (Avisar et al., 2008; Natesan et al., 2009). Each construct encodes the globular tail domain of the corresponding myosin XI. Expression of the dominant negative constructs in pCB302 is regulated in planta by the nopaline synthase promoter (Xiang et al., 1999; Avisar et al., 2008).
Agroinfiltration of tobacco leaves
Nicotiana benthamiana and A. thaliana were transiently transformed by pressure-infiltrating cultures of A. tumefaciens (Wroblewski et al., 2005; Wydro et al., 2006; Conley et al., 2009b). Agrobacterium tumefaciens were grown overnight in 3 ml of YEB medium with the appropriate antibiotics. Then 50 μl of the overnight culture was transferred to 50 ml of YEB containing 25 μl of 200 mM acetosyringone and 500 μl of 1 M MES, and grown until cell density reached an OD600 of 0.5–0.8. Cells were collected by centrifugation, resuspended in Gamborg’s solution to a final OD600 of 1.0, and incubated for an additional hour prior to infiltration. For co-infiltration, equal volumes of A. tumefaciens cultures containing different vectors were combined to maintain a final OD600 of 1.0. The p19 vector encodes a 19 kDa protein from Tomato bushy stunt virus, which has been shown to enhance transgene expression through suppression of post-transcriptional gene silencing (PTGS; Silhavy et al., 2002; Voinnet et al., 2003) and was added to all transient transformations.
A minor variation of this protocol was used for co-expression with TP-ssRuBisCO–YFP as this construct produced a very strong fluorescence signal compared with that of ADT–CFP. Therefore, A. tumefaciens strains containing ADT–CFP and p19 constructs (1:1) were infiltrated 1 d before the infiltration of TP-ssRuBisCO–YFP and p19 (1:1) constructs. In addition, A. tumefaciens containing the TP-ssRuBisCO–YFP construct were infiltrated at a lower OD600 of ~0.5. Hence visualization of subcellular localization was performed 4 days post-infiltration (dpi) with TP-ssRuBisCO–YFP (which equals 5 dpi, with an ADT–CFP).
Transient transformants were assayed 5 dpi using a Leica SP2 confocal laser scanning microscope equipped with a ×63 water immersion objective. The abaxial surface of leaf tissue was viewed to observe the localization pattern of fluorescent proteins in the lower epidermis and in mesophyll cells. CFP and chlorophyll were excited with a blue diode laser (405 nm), and emission was collected from 440 nm to 485 nm and from 630 nm to 690 nm, respectively. YFP was excited with a 514 nm argon laser and its emission was collected from 540 nm to 550 nm. For co-localization experiments, CFP and YFP emissions were collected sequentially to avoid emission crosstalk of the fluorophore pair (Shaner et al., 2005; Conley et al., 2009b). Images were analyzed using Leica Confocal Software (Leica, V2.61) or ImageJ 1.45s (Schneider et al., 2012). Chlorophyll, CFP, and YFP fluorescence was false colored red, cyan, and yellow, respectively.
Western blots
For one protein extract, three leaf disc samples (9 mm) were collected from transiently transformed N. benthamiana plants at 4 dpi. Total soluble protein (TSP) was extracted and quantified as described by Conley et al. (2009a). For each sample, 10 μg of TSP were size separated by 10% SDS–PAGE. The proteins were probed with a primary anti-GFP antibody (Clontech catalog no. 632380; designed to recognize GFP and fluorescent variants including CFP) at a 1:5000 dilution. Subsequently, a secondary goat anti-mouse IgG (H+L) horseradish peroxidase-conjugated antibody (Bio-Rad catalog no. 170-6516) was used at a 1:3000 dilution. CFP fusion proteins were visualized with the enhanced chemiluminescence (ECL) detection kit (GE Healthcare, Mississauga, ON, Canada).
Measurements and statistics
Stromule and chloroplast lengths were determined using the measuring tool from ImageJ 1.45s (Schneider et al., 2012). Chloroplasts were measured in a straight line across their longest axis. Stromules were considered to be any extensions from chloroplasts that were >1 μm in length. Chloroplasts were analyzed for stromules only if they contained detectable TP-ADT2–CFP fluorescence. For non-linear stromules, several linear measurements were taken to account for bends and curves, and subsequently added together to provide a more accurate measurement of stromule length. Nuclear-localized ADT5–CFP was measured as a proportion of total cells exhibiting ADT5–CFP fluorescence. To determine the proportion of cells having ADT5–CFP within the nucleus, cells were analyzed only if ADT5–CFP was present in the cell/chloroplast. The proportion of chloroplasts with stromules, average stromule and chloroplast lengths, and proportion of cells with ADT5 in the nucleus were analyzed using one-way ANOVA (multiple comparisons) on GraphPad Prism 7.0.
Results
ADTs localize to stroma and stromules of chloroplasts
To determine the subcellular localization of ADTs, full-length A. thaliana ADTs were transiently expressed as CFP fusions in N. benthamiana leaves. ADT genes were cloned (Fig. 1B) upstream of the CFP-coding sequence to maintain the fluorescent tag even if the putative TP is cleaved in planta (Li and Chiu, 2010). ADTs were tagged with CFP, instead of GFP, to avoid emission spectra overlap with organelle markers tagged with YFP (Shaner et al., 2005; Nelson et al., 2007; Lu et al., 2010). Leaves of 3- to 5-week-old N. benthamiana leaves were co-transformed with an A. tumefaciens strain harboring an ADT–CFP construct, and a strain encoding p19 to enhance recombinant protein expression (Voinnet et al., 2003). For co-localization experiments, leaves were also co-transformed with A. tumefaciens carrying a plasmid-encoded organelle marker. ADT–CFP expression was confirmed by isolating total protein and performing western blots (Supplementary Fig. S2 at JXB online).
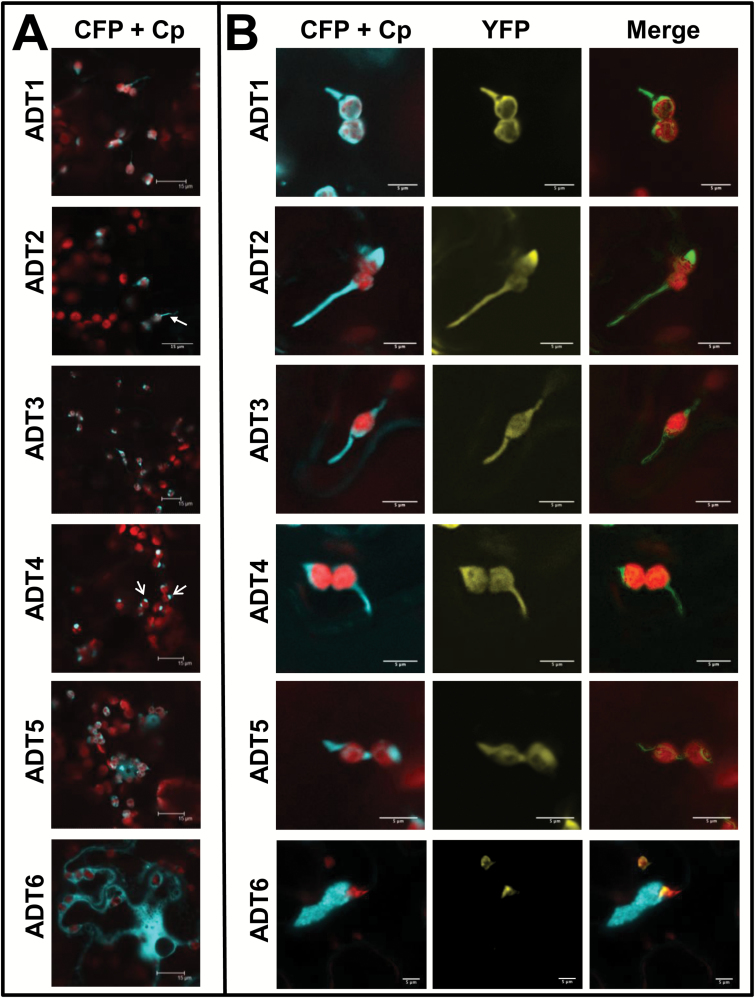
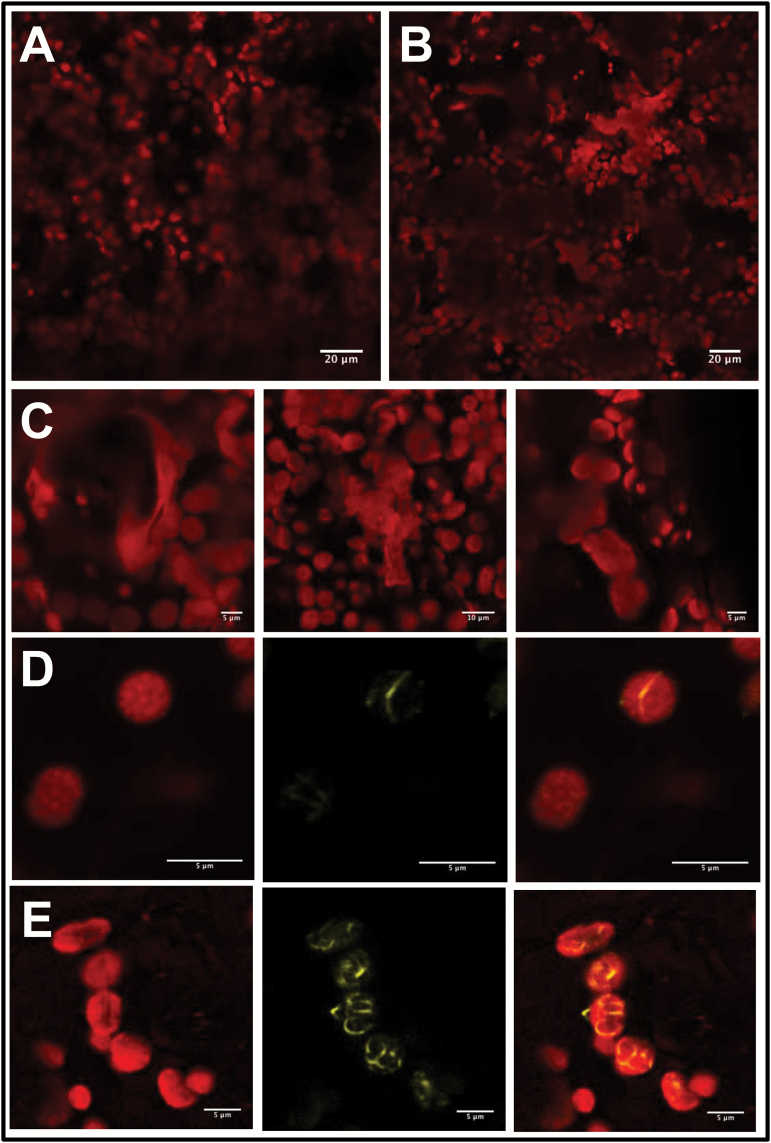
As transformation controls, non-infiltrated tissue, pCB without an insert (empty vector; EV), and pCB infiltrated only with p19 were observed by confocal laser scanning microscopy. For all controls, only chlorophyll autofluorescence was detected and no fluorescence was observed in the CFP and YFP channels in the absence of fusion proteins (Supplementary Fig. S1). ADTs localized to the chloroplast stroma, but were also seen within thread-like structures (e.g. the arrow in ADT2) or as globular structures (e.g. arrows in ADT4) near the chloroplast, but the CFP signal did not directly overlap with chlorophyll autofluorescence (Fig. 2A). The shape and length of these structures were variable, ranging from short and globular to long and narrow protrusions from the chloroplast body. Unlike ADT1–ADT5, ADT6 did not localize to chloroplasts, but was mostly present in the cytosol (Fig. 2A, bottom panel).
Fig. 2.
Subcellular localization of ADT–FP fusion proteins and co-localization with TP-ssRuBisCO–YFP. (A) ADT–CFP subcellular localization patterns. ADT1–ADT5 localized to stroma and to areas seemingly close to the chloroplast just outside of the autofluorescence signal generated by chlorophyll. They often appear either in thread-like structures (e.g. the arrow in ADT2) or globular structures (e.g. the arrows in ADT4). The ADT6–CFP pattern is distinctly different, showing a cytosolic distribution. Images were taken at a lower magnification to allow observation of the CFP signal relative to several chloroplasts. (B) Close-ups of ADT–CFP subcellular localization patterns in relation to TP-ssRuBisCO–YFP. In contrast to the chlorophyll autofluorescence, the TP-ssRuBisCO–YFP is a stroma-specific marker that visualizes all stroma-filled areas within the chloroplast including stromules. ADT1–ADT5 are found within the main body of chloroplast and in stromules, while ADT6 is found within the cytosol and does not co-localize with TP-ssRuBisCO–YFP.
We hypothesized that the thread-like and globular structures were stromules (Köhler and Hanson, 2000). To confirm that ADTs do localize to stromules, the ADT–CFP fusion constructs were co-expressed with the TP of the small subunit of RuBisCO fused to YFP (TP-ssRuBisCO–YFP). This construct is known to localize to the stroma, and therefore can be used to identify stromules (Nelson et al., 2007). The fluorescence of CFP fusion proteins with ADT1–ADT5 overlapped with the fluorescence of TP-ssRuBisCO–YFP, which confirms that these ADTs are targeted to stromules within the chloroplasts (Fig. 2B). The fluorescence of ADT6–CFP did not overlap with the fluorescence of TP-ssRuBisCO–YFP as ADT6 is mostly found within the cytosol (Fig. 2B, bottom panel).
To determine if the TP domain of ADTs is responsible for chloroplast and stromule localization, the TP sequence was expressed as a TP-ADT2–CFP fusion protein and was detected in chloroplasts and stromules (Fig. 3). These data are consistent with the TP being sufficient to target ADT sequences to chloroplasts and stromules.
Fig. 3.
Localization of ADTs to the chloroplast is dependent on the transit peptide sequences. To test if the transit peptide sequences are sufficient for the transport of ADTs to the chloroplast, the first 99 amino acids of ADT2 (TP-ADT2–CFP) were expressed transiently in N. benthamiana leaves.
ADT2 localizes to chloroplasts in a ring structure
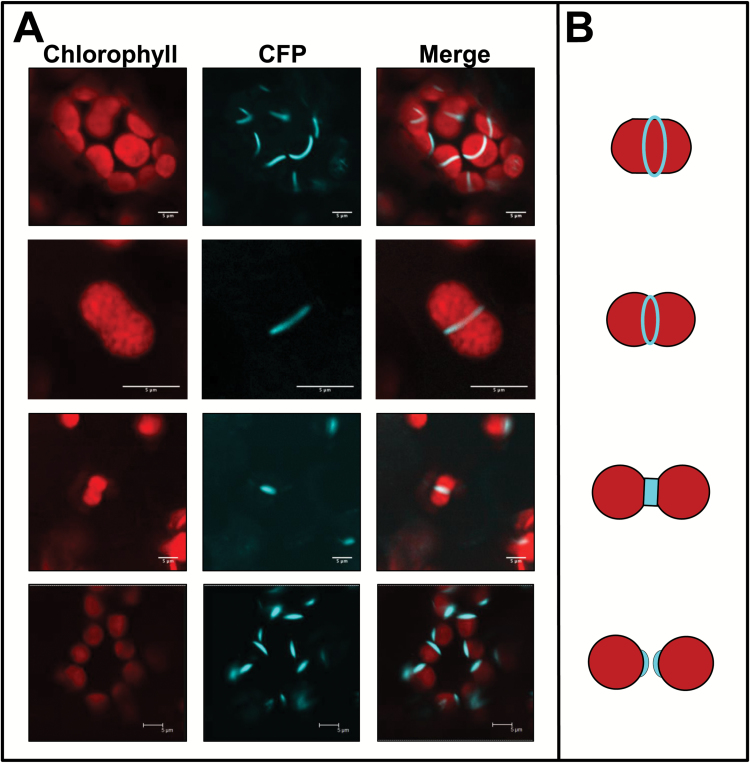
ADT2–CFP displayed a unique localization pattern compared with the other ADTs (Fig. 4A). In chloroplasts with no apparent central constriction, ADT2–CFP localized to a band at the equatorial plane. Stacked confocal images showed that ADT2 formed a ring around the center of the chloroplast (data not shown). In elongated chloroplasts with a slight indentation, suggestive of an early chloroplast division stage, ADT2–CFP localized as a band around the middle of the elongation exactly at the point of indentation. In chloroplasts with a clear indentation, indicative of a later stage of division, ADT2–CFP was found at the site of constriction. ADT2 localization to the poles of chloroplasts was consistent with remnants of the division ring on daughter chloroplasts (Miyagishima, 2011). These ADT2 localization patterns are strikingly similar to those of proteins that are involved in chloroplast division, a process requiring placement of multiple proteinaceous rings followed by constriction that partitions the chloroplast into two equal-sized daughter chloroplasts (Fig. 4B; Miyagishima, 2011).
Fig. 4.
ADT2 forms structures consistent with chloroplast division rings. (A) In addition to being expressed within chloroplasts and stromules, ADT2 was also found to accumulate in places consistent with chloroplast division rings. The top panel shows ADT2 forming rings at the equatorial plate of the chloroplast. On occasion, ADT2 was found in the constriction zone of chloroplasts (two middle panels). In these cases, the chloroplasts have a distinct dumb-bell shape and the degree of indentation depends on how far the division process has proceeded. In addition, ADT2 accumulated in a spindle-like shape that tapers at chloroplast poles (bottom panel). This fusiform ADT2 accumulation was only found at one pole of the chloroplast and is distinct from a stromule pattern shown in Fig. 2. (B) Schematic of chloroplast division stages: from top to bottom, positioning of chloroplast division rings; slightly constricted chloroplast just prior to division; two daughter chloroplast following division. Analogous to the fluorescent images, chloroplasts are shown in red and the position of ring proteins in blue (adapted from Miyagishima, 2011).
To initiate division, chloroplasts have to reach a certain size (Pyke, 1999). Therefore, we argue that chloroplasts with ADT2–CFP at the equatorial plane should be the largest as they are in the process of dividing. Conversely, chloroplasts with ADT2–CFP at their pole should be the smallest as they have just recently divided. In addition, these two classes should have little variation in size as they represent distinct stages in chloroplast development. In contrast, growing chloroplasts should vary in size as they encompass all division stages. While chloroplast volume would be the most accurate way to measure chloroplast size, it is difficult to determine. Therefore, chloroplast size was measured as the length of a chloroplast across its longest axis. Chloroplasts from uninfiltrated N. benthamiana plants were used to determine the average size of a chloroplast because they should contain chloroplasts at many different developmental stages, and thus different sizes. Chloroplast sizes measured in three uninfiltrated plants had an average length of 5.1 μm (Table 2). In chloroplasts with ADT2–CFP present on a pole, the average length of these chloroplasts was significantly shorter (P<0.05), at 4.2 μm (Table 2). Lastly, chloroplasts with ADT2–CFP localized at the equatorial plane were significantly longer, at 6.7 μm (P<0.05; Table 2). As predicted, the SD from the mean was larger for chloroplasts from uninfiltrated plants, consistent with a mixed population of chloroplasts, compared with chloroplasts with ADT2–CFP at a pole or at the equatorial plane (Table 2). These data support the hypothesis that the ADT2 localization patterns we observed are consistent with different chloroplast division stages.
Table 2.
Comparison of chloroplast lengths
| Type of chloroplast | No. of plants | No. of chloroplasts | Length (µm)a | SD |
|---|---|---|---|---|
| Uninfiltrated | 3 | 68 | 5.1 | 1.12 |
| With polar ADT2 | 3 | 75 | 4.2* | 0.60 |
| With equatorial ADT2 ring | 5 | 35 | 6.7* | 0.85 |
a The length of the chloroplast was measured at its longest axis.
* Significantly different from the chloroplasts in the uninfiltrated control (P<0.05) as determined by a t-test.
A single amino acid change in ADT2 affects chloroplast morphology and FtsZ2 localization in A. thaliana
Mutations in genes encoding components of the chloroplast division machinery result in changes to chloroplast morphology (Pyke, 1999). If ADT2 has a role in chloroplast division, it is reasonable to expect that an adt2 mutant will have distorted chloroplasts. Unlike for other ADT genes, no A. thaliana T-DNA insertion line that abolishes ADT2 mRNA production exists (Corea et al., 2012b). However, a point mutation within the coding sequence of the ACT regulatory domain of ADT2 (adt2-1D) has been documented in which conversion of a serine to an alanine prevents allosteric inhibition of the enzyme (Huang et al., 2010). To determine if adt2-1D affects chloroplast morphology, homozygous adt2-1D plants were examined by confocal microscopy and compared with wild-type Col-0 plants of identical age (Fig. 5A, B). Chloroplasts in adt2-1D plants differed greatly in appearance, and were highly heterogeneous in size and shape (Fig. 5C). This contrasted with chloroplasts in wild-type Col-0, which were ovoid in shape and relatively uniform in size. Although many adt2-1D chloroplasts were clearly affected by the mutation, wild-type appearing chloroplasts can still be observed, suggesting a partial loss of ADT2 function.
Fig. 5.
Chloroplast morphology and FtsZ2–YFP localization is affected by a point mutation in ADT2. (A) Chloroplasts in wild-type A. thaliana Col-0. (B) Chloroplasts in adt2-1D A. thaliana mutants. (C) Close-ups of chloroplasts observed in adt2-1D to show the heterogeneity in shape and size. (D) Transiently expressed FtsZ–YFP in wild-type Col-0 localizes as expected to a single ring at the equatorial plane. (E) In contrast, FtsZ2–YFP localizes as long spiralling filaments within adt2-1D chloroplasts. (D, E) Images of chlorophyll fluorescence (left) and FtsZ2–YFP (middle) are shown separately and merged (right).
FtsZ is a tubulin-like protein that is a central component of the chloroplast division apparatus (Vitha et al., 2001; TerBush et al., 2013). There are two FtsZ proteins (FtsZ1 and FtsZ2), which assemble to form the earliest known division ring, the Z-ring within the stroma. As the localization of FtsZ in chloroplast division mutants has been used to provide insight into the function of putative division proteins (Vitha et al., 2003; Glynn et al., 2007; Fujiwara et al., 2008; Glynn et al., 2009; Nakanishi et al., 2009a), we were interested to determine if FtsZ localization was affected in adt2-1D plants. Thus we generated an FtsZ2–YFP fusion construct and expressed it in adt2-1D plants. Expression of FtsZ2–YFP in wild-type Col-0 leaves led to the formation of the expected single ring (Fig. 5D). In contrast, FtsZ2–YFP in adt2-1D plants was less organized and formed what appeared to be spirals or multiple rings (Fig. 5E). These results demonstrate that deregulation of ADT2 abolishes proper placement of FtsZ2, further supporting an involvement of ADT2 in chloroplast division.
ADT5 is found in the nucleus
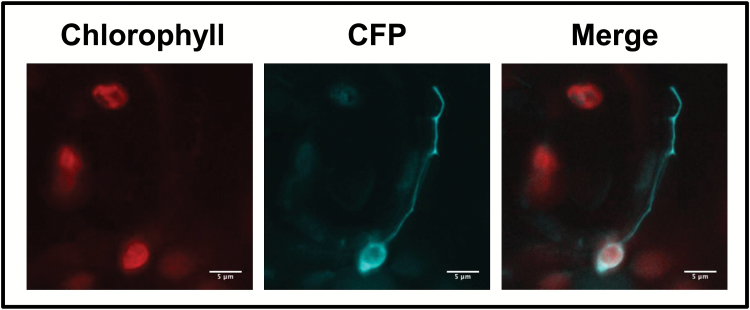
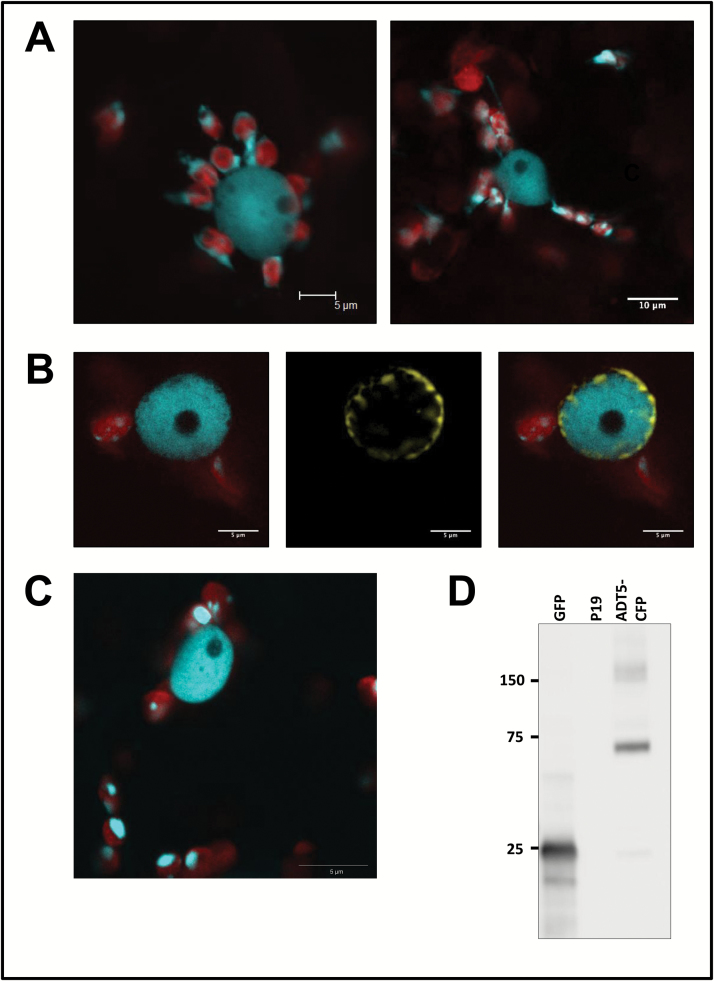
In addition to its chloroplast localization, only ADT5–CFP was also detected in nuclei at ~4–5 dpi (Figs 2A, 6A). To confirm this finding, ADT5–CFP was co-infiltrated with a YFP fusion to the nuclear marker NUP1, a component of the nuclear pore complex in A. thaliana that was previously shown to localize to the nuclear membrane (Lu et al., 2010). Confocal imaging determined that NUP1–YFP localized around ADT5–CFP (Fig. 6B). As NUP1–YFP localizes to the nuclear membrane, this result confirms that ADT5–CFP is contained within the nucleus and localizes uniformly throughout the nucleoplasm. As these results were obtained with constructs using a CaMV 35S promoter, we repeated the experiment and expressed ADT5–CFP under control of its native promoter (Fig. 6C) and confirmed the nuclear localization pattern. To ensure that the observed nuclear localization is not due to a smaller diffusible cleavage product or to a particularly high level of ADT5 compared with other ADTs, we performed western blots (Fig. 6D; Supplementary Fig. S2). The blot containing all ADTs expressed under the control of the CaMV 35S promoter shows a low level of cleavage in all lanes. However, more cleavage product is seen for ADT1 and ADT3 compared with ADT5, while no nuclear localization is observed with either ADT1 or ADT3. The blot showing ADT5–CFP under the control of its native promoter (Fig. 6D) shows a shadow band of the size of CFP, and yet a very clear nuclear localization is evident (Fig. 6C). These results indicate that the nuclear localization pattern is a bona fide ADT5 localization. There are additional bands of higher molecular weight (Fig. 5D; Supplementary Fig. S2) that might correspond to ADT dimers or even higher multimers.
Fig. 6.
ADT5 is found in the nucleus. ADT5–CFP proteins are unique as they are the only full-length ADT proteins that were found in the nucleus. (A) Nuclei show a close association with chloroplasts (left) or with stromules of chloroplasts (right). Both images show ADT5–CFP within nuclei. (B) Co-localization of ADT5–CFP with NUP1–YFP. To determine if ADT5–CFP localizes to the nucleus, it was co-expressed with NUP1–YFP in N. benthamiana. Images of chlorophyll fluorescence and ADT5–CFP are shown merged (left). NUP1–YFP is shown alone (middle) and merged with ADT5–CFP and chlorophyll fluorescence (right). NUP1–YFP localizes to the nuclear membrane and surrounds ADT5–CFP, confirming that it localizes to the nucleus. (C) ADT5–CFP transiently expressed with its native ADT5 promoter also localizes to the nucleus. (D) Western blot of ADT5–CFP (calculated size 73.9 kDa) expressed with its native promoter and visualized with a GFP antibody is detected at its expected size. As negative controls, proteins isolated from leaves transformed with GFP (25 kDa) and p19 are shown. Total soluble protein was isolated from transiently transformed leaves, and 10 μg of total soluble protein was size separated by 10% SDS–PAGE. Sizes of the protein ladder are given in kDa.
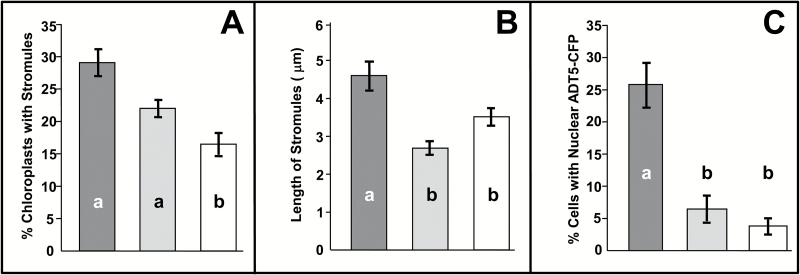
Furthermore, we often observed ADT5–CFP-containing nuclei surrounded by chloroplasts that appeared to be connected to the nucleus through stromules (Fig. 6A), suggesting that ADT5 nuclear localization depends on stromule-mediated transport. To test if stromule formation affects the nuclear localization of ADT5, we transiently expressed myosin XI tail domains (dnMyoXI-2 and dnMyoXI-K/GTD) as they were previously shown to inhibit stromule formation through a dominant negative effect on wild-type myosin XI (Avisar et al., 2008; Natesan et al., 2009). To ensure that these treatments inhibit stromules, several control transformations were performed. To visualize stromules in control transformations, TP-ADT2–CFP was used, as it easily visualizes stromules and therefore provides a very sensitive marker to detect stromule inhibition. TP-ADT2–CFP was co-infiltrated into N. benthamiana leaves with an empty pCB vector, a dnMyoXI-2 construct, or a dnMyoXI-K/GTD construct (Avisar et al., 2008), and the number of chloroplasts having stromules (Fig. 7A) and the length of stromules formed (Fig. 7B) were determined. In plants transformed with an empty vector, 28.7% of chloroplasts had stromules (Fig. 7A) and the average length of these was 4.6 μm (Fig. 7B). Infiltration with dnMyoXI-2 decreased the percentage of chloroplasts with stromules (22.2%; Fig. 7A) compared with that of the control, and significantly reduced (P<0.001) the average length of stromules to 2.7 μm (Fig. 7B). Treatment with dnMyoXI-K/GTD also caused a significant decrease (P<0.001) in the percentage of chloroplasts with stromules (17.3%; Fig. 7A) and significantly decreased (P<0.001) the average length of stromules to 3.5 μm (Fig. 7B). These results confirm that both myosin domains affect stromule formation.
Fig. 7.
The presence of ADT5 in the nucleus is affected by the ability to form stromules. To determine if nuclear localization of ADT5 is dependent on stromules, plants were co-infiltrated with TP-ADT2–CFP (A and B) as a control or ADT5–CFP (C) and an empty vector (dark gray), dominant negative myosin XI-2 (dnMyoXI-2; light gray) and myosin XI-K (dnMyoXI-K/GTD; white), respectively. (A) Percentage of chloroplasts having stromules. Chloroplasts were analyzed if they contained any visible TP-ADT2–CFP fluorescence and were determined to have a stromule if the projection was longer than 1 μm. In total 554, 395, and 579 chloroplasts were analyzed from plants transformed with an empty vector, dnMyoXI-2, and dnMyoXI-K/GTD, respectively. (B) Average length of stromules. A total of 166, 93, and 91 stromules were measured from plants transformed with an empty vector, dnMyoXI-2, and dnMyoXI-K/GTD, respectively. (C) Nuclear localization of ADT5–CFP. Cells were analyzed for CFP fluorescence in the nucleus only if any ADT5–CFP fluorescence was detectable. A total of 131, 190, and 358 cells were analyzed from plants transformed with an empty vector, dnMyoXI-2, and dnMyoXI-K/GTD, respectively. Each experiment was performed on three independent occasions. Significant differences (P<0.001) as determined by a one-way ANOVA (multiple comparisons) are indicated by different letters. Averages ± SE of the mean are plotted.
To determine if the ability to form stromules affects nuclear localization of ADT5, ADT5–CFP was co-expressed with the empty pCB vector, dnMyoXI-2, or dnMyoXI-K/GTD (Fig. 7C). The extent of ADT5–CFP nuclear localization was expressed as a percentage of cells containing CFP fluorescence. Co-infiltration with the empty vector control showed that 25.9% of cells had ADT5–CFP fluorescence visible in the nucleus (Fig. 7C). In contrast, co-infiltration with dnMyoXI-2 or dnMyoXI-K/GTD showed that ADT5–CFP was detected in the nucleus in only 7.1% and 4.3% of cells, respectively (Fig. 7C), both significant reductions (P<0.001) from the control. These data demonstrate that ADT5–CFP nuclear localization is decreased by the same conditions shown to decrease stromule formation.
ADTs in A. thaliana
Transient transformations using agroinfiltration are widely used in N. benthamiana but have traditionally been difficult in A. thaliana (Wroblewski et al., 2005). We were able to transform A. thaliana reliably by growing plants with 20 mM l-ascorbic acid, which seemed to decrease necrosis of leaves associated with agroinfiltration. As it was not the focus of this study, the reason for this was not addressed. However, as l-ascorbic acid can scavenge damaging reactive oxygen species (Gallie, 2013), the increased tolerance of A. thaliana to agroinfiltration may be due to a decrease in oxidative stress.
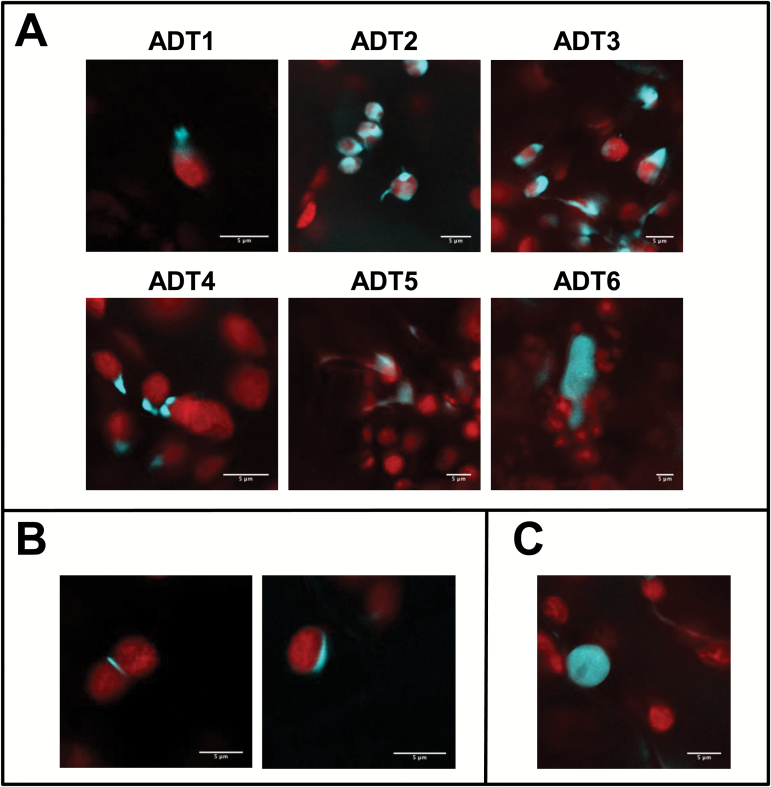
ADT–CFP fusion genes were transiently expressed in A. thaliana to confirm that localization in a heterologous host reflected the situation in the native environment (Fig. 8). As in N. benthamiana, all ADT–CFPs in A. thaliana, with the exception of ADT6–CFP, which appeared in the cytosol, exhibited stroma and stromule-like patterns (Fig. 8A). Similarly, the unique localization patterns of ADT2–CFP and ADT5–CFP were observed in A. thaliana, localizing to the equatorial plane and poles of chloroplasts (Fig. 8B), and to nuclei (Fig. 8C), respectively. This finding was important as it verifies that the findings in N. benthamiana are not artifacts.
Fig. 8.
ADT localization to the stroma and stromules, the chloroplast equatorial plane, and the nucleus can also be detected in A. thaliana. To test if the ADT patterns determined in N. benthamiana reflect expression in A. thaliana, all six ADT–CFP fusion proteins were transiently expressed in A. thaliana Col-0. All images show a merge of chlorophyll and CFP fluorescence. (A) ADT1–CFP through ADT5–CFP localize to stroma and structures resembling stromules of varying shapes and lengths, with varying levels of fluorescence in the stroma. ADT6–CFP localizes outside of chloroplasts in the cytosol. (B) Chloroplast division patterns for ADT2–CFP. (C) Nuclear localization of ADT5–CFP.
Discussion
Phenylalanine biosynthesis and stromules
ADTs were seen to localize to thread-like structures seemingly outside of the region of chlorophyll autofluorescence (Fig. 2A). Co-localization of ADT–CFP fluorescence with the stromule marker TP-ssRuBisCO–YFP (Nelson et al., 2007) confirmed that all ADTs except ADT6 localized to stromules (Fig. 2B). Prior to our study, A. thaliana ADTs were found uniformly throughout the stroma of chloroplasts, with no indication of stromule localization (Rippert et al., 2009). However, the said study used protoplasts, which represent dedifferentiated cells that are also in a state of stress (Genschik et al., 1992; Reyes et al., 2010).
Stromules are dynamic structures that range in size and shape from short beak-like projections to long and elaborate tubules (Köhler and Hanson, 2000; Gunning, 2005; Gray, 2013). The function of stromules has been the subject of debate, but the idea that they increase transport of compounds synthesized within plastids to other areas of the cell is now generally accepted (Hanson and Sattarzadeh, 2013). The presence of biosynthetic enzymes, such as ADTs, in stromules is consistent with this hypothesis. Phenylalanine, the product of ADT enzymatic activity, is required within the cytosol for the synthesis of proteins and as a precursor for phenylpropanoids such as lignins and flavonoids (Fraser and Chapple, 2011). It stands to reason that stromules could have a high concentration of phenylalanine as a result of ADT activity, providing an effective means of increasing phenylalanine export into the cytosol. Interestingly, abiotic stressors, such as drought and salt stress, known to induce stromules (Gray et al., 2012), are also associated with increased flavonoid levels in leaves (Agati et al., 2011; Mewis et al., 2012), and genes encoding flavonoid biosynthetic enzymes are up-regulated in response to salinity-induced stress (Walia et al., 2005). For example, PHENYLALANINE AMMONIA LYASE1 (PAL1) uses phenylalanine as a substrate to catalyze the first step of phenylpropanoid biosynthesis in the cytosol (Walia et al., 2005; Fraser and Chapple, 2011). Under conditions of salt stress, PAL1 up-regulation coincides with the formation of stromules, suggesting that these two events might be linked and that both events contribute to an increased transport of phenylalanine to the cytosol.
ADTs are not the only enzymes that have been associated with stromules. Other examples include geranylgeranyl diphosphate synthase (GGPS), which synthesizes geranylgeranyl diphosphate in the stromules of chloroplasts, and the compound is required in the cytosol as part of isoprenoid metabolism (Thabet et al., 2012), or RuBisCO and an aspartate aminotransferase (ASP5), which are present in stromules and capable of moving between plastids via stromules when expressed as GFP fusion proteins (Kwok and Hanson, 2004). This allows for the speculation that chloroplastic enzymes that synthesize molecules required in the cytosol preferentially localize to stromules as these might facilitate metabolite export.
In silico analysis of A. thaliana ADT sequences using ChloroP (Emanuelsson et al., 1999) predicted that their N-terminal sequences were likely to encode TPs directing the enzymes to the chloroplast. We present data that confirm the in silico prediction for ADT2, as the N-terminal portion was necessary and sufficient to allow direct import into chloroplasts and specifically stromules (Fig. 3A, B). These data also corroborate observations made for all three petunia ADTs, where the N-terminal portion of ADTs directed GFP fusion proteins to chloroplasts (Maeda et al., 2010).
ADT2 and a role in chloroplast division
Aside from its inclusion in stroma and stromules, ADT2 localized as a ring around the equatorial plane or at the poles of chloroplasts (Figs 4A, 8B). The similarity of these patterns to those of chloroplast division proteins (Miyagishima, 2011) during and after division led to an investigation of a possible second, non-enzymatic role for ADT2 in chloroplast division. Since chloroplast division is regulated by size (Pyke, 1999), we reasoned that chloroplasts with either the equatorial or polar localization patterns would be larger and smaller, respectively, than average sized chloroplasts. This was confirmed upon comparing chloroplast lengths across their longest axis (Table 2). Additionally, the SD of chloroplast lengths varied. It was lowest in chloroplasts with ADT2 at the equator or at a pole, in agreement with these chloroplasts being in very distinct phases, either just prior to or post-division, and therefore very similar in size. This was in contrast to chloroplasts from uninfiltrated plants, which are comprised of chloroplasts in all division states. Therefore, our results suggest that ADT2 localizes to the division plane early in the division process and remains there throughout the duration of constriction and separation into daughter organelles. Similarly, known division proteins such as FtsZ and ACCUMULATION AND REPLICATION OF CHLOROPLASTS 6 (ARC6) assemble at the equatorial plane in the process leading to constriction and division (Vitha et al., 2001; 2003).
The striking similarities between ADT2 and other chloroplast division proteins prompted observation of chloroplasts in adt2 mutant plants. Interestingly, no T-DNA insertion knockout lines that abolish ADT2 mRNA are available (Corea et al., 2012b). This makes ADT2 unique and raises the possibility that an adt2 knockout is lethal. However, plants homozygous for the adt2-1D point mutation have been documented (Huang et al., 2010). The appearance of adt2-1D chloroplasts was variable and the presence of misshapen and heterogeneous chloroplasts is consistent with previous descriptions of chloroplast morphology in division mutants (Pyke and Leech, 1992; Colletti et al., 2000; Glynn et al., 2009; Nakanishi et al., 2009b). In the adt2-1D plants, chloroplasts that appear wild type can still be observed. This infers that the single amino acid substitution probably does not abolish ADT2’s function, but impairs it. During chloroplast division, FtsZ forms the first known division ring within the stroma (TerBush et al., 2013). Given the central role that FtsZ proteins play in division, we reasoned that FtsZ localization should be affected in adt2-1D chloroplasts if ADT2 is a chloroplast division protein. Expression of an FtsZ2–YFP fusion construct in adt2-1D plants revealed long and spiraling FtsZ2–YFP filaments throughout the chloroplast stroma. Although FtsZ2–YFP was overexpressed in our study, in wild-type Col-0 chloroplasts the fusion protein localized as expected as a single equatorial ring. We propose that the abnormal appearance of FtsZ2–YFP within adt2-1D chloroplasts suggests that ADT2 regulates FtsZ positioning. However, we cannot ignore the possibility that elevated phenylalanine levels in adt2-1D (up to 160-fold compared with the wild type; Huang et al., 2010) are at least in part responsible for the observed changes.
It is intriguing to note that not all ring proteins have been identified. Although FtsZ, one of the inner rings, and ARC5 (dynamin), one of the outer rings, have been known for some time (Vitha et al., 2001; Gao et al., 2003), the identity of the outer plastid-dividing (PD) ring (Yoshida et al., 2010), polyglucan filaments, has just recently been suggested, and the composition of the inner PD ring is still unknown. The realization that enzymes can have a second unrelated, non-enzymatic function, even as part of cellular structural components, is discovered more and more frequently (Huberts and van der Klei, 2010; MoonProt Database: www.moonlightingproteins.org, last accessed 13 February 2017). A good example of this is the Physcomitrella patens enzyme presenilin, the catalytic unit for γ-secretase, which has an independent function in the cytoskeletal network (Khandelwal et al., 2007). However, additional studies will be required to determine the precise relationship between ADT2, phenylalanine levels, chloroplast division, and other components of the chloroplast division machinery.
ADT5 and the nucleus
Similar to ADT2, ADT5 has an additional unique localization pattern and was clearly observed in the nuclei of both N. benthamiana and A. thaliana (Fig. 6A, C). Proteins with dual plastid and nuclear localization may be significant in the context of retrograde signaling. While retrograde signaling traditionally refers to chemical messengers that are released from plastids and affect nuclear gene expression (Inaba et al., 2011), it is becoming apparent that proteins within the chloroplast can also act as retrograde signals (Isemer et al., 2012; Krause et al., 2012). One such protein is WHIRLY1 from A. thaliana, which can move directly from plastids to the nucleus (Isemer et al., 2012). In plastids, WHIRLY1 contributes to plastid genome stability by preventing illegitimate recombination (Maréchal et al., 2009). In the nucleus, it acts as a transcriptional activator of pathogen response genes (Isemer et al., 2012), consistent with the increased pathogen susceptibility associated with decreased WHIRLY1 DNA binding ability (Desveaux et al., 2005). Whether ADT5 has a role in retrograde signaling is currently unknown. In addition, many other enzymes with diverse functions have been reported in both plastids and nuclei, such as phosphate-isopentyltransferase 3, an enzyme involved in cytokinin biosynthesis (Galichet et al., 2008; Krause et al., 2012), CDT1, a kinase required in cell cycle regulation (Raynaud et al., 2005), and a dihydrofolate reductase required for nucleotide metabolism (Luo et al., 1997), but often the nuclear role of these enzymes is ill defined.
While a direct mechanism of protein transport between plastids and the nucleus through stromules is a hypothetical mode (Krause et al., 2012), they have been shown to interconnect plastids (Köhler et al., 1997; Kwok and Hanson, 2004; Hanson and Sattarzadeh, 2013). Chloroplasts with stromules containing ADT5–CFP often appeared to connect directly with the nucleus (Fig. 6A). Expression of dominant negative forms of myosin XI were found to inhibit stromules (Fig. 7A, B) and also significantly reduce ADT5–CFP localization to the nucleus (Fig. 7C), providing indirect evidence of stromule-mediated nuclear transport. We are aware that there are alternative interpretations of these results, as myosin XI is involved in other processes including movement of organelles (Avisar et al., 2008) and cytoplasmic streaming (Shimmen and Yokota, 2004). Regardless of this, the appearance of stromules directly connecting to nuclei (Fig. 6A) makes the possibility of a stromule-mediated nuclear transport system intriguing.
Currently, the role that ADT5 plays in the nucleus is unknown. It is conceivable that it acts as a transcriptional regulator of ADT genes or other genes within the same biosynthetic pathway. There is precedence for an enzyme to act as a transcriptional regulator of functionally related genes. For example, A. thaliana HEXOKINASE1 (HXK1) is involved in glucose metabolism in mitochondria, but also localizes to the nucleus where it forms part of a protein complex affecting transcription of genes involved in glucose signaling (Cho et al., 2006). Furthermore, in the budding yeast Saccharomyces cerevisiae, ARG5,6, an enzyme involved in arginine biosynthesis, is able to bind DNA directly and regulate gene expression (Hall et al., 2004). Interestingly, mutant analysis has shown that loss of ADT5 activity cannot be compensated for by other ADTs and is the only single ADT knockout with a visible phenotype (Corea et al., 2012b), consistent with a unique nuclear role for ADT5.
ADT2 and ADT5: moonlighting proteins
The term ‘moonlighting protein’ was coined to describe proteins that perform multiple autonomous and often unrelated functions without these functions being partitioned into different domains of the protein or resulting from alternative splicing or gene fusion (reviewed in Jeffery, 1999, 2013). Since the description of the first moonlighting proteins, the number of documented moonlighting proteins has increased to ~300 (Jeffery, 2013; MoonProt Database: www.moonlightingproteins.org). Enzymes are common among moonlighting proteins and many have additional non-enzymatic functions, including roles as structural components and as regulators of transcription or translation (Jeffery, 2013). For example, in Tetrahymena, a citrate synthase acts as an enzyme in mitochondria while in the cytosol it can polymerize to form 14 nm filaments and then act as a cytoskeletal protein (Kojima et al., 1997). We propose that ADT2 and ADT5 are moonlighting proteins. In the current study we provide evidence that ADT2, with demonstrated arogenate dehydratase activity (Cho et al., 2007), can form rings around chloroplasts similar to FtsZ or ARC5 as part of their role in chloroplast division (Vitha et al., 2001; Gao et al., 2003). The dual localization of ADT5 to chloroplasts and nuclei suggests that ADT5 has an additional role in the nucleus, possibly in transcriptional regulation. As the entire ADT enzyme family catalyzes the final step in phenylalanine biosynthesis, we expect this to be a key regulatory step.
Moonlighting proteins appear to be ubiquitous in nature, with documented examples in simple single-cell organisms, such as archaea and bacteria, and complex eukaryotes including plants and animals. It seems likely that moonlighting proteins are evolutionarily advantageous, ensuring that the number of genes in a genome does not limit the number of functions they are capable of performing. Many moonlighting proteins are ancient in terms of their evolutionary history, giving them ample time to adapt to a second role (Jeffery, 2013). Analyzing the subcellular localization of A. thaliana ADTs shows that the function of enzymes is far more complex than previously realized.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Negative controls.
Fig. S2. Western blots showing expression of transiently expressed ADTs.
Supplementary Material
Acknowledgments
We would like to thank Lisa Amyot and Reza Saberianfar for their technical support when using the confocal microscope, and Drs Gary Tian and Yuhai Cui, for the donation of the NUCLEOPORIN1–YFP nuclear marker. We are grateful to Agriculture and Agri-Food Canada (AAFC) in London for providing generous access to their confocal microscope. Nicotiana benthamiana seeds and Agrobacterium strains were provided by Jamie McNeil and Dr R. Menassa, AAFC, London, Ontario. Many thanks to Dr Tengfang Huang and Dr Georg Jander (Boyce Thompson Institute for Plant Research (Ithaca, NY) for generously providing us with adt2-1D seeds. We would like to express our thanks to Drs R. Menassa and K. Dobinson, AAFC, London, Ontario, Emily Clayton, and Emily Cornelius for diligently proofreading the manuscript. This work was supported in part by a grant from MITACS to CDB and SEK, and an NSERC Discovery grant to SEK.
Glossary
Abbreviations:
- ACT
aspartokinase–chorismate mutase–TyrA
- ADT
arogenate dehydratase
- CFP
cyan fluorescent protein
- dpi
days post-infiltration
- EV
empty vector
- GFP
green fluorescent protein
- FtsZ
filamentous temperature sensitive Z
- PAT
prephenate aminotransferase
- PDT
prephenate dehydratase
- PP
phenylpyruvate
- PPAT
phenylpyruvate aminotransferase
- PTGS
post-transcriptional gene silencing
- TP
transit peptide
- YFP
yellow fluorescent protein.
References
- Agati G, Biricolti S, Guidi L, Ferrini F, Fini A, Tattini M. 2011. The biosynthesis of flavonoids is enhanced similarly by UV radiation and root zone salinity in L. vulgare leaves. Journal of Plant Physiology 168, 204–212. [DOI] [PubMed] [Google Scholar]
- Avisar D, Prokhnevsky AI, Makarova KS, Koonin EV, Dolja VV. 2008. Myosin XI-K is required for rapid trafficking of Golgi stacks, peroxisomes, and mitochondria in leaf cells of Nicotiana benthamiana. Plant Physiology 146, 1098–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertani G. 2004. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. Journal of Bacteriology 186, 595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bross CD, Corea ORA, Kaldis A, Menassa R, Bernards MA, Kohalmi SE. 2011. Complementation of the pha2 yeast mutant suggests functional differences for arogenate dehydratases from Arabidopsis thaliana. Plant Physiology and Biochemistry 49, 882–890. [DOI] [PubMed] [Google Scholar]
- Cho MH, Corea ORA, Yang H, et al. 2007. Phenylalanine biosynthesis in Arabidopsis thaliana. Identification and characterization of arogenate dehydratases. Journal of Biological Chemistry 282, 30827–30835. [DOI] [PubMed] [Google Scholar]
- Cho YH, Yoo SD, Sheen J. 2006. Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell 127, 579–589. [DOI] [PubMed] [Google Scholar]
- Colletti KS, Tattersall EA, Pyke KA, Froelich JE, Stokes KD, Osteryoung KW. 2000. A homologue of the bacterial cell division site-determining factor MinD mediates placement of the chloroplast division apparatus. Current Biology 10, 507–516. [DOI] [PubMed] [Google Scholar]
- Conley AJ, Joensuu JJ, Jevnikar AM, Menassa R, Brandle JE. 2009a . Optimization of elastin-like polypeptide fusions for expression and purification of recombinant proteins in plants. Biotechnology and Bioengineering 103, 562–573. [DOI] [PubMed] [Google Scholar]
- Conley AJ, Joensuu JJ, Menassa R, Brandle JE. 2009b . Induction of protein body formation in plant leaves by elastin-like polypeptide fusions. BMC Biology 7, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corea ORA, Bedgar DL, Davin LB, Lewis NG. 2012a . The arogenate dehydratase gene family: towards understanding differential regulation of carbon flux through phenylalanine into primary versus secondary metabolic pathways. Phytochemistry 82, 22–37. [DOI] [PubMed] [Google Scholar]
- Corea ORA, Ki C, Cardenas CL, Kim SJ, Brewer SE, Patten AM, Davin LB, Lewis NG. 2012b . Arogenate dehydratase isoenzymes profoundly and differentially modulate carbon flux into lignins. Journal of Biological Chemistry 287, 11446–11459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desveaux D, Maréchal A, Brisson N. 2005. Whirly transcription factors: defense gene regulation and beyond. Trends in Plant Science 10, 95–102. [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. 2006. Gateway-compatible vectors for plant functional genomics and proteomics. Plant Journal 45, 616–629. [DOI] [PubMed] [Google Scholar]
- Ehlting J, Mattheus N, Aeschliman DS, et al. 2005. Global transcript profiling of primary stems from Arabidopsis thaliana identifies candidate genes for missing links in lignin biosynthesis and transcriptional regulators of fiber differentiation. The Plant Journal 42, 618–640. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G. 1999. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Science 8, 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CM, Chapple C. 2011. The phenylpropanoid pathway in Arabidopsis. Arabidopsis Book 9, e0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara MT, Hashimoto H, Kazama Y, Abe T, Yoshida S, Sato N, Itoh RD. 2008. The assembly of the FtsZ ring at the mid-chloroplast division site depends on a balance between the activities of AtMinE1 and ARC11/AtMinD1. Plant and Cell Physiology 49, 345–361. [DOI] [PubMed] [Google Scholar]
- Galichet A, Hoyerová K, Kamínek M, Gruissem W. 2008. Farnesylation directs AtIPT3 subcellular localization and modulates cytokinin biosynthesis in Arabidopsis. Plant Physiology 146, 1155–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR. 2013. The role ofl-ascorbic acid recycling in responding to environmental stress and in promoting plant growth. Journal of Experimental Botany 64, 433–443. [DOI] [PubMed] [Google Scholar]
- Gao H, Kadirjan-Kalbach D, Froehlicht JE, Osteryoung KW. 2003. ARC5, a cytosolic dynamin-like protein from plants, is part of the chloroplast division machinery. Proceedings of the National Academy of Sciences, USA 100, 4328–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschik P, Criqui MC, Parmentier Y, Marbach J, Durr A, Fleck J, Jamet E. 1992. Isolation and characterization of a cDNA encoding a 3-hydroxy-3-methylglutaryl coenzyme A reductase from Nicotiana sylvestris. Plant Molecular Biology 20, 337–341. [DOI] [PubMed] [Google Scholar]
- Glynn JM, Miyagishima SY, Yoder DW, Osteryoung KW, Vitha S. 2007. Chloroplast division. Traffic 8, 451–461. [DOI] [PubMed] [Google Scholar]
- Glynn JM, Yang Y, Vitha S, Schmitz AJ, Hemmes M, Miyagishima SY, Osteryoung KW. 2009. PARC6, a novel chloroplast division factor, influences FtsZ assembly and is required for recruitment of PDV1 during chloroplast division in Arabidopsis. The Plant Journal 59, 700–711. [DOI] [PubMed] [Google Scholar]
- Gray JC. 2013. Stromule formation. In: Biswal B, Krupinska K, Biswal UC, eds. Plastid development in leaves during growth and senescence. Dordrecht, The Netherlands: Springer, 169–186. [Google Scholar]
- Gray JC, Hansen MR, Shaw DJ, Graham K, Dale R, Smallman P, Natesan SK, Newell CA. 2012. Plastid stromules are induced by stress treatments acting through abscisic acid. The Plant Journal 69, 387–398. [DOI] [PubMed] [Google Scholar]
- Gunning BE. 2005. Plastid stromules: video microscopy of their outgrowth, retraction, tensioning, anchoring, branching, bridging, and tip-shedding. Protoplasma 225, 33–42. [DOI] [PubMed] [Google Scholar]
- Hall DA, Zhu H, Zhu X, Royce T, Gerstein M, Snyder M. 2004. Regulation of gene expression by a metabolic enzyme. Science 306, 482–484. [DOI] [PubMed] [Google Scholar]
- Hanson MR, Sattarzadeh A. 2013. Trafficking of proteins through plastid stromules. The Plant Cell 25, 2774–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley JL, Temple GF, Brasch MA. 2000. DNA cloning using in vitro site-specific recombination. Genome Research 10, 1788–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hébrard C, Trap-Gentil MV, Lafon-Placette C, Delaunay A, Joseph C, Lefèbvre M, Barnes S, Maury S. 2013. Identification of differentially methylated regions during vernalization revealed a role for RNA methyltransferases in bolting. Journal of Experimental Botany 64, 651–663. [DOI] [PubMed] [Google Scholar]
- Hellens R, Mullineaux P, Klee H. 2000. Technical Focus: a guide to Agrobacterium binary Ti vectors. Trends in Plant Science 5, 446–451. [DOI] [PubMed] [Google Scholar]
- Herrmann KM, Weaver LM. 1999. The shikimate pathway. Annual Review of Plant Physiology and Plant Molecular Biology 50, 473–503. [DOI] [PubMed] [Google Scholar]
- Hoekema A, Hirsch PR, Hooykaas PJJ, Schilperoort RA. 1983. A binary plant vector strategy based on separation of vir- and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature 303, 179–180. [Google Scholar]
- Huang T, Tohge T, Lytovchenko A, Fernie AR, Jander G. 2010. Pleiotropic physiological consequences of feedback-insensitive phenylalanine biosynthesis in Arabidopsis thaliana. The Plant Journal 63, 823–835. [DOI] [PubMed] [Google Scholar]
- Huberts DH, van der Klei IJ. 2010. Moonlighting proteins: an intriguing mode of multitasking. Biochimica et Biophysica Acta 1803, 520–525. [DOI] [PubMed] [Google Scholar]
- Inaba T, Yazu F, Ito-Inaba Y, Kakizaki T, Nakayama K. 2011. Retrograde signaling pathway from plastid to nucleus. International Review of Cell and Molecular Biology 290, 167–204. [DOI] [PubMed] [Google Scholar]
- Isemer R, Mulisch M, Schäfer A, Kirchner S, Koop HU, Krupinska K. 2012. Recombinant Whirly1 translocates from transplastomic chloroplasts to the nucleus. FEBS Letters 586, 85–88. [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D, Burke JF. 1981. Rapid and efficient cosmid cloning. Nucleic Acids Research 9, 2989–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery CJ. 1999. Moonlighting proteins. Trends in Biochemical Sciences 24, 8–11. [DOI] [PubMed] [Google Scholar]
- Jeffery CJ. 2013. New ideas on protein moonlighting. In: Henderson B, ed. Moonlighting cell stress proteins in microbial infections. Dordrecht, The Netherlands: Springer, 51–66. [Google Scholar]
- Jung E, Zamir LO, Jensen RA. 1986. Chloroplasts of higher plants synthesizel-phenylalanine vial-arogenate. Proceedings of the National Academy of Sciences, USA 83, 7231–7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karve A, Rauh BL, Xia X, Kandasamy M, Meagher RB, Sheen J, Moore BD. 2008. Expression and evolutionary features of the hexokinase gene family in Arabidopsis. Planta 228, 411–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandelwal A, Chandu D, Roe CM, Kopan R, Quatrano RS. 2007. Moonlighting activity of presenilin in plants is independent of γ-secretase and evolutionary conserved. Proceedings of the National Academy of Sciences, USA 104, 13337–13342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaggs AR. 2003. The biosynthesis of shikimate metabolites. Natural Product Reports 20, 119–136. [DOI] [PubMed] [Google Scholar]
- Köhler RH, Cao J, Zipfel WR, Webb WW, Hanson MR. 1997. Exchange of protein molecules through connections between higher plant plastids. Science 276, 2039–2042. [DOI] [PubMed] [Google Scholar]
- Köhler RH, Hanson MR. 2000. Plastid tubules of higher plants are tissue-specific and developmentally regulated. Journal of Cell Science 113, 81–89. [DOI] [PubMed] [Google Scholar]
- Kojima H, Watanabe Y, Numata O. 1997. The dual functions of Tetrahymena citrate synthase are due to the polymorphism of its isoforms. Journal of Biochemistry 122, 998–1003. [DOI] [PubMed] [Google Scholar]
- Koncz C, Schell J. 1986. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Molecular and General Genetics 204, 383–396. [Google Scholar]
- Krause K, Oetke S, Krupinska K. 2012. Dual targeting and retrograde translocation: regulators of plant nuclear gene expression can be sequestered by plastids. International Journal of Molecular Sciences 13, 11085–11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok EY, Hanson MR. 2004. GFP-labelled Rubisco and aspartate aminotransferase are present in plastid stromules and traffic between plastids. Journal of Experimental Botany 55, 595–604. [DOI] [PubMed] [Google Scholar]
- Laskar DD, Corea ORA, Patten AM, Kang CH, Davin LB, Lewis NG. 2010. Vascular plant lignification: biochemical/structural biology considerations of upstream aromatic amino acid and monolignol pathways. In: Mander L, Liu HW, eds. Comprehensive natural products II: chemistry and biology, Vol 6, carbohydrates, nucleosides and nucleic acids. Amsterdam: Elsevier, 541–604 [Google Scholar]
- Li HM, Chiu CC. 2010. Protein transport into chloroplasts. Annual Review of Plant Biology 61, 157–180. [DOI] [PubMed] [Google Scholar]
- Lu Q, Tang X, Tian G, et al. 2010. Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. The Plant Journal 61, 259–270. [DOI] [PubMed] [Google Scholar]
- Luo M, Orsi R, Patrucco E, Pancaldi S, Cella R. 1997. Multiple transcription start sites of the carrot dihydrofolate reductase-thymidylate synthase gene, and sub-cellular localization of the bifunctional protein. Plant Molecular Biology 33, 709–722. [DOI] [PubMed] [Google Scholar]
- Maeda H, Dudareva N. 2012. The shikimate pathway and aromatic amino acid biosynthesis in plants. Annual Review of Plant Biology 63, 73–105. [DOI] [PubMed] [Google Scholar]
- Maeda H, Shasany AK, Schnepp J, Orlova I, Taguchi G, Cooper BR, Rhodes D, Pichersky E, Dudareva N. 2010. RNAi suppression of Arogenate Dehydratase1 reveals that phenylalanine is synthesized predominantly via the arogenate pathway in petunia petals. The Plant Cell 22, 832–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H, Yoo H, Dudareva N. 2011. Prephenate aminotransferase directs plant phenylalanine biosynthesis via arogenate. Nature Chemical Biology 7, 19–21. [DOI] [PubMed] [Google Scholar]
- Maréchal A, Parent JS, Véronneau-Lafortune F, Joyeux A, Lang BF, Brisson N. 2009. WHIRLY proteins maintain plastid genome stability in Arabidopsis. Proceedings of the National Academy of Sciences, USA 106, 14693–14698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mewis I, Khan MA, Glawischnig E, Schreiner M, Ulrichs C. 2012. Water stress and aphid feeding differentially influence metabolite composition in Arabidopsis thaliana (L.). PLoS One 7, e48661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishima SY. 2011. Mechanism of plastid division: from a bacterium to an organelle. Plant Physiology 155, 1533–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi H, Suzuki K, Kabeya Y, Miyagishima SY. 2009a . Plant-specific protein MCD1 determines the site of chloroplast division in concert with bacteria-derived MinD. Current Biology 19, 151–156. [DOI] [PubMed] [Google Scholar]
- Nakanishi H, Suzuki K, Kabeya Y, Okazaki K, Miyagishima SY. 2009b . Conservation and differences of the Min system in the chloroplast and bacterial division site placement. Communicative and Integrative Biology 2, 400–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natesan SK, Sullivan JA, Gray JC. 2005. Stromules: a characteristic cell-specific feature of plastid morphology. Journal of Experimental Botany 56, 787–797. [DOI] [PubMed] [Google Scholar]
- Natesan SK, Sullivan JA, Gray JC. 2009. Myosin XI is required for actin-associated movement of plastid stromules. Molecular Plant 2, 1262–1272. [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. 2007. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. The Plant Journal 51, 1126–1136. [DOI] [PubMed] [Google Scholar]
- Popescu SC, Popescu GV, Bachan S, Zhang Z, Seay M, Gerstein M, Snyder M, Dinesh-Kumar SP. 2007. Differential binding of calmodulin-related proteins to their targets revealed through high-density Arabidopsis protein microarrays. Proceedings of the National Academy of Sciences, USA 104, 4730–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke KA. 1999. Plastid division and development. The Plant Cell 11, 549–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke KA, Leech RM. 1992. Chloroplast division and expansion is radically altered by nuclear mutations in Arabidopsis thaliana. Plant Physiology 99, 1005–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud C, Perennes C, Reuzeau C, Olivier C, Brown S. 2005. Cell and plastid division are coordinated through the prereplication factor AtCDT. Proceedings of the National Academy of Sciences, USA 102, 8216–8221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes FC, Sun B, Guo H, Gruis DF, Otegui MS. 2010. Agrobacterium tumefaciens-mediated transformation of maize endosperm as a tool to study endosperm cell biology. Plant Physiology 153, 624–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippert P, Puyaubert J, Grisollet D, Derrier L, Matringe M. 2009. Tyrosine and phenylalanine are synthesized within the plastids in Arabidopsis. Plant Physiology 149, 1251–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell D. 2001. Molecular cloning: a laboratory manual, 3rd edn Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nature Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Steinbach PA, Tsien RY. 2005. A guide to choosing fluorescent proteins. Nature Methods 2, 905–909. [DOI] [PubMed] [Google Scholar]
- Shimmen T, Yokota E. 2004. Cytoplasmic streaming in plants. Current Opinion in Cell Biology 16, 68–72. [DOI] [PubMed] [Google Scholar]
- Silhavy D, Molnár A, Lucioli A, Szittya G, Hornyik C, Tavazza M, Burgyán J. 2002. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO Journal 21, 3070–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I, Brandizzi F. 2012. Fluorescent protein-based technologies: shedding new light on the plant endomembrane system. The Plant Journal 70, 96–107. [DOI] [PubMed] [Google Scholar]
- Tan K, Li H, Zhang R, Gu M, Clancy ST, Joachimiak A. 2008. Structures of open (R) and close (T) states of prephenate dehydratase (PDT)—implication of allosteric regulation byl-phenylalanine. Journal of Structural Biology 162, 94–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TerBush AD, Yoshida Y, Osteryoung KW. 2013. FtsZ in chloroplast division: structure, function and evolution. Current Opinion in Cell Biology 25, 461–470. [DOI] [PubMed] [Google Scholar]
- Thabet I, Guirimand G, Guihur A, Lanoue A, Courdavault V, Papon N, Bouzid S, Giglioli-Guivarc’h N, Simkin AJ, Clastre M. 2012. Characterization and subcellular localization of geranylgeranyl diphosphate synthase from Catharanthus roseus. Molecular Biology Reports 39, 3235–3243. [DOI] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, et al. 2006. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604. [DOI] [PubMed] [Google Scholar]
- Tzin V, Galili G. 2010. New insights into the shikimate and aromatic amino acids biosynthesis pathways in plants. Molecular Plant 3, 956–972. [DOI] [PubMed] [Google Scholar]
- Vervliet G, Holsters M, Teuchy H, Van Montagu M, Schell J. 1975. Characterization of different plaque-forming and defective temperate phages in Agrobacterium. Journal of General Virology 26, 33–48. [DOI] [PubMed] [Google Scholar]
- Vitha S, Froehlich JE, Koksharova O, Pyke KA, van Erp H, Osteryoung KW. 2003. ARC6 is a J-domain plastid division protein and an evolutionary descendant of the cyanobacterial cell division protein Ftn2. The Plant Cell 15, 1918–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitha S, McAndrew RS, Osteryoung KW. 2001. FtsZ ring formation at the chloroplast division site in plants. Journal of Cell Biology 153, 111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivan AL, Caceres RA, Abrego JR, Borges JC, Ruggiero Neto J, Ramos CH, de Azevedo WF, Jr , Basso LA, Santos DS. 2008. Structural studies of prephenate dehydratase from Mycobacterium tuberculosis H37Rv by SAXS, ultracentrifugation, and computational analysis. Proteins 72, 1352–1362. [DOI] [PubMed] [Google Scholar]
- Vogt T. 2010. Phenylpropanoid biosynthesis. Molecular Plant 3, 2–20. [DOI] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D. 2003. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. The Plant Journal 33, 949–956. [DOI] [PubMed] [Google Scholar]
- Walia H, Wilson C, Condamine P, et al. 2005. Comparative transcriptional profiling of two contrasting rice genotypes under salinity stress during the vegetative growth stage. Plant Physiology 139, 822–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise AA, Liu Z, Binns AN. 2006. Three methods for the introduction of foreign DNA into Agrobacterium. Methods in Molecular Biology 343, 43–53. [DOI] [PubMed] [Google Scholar]
- Wroblewski T, Tomczak A, Michelmore R. 2005. Optimization of Agrobacterium-mediated transient assays of gene expression in lettuce, tomato and Arabidopsis. Plant Biotechnology Journal 3, 259–273. [DOI] [PubMed] [Google Scholar]
- Wydro M, Kozubek E, Lehmann P. 2006. Optimization of transient Agrobacterium-mediated gene expression system in leaves of Nicotiana benthamiana. Acta Biochimica Polonica 53, 289–298. [PubMed] [Google Scholar]
- Xiang C, Han P, Lutziger I, Wang K, Oliver DJ. 1999. A mini binary vector series for plant transformation. Plant Molecular Biology 40, 711–717. [DOI] [PubMed] [Google Scholar]
- Yamada T, Matsuda F, Kasai K, Fukuoka S, Kitamura K, Tozawa Y, Miyagawa H, Wakasa K. 2008. Mutation of a rice gene encoding a phenylalanine biosynthetic enzyme results in accumulation of phenylalanine and tryptophan. The Plant Cell 20, 1316–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Kuroiwa H, Misumi O, et al. 2010. Chloroplasts divide by contraction of a bundle of nanofilaments consisting of polyglucan. Science 329, 949–953. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.