Highlight
During Pseudomonas syringae infection the MAPK phosphatase AP2C1 negatively regulates plant resistance and modulates defense responses, such as changing hormone levels, callose deposition and transcriptional reprogramming.
Keywords: Callose, defense genes, MAPK, MAPK phosphatase, PAMP, PP2C phosphatase, Pseudomonas syringae, salicylic acid, transcription factors.
Abstract
Mitogen-activated protein kinases (MAPKs) mediate plant immune responses to pathogenic bacteria. However, less is known about the cell autonomous negative regulatory mechanism controlling basal plant immunity. We report the biological role of Arabidopsis thaliana MAPK phosphatase AP2C1 as a negative regulator of plant basal resistance and defense responses to Pseudomonas syringae. AP2C2, a closely related MAPK phosphatase, also negatively controls plant resistance. Loss of AP2C1 leads to enhanced pathogen-induced MAPK activities, increased callose deposition in response to pathogen-associated molecular patterns or to P. syringae pv. tomato (Pto) DC3000, and enhanced resistance to bacterial infection with Pto. We also reveal the impact of AP2C1 on the global transcriptional reprogramming of transcription factors during Pto infection. Importantly, ap2c1 plants show salicylic acid-independent transcriptional reprogramming of several defense genes and enhanced ethylene production in response to Pto. This study pinpoints the specificity of MAPK regulation by the different MAPK phosphatases AP2C1 and MKP1, which control the same MAPK substrates, nevertheless leading to different downstream events. We suggest that precise and specific control of defined MAPKs by MAPK phosphatases during plant challenge with pathogenic bacteria can strongly influence plant resistance.
Introduction
The ability to grow in an environment full of potentially pathogenic microbes is very important for plant survival. Plants recognize pathogen-associated molecular patterns (PAMPs), such as flagellin or elongation factor Tu (EF-Tu), via plasma membrane-localized pattern recognition receptors (PRRs) (Macho and Zipfel, 2014). This recognition rapidly activates a signaling network of mitogen-activated protein kinases (MAPKs) (Meng and Zhang, 2013), which induces a basal level of immunity called PRR-triggered immunity (PTI, also known as pattern- or PAMP-triggered immunity) (Tena et al., 2011; Rasmussen et al., 2012; Meng and Zhang, 2013; Couto and Zipfel, 2016). MAPKs play an important role in PTI, but how they are negatively regulated by phosphatases is less understood.
Arabidopsis MAPKs MPK3, 4 and 6 are central for the regulation of several basal defense responses, including pathogen-induced gene expression and the production of plant stress hormones, as well as antimicrobial compounds (Tena et al., 2011; Rasmussen et al., 2012; Meng and Zhang, 2013). Phosphorylation of the MAPK motif ‘pTEpY’ by MAPK kinases (MAPKKs) is essential for their activation, whereas MAPK inactivation relies on different types of protein phosphatases to dephosphorylate this motif. Protein tyrosine (PTP) and dual specificity (DSP) phosphatases dephosphorylate phospho-tyrosines (pY) or both phospho-threonines (pT) and pY, respectively (Bartels et al., 2010; Caunt and Keyse, 2013). Ser/Thr protein phosphatases of type 2C (PP2C) target pT in the ‘pTEpY’ loop (Meskiene et al., 2003; Fuchs et al., 2013). These types of phosphatases have been found to control MAPK signaling during plant defense. PTP1 and the DSP MKP1 have genetically overlapping roles in suppressing plant defense responses via inactivation of MPK3 and MPK6 (Bartels et al., 2009). MKP1 negatively regulates a subset of PAMP-regulated genes, and MPK6-dependent resistance to the virulent bacterial pathogen Pseudomonas syringae pv. tomato (Pto) DC3000 (Bartels et al., 2009; Anderson et al., 2011, 2014). MKP2 interacts with MPK6 and controls the hypersensitive response in plants (Lumbreras et al., 2010). Structurally and functionally different from PTP/DSP, phosphatases of the PP2C type such as Arabidopsis AP2C1, 2, 3 and 4 (Schweighofer et al., 2004) also dephosphorylate MAPKs (Umbrasaite et al., 2010). The alfalfa AP2C1 ortholog MP2C inactivates the MPK6 alfalfa ortholog SIMK by dephosphorylating pT in the ‘pTEpY’ motif (Meskiene et al., 2003).
Among the 80 Arabidopsis PP2C family members, AP2C proteins feature a kinase interaction motif (KIM) (Fuchs et al., 2013), which mediates interaction with MAPKs and is highly similar to the KIM motif found in mammalian MAPK phosphatases (Farooq and Zhou, 2004). The specificities for interactions between AP2Cs and MAPKs have been demonstrated, and extensive yeast two-hybrid screenings with AP2Cs repeatedly isolated MPK6 (Schweighofer et al., 2007; Umbrasaite et al., 2010). Interaction between AP2Cs and MPK3, 4 and 6 has been confirmed by bimolecular fluorescence complementation (BiFC) (Schweighofer et al., 2007; Brock et al., 2010; Umbrasaite et al., 2010) and co-immunoprecipitation (Schweighofer et al., 2007). AP2C1 modulates wound- and PAMP-induced MAPK activities (Schweighofer et al., 2007; Galletti et al., 2011), and its overexpression impairs wound-induced ethylene (ET) production (Schweighofer et al., 2007) and plant immunity to the necrotrophic fungus Botrytis cinerea (Schweighofer et al., 2007; Galletti et al., 2011). PAMP perception highly upregulates the expression of AP2C1 and its closest homolog AP2C2 in Arabidopsis plants (Navarro et al., 2004; Zipfel et al., 2006). However, their role in plant responses to hemibiotrophic pathogens such as Pseudomonas syringae remains unclear. We approach this question in the current study.
Many of the PAMP-induced transcriptional alterations (Zipfel et al., 2004, 2006) are downstream of MAPKs projected for resetting the cellular processes for defense responses (Meng and Zhang, 2013). PAMPs and pathogens transiently activate MPK3, 4, 6, and 11 (Nühse et al., 2000; Asai et al., 2002; Bethke et al., 2012), which leads to phosphorylation of proteins, including transcription factors (TFs) (Liu and Zhang, 2004; Bethke et al., 2009; Popescu et al., 2009; Andreasson and Ellis, 2010; Mao et al., 2011; Lassowskat et al., 2014). The ability of MAPKs to phosphorylate different TFs, which are also partially shared between MAPKs (Popescu et al., 2009), adds to the complexity in dissecting defined roles of MAPKs. Direct targets of MAPKs are WRKY TFs, which regulate the expression of many pathogen-responsive genes. MPK3 and MPK6 phosphorylate WRKY33, which stimulates the expression of PHYTOALEXIN DEFICIENT 3 (PAD3) (Mao et al., 2011). PAD3 encodes an enzyme required for the synthesis of the antimicrobial compound camalexin (Qiu et al., 2008). WRKY33 is also involved in MPK3- and MPK6-induced expression of ACS2 and ACS6, which encode enzymes involved in ET biosynthesis (Li et al., 2012). Direct phosphorylation of ACS2 and ACS6 by MPK3 and MPK6 and ensuing stabilization of these proteins enhances ET biosynthesis (Liu and Zhang, 2004; Joo et al., 2008; Han et al., 2010). ET is important for PTI amplification and maintenance by replenishment of ligand-free FLS2 and also by activation of signaling mediated by PEPR1/2 to induce immunity to pathogens (Boutrot et al., 2010; Liu et al., 2013; Tintor et al., 2013; Zipfel, 2013). ET promotes the release of ERF104 from MPK6, presumably to access target genes for plant defense (Bethke et al., 2009). PAMP perception by PRRs leads to production of salicylic acid (SA) (Mishina and Zeier, 2007), which plays a central role in PTI (Dempsey et al., 2011). MPK3 and MPK6 positively regulate SA signaling (Zhang et al., 2007b), and MPK4 has been identified as a positive regulator of PTI (Zhang et al., 2012). An SA marker gene, PR1 (Ward et al., 1991), is also regulated by alternative SA-independent mechanisms, such as the one induced by sustained MPK3 activation (Tsuda et al., 2013).
Transcriptional regulation plays an important role in plants (Mitsuda and Ohme-Takagi, 2009). For global TF analysis, qRT-PCR is a sensitive and preferred method (Czechowski et al., 2004; Rauf et al., 2013), but TF profiling by qRT-PCR has not yet been applied to plant–pathogen studies in respect to cell signaling. In our study, we employed an Arabidopsis whole genome qRT-PCR platform for 1880 TFs and 137 defense response genes (Brotman et al., 2012) aiming to identify genes involved in AP2C1 regulated cell signaling ensuing infection by pathogen Pto DC3000.
The present study demonstrates a strongly enhanced resistance to Pto DC3000 in the ap2c1 mutant, providing an important genetic model for investigating the basis for the induction of enhanced plant resistance. We address the regulatory mechanism of MAPK activities by AP2C1 and relate activation of MAPKs with distinct cellular responses during pathogen/PAMP-induced signaling, expression of TFs and defense genes, as well as ethylene and callose accumulation. These findings highlight the significance of the MAPK pathway regulation by AP2C1 phosphatase for plant defense.
Materials and methods
Plant material
For this study the following mutant lines of Arabidopsis thaliana (L.) Heynh. (Col-0 accession) were used: ap2c1 (SALK_065126) (Schweighofer et al., 2007), ap2c2 (GABI-Kat_316F11) (Umbrasaite et al., 2010), mkp1 (Bartels et al., 2009), mpk3-1 (SALK_151594), mpk6-2 (SALK_073907), fls2c (SAIL_691_C4) (Zipfel et al., 2004), efr-1 (SALK_044334) (Zipfel et al., 2006), fls2c efr-1 cerk1-2 (fec) (Gimenez-Ibanez et al., 2009b), 35S-AP2C1-GFP, and ap2c1 complemented line (644) (Schweighofer et al., 2007). ap2c1 mpk3 and ap2c1 mpk6 double mutants were created by crossing single mutants and verified by PCR in the F2 generation [ap2c1 T-DNA (forward: 5′-TGGTTCACGTAGTGGGCCATCG-3′ and reverse: 5′-CATCAGACGAGCCTCGTGAAGCAGATAAATCG-3′); AP2C1 (forward: 5′-TCGGCCGCTGTGGCTGCG -3′ and reverse: 5′-CATCAGACGAGCCTCGTGAAGCAGATAAATCG-3′; mpk3 T-DNA (forward: 5′-GCTTGGCACACCGACAGAATCT-3′ and reverse: 5′-TGGTTCACGTAGTGGGCCATCG-3′); MPK3 (forward: 5′-GCTTGGCACACCGACAGAATCT-3′ and reverse: 5′-ACCGTATGTTGGATTGAGTGCTATG-3′); mpk6 T-DNA (forward: 5′-GGCATCGTTTGTTCGGCTATG-3′ and reverse: 5′- TGGTTCACGTAGTGGGCCATCG -3′); MPK6 (forward: 5′-GGCATCGTTTGTTCGGCTATG-3′ and reverse: 5′- GATCT CGTCCAGGGAAGAGTG-3′)]. Plants on soil were grown at 21–22 °C with an 8 h light/16 h dark photoperiod in environmentally controlled growth chambers. For growth under sterile conditions seeds were surface sterilized (1 min in 96% ethanol, 5 min in 7.5% NaOCl–0.01% Triton X-100, five washes with sterile water). Seedlings were grown on plates containing half-strength Murashige and Skoog (MS) medium (Duchefa), 1% sucrose, and 0.7% Bacto agar (Invitrogen) at 22 °C with a 10 h light/14 h dark photoperiod. For ethylene measurements seeds were spread in glass vials containing 15 ml half-strength MS medium (Duchefa), 1% sucrose, and 0.7% Bacto agar (Invitrogen), and the glass vials were transferred to a growth chamber and kept in long day photoperiod (16 h light/8 h dark), at 50% humidity and 23 °C temperature.
Pseudomonas infection and growth assay
Bacterial strains used in this study were Pseudomonas syringae pv. tomato DC3000 (Pto DC3000), Pseudomonas syringae pv. tomato DC3000 ΔavrPto/ΔavrPtoB (Pto DC3000 ΔavrPto/ΔavrPtoB), Pseudomonas syringae pv. tomato DC3000 COR− (Pto DC3000 COR−). For bacterial enumeration assays, plants were sprayed with the strains (inoculum: 107 cfu ml−1, OD600=0.02), in the presence of 0.001% (v/v) Silwet L-77, as described (Zipfel et al., 2004). Sprayed plants were covered with a transparent plastic lid for the remaining time of the experiment. Bacterial titer was estimated 3 or 4 d after infection.
PAMP treatment, protein extraction, SDS-PAGE and western blot
PAMP treatments were performed by spraying flg22 and elf18 elicitor peptides (Peptron, South Korea), which were dissolved in water to a concentration of 0.1 or 1 µM.
Total protein was extracted from frozen leaf tissue, subjected to SDS-PAGE, transferred to a polyvinylidene fluoride membrane (Millipore), and used for immunodetection as described (Meskiene et al., 2003). Equal loading was checked by Ponceau S staining. Anti-phospho-p44/42 MPK (Thr202/Tyr204) antibody (Cell Signaling Technology), was used to detect doubly phosphorylated MAPKs. Antigen–antibody complexes were detected with horseradish peroxidase-conjugated anti-rabbit secondary antibody (Cell Signaling Technology) followed by chemiluminescence detection with SuperSignal West Pico chemiluminescent substrate (Pierce).
Quantitative RT-PCR
Plants were grown for 4 weeks on soil and treated for 0, 4, and 48 h with Pto DC3000 (OD600=0.02). Total RNA was isolated with the RNeasy Plant Mini kit (Qiagen). DNase Turbo DNA-free (Ambion) was used for genomic DNA removal. Absence of genomic DNA was confirmed by qRT-PCR using intron-specific primers for the gene At5g65080 (forward: 5′-TTTTTTGCCCCCTTCGAATC-3′ and reverse: 5′-ATCTTCCGCCACCACATTGTAC-3′). RNA integrity was checked on 1% (w/v) agarose gel and concentration measured with a Nanodrop ND-1000 spectrophotometer (Thermo Scientific) before and after DNAse I digestion. cDNA was synthesized from 2 µg of total RNA using RevertAid-First Strand cDNA Synthesis Kit (Fermentas) with oligo-dT primers, according to the manufacturer’s instructions. The efficiency of cDNA synthesis was determined by qRT-PCR amplifications of control transcripts of the genes ACTIN2 (At3g18780; forward: 5′-TTCCTCAGCACATTCCAGCAGAT-3′ and reverse: 5′-AACGATTCCTGGACCTGCCTCATC-3′), GAPDH 5′ region (At1g13440; forward: 5′-TCTCGATCTCAATTTCGCAAAA-3′ and reverse: 5′-CGAAACCGTTGATTCCGATTC-3′) and GAPDH 3′ region (At1g13440; forward: 5′-TTGGTGACAACAGGTCA AGCA-3′ and reverse: 5′-AAACTTGTCGCTCAATGCAATC-3′). qPCRs were performed either with SyberGreen master mix (Sigma-Aldrich) or with Power SYBER Green PCR Master Mix (Applied Biosystems). Gene expression was normalized to ACTIN2.
TF and defense-related expression profiling platforms were used as described previously (Czechowski et al., 2004; Brotman et al., 2012).
ET measurements
Thirteen-day-old seedlings were treated with 1 µM elf18, 1 µM flg22, Pto DC3000 (OD600=0.04) or 10 mM MgCl2 solution. Two hundred microliters of solution was carefully placed on plant leaves as droplets. Glass vials with treated seedlings were capped air-tight and transferred to a plant growth chamber. Next day accumulated ethylene was measured using a Thermo Scientific FOCUS GC gas chromatograph with flame ionization detector and Supelco column (length 1.8 m, external diameter 3.2 mm, internal diameter 2.1 mm): 500 µl of gas sample was taken using a gas-tight micro syringe and loaded into the machine. Chromatograms were analysed using the Chrom-card program.
SA and camalexin measurements
For the analysis of free and conjugated SA and camalexin, 0.25 g of leaves (with 200 ng of o-anisic acid) was extracted once with 2 ml of 70% methanol and once with 2 ml of 90% methanol by using a homogenizer (Polytron; Kinematica, Littau, Switzerland). After evaporation of the methanol from the combined extracts, trichloroacetic acid precipitation was performed. Free phenols and camalexin were extracted into cyclohexane–ethyl acetate (1:1). The remaining aqueous phase was submitted to acid hydrolysis in the presence of 4 M HCl at 80 °C for 1 h, and the liberated phenols were extracted into cyclohexane–ethyl acetate, as described above. For HPLC, the organic phase was evaporated, and the samples were resuspended in 85% phosphate buffer–15% acetonitrile. Chromatography was performed on a reverse phase HPLC column (ABZ+, 25 cm×4.6 mm; Supelco, Buchs, Switzerland) as described (Meuwly and Métraux, 1993).
Callose deposition
Callose deposition was observed after spraying with a solution of 1 µM elf18 or Pto DC3000 (OD600=0.02) for 24 h as previously described (Luna et al., 2011). Callose deposits were quantified using ImageJ software.
Analysis of promoter motifs
Promoter motifs were identified using the Athena web-based research tool (http://www.bioinformatics2.wsu.edu/Athena). TF-binding frequency and enrichment for sub-selected promoters and TFs were calculated.
Results
Disease resistance to P. syringae is strongly enhanced in the Arabidopsis protein phosphatase mutant ap2c1
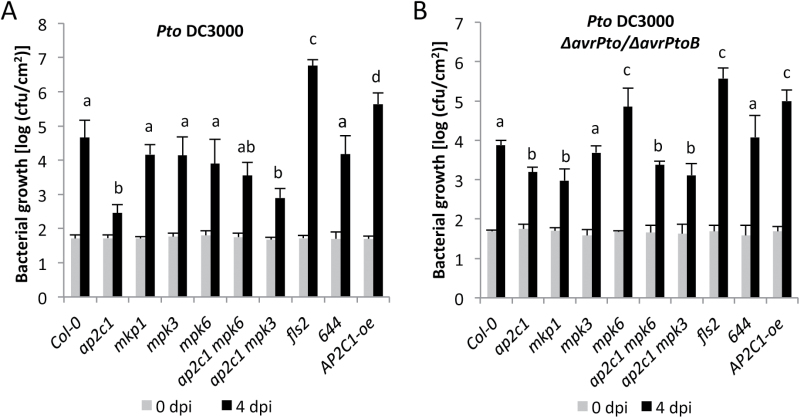
Previously, we demonstrated that the T-DNA knock-out mutant of the MAPK phosphatase ap2c1 produces higher amounts of wound-induced jasmonic acid (JA) and is more resistant to phytophagous mites (Tetranychus urticae) (Schweighofer et al., 2007). Plants with increased AP2C1 levels are compromised in basal and PAMP-induced resistance against the necrotrophic fungal pathogen Botrytis cinerea (Schweighofer et al., 2007; Galletti et al., 2011). To test whether AP2C1 regulates plant defense against bacterial hemibiotrophs, we infected ap2c1 plants by spray-inoculation with the virulent bacterium Pseudomonas syringae pv. tomato DC3000 (Pto DC3000). Remarkably, we observed a strongly increased resistance in ap2c1 compared with the wild-type Col-0 at 4 days post-inoculation (dpi) (Fig. 1A). This phenotype was observed neither in the null mutants mpk3 or mpk6, nor in mkp1, whereas fls2 plants were significantly more susceptible to Pto DC3000 (Fig. 1A and Supplementary Fig. S1 at JXB online), as reported previously (Zipfel et al., 2004). The enhanced resistance to bacteria observed in ap2c1 was reversed in transgenic lines complemented with AP2C1-GFP expressed under the native promoter (line 644) (Schweighofer et al., 2007) (see Supplementary Fig. S1), indicating that the phenotype observed in ap2c1 is caused by the lack of AP2C1. Moreover, plants overexpressing AP2C1 (AP2C1-oe) were more susceptible to Pto DC3000 (Fig. 1). Ap2c1 mutants were also more resistant to strains with attenuated virulence such as Pto DC3000 ΔavrPto/ΔavrPtoB (Fig. 1B), which lack the type III-secreted effectors AvrPto and AvrPtoB (Lin and Martin, 2005).
Fig. 1.
Susceptibility of plants to the pathogen P. syringae. Four-week-old plants were spray-infected with P. syringae pv. tomato (Pto) DC3000 (A) or Pto DC3000 ΔavrPto/ΔavrPtoB (B) and bacterial count measured at 4 days post-infection (dpi). Values shown are means±standard deviation (n=8) of one representative experiment from three independent repetitions. One-way ANOVA/Holm–Sidak: a≠b, P<0.01; a≠c, P<0.01; a≠d, P<0.01.
To assess the specificity of the enhanced plant resistance detected in ap2c1, we tested whether a similar phenotype could be observed in a T-DNA knock-out mutant line of the related MAPK phosphatase AP2C2 (Umbrasaite et al., 2010), the closest paralog to AP2C1. Although we did not find significant changes in response to Pto DC3000 in ap2c2 plants at 3 and 4 dpi (see Supplementary Fig. S1A), these plants, as well as ap2c1 mutants, were more resistant to the weakly virulent Pto DC3000 COR– (Supplementary Fig. S1B), which lack the jasmonic acid mimic coronatine (COR) (Brooks et al., 2004).
Taken together, our data show that AP2C1 and to some extent AP2C2 are negative regulators of plant resistance to pathogenic bacteria.
Activation of MAPKs by PAMPs and Pto DC3000 is enhanced in the ap2c1 mutant
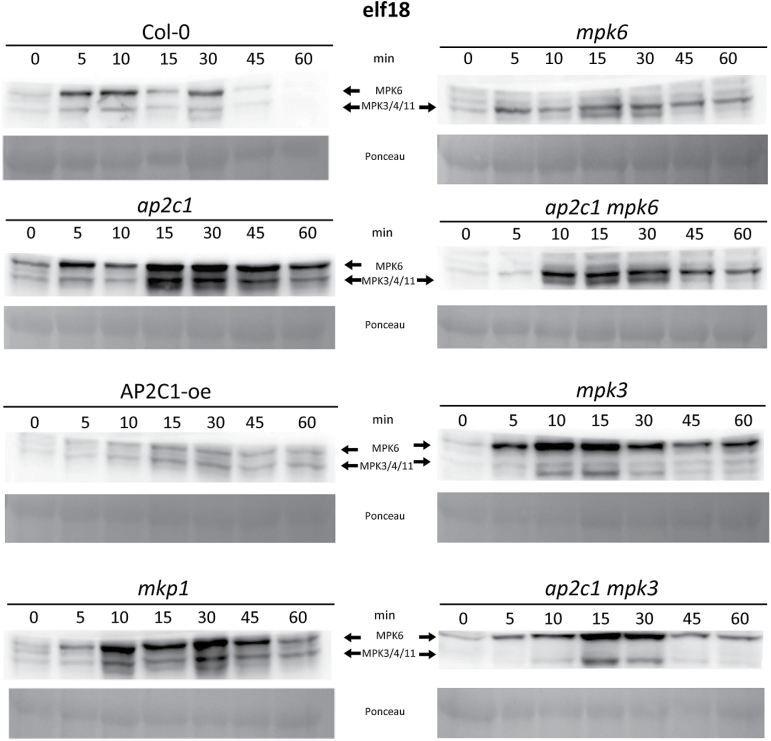
To better understand the underlying mechanism of the enhanced bacterial resistance observed in ap2c1 plants, we analysed the PAMP-induced activation of MPK3, MPK4 and MPK6, which are the substrates of AP2C1 (Schweighofer et al., 2007; Galletti et al., 2011). Our data show more pronounced and prolonged elf18- and flg22-induced MAPK activation in ap2c1 than in Col-0 plants (Fig. 2 and Supplementary Fig. S2). Conversely, ectopic expression of AP2C1 strongly inhibited elf18-induced MAPK phosphorylation (Fig. 2). Remarkably, the double mutant ap2c1 mpk6 showed an MPK3/MPK4 phosphorylation that is much more pronounced than Col-0 and mpk6 lines (Fig. 2). Similarly, the ap2c1 mpk3 displayed a higher PAMP-induced MPK6 activity compared with the single mutants ap2c1 or mpk3. The activation of MAPKs in mkp1 plants was also enhanced and prolonged compared with Col-0 plants, but showed different kinetics and activity maximum.
Fig. 2.
AP2C1 controls elf18-induced MAPK activation. Western blotting with p44/42 antibodies after application of 1 µM elf18 on seedlings. Profiling of MAPK activation by an immunological assay that detects phosphorylation of the MAPKs. MPK6 and MPK3/4/11 corresponding immunoreactive bands are indicated by arrows in the top panels. Ponceau staining was used to estimate equal loading (bottom panels).
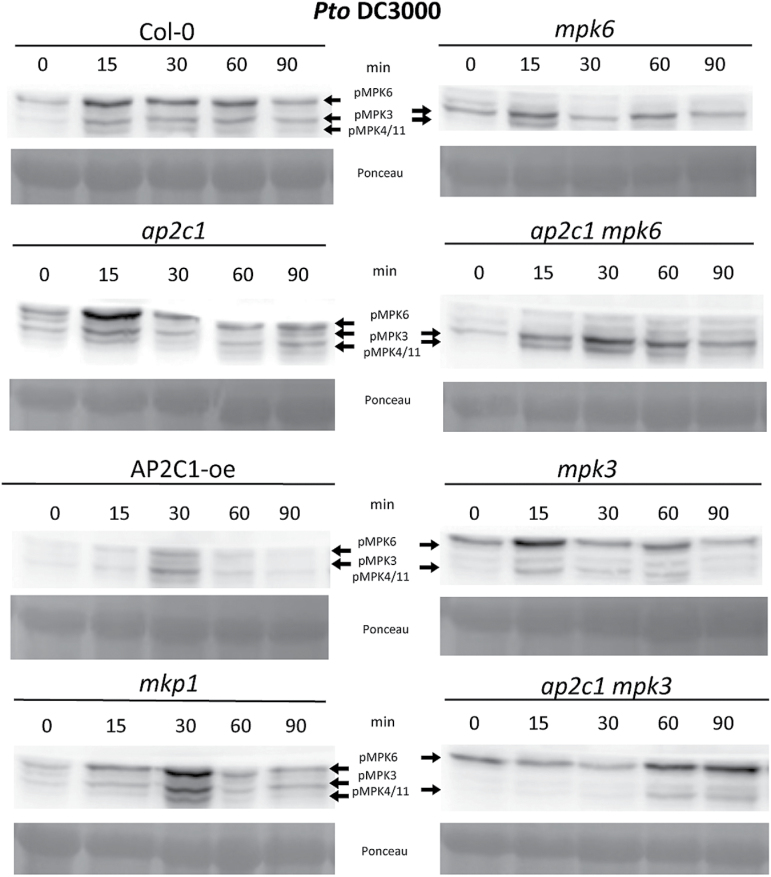
To assess the effect of bacterial infection on MAPK activation, we characterized MAPK phosphorylation in response to Pto DC3000. In the ap2c1 line, Pto DC3000 triggered higher MPK6 activation, with its maximum activity at an earlier time point compared with Col-0 (Fig. 3), whereas MAPK activities were abolished in the AP2C1-oe line. MAPK activities in the mkp1 mutant were also more pronounced than in Col-0, but their kinetics was different than in ap2c1 plants. Double mutant plants ap2c1 mpk6 showed stronger activation of MPK3/4/11, and ap2c1 mpk3 stronger activation of MPK6 compared with Col-0 (Fig. 3).
Fig. 3.
Analysis of MAPK activation in response to infection with Pto DC3000. Western blotting with anti-p44/42 antibodies. Bacteria induced activation of MAPKs in plants after treatment with Pto DC3000; OD600=0.02. The immunoreactive protein bands corresponding to respective MAPKs are indicated in the top panels. Ponceau staining was used to estimate equal loading (bottom panels).
Taken together, our data demonstrate that during PAMP- and pathogen-induced responses AP2C1 plays a significant role in the negative regulation of MPK3, MPK4/11 and predominantly MPK6. AP2C1 deactivates MPK6 during early stages of bacterial invasion, whereas in the absence of AP2C1 a different phosphatase, such as MKP1, can effectively inactivate these kinases.
To investigate whether MPK3 or MPK6 plays a role in increased resistance observed in ap2c1, we analysed the double mutant lines ap2c1 mpk3 and ap2c1 mpk6 for plant resistance to Pto DC3000 and Pto DC3000 ΔavrPto/ΔavrPtoB. Absence of each of the MAPKs did affect the disease resistance as both double mutant lines demonstrated significantly enhanced resistance compared with the Col-0 plants (Fig. 1A, B). In response to Pto DC3000, ap2c1 mpk6 plants showed significantly higher bacterial growth than the ap2c1 mpk3 mutant, indicating a more substantial contribution of MPK6 to plant basal resistance in the ap2c1 background than of MPK3 (Fig. 1A, B).
Defense-related genes are deregulated in ap2c1 and MAPK mutant plants
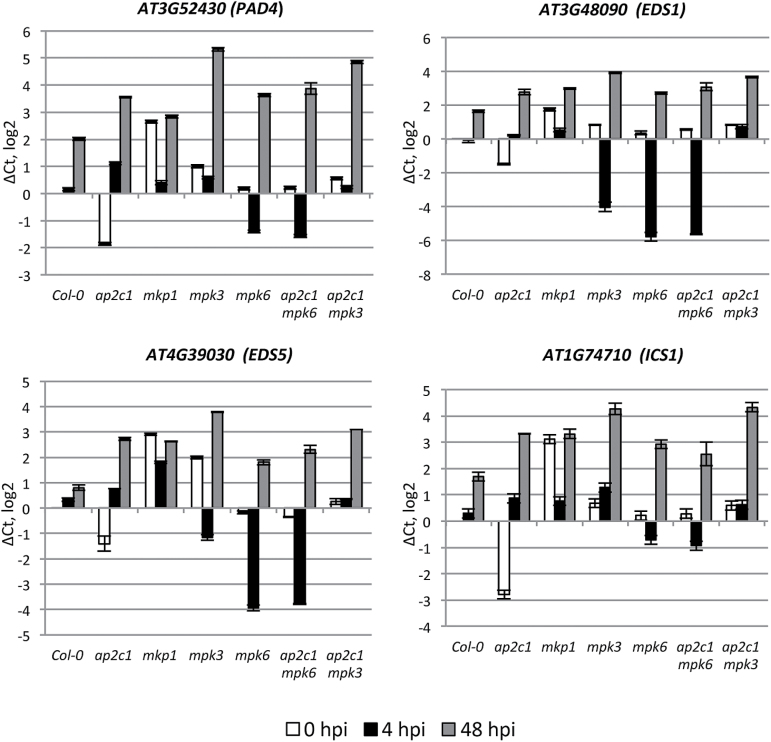
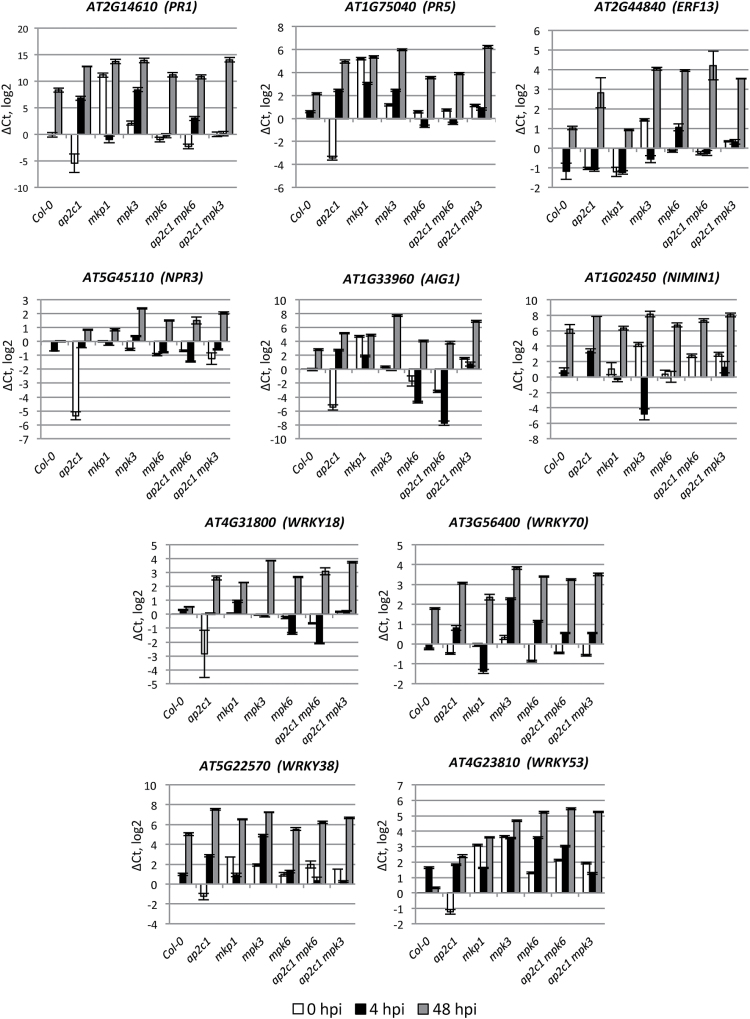
To evaluate the impact of AP2C1 on pathogen responsive gene expression, we performed qRT-PCR of 137 defense-related genes (Brotman et al., 2012). To cover ‘early’ and ‘late’ pathogen-induced gene expression, leaves were collected at 4 and 48 h post-infection (hpi) with Pto DC3000, respectively. Our data demonstrate significant deregulation (>1 on a log2 scale, which equals >2-fold) of genes involved in SA production (EDS1, PAD4, EDS5, and ICS1) and SA-induced genes, including PR1, PR5, ERF13, PAD3, NPR3, AIG1, and NIMIN1 as well as transcription factors WRKY18, WRKY70, WRKY53, and WRKY38, in single and double ap2c1 mpk3 and ap2c1 mpk6 mutants compared with Col-0 (Figs 4–7 and Supplementary Fig. S3). Importantly, we observed a significant down-regulation of almost all genes listed above in untreated ap2c1 plants. At 4 hpi we observed strong up-regulation of PR1, PR5, PAD4, AIG1, WRKY70, WRKY38, and NIMIN1 genes specifically in ap2c1 plants (Figs 4 and 5, and Supplementary Fig. S3).
Fig. 4.
qRT-PCR analysis of SA-related gene expression. Adult 4-week-old plants were sprayed with Pto DC3000 or water as a mock control and harvested at 0 h (white bars), 4 h (black bars) and 48 h (2 dpi; gray bars) post-infection. The relative gene expression was normalized to the reference gene, ACTIN2. Results are from three biological and two technical replicates for each experiment. Error bars indicate SE. Values on the Y-axis are given on a log2 scale.
Fig. 5.
qRT-PCR analysis of defense-related gene expression. Adult 4-week-old plants were sprayed with Pto DC3000 or water as a mock control and harvested at 0 h (white bars), 4 h (black bars), and 48 h (2 dpi; gray bars) post-infection. The relative gene expression was normalized to the reference gene, ACTIN2. Results are mean of three biological and two technical replicates for each experiment. Error bars indicate SE. Values on the Y-axis are given on a log2 scale.
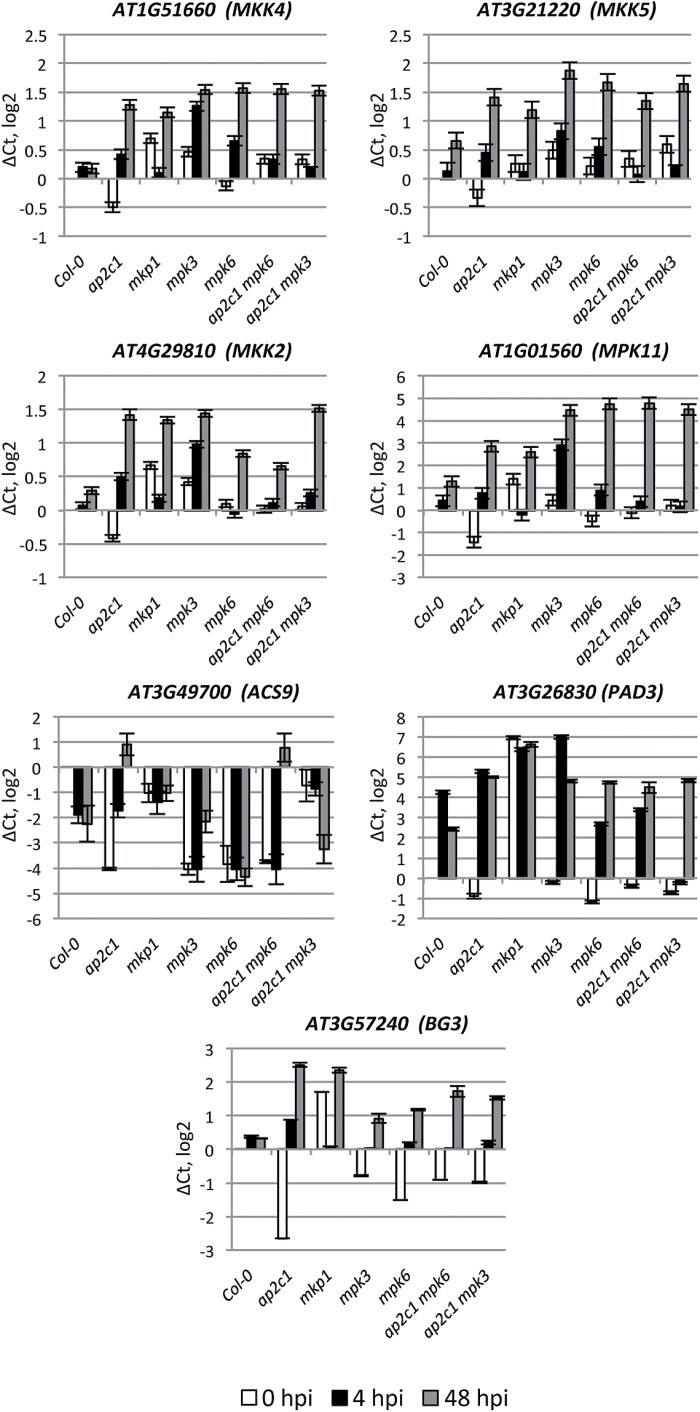
In ap2c1, we also observed strong up-regulation of genes encoding components of the MAPK signaling cascade, such as MPK11, which is a flg22-activated gene (Bethke et al., 2012), MKK5 and MKK4, which are MAPKKs upstream of MPK3/MPK6 (Asai et al., 2002), and MKK2, which is a positive regulator of plant immunity against P. syringae, acting upstream of MPK4/MPK6 (Brader et al., 2007). BETA-1,3-GLUCANASE 3 (BG3) (Dong et al., 1991) and ACS9, which is involved in ET biosynthesis (Gomi et al., 2005; Christians et al., 2009; Tsuchisaka et al., 2009), were also upregulated in comparison with Col-0 plants (Fig. 6 and Supplementary Fig. S3). However, ACS9 expression was repressed in the ap2c1 mpk3 line.
Fig. 6.
qRT-PCR analysis of pathogen-related gene expression (MAPK cascade components; ET-, camalexin- and callose-related genes). Adult 4-week-old plants were sprayed with Pto DC3000 or water as a mock control and harvested at 0 h (white bars), 4 h (black bars), and 48 h (gray bars) post-infection. The relative gene expression was normalized to the reference gene, ACTIN2. Results are mean of three biological and two technical replicates for each experiment. Error bars indicate SE. Values on the Y-axis are given on a log2 scale.
Identification of AP2C1-regulated genes by genome-wide TF expression analysis
In order to identify possible links between enhanced activity of MAPKs and up-regulation of defense genes, we performed a genome-wide analysis of the expression of 1880 TF genes in Col-0 and ap2c1, using qRT-PCR. We found that 88 TF genes from 21 families, including basic helix–loop–helix (bHLH), WRKY, NAC, and AP2/ERF were differently regulated (>4 on a log2 scale) after 4 hpi with Pto DC3000 in ap2c1 vs Col-0 (Supplementary Table S1 and Fig. 7).
Fig. 7.
Heat map of 88 TF genes differentially regulated during Pto DC3000 immune response in ap2c1 compared with WT plants. Expression levels were determined in leaves of treated plants by multi-parallel qRT-PCR analysis. Red and green indicate higher and lower expression values, respectively. Intensity of the colors is proportional to the absolute value of log2 of the difference in gene expression between ap2c1 and WT. Black indicates no change in gene expression. Two biological replicates with two technical replicates in each were analysed.
A promoter analysis of the 88 TF genes revealed enrichment of several motifs (Supplementary Table S2), such as CARGCW8GAT (8% increase over background), ARF binding site (8%), AtMYC2BSinRD22 (6%), MYCATERD1 (6%), L1-box motif (6%) and ATHB2 binding site (9%), suggesting these motifs as potential target sites for regulation by activated AP2C1-determined (MAPK) signaling. These results reveal the importance of AP2C1 as a regulator of many transcriptional responses, and suggest that many target genes are deregulated in the ap2c1 mutant, thus altering plant immune responses after pathogen application.
Induction of AP2C1 and AP2C2 expression by PAMPs and Pto DC3000
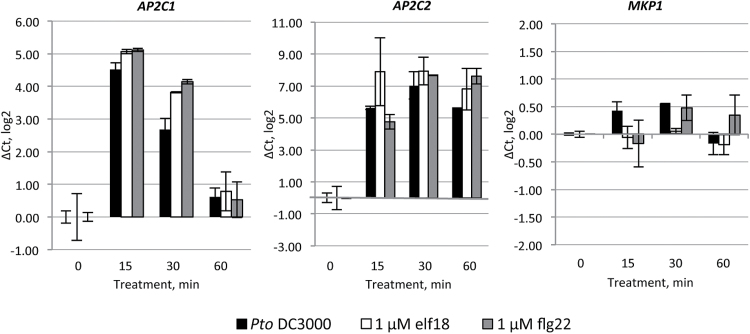
AP2C1 and AP2C2 have been detected as early PAMP-induced genes (Zipfel et al., 2006) and the Genevestigator expression database (Hruz et al., 2008) reports enhancement of their expression by PAMP treatment or during bacterial/fungal infection (see Supplementary Fig. S4). Monitoring the expression of AP2C1, AP2C2 and other known or potential MAPK phosphatase genes in response to elf18, we observed differences in expression levels of these genes in seedlings upon a 180 min treatment with elf18 (Supplementary Fig. S5). AP2C2 and AP2C3 were significantly up-regulated upon treatment with elf18, whereas expression levels of other MAPK phosphatases, such as AP2C1 and AP2C4, as well as of members from the PTP/DSP class were not significantly altered (Supplementary Fig. S5). To examine AP2C1 and AP2C2 expression at earlier time points, we performed qRT-PCR at 15, 30 and 60 min after application of Pto DC3000, elf18 or flg22 (Fig. 8). Strong AP2C1 and AP2C2 expression was detected already at 15 min after treatments. Interestingly, AP2C1 transcript was no longer detectable at 60 min, whereas AP2C2 demonstrated sustained expression, but MKP1 expression was not altered (Fig. 8).
Fig. 8.
Gene expression of AP2C1, AP2C2, and MKP1 after treatment with Pto DC3000, elf18 or flg22. Fourteen-day-old seedlings were treated with Pto DC3000, 1 µM elf18 or 1 µM flg22 and harvested at 0, 15, 30, and 60 min. The relative gene expression was normalized to the reference gene, ACTIN2. Results are mean of two biological and two technical replicates for each experiment. Error bars indicate SE. Values on the Y-axis are given on a log2 scale.
Collectively, the observation that both PAMPs and bacteria increase AP2C1 and AP2C2 expression in plants substantiates the idea of AP2C1 and AP2C2 as important players in plant response to PAMP and Pto DC3000.
AP2C1 negatively controls accumulation of ET, but not camalexin or SA in response to PAMP or Pto DC3000
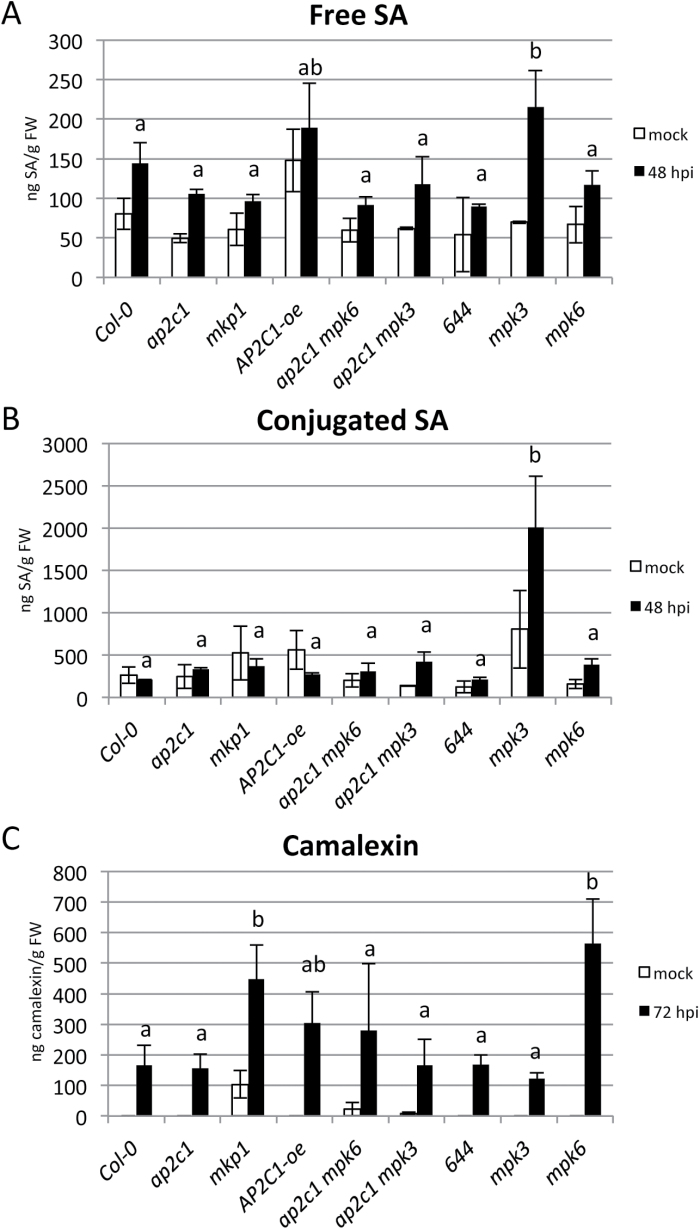
Our analysis of defense gene expression indicated that SA and/or ET homeostasis might be modulated in the ap2c1 mutant. Thus, we measured free and conjugated SA in plants at 24, 48 and 72 hpi with Pto DC3000. Our results show no significant differences in amounts of free or conjugated SA after pathogen application in ap2c1 or mkp1 mutant lines compared with Col-0, while these amounts increase in mpk3 plants (Fig. 9A, B).
Fig. 9.
Analysis of SA and camalexin accumulation in plants treated with Pto DC3000. Levels of total free (A) and conjugated (B) SA, or total camalexin (C) as determined by HPLC in leaves of 4-week-old soil-grown plants mock spayed or treated with Pto DC3000 (OD600=0.02). (Results shown are mean±SE; n=4). One-way ANOVA/Holm–Sidak: a≠b, P<0.05.
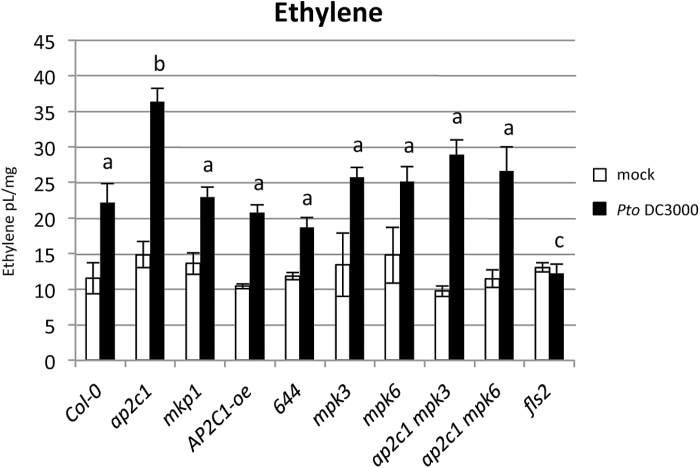
We measured ET levels after elf18, flg22 or Pto DC3000 treatments. Our results show no significant differences in ET amounts in all tested lines, except efr and fls2 during elf18 or flg22 application, respectively (see Supplementary Fig. S6). However, in response to bacteria, ap2c1 plants produce almost double the amount of ET as Col-0 and other lines tested (Fig. 10). The ap2c1 complemented line 644 produced ET amounts similar to Col-0, suggesting that lack of AP2C1 leads to enhanced ET production.
Fig. 10.
Pto DC3000-induced ethylene production in seedlings. Two-week-old seedlings of Col-0 and corresponding mutant lines were treated with Pto DC3000 (OD600=0.02) and total amount of ethylene produced by treated plants in 24 h was measured. (Results shown are mean±SE; n=6). One-way ANOVA/Holm–Sidak between treated line samples: a≠b, P<0.05; a≠c, P<0.02.
Since an important part of plant pathogen defense responses is the production of the antimicrobial compound camalexin (3-thiazolylindole) (Meng and Zhang, 2013), we studied its accumulation in plant leaves after Pto DC3000 infection. Camalexin accumulation after bacterial inoculation was similar in Col-0 and ap2c1, while higher amounts of camalexin were produced in mkp1 and mpk6 in comparison to Col-0, mpk3 and the corresponding double mutant lines (Fig. 9C).
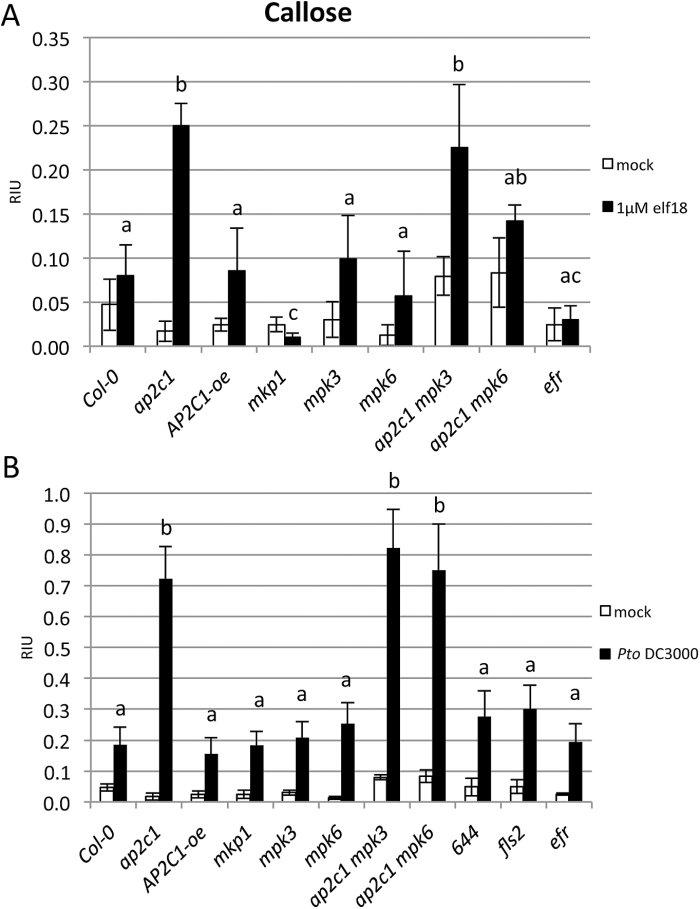
Callose deposition is enhanced in ap2c1 seedlings in response to PAMP and Pto DC3000
To measure callose accumulation in plants, we treated 2-week-old seedlings with 1 μM elf18 or Pto DC3000 for 24 h. Interestingly, plants lacking AP2C1 responded strongly to elf18 or Pto DC3000 by accumulating several times higher amounts of callose compared with Col-0 or AP2C1-oe plants (Fig. 11 and Supplementary Fig. S7). The enhancement of callose accumulation observed in ap2c1 remained in ap2c1 mpk3 and ap2c1 mpk6 plants (Fig. 11). At the same time, the complementation of ap2c1 mutation (line 644) showing a response similar to Col-0 indicated the reversion to the wild type phenotype (Fig. 11B). Corresponding to the previously published data (Anderson et al., 2011, 2014) mkp1 showed reduced callose deposition after PAMP treatment, and in response to Pto these plants responded similarly to Col-0.
Fig. 11.
Callose deposition in cotyledons in response to elf18 or Pto DC3000. Ten-day-old seedlings were treated for 24 h with 1 µM elf18 (A) or Pto DC3000 (OD=0.02) (B). Photographs of aniline blue-stained cotyledons under UV epifluorescence were quantified with ImageJ. Data shown are mean values±SE (n>10) of relative callose intensities (RIU, relative intensity units) as measured at 24 h after pathogen or PAMP treatment. One-way ANOVA/Holm–Sidak between treated lines samples: a≠b, P<0.03; a≠c, P<0.05.
Discussion
Despite recent progress in understanding the role of signaling by MAPKs in plant immunity, less is known about their negative regulation by MAPK phosphatases. Focusing on the role of the MAPK phosphatase AP2C1, we have gained insights about the mechanism of negative regulation of MAPK signaling and its association with plant defense responses during PTI as well as plant resistance to pathogenic bacteria. We found that loss of AP2C1 leads to strongly enhanced resistance to Pto DC3000 that correlates with enhanced PAMP- and Pto-induced MAPK activities displaying particular kinetics, specific transcriptional reprogramming after Pto inoculation, and enhanced accumulation of ET and callose in infected plant tissues.
Additionally, our study pinpoint the specificity of MAPK regulation by the MAPK phosphatases AP2C1 and MKP1, which control the same substrate MAPKs, but differentially regulate the amplitude or timing of the kinase activities and related downstream events. These MAPK phosphatases evolved independently and utilize different dephosphorylation mechanisms (Brautigan, 2013). Importantly, the distinctive control of MAPKs by AP2C1 and MKP1 clearly correlates with the differences in plant pathogen responses. We suggest that, during challenge with pathogenic bacteria, individual negative regulators precisely and specifically control defined MAPKs to influence plant resistance. This is supported by our results demonstrating stronger resistance to Pto DC3000 in ap2c1 compared with mkp1 plants, which is likely due to a different mode of MAPK activation. We found that AP2C1 is a major determinant for plant sensitivity to Pto, acting in the earlier phase of kinase activation by flg22 or Pto, whereas the later phase is controlled by MKP1. Although enhanced resistance to Pto DC3000 was reported for mkp1 (Col-0) (Bartels et al., 2009; Anderson et al., 2011, 2014), in our conditions these plants showed a similar disease resistance phenotype as Col-0. Pto DC3000 effectors AvrPto and AvrPtoB intercept signaling upstream of MAPKKK (He et al., 2006) by interaction with FLS2, EFR, and BAK1 receptors (Göhre et al., 2008; Shan et al., 2008; Xiang et al., 2008; Gimenez-Ibanez et al., 2009a; Cheng et al., 2011), and thus bacteria lacking these factors are less virulent. Interestingly, strong resistance of ap2c1 plants to Pto DC3000 was reduced but still significant to isogenic hypovirulent strain ΔavrPto/ΔavrPtoB, whereas mkp1 resistance was significantly enhanced, suggesting differences in control of plant resistance to bacterial infection for these MAPK phosphatases.
The closely related PP2Cs AP2C1 and AP2C2 are induced by PAMPs and pathogens, but their expression pattern differs: AP2C1 expression is transient, whereas that of AP2C2 is more sustained. AP2C1 plays a master role in controlling resistance to Pto DC3000; however, both AP2C1 and AP2C2 negatively regulate plant resistance to the less virulent Pto DC3000 strain COR–.
Strongly overlapping functions of MPK3 and MPK6, and lethality of their double mutants (Wang et al., 2007) makes it difficult to decipher their individual roles in pathogen-induced cell signaling. MAPKK gain-of-function (GOF) approach helps to uncover the roles of MAPKs in defense responses (Meng and Zhang, 2013). The possibility of studying the mechanism of actions of specific MAPKs activated in their own domain is provided by T-DNA knock-out mutant plants of MAPK phosphatases (Schweighofer et al., 2007; Bartels et al., 2009; Anderson et al., 2011, 2014). Here, studying the ap2c1 mutant enabled us to reveal the roles of MPK3 and MPK6 in PTI. Genetic evidence emphasizes a strong positive role of MPK6 and also a contribution of MPK3 to PTI. Our results support the idea that activity of MAPKs, and mostly MPK6, which is regulated by AP2C1, is important for enhancement of plant resistance to Pto DC3000. Given the central role of MAPKs in signal transduction in response to PAMPs and Pto we showed that the precisely attenuated activity of MAPKs by a specific negative regulator in their native domain leads to a reduction of plant resistance. In addition, suppressed MAPK activities in response to PAMP and Pto and enhanced disease susceptibility due to AP2C1 overexpression underline AP2C1 as a negative regulator of PTI and basal resistance. It correlates with the previous finding that AP2C1 overexpression enhances plants sensitivity to the necrotrophic fungus Botrytis cinerea (Schweighofer et al., 2007). In another report AP2C1 over-expression revealed the major contribution of MPK6 in flg22-triggered resistance and contribution of MPK3 in basal resistance to B. cinerea, whereas no significant phenotypes were observed in ap2c1 (Galletti et al., 2011). In this study the robust plant resistance in the ap2c1 and the enhanced sensitivity due to AP2C1 overexpression demonstrate the importance of AP2C1 for negative regulation of plant immunity to the hemibiotrophic bacterium Pto DC3000.
Regulation of callose accumulation by cellular signaling is not well understood. It has been proposed that the activity of the callose synthase PMR4 might be regulated by post-translational mechanisms (Jacobs et al., 2003; Ellinger et al., 2013). Correlation of enhanced MAPK activation in ap2c1 with enhanced deposition of Pto- and PAMP-induced callose suggests that the control of MAPK activation defines a possible mechanism in callose accumulation. Our findings are consistent with a previous report on enhanced callose accumulation by GOF MKK5, which activates MPK3/MPK6 (Zhang et al., 2007a). It is tempting to speculate that PAMP- and Pto-triggered callose accumulation may result from the activation of PMR4 by AP2C1-defined MAPK signaling, since our results suggest a specific signal derived from MAPK activation towards callose accumulation. This feature is characteristic for AP2C1-controlled MAPK pathways, as mkp1 shows strongly reduced callose accumulation in response to elf18, even though the mkp1 mutant exhibits PAMP/Pto-induced MAPK activities. It can be speculated that higher activities of both MPK3 and MPK6 are required, possibly at specific locations in plant tissues or intracellular pools. In summary, our findings suggest that MAPK activities, which are enhanced due to the lack of AP2C1 in response to PAMP or Pto, control callose accumulation. Since callose is thought to act as a physical barrier to pathogen invasion (Ham et al., 2007; Luna et al., 2011), enhanced deposition of callose in ap2c1 may explain the increased plant resistance to Pto DC3000 in this mutant.
Previously we reported reduced wound-induced ET production due to AP2C1 overexpression, whereas ap2c1 plants showed no significant difference to Col-0 (Schweighofer et al., 2007). Here, in response to Pto, strongly upregulated ET amounts and enhanced kinase activities in ap2c1 corroborate MPK6/MPK3-coordinated ET biosynthesis (Meng and Zhang, 2013). Intriguingly, elf18 and flg22 do not additionally enhance ET amounts in ap2c1 plants, but Pto does, suggesting that recognition of bacteria by other plant receptors is integrated at the MPK3/MPK6–AP2C1 module. Notably, mkp1 shows no additional enhancement of ET in response to Pto suggesting that an AP2C1-specific process of MAPK control is involved in regulation of ET amounts in response to bacterial infection. Importantly, study of ap2c1 plants indicates that enhanced ET production and plant resistance to pathogenic bacteria are connected events. The observed repression of Pto-induced ACS9 expression (Tsuchisaka et al., 2009) in the ap2c1 mpk3 line is evidence for a potential link for the proposed role of AP2C1 in MPK6-mediated regulation of ET biosynthesis (Liu and Zhang, 2004; Schweighofer et al., 2007).
High-throughput methods for gene expression indicated a number of TFs playing key regulatory functions in pathogen responses (Chen et al., 2002; Czechowski et al., 2004; Caldana et al., 2007); however, relatively few have been characterized as downstream targets of MAPK signaling (Andreasson and Ellis, 2010; Meng and Zhang, 2013). During Pto DC3000 infection, a contribution of AP2C1 was shown for 88 out of 1880 Arabidopsis TFs. This enables us to link transcriptional changes to the modulation of MAPK signaling and plant responses to pathogens. We observed a substantial and prevalent increase in expression of bHLH, basic region/leucine zipper motif (bZIP), AP2/ERF, NAC, and WRKY family members, whereas a predominant reduction for LOB(AS2), MADS, and MYB TFs indicates that loss of AP2C1 strongly affects TF transcript abundance during pathogen-induced signaling. Correlation of enhanced plant resistance with alterations in the expression of 88 TFs suggests their role in PTI and an impact on disease resistance. The identified TFs might be indirect downstream targets of MAPK signaling; a possible scenario is that the MAPKs control regulatory proteins, including perhaps TFs directly binding to the cis-regulatory elements enriched in the promoters of the TF genes affected in the ap2c1 mutant (see Results), and that this control is affected by AP2C1. Significant transcriptional changes of defense-related genes indicate the involvement of a cell signaling cascade in their regulation. Strong downregulation of PR1 and PR5 in untreated ap2c1 plants, and high upregulation at 4 hpi with Pto is remarkable, even though after infection a negative regulator of NPR1-mediated PR gene induction, namely NIMIN1 (Weigel et al., 2005), is upregulated. Since no significant changes of SA amounts were observed in pathogen-treated plants, it is likely that SA-independent mechanisms are activated in ap2c1, such as the one suggested to be induced by sustained MPK3/MPK6 activation (Tsuda et al., 2013). Different roles of AP2C1 and MKP1 regarding the control of PR1 expression are indicated by very high PR1 levels in untreated mkp1 confirming a previous report (Bartels et al., 2009), although in our conditions the SA amounts were similar to Col-0. AP2C1 is required to maintain the basal expression levels of several defense-related genes. Downregulation of AIG1, BG3, EDS1, EDS5, ICS1, PAD3, PAD4, PR1, PR5, WRKY38, and WRKY53 during ambient conditions in ap2c1 and their upregulation in mkp1 compared with Col-0 plants indicate different cellular roles of AP2C1 and MKP1. The upregulation of WRKY38, known to function as a negative regulator of plant basal defense (Kim et al., 2008), in ap2c1 compared with Col-0 or mkp1 mutant plants, and the different regulation of WRKY70 demonstrate that these MAPK phosphatases control PTI responses differentially. AP2C1 also plays a different role from MKP1 as part of a regulatory mechanism for several MAPK(K)s, since their expression is downregulated in untreated conditions in ap2c1 plants. Changes in TF and defense responsive gene expression may contribute to or trigger enhanced Pto resistance of the ap2c1 plants.
Analyses of the single MAPK mutants revealed differential control by MPK3 and MPK6 of the expression of several defense genes and the double mutants ap2c1 mpk3 and ap2c1 mpk6 indicated the expression of PAD4, ICS1, PR5, NPR3, AIG1, WRKY18, and BG3 as MPK3 and MPK6 dependent, but epistatic to AP2C1.
In summary, our data reveal that MPK3 negatively regulates PR5, PR1, WRKY38, WRKY53, PAD4, MPK11, and ICS1, most probably via an AP2C1-controlled pathway. We also found that expression of PR5, PR1, and NIMIN1 is redundantly controlled by MPK3/MPK6, while MPK6 is not regulated by AP2C1 to control their expression. WRKY18 is controlled by AP2C1 but is independent of MPK3 and MPK6. After Pto infection the pattern of gene expression becomes even more complex, making it challenging to dissect dependencies on the action of single genes.
Specific regulation of camalexin by MKP1-, but not by AP2C1-controlled MAPK pathways is indicated by enhanced camalexin amounts in mkp1 but not in ap2c1, even though PAD3 (Schuhegger et al., 2006) and PAD4 (Zhou et al., 1998) required for camalexin biosynthesis are upregulated in both lines after Pto infection. Our data support a previous report that MKP1 negatively regulates camalexin biosynthesis by controlling MPK3 and MPK6 (Bartels et al., 2009). Intriguingly, high upregulation of PAD3 expression and SA amount in response to Pto in mpk3 plants, and enhanced camalexin amounts in mpk6 plants in comparison with Col-0 underline defined roles of these MAPKs in PTI. PAD4 and its interacting partner EDS1 function together to promote SA-dependent and -independent defenses (Feys et al., 2001), and their regulation in ap2c1 is MPK3 and MPK6 dependent. The fact that mutations in either of these MAPKs did not, or only partially, suppress enhancement of several defense responses implies their redundancy and suggests specific functions in signaling pathogen attack. This redundancy is illustrated by strongly enhanced remaining MPK6 activity in plants after elimination of MPK3 as it was observed previously in wound-induced signaling (Menke et al., 2004).
In plant immunity the integration of perception of multiple non-self signals on the same conservative MAPK (Tena et al., 2011; Rasmussen et al., 2012; Meng and Zhang, 2013) indicates that regulation of specificity has to be ensured. Although RACK1 was recently identified as the first plant scaffold protein in protease-mediated but not flg22-mediated defense responses (Cheng et al., 2015), regulation of both defense and developmental pathways by a MKK4/MKK5–MPK3/MPK6 module is puzzling, especially as MPK6 and MPK3 share a large percentage (40%) of tested substrates (Popescu et al., 2009). Our results suggest that protein phosphatases bring precision in pathogen-activated MAPK control and this regulation may to some extent provide the specificity towards the downstream responses. An interesting question for future research is whether these negative regulators of MAPK activities act on the same pool of MAPKs or target MAPKs at different subcellular locations. Our previous results showed cytoplasmic and nuclear AP2C1 interaction with MPK6 (Schweighofer et al., 2007), whereas Bartels et al. (2009) demonstrated an interaction of MKP1 with MPK6 predominantly in the cytoplasm. These phosphatases may therefore act on different subcellular pools of MAPKs.
It is obvious that the regulation of signaling pathways by protein phosphatases is complex. MAPK activities have to be kept under control by different MAPK phosphatases that exhibit different tissue expression patterns and subcellular localizations thereby collectively regulating signaling. It was shown that a coordinated action of MKP1 and PTP1 phosphatases is crucial for repression of plant SA-dependent autoimmune-like responses (Bartels et al., 2009). A negative control mechanism of MAPKs maintains a highly conserved property to balance the activity of the upstream components with the downstream responses, as the duration and magnitude of MAPK activation play a major role in determining the biological outcome of signaling in animal cells (Caunt and Keyse, 2013).
Taken together, studying ap2c1 plants has proved useful to connect this phosphatase with MPK3/MPK6-dependent endpoints, such as the activation of TF and pathogen-related gene expression, and ET and callose accumulation. Consequently, the proper spatial and temporal regulation of MAPK pathways by protein phosphatases may significantly contribute to specificity of signaling outcome. The future challenge is to elucidate the precise biological mechanism conferred by AP2C1 regulation of MAPK signaling and related plant responses.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Susceptibility of plants to P. syringae.
Fig. S2. Analysis of MAPK activation in plants by flg22.
Fig. S3. Heat map of pathogen-related gene expression during the immune response to Pto DC3000 infection.
Fig. S4. Expression analysis of AP2C1 and AP2C2 in response to pathogens and PAMPs using Genevestigator.
Fig. S5. Induction of MAPK-phosphatases upon elf18 treatment.
Fig. S6. PAMP-induced ethylene production in seedlings.
Fig. S7. Callose deposition in cotyledons in response to the PAMP elf18 or to Pto DC3000.
Table S1. Expression of transcription factors (TFs) in plants in response to Pto DC3000 treatment.
Table S2. Promoter region analysis of 88 selected TFs, which are significantly altered in ap2c1 plants treated with Pto DC3000 in comparison to WT.
Author contributions
VS, JB, JS, KK, MS, SB, and FB performed experiments; VS, AS, MS, FM, SB, BMR, FB, CZ, and IM designed experiments; and VS, JB, AS, FM, SB, BMR, FB, CZ, and IM wrote the manuscript.
Supplementary Material
Acknowledgements
We thank Roman Ulm for providing mkp1 (Col-0) seeds, Muhammad Arif for help with gene expression analyses and the Nottingham Arabidopsis Stock Centre for SALK mutant lines. This work was supported by the Lithuanian-Swiss Cooperation Program with the grant CH-3-ŠMM-01/10 and by the Austrian Science Fund (FWF) with the grants I-255 and W1220-B09 to IM. AS was supported by the Research Council of Lithuania with the grant MIP 003/2014. CZ was supported by The Gatsby Charitable Foundation, the UK Biotechnology and Biological Sciences Research Council with the grants BB/G024944/1(ERA-PG ‘Pathonet’) and BB/G024936/1 (ERA-PG ‘PRR-CROP’), and the European Research Council (‘PHOSPHinnATE’). JB was supported by a short term EMBO fellowship. SB and BMR gratefully acknowledge the financial support by the MPI of Molecular Plant Physiology. The authors declare no competing interests.
References
- Anderson JC, Bartels S, González Besteiro MA, Shahollari B, Ulm R, Peck SC. 2011. Arabidopsis MAP Kinase Phosphatase 1 (AtMKP1) negatively regulates MPK6-mediated PAMP responses and resistance against bacteria. The Plant Journal 67, 258–268. [DOI] [PubMed] [Google Scholar]
- Anderson JC, Wan Y, Kim YM, Pasa-Tolic L, Metz TO, Peck SC. 2014. Decreased abundance of type III secretion system-inducing signals in Arabidopsis mkp1 enhances resistance against Pseudomonas syringae. Proceedings of the National Academy of Sciences of the United States of America 111, 6846–6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasson E, Ellis B. 2010. Convergence and specificity in the Arabidopsis MAPK nexus. Trends in Plant Science 15, 106–113. [DOI] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J. 2002. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983. [DOI] [PubMed] [Google Scholar]
- Bartels S, Anderson JC, González Besteiro MA, Carreri A, Hirt H, Buchala A, Métraux JP, Peck SC, Ulm R. 2009. MAP kinase phosphatase1 and protein tyrosine phosphatase1 are repressors of salicylic acid synthesis and SNC1-mediated responses in Arabidopsis. The Plant Cell 21, 2884–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels S, González Besteiro MA, Lang D, Ulm R. 2010. Emerging functions for plant MAP kinase phosphatases. Trends in Plant Science 15, 322–329. [DOI] [PubMed] [Google Scholar]
- Bethke G, Pecher P, Eschen-Lippold L, Tsuda K, Katagiri F, Glazebrook J, Scheel D, Lee J. 2012. Activation of the Arabidopsis thaliana mitogen-activated protein kinase MPK11 by the flagellin-derived elicitor peptide, flg22. Molecular Plant-Microbe Interactions 25, 471–480. [DOI] [PubMed] [Google Scholar]
- Bethke G, Unthan T, Uhrig JF, Poschl Y, Gust AA, Scheel D, Lee J. 2009. Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proceedings of the National Academy of Sciences of the United States of America 106, 8067–8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutrot F, Segonzac C, Chang KN, Qiao H, Ecker JR, Zipfel C, Rathjen JP. 2010. Direct transcriptional control of the Arabidopsis immune receptor FLS2 by the ethylene-dependent transcription factors EIN3 and EIL1. Proceedings of the National Academy of Sciences of the United States of America 107, 14502–14507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brader G, Djamei A, Teige M, Palva ET, Hirt H. 2007. The MAP kinase kinase MKK2 affects disease resistance in Arabidopsis. Molecular Plant-Microbe Interactions 20, 589–596. [DOI] [PubMed] [Google Scholar]
- Brautigan DL. 2013. Protein Ser/Thr phosphatases—the ugly ducklings of cell signalling. The FEBS Journal 280, 324–345. [DOI] [PubMed] [Google Scholar]
- Brock AK, Willmann R, Kolb D, Grefen L, Lajunen HM, Bethke G, Lee J, Nürnberger T, Gust AA. 2010. The Arabidopsis mitogen-activated protein kinase phosphatase PP2C5 affects seed germination, stomatal aperture, and abscisic acid-inducible gene expression. Plant Physiology 153, 1098–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks DM, Hernández-Guzmán G, Kloek AP, Alarcón-Chaidez F, Sreedharan A, Rangaswamy V, Peñaloza-Vázquez A, Bender CL, Kunkel BN. 2004. Identification and characterization of a well-defined series of coronatine biosynthetic mutants of Pseudomonas syringae pv. tomato DC3000. Molecular Plant-Microbe Interactions 17, 162–174. [DOI] [PubMed] [Google Scholar]
- Brotman Y, Lisec J, Méret M, Chet I, Willmitzer L, Viterbo A. 2012. Transcript and metabolite analysis of the Trichoderma-induced systemic resistance response to Pseudomonas syringae in Arabidopsis thaliana. Microbiology 158, 139–146. [DOI] [PubMed] [Google Scholar]
- Caldana C, Scheible WR, Mueller-Roeber B, Ruzicic S. 2007. A quantitative RT-PCR platform for high-throughput expression profiling of 2500 rice transcription factors. Plant Methods 3, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caunt CJ, Keyse SM. 2013. Dual-specificity MAP kinase phosphatases (MKPs): shaping the outcome of MAP kinase signalling. The FEBS Journal 280, 489–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Provart NJ, Glazebrook J, et al. 2002. Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. The Plant Cell 14, 559–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W, Munkvold KR, Gao H, et al. 2011. Structural analysis of Pseudomonas syringae AvrPtoB bound to host BAK1 reveals two similar kinase-interacting domains in a type III Effector. Cell Host and Microbe 10, 616–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Li JF, Niu Y, et al. 2015. Pathogen-secreted proteases activate a novel plant immune pathway. Nature 521, 213–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christians MJ, Gingerich DJ, Hansen M, Binder BM, Kieber JJ, Vierstra RD. 2009. The BTB ubiquitin ligases ETO1, EOL1 and EOL2 act collectively to regulate ethylene biosynthesis in Arabidopsis by controlling type-2 ACC synthase levels. The Plant Journal 57, 332–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couto D, Zipfel C. 2016. Regulation of pattern recognition receptor signalling in plants. Nature Reviews. Immunology 16, 537–552. [DOI] [PubMed] [Google Scholar]
- Czechowski T, Bari RP, Stitt M, Scheible WR, Udvardi MK. 2004. Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. The Plant Journal 38, 366–379. [DOI] [PubMed] [Google Scholar]
- Dempsey DA, Vlot AC, Wildermuth MC, Klessig DF. 2011. Salicylic Acid biosynthesis and metabolism. The Arabidopsis Book 9, e0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Mindrinos M, Davis KR, Ausubel FM. 1991. Induction of Arabidopsis defense genes by virulent and avirulent Pseudomonas syringae strains and by a cloned avirulence gene. The Plant Cell 3, 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinger D, Naumann M, Falter C, Zwikowics C, Jamrow T, Manisseri C, Somerville SC, Voigt CA. 2013. Elevated early callose deposition results in complete penetration resistance to powdery mildew in Arabidopsis. Plant Physiology 161, 1433–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooq A, Zhou MM. 2004. Structure and regulation of MAPK phosphatases. Cellular Signalling 16, 769–779. [DOI] [PubMed] [Google Scholar]
- Feys BJ, Moisan LJ, Newman MA, Parker JE. 2001. Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. The EMBO Journal 20, 5400–5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs S, Grill E, Meskiene I, Schweighofer A. 2013. Type 2C protein phosphatases in plants. The FEBS Journal 280, 681–693. [DOI] [PubMed] [Google Scholar]
- Galletti R, Ferrari S, De Lorenzo G. 2011. Arabidopsis MPK3 and MPK6 play different roles in basal and oligogalacturonide- or flagellin-induced resistance against Botrytis cinerea. Plant Physiology 157, 804–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez-Ibanez S, Hann DR, Ntoukakis V, Petutschnig E, Lipka V, Rathjen JP. 2009a AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Current Biology 19, 423–429. [DOI] [PubMed] [Google Scholar]
- Gimenez-Ibanez S, Ntoukakis V, Rathjen JP. 2009b The LysM receptor kinase CERK1 mediates bacterial perception in Arabidopsis. Plant Signaling and Behavior 4, 539–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göhre V, Spallek T, Häweker H, Mersmann S, Mentzel T, Boller T, de Torres M, Mansfield JW, Robatzek S. 2008. Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Current Biology 18, 1824–1832. [DOI] [PubMed] [Google Scholar]
- Gomi K, Ogawa D, Katou S, et al. 2005. A mitogen-activated protein kinase NtMPK4 activated by SIPKK is required for jasmonic acid signaling and involved in ozone tolerance via stomatal movement in tobacco. Plant and Cell Physiology 46, 1902–1914. [DOI] [PubMed] [Google Scholar]
- Ham JH, Kim MG, Lee SY, Mackey D. 2007. Layered basal defenses underlie non-host resistance of Arabidopsis to Pseudomonas syringae pv. phaseolicola. The Plant Journal 51, 604–616. [DOI] [PubMed] [Google Scholar]
- Han L, Li GJ, Yang KY, Mao G, Wang R, Liu Y, Zhang S. 2010. Mitogen-activated protein kinase 3 and 6 regulate Botrytis cinerea-induced ethylene production in Arabidopsis. The Plant Journal 64, 114–127. [DOI] [PubMed] [Google Scholar]
- He P, Shan L, Lin NC, Martin GB, Kemmerling B, Nürnberger T, Sheen J. 2006. Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell 125, 563–575. [DOI] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. 2008. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Advances in Bioinformatics 2008, 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs AK, Lipka V, Burton RA, Panstruga R, Strizhov N, Schulze-Lefert P, Fincher GB. 2003. An Arabidopsis callose synthase, GSL5, is required for wound and papillary callose formation. The Plant Cell 15, 2503–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo S, Liu Y, Lueth A, Zhang S. 2008. MAPK phosphorylation-induced stabilization of ACS6 protein is mediated by the non-catalytic C-terminal domain, which also contains the cis-determinant for rapid degradation by the 26S proteasome pathway. The Plant Journal 54, 129–140. [DOI] [PubMed] [Google Scholar]
- Kim KC, Lai Z, Fan B, Chen Z. 2008. Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. The Plant Cell 20, 2357–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassowskat I, Böttcher C, Eschen-Lippold L, Scheel D, Lee J. 2014. Sustained mitogen-activated protein kinase activation reprograms defense metabolism and phosphoprotein profile in Arabidopsis thaliana. Frontiers in Plant Science 5, 554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Meng X, Wang R, Mao G, Han L, Liu Y, Zhang S. 2012. Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in Arabidopsis. PLoS Genetics 8, e1002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin NC, Martin GB. 2005. An avrPto/avrPtoB mutant of Pseudomonas syringae pv. tomato DC3000 does not elicit Pto-mediated resistance and is less virulent on tomato. Molecular Plant-Microbe Interactions 18, 43–51. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang S. 2004. Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. The Plant Cell 16, 3386–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Wu Y, Yang F, Zhang Y, Chen S, Xie Q, Tian X, Zhou JM. 2013. BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proceedings of the National Academy of Sciences of the United States of America 110, 6205–6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumbreras V, Vilela B, Irar S, Sole M, Capellades M, Valls M, Coca M, Pages M. 2010. MAPK phosphatase MKP2 mediates disease responses in Arabidopsis and functionally interacts with MPK3 and MPK6. The Plant Journal 63, 1017–1030. [DOI] [PubMed] [Google Scholar]
- Luna E, Pastor V, Robert J, Flors V, Mauch-Mani B, Ton J. 2011. Callose deposition: a multifaceted plant defense response. Molecular Plant-Microbe Interactions 24, 183–193. [DOI] [PubMed] [Google Scholar]
- Macho AP, Zipfel C. 2014. Plant PRRs and the activation of innate immune signaling. Molecular Cell 54, 263–272. [DOI] [PubMed] [Google Scholar]
- Mao G, Meng X, Liu Y, Zheng Z, Chen Z, Zhang S. 2011. Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. The Plant Cell 23, 1639–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Zhang S. 2013. MAPK cascades in plant disease resistance signaling. Annual Review of Phytopathology 51, 245–266. [DOI] [PubMed] [Google Scholar]
- Menke FL, van Pelt JA, Pieterse CM, Klessig DF. 2004. Silencing of the mitogen-activated protein kinase MPK6 compromises disease resistance in Arabidopsis. The Plant Cell 16, 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meskiene I, Baudouin E, Schweighofer A, Liwosz A, Jonak C, Rodriguez PL, Jelinek H, Hirt H. 2003. Stress-induced protein phosphatase 2C is a negative regulator of a mitogen-activated protein kinase. The Journal of Biological Chemistry 278, 18945–18952. [DOI] [PubMed] [Google Scholar]
- Meuwly P, Métraux JP. 1993. Ortho-anisic acid as internal standard for the simultaneous quantitation of salicylic acid and its putative biosynthetic precursors in cucumber leaves. Analytical Biochemistry 214, 500–505. [DOI] [PubMed] [Google Scholar]
- Mishina TE, Zeier J. 2007. Pathogen-associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. The Plant Journal 50, 500–513. [DOI] [PubMed] [Google Scholar]
- Mitsuda N, Ohme-Takagi M. 2009. Functional analysis of transcription factors in Arabidopsis. Plant and Cell Physiology 50, 1232–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Zipfel C, Rowland O, Keller I, Robatzek S, Boller T, Jones JD. 2004. The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiology 135, 1113–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nühse TS, Peck SC, Hirt H, Boller T. 2000. Microbial elicitors induce activation and dual phosphorylation of the Arabidopsis thaliana MAPK 6. The Journal of Biological Chemistry 275, 7521–7526. [DOI] [PubMed] [Google Scholar]
- Popescu SC, Popescu GV, Bachan S, Zhang Z, Gerstein M, Snyder M, Dinesh-Kumar SP. 2009. MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes and Development 23, 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu JL, Fiil BK, Petersen K, et al. 2008. Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. The EMBO Journal 27, 2214–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen MW, Roux M, Petersen M, Mundy J. 2012. MAP kinase cascades in Arabidopsis innate immunity. Frontiers in Plant Science 3, 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauf M, Arif M, Fisahn J, Xue GP, Balazadeh S, Mueller-Roeber B. 2013. NAC transcription factor speedy hyponastic growth regulates flooding-induced leaf movement in Arabidopsis. The Plant Cell 25, 4941–4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuhegger R, Nafisi M, Mansourova M, Petersen BL, Olsen CE, Svatos A, Halkier BA, Glawischnig E. 2006. CYP71B15 (PAD3) catalyzes the final step in camalexin biosynthesis. Plant Physiology 141, 1248–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweighofer A, Hirt H, Meskiene I. 2004. Plant PP2C phosphatases: emerging functions in stress signaling. Trends in Plant Science 9, 236–243. [DOI] [PubMed] [Google Scholar]
- Schweighofer A, Kazanaviciute V, Scheikl E, et al. 2007. The PP2C-type phosphatase AP2C1, which negatively regulates MPK4 and MPK6, modulates innate immunity, jasmonic acid, and ethylene levels in Arabidopsis. The Plant Cell 19, 2213–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L, He P, Li J, Heese A, Peck SC, Nürnberger T, Martin GB, Sheen J. 2008. Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host and Microbe 4, 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tena G, Boudsocq M, Sheen J. 2011. Protein kinase signaling networks in plant innate immunity. Current Opinion in Plant Biology 14, 519–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tintor N, Ross A, Kanehara K, Yamada K, Fan L, Kemmerling B, Nurnberger T, Tsuda K, Saijo Y. 2013. Layered pattern receptor signaling via ethylene and endogenous elicitor peptides during Arabidopsis immunity to bacterial infection. Proceedings of the National Academy of Sciences of the United States of America 110, 6211–6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchisaka A, Yu G, Jin H, Alonso JM, Ecker JR, Zhang X, Gao S, Theologis A. 2009. A combinatorial interplay among the 1-aminocyclopropane-1-carboxylate isoforms regulates ethylene biosynthesis in Arabidopsis thaliana. Genetics 183, 979–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Mine A, Bethke G, Igarashi D, Botanga CJ, Tsuda Y, Glazebrook J, Sato M, Katagiri F. 2013. Dual regulation of gene expression mediated by extended MAPK activation and salicylic acid contributes to robust innate immunity in Arabidopsis thaliana. PLoS Genetics 9, e1004015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbrasaite J, Schweighofer A, Kazanaviciute V, et al. 2010. MAPK phosphatase AP2C3 induces ectopic proliferation of epidermal cells leading to stomata development in Arabidopsis. PloS One 5, e15357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ngwenyama N, Liu Y, Walker JC, Zhang S. 2007. Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. The Plant Cell 19, 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward ER, Uknes SJ, Williams SC, Dincher SS, Wiederhold DL, Alexander DC, Ahl-Goy P, Metraux JP, Ryals JA. 1991. Coordinate gene activity in response to agents that induce systemic acquired resistance. The Plant Cell 3, 1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel RR, Pfitzner UM, Gatz C. 2005. Interaction of NIMIN1 with NPR1 modulates PR gene expression in Arabidopsis. The Plant Cell 17, 1279–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang T, Zong N, Zou Y, et al. 2008. Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Current Biology 18, 74–80. [DOI] [PubMed] [Google Scholar]
- Zhang J, Shao F, Li Y, et al. 2007a A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host and Microbe 1, 175–185. [DOI] [PubMed] [Google Scholar]
- Zhang X, Dai Y, Xiong Y, DeFraia C, Li J, Dong X, Mou Z. 2007b Overexpression of Arabidopsis MAP kinase kinase 7 leads to activation of plant basal and systemic acquired resistance. The Plant Journal 52, 1066–1079. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Wu Y, Gao M, Zhang J, Kong Q, Liu Y, Ba H, Zhou J, Zhang Y. 2012. Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host and Microbe 11, 253–263. [DOI] [PubMed] [Google Scholar]
- Zhou N, Tootle TL, Tsui F, Klessig DF, Glazebrook J. 1998. PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. The Plant Cell 10, 1021–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C. 2013. Combined roles of ethylene and endogenous peptides in regulating plant immunity and growth. Proceedings of the National Academy of Sciences of the United States of America 110, 5748–5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G. 2006. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125, 749–760. [DOI] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T. 2004. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428, 764–767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.