Highlight
RTCS regulates the expression of non-syntenic genes and genes associated with phytohormone action, transcription factors, and cell cycle regulators to control seminal root initiation during embryogenesis in maize.
Keywords: Embryo, primordia, RNA-Seq, rtcs, seminal root, transcriptome, Zea mays.
Abstract
Seminal roots of maize are pivotal for early seedling establishment. The maize mutant rootless concerning crown and seminal roots (rtcs) is defective in seminal root initiation during embryogenesis. In this study, the transcriptomes of wild-type and rtcs embryos were analyzed by RNA-Seq based on histological results at three stages of seminal root primordia formation. Hierarchical clustering highlighted that samples of each genotype grouped together along development. Determination of their gene activity status revealed hundreds of genes specifically transcribed in wild-type or rtcs embryos, while K-mean clustering revealed changes in gene expression dynamics between wild-type and rtcs during embryo development. Pairwise comparisons of rtcs and wild-type embryo transcriptomes identified 131 transcription factors among 3526 differentially expressed genes [false discovery rate (FDR) <5% and |log2Fc|≥1]. Among those, functional annotation highlighted genes involved in cell cycle control and phytohormone action, particularly auxin signaling. Moreover, in silico promoter analyses identified putative RTCS target genes associated with transcription factor action and hormone metabolism and signaling. Significantly, non-syntenic genes that emerged after the separation of maize and sorghum were over-represented among genes displaying RTCS-dependent expression during seminal root primordia formation. This might suggest that these non-syntenic genes came under the transcriptional control of the syntenic gene rtcs during seminal root evolution. Taken together, this study provides first insights into the molecular framework underlying seminal root initiation in maize and provides a starting point for further investigations of the molecular networks underlying RTCS-dependent seminal root initiation.
Introduction
Specialized root systems in plants are vital for growth and adaptation because they provide mechanical support, mediate water and nutrient uptake, and interact with the rhizosphere (reviewed in Hawkes et al., 2007; Lynch, 2013; Villordon et al., 2014). The complex maize root system comprises roots that are laid down during embryogenesis and post-embryonic roots that are formed after germination. The embryonic root system includes a primary root and a variable number of seminal roots, while shoot-borne and lateral roots are formed post-embryonically (reviewed in Hochholdinger et al., 2004; Yu et al., 2016). Primary roots are initiated at the basal pole of the embryo and emerge 2–3 days after germination. Seminal roots are initiated at the scutellar node between 22 and 40 days after pollination and emerge in germinating seeds soon after the primary root (Sass, 1977; Erdelska and Vidovencova, 1993). Seminal roots are formed primarily by dorsal primordia (Salvi et al., 2016). The number of seminal roots in maize varies within a genotype and between different genotypes, and ranges from 0 to 13 (Erdeska and Vidovencova, 1993; Burton et al., 2013; Tai et al., 2016).
While maize does form seminal roots, its close relative sorghum (Sorghum bicolor) does not (Singh et al., 2010). The number of seminal roots has increased during maize domestication in landraces and modern varieties compared with their ancestor teosinte, suggesting that seminal root number has probably been selected inadvertently as an adaptive trait during maize domestication (Burton et al., 2013, Tai et al., 2016). Seminal roots are essential in the establishment and early development of maize seedlings (Sanguineti et al., 1998; Lynch, 2011). Moreover, at later developmental stages, seminal root length and number are positively correlated with shoot biomass at low phosphorus levels (Zhu et al., 2006). Furthermore, seminal root length correlates with grain yield under both low and high nitrogen levels (Abdel-Ghani et al., 2015). Other seminal root traits such as number, angle, and diameter have been considered in designing hypothetical root ideotypes optimized for maize adaptation to a range of environmental stresses and increased sustainability of water and nutrient acquisition (Lynch, 2013). QTL mapping studies have suggested that a small number of major loci controls seminal root formation (Tuberosa et al., 2002; Hund et al., 2004; Zhu et al., 2006; Burton et al., 2013). This would facilitate a precise manipulation of root ideotypes to obtain optimized seminal root traits (Lynch, 2013; Salvi et al., 2016).
In maize, two monogenic mutants defective in seminal root initiation have been described: rootless concerning crown and seminal roots (rtcs) and rootless with undetectable meristems 1 (rum1). These two genes have been co-mapped with the two major QTLs controlling seminal root number (Salvi et al., 2016) and are key components of auxin signaling (Taramino et al., 2007; von Behrens et al., 2011). The rum1 gene encodes the monocot-specific Aux/IAA10 protein (von Behrens et al., 2011), while rtcs encodes a member of the plant-specific LATERAL ORGAN BOUNDARIES DOMAIN (LBD) transcription factor family (Taramino et al., 2007; Majer et al., 2012). In maize, the LBD family comprises 43 members (Majer and Hochholdinger, 2011). LBD proteins have crucial functions in defining lateral organ boundaries and are involved in various root-related developmental processes, including lateral root formation in Arabidopsis (Péret et al., 2009), lateral and shoot-borne root formation in rice (Inukai et al., 2005), and shoot-borne and seminal root formation in maize (Taramino et al., 2007). LBD proteins contain a characteristic N-terminal LOB domain (Shuai et al., 2002) that can bind to the LBD motif (5′-GCGGCG-3′) in promoters of target genes (Husbands et al., 2007). RTCS can bind to the LBD motifs of the auxin response factor arf34, and the ARF34 protein can, vice versa, bind to rtcs (Majer et al., 2012).
In the present study, RTCS-mediated transcriptional regulation of maize seminal root initiation during embryogenesis was studied by RNA-Seq. The objective of this study was to identify genes regulated by RTCS during maize embryogenesis and to gain novel insights into the molecular framework underlying seminal root primordia formation.
Materials and methods
Plant material and growth conditions
Surface-sterilized seeds of maize families segregating 3:1 for wild-type versus mutant rtcs seedlings were germinated in distilled water in paper rolls (Hetz et al., 1996) and grown for 16 h in light at 28 °C and 8 h in dark at 22 °C at a constant humidity of 60% in a growth chamber. Seedlings at 10 d old were phenotyped and genotyped according to Xu et al. (2015) to select homozygous rtcs and homozygous wild-type siblings. Subsequently, homozygous wild-type and mutant seedlings were transferred to soil pots in a growth chamber and grown under the same conditions as the paper rolls. After selfing, embryos were harvested from homozygous wild-type and mutant rtcs plants at 25, 30, and 35 d after pollination. Identification of homozygous wild-type and mutant plants from segregating families ensured a very close genetic relationship of these genotypes. The segregating families from which these plants were selected had been previously selfed for more than seven generations and were thus highly isogenic.
Histology of maize embryos
Embryos were fixed in 4% paraformaldehyde for 12 h at 4 °C and subsequently embedded in paraffin as described by Lim et al. (2000). Cross-sections of 14 µm were prepared with a Leica 2035 biocut-microtome (Leica, Nussloch, Germany). Sections were then deparaffinized and stained with Safranin O (AppliChen, Darmstadt, Germany) and Fast Green (Sigma-Aldrich, Taufkirchen, Germany) as previously described (Hetz et al., 1996). Stained embryo sections were examined under a Zeiss-Axioskop HBO 100W/2 microscope (Zeiss, Jena, Germany) and photographed using a bright field camera (PixCell IIe System, Krefeld, Germany).
Embryo RNA isolation and RNA-Seq
At 25, 30, and 35 d after pollination, embryos of similar size of the two genotypes were subjected to RNA-Seq analyses. Pools of 10 embryos were sampled per biological replicate. Four independent biological replicates were analyzed for each genotype and developmental stage. After harvest, embryos were immediately frozen in liquid nitrogen and stored at −80 °C until RNA isolation. Total RNA was extracted with the RNeasy Plant Mini Kit (Qiagen, Venlo, Netherlands). RNA integrity and quality were assessed by agarose gel electrophoresis and on an Agilent RNA 6000 Nano Chip in an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). High-quality samples with RIN (RNA integrity number) values >8 were subjected to RNA-Seq (Illumina, San Diego, CA, USA). cDNA libraries were prepared as described by the manufacturer. Finally, the Illumina HiSeq 4000 platform was used to generate 100-bp paired-end reads.
Processing and mapping of reads
Raw sequencing reads were trimmed by adapter removal. Low-quality reads containing ˃50% low-quality bases were excluded. Read mapping to the maize reference genome sequence was performed with CLC Genomics Workbench software (Version 8.0.1; https://www.qiagenbioinformatics.com/products/clc-genomicsworkbench/). Reads were first mapped to the maize B73 reference genome sequence (RefGen_v2; ftp://ftp.gramene.org/pub/gramene/maizesequence.org/release-5b/assembly/). If at least 90% of a read matched with 90% similarity to the reference, it was considered as mapped. Stacked reads that share the same start and end coordinate were considered as one read. Gaps up to 50 kb were allowed within reads to span introns. Mapped reads were further projected to the filtered gene set (FGS v2; Release 5b; ftp://ftp.gramene.org/pub/gramene/maizesequence.org/release-5b/filtered-set/). Only reads that mapped to unique positions in the filtered gene set with 80% of their bases displaying 90% similarity to the reference were selected for subsequent expression analyses. RNA-Seq data have been deposited in the NCBI sequencing read archive (SRA; http://www.ncbi.nlm.nih.gov/sra) at AC: SRP079373.
Sample relationships were analyzed by a Principal component analysis (PCA) and hierarchical clustering. PCA was conducted by using the prcomp function in R with default settings. Hierarchical clustering of all samples was generated based on Pearson correlations in the CLC Genomics Workbench package.
Statistical procedures to determine gene activity
The transcriptional activity status of all genes (active/inactive) in wild-type and rtcs embryos at each developmental stage was determined using a generalized linear mixed model with a negative binomial response. The log of the mean was assumed to be a linear combination of fixed and random effects, plus sample and gene-specific normalization factors (described below). Each combination of genotype and stage was represented by a fixed effect, and random effects accounting for additional variation from sequencing lanes were also included. The log of the TMM normalization factor (Robinson and Oshlack, 2010) was added to normalize across samples, and a smooth function of gene length and GC content was used to normalize across genes.
The vector of fixed effects for each gene was assumed to be a draw from a multivariate normal distribution with an unknown and unrestricted mean and an unknown diagonal variance–covariance matrix. Within a gene, the log of the negative binomial dispersion parameter was assumed to be constant and a draw from a normal distribution with unknown mean and variance. The random effects were assumed to follow gamma distributions, where the parameters for the lane effects were specified to create a vague distribution. An empirical Bayes procedure via the R package ‘ShrinkBayes’ (Van De Wiel et al., 2012) was used to estimate the unknown parameters, and to approximate the posterior distribution for the fixed effect associated with the gene (g), genotype (t), and stage (s) using the integrated nested Laplace approximation (Rue et al., 2009).
Activity of a gene was determined by computing P gts(T), the posterior probability that the fixed effect for gene (g), genotype (t), and stage (s) was larger than a given threshold T. A gene (g) was called active for genotype (t) and stage (s) if P gts(T)>0.5 and otherwise inactive. This method classifies genes as active or inactive based on the posterior distribution of fixed effects considering raw read count, sequencing differences from sample to sample, gene length, and GC content differences, and therefore is more accurate than classifications based only on a single raw read count threshold applied to all genes.
Statistical analysis of differential gene expression
To determine differential expression, only genes with a minimum of five mapped reads in all four biological replicates of at least one genotype were taken into account. Gene expression was normalized as FPKM (fragments per kilobase of exon model per million mapped reads) values. The R package limma based on linear models (Ritchie et al., 2015) was used to identify differentially expressed genes in pairwise contrasts. The model takes the lane effect into account, considering it as a random effect. The number of differentially expressed genes was controlled by FDR<5% and |log2Fc|≥1. Euclidean algorithm-based K-means clustering was performed to generate the expression clusters of gene expression dynamics along three stages using the OmicShare tools (www.omicshare.com/tools). The eight clusters of the identified gene dynamic expression were based on the formula 3(t−1)–1 for theoretical possible dynamic expression patterns. ‘t–1’ indicates two transitions across three time points in this study. The number ‘3’ indicates the three expression patterns of a gene: increase, decrease, or stay the same between consecutive time points. The number ‘1’ indicates the cluster of which the expression of genes has no change through time points. Genes were assigned to the different clusters with FDR<5% and |log2Fc|≥1 for each of the pairwise comparisons to identify robust expression patterns.
Functional annotation of genes and metabolic pathway analyses
The function of genes was annotated using MapMan (http://mapman.gabipd.org/web/guest/mapman) based on the functional annotation file ZmB73_5b_FGS_2011 and their homologs in Arabidopsis and rice, and a list of maize curated genes (Thimm et al., 2004). Identification of significantly over- or under-represented categories was determined by a χ 2 test (P<0.01) with Yate’s continuity correction. A category was declared over-represented if significantly more genes than expected were assigned to it. The expected gene number in each of the 32 major annotated categories was calculated based on the distribution of all expressed genes to the categories. Annotation of transcription factors (TFs) and assignment into TF families was based on the Plant Transcription Factor Database (v3.0; http://planttfdb.cbi.pku.edu.cn/; Jin et al., 2013).
Results
Histological analyses of seminal root primordia formation in wild-type and rtcs embryos
To study the process of formation of seminal root primordia, the anatomical structure of wild-type embryos in the region of the scutellar node was examined in transverse sections 25, 30, and 35 d after pollination (dap) and compared with rtcs cross-sections at the same developmental stages. At 25 dap, seminal root primordia were neither detectable in cross-sections of wild-type nor in rtcs embryos (Fig. 1A). At 30 dap, primordia were initiated in wild-type but not in rtcs embryos (Fig. 1A). Finally, at 35 dap wild-type embryos displayed fully developed seminal root primordia while these structures were entirely absent in rtcs embryos (Fig. 1A). As expected, embryo length and weight significantly increased during development (Fig. 1B). Embryos of both genotypes that displayed a similar length and weight at each stage (Fig. 1B) were selected for transcriptomic analyses.
Fig. 1.
Histological and phenotypic analyses of wild-type (WT) and rtcs embryos during formation of seminal root primordia at three developmental stages. (A) Transverse sections of Safranin-/Fast Green-stained WT and rtcs embryos 25, 30, and 35 d after pollination (dap). Seminal root primordia are indicated by black arrows. The main strand of the scutellar bundle is indicated by black asterisks. Scale bars are 300 µm. (B) Quantification of embryo length and weight in WT and rtcs at 25, 30 and 35 dap. *, P<0.01; **, P<0.001; n=10; error bars indicate ±SD.
RNA-Seq and transcriptome relationships of wild-type and rtcs embryos
Global gene expression profiles of wild-type and rtcs embryos at 25, 30, and 35 dap were analyzed using an Illumina HiSeq 4000 platform in four biological replicates per genotype/stage combination. On average, ~32 million 100-bp paired-end reads were obtained per sample (see Supplementary Table S1 at JXB online). The sequencing reads were quality checked and ~28 million (88%) of the remaining high-quality reads were mapped to unique positions of the maize reference genome (ZmB73 FefGen_v2; Supplementary Table S1). Finally, after removal of stacked reads that share the same start and end coordinates, ~67% of the remaining reads were mapped uniquely to the 39656 maize high-confidence gene models of the filtered gene set (FGSv2, release 5b.60; Supplementary Table S1).
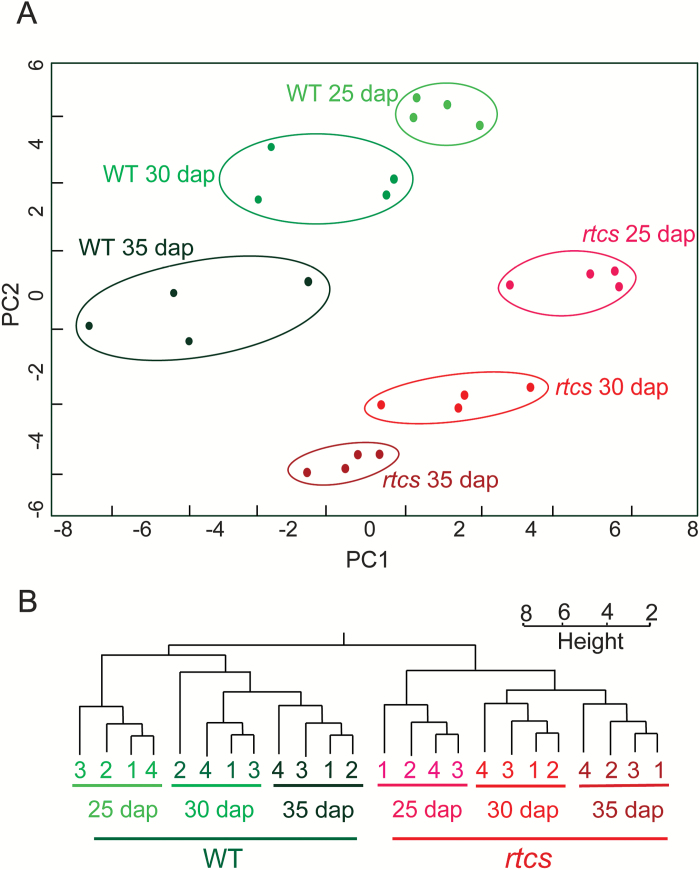
The relationship of transcriptome samples was assessed by a principal component analysis (PCA) (Fig. 2A) and by hierarchical clustering (Fig. 2B). Both analyses revealed a close relationship of the four biological replicates of each genotype/stage combination. Moreover, the hierarchical clustering analysis revealed that transcriptomes representing the three developmental stages of each of the two genotypes were more closely related to each other than samples of the same developmental stage of different genotypes (Fig. 2B).
Fig. 2.
Transcriptome relationships of wild-type and rtcs embryos. (A) Principal component analysis (PCA) of wild-type and rtcs embryo RNA-Seq samples during development. (B) Hierarchical clustering of the RNA-Seq samples based on Pearson correlation. Height indicates the degree of variance of the y-axis.
Transcriptome-wide gene activity status in wild-type and rtcs embryos
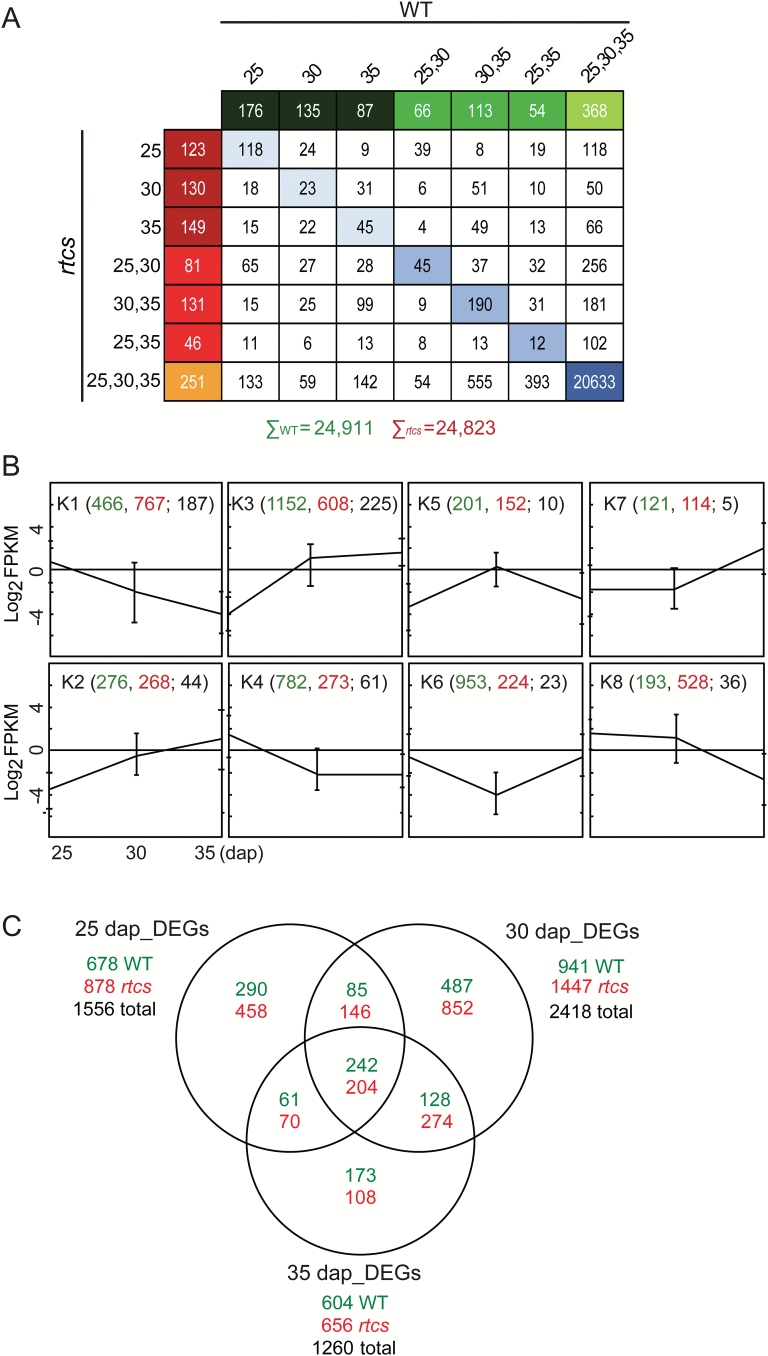
A generalized linear mixed model was applied to determine the activity status (active/inactive) of all genes at each of the six genotype/stage combinations (see Methods). In total, 25822 genes (65% of the FGSv2; Fig. 3A; see Supplementary Table S2) were declared active in at least one genotype/stage combination. Wild-type embryos expressed 24911 genes and rtcs embryos 24823. A substantial subset of these genes (20633; 80%) was constitutively active in all six genotype/stage combinations, whereas among the genes that were not constitutively active, only between 12 and 190 genes were active at the same developmental stages in both genotypes (blue boxes in Fig. 3A). Notably, hundreds of genes were only active either in embryos of wild-type (Fig. 3A green boxes: 999 genes) or in rtcs (Fig. 3A red boxes: 911 genes), suggesting genotype-specific functions of these genes during development. Genes active in only one genotype were functionally annotated based on a set of 3599 classical maize genes with manually curated functions (Schnable et al., 2011). This analysis revealed that 92 classical maize genes were exclusively active in wild-type embryos (e.g. the highly abundant ABA signaling gene ABI3 and the auxin signaling gene Aux/IAA33), while 85 classical maize genes were specifically active in rtcs embryos. Many of these classical genotype-specific genes are suggested to encode transcription factors (Supplementary Table S3).
Fig. 3.
Differential gene expression during embryo development and between wild-type (WT) and rtcs embryos. (A) Stage and genotype specificity of gene activity in wild-type and rtcs embryos. Cells in green indicate genes exclusively active in wild-type and cells in red indicate genes only active in rtcs embryos. Cells in blue indicate genes active in both genotypes at the same stage or stages. (B) K-means clustering showing the pattern changes of gene expression across the three stages in wild-type and rtcs embryos. Error bars indicate SD of gene expression in wild-type embryos. Numbers in green and red indicate the genes following the respective pattern in wild-type and rtcs embryos, respectively. Numbers in black indicate genes displaying identical patterns in both rtcs and wild-type embryos. (C) Venn diagram displaying the number of genes differentially expressed between wild-type and rtcs and their overlap during development. For each developmental stage, the total number of differentially expressed genes (black), and the numbers of genes preferential expressed in wild-type (green) and rtcs (red) are indicated. DEGs, differentially expressed genes; dap, days after pollination.
Expression dynamics of wild-type and rtcs embryos during development
To explore the dynamics of gene expression during different stages of formation of seminal root primordia in maize embryos, eight patterns of gene expression along the three developmental stages were identified in wild-type and rtcs by K-means clustering (Fig. 3B; see Supplementary Table S4). In total, 4144 genes were assigned to the eight clusters (K1 to K8) in wild-type embryos (green numbers in Fig. 3B), while 2934 genes were assigned to rtcs embryos (red numbers). This implies less dynamic gene expression patterns in rtcs embryos along the three developmental stages than in wild-type embryos. We further compared overlapping gene expression patterns between wild-type and rtcs for each of the eight clusters. In total, 591 genes exhibited the same expression pattern in both genotypes (Fig. 3B, black numbers). This suggests that during the formation of seminal root primordia in maize embryos, expression patterns of a large number of genes change in rtcs versus wild-type embryos.
Pairwise contrasts were further determined to identify genes differentially expressed between wild-type and rtcs embryos (Fig. 3C). Overall, 3526 genes (FDR<5%, |log2Fc|≥1) exhibited differential expression at least at one developmental stage in rtcs versus wild-type embryos, including 1556 at 25 dap, 2418 at 30 dap, and 1260 at 35 dap (see Supplementary Table S5). In total, 13% (446/3526) of differentially expressed genes were significantly different between wild-type and rtcs embryos at all three developmental stages, whereas 67% (2368/3526) genes were differentially expressed at only one developmental stage. Among the differentially expressed genes, 131 genes encoding transcription factors were identified via searches in the maize TF database PlantTFDB v3.0 (http://planttfdb.cbi.pku.edu.cn/; Jin et al., 2013; Supplementary Table S5).
Subsequently, genes with dynamic (Fig. 3B) and differential expression (Fig. 3C) patterns were assigned to MapMan functional categories (Thimm et al., 2004) to identify enriched molecular processes. Enriched functional categories (P<0.01) were only found among differentially expressed genes (see Supplementary Table S6). Genes differentially expressed at 25 dap were enriched for the categories ‘hormone’, ‘minor CHO metabolism’, ‘secondary metabolism’, ‘stress’, and ‘redox regulation’. The categories ‘minor CHO metabolism’ and ‘stress’ were enriched among genes differentially expressed at 30 dap. Finally, genes exhibiting differential expression at 35 dap were enriched for ‘minor CHO metabolism’, ‘stress’, and ‘cell wall’.
Differential expression of genes involved in cell proliferation
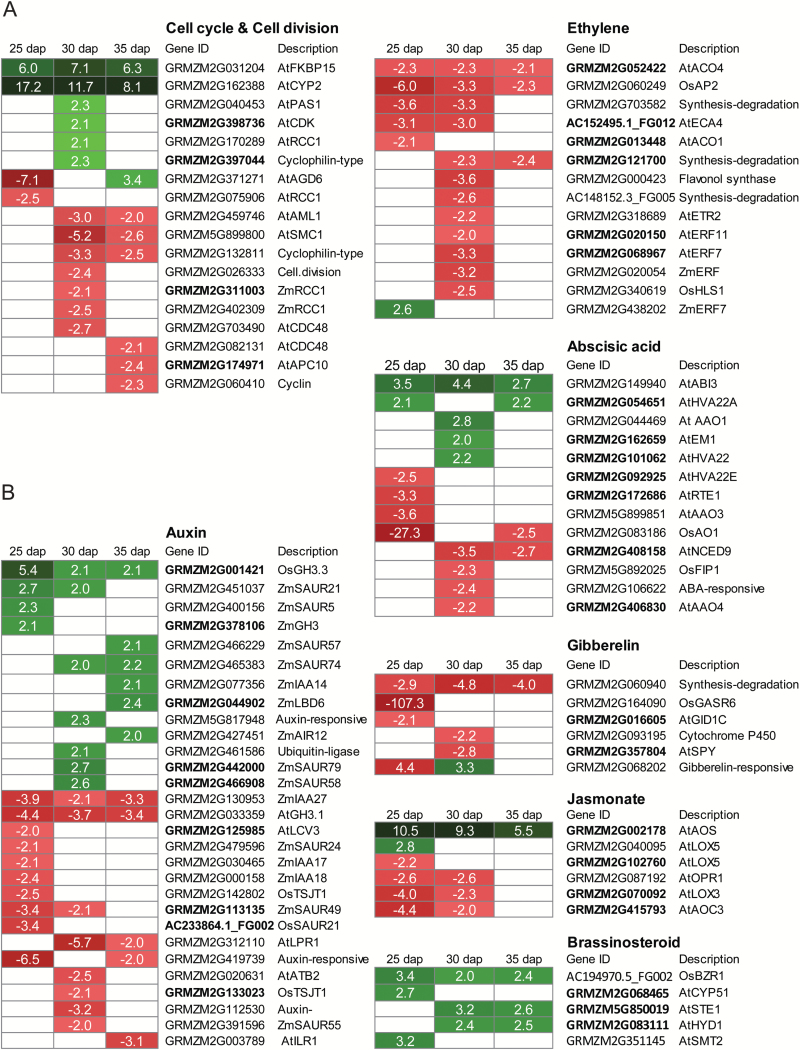
Cell proliferation and differentiation are processes regulating seminal root primordia formation. Accordingly, 18 genes related to cell cycle and cell division exhibited differential expression in at least one developmental stage (Fig. 4A; see Supplementary Table S7). Most genes (13) were differentially expressed at 30 dap, while only four genes were differentially expressed 25 dap, implying active cell division progression from 30 dap onwards. The expression levels of six cell cycle-related genes were up to 17-fold higher in wild-type embryos, including two genes with homology to AtFKBP15 and AtCYP2 that were consistently up-regulated >6-fold at all three stages. Decreased expression of cell cycle genes in wild-type embryos, as for instance illustrated by two regulators of cell division cycle protein (CDC48), was also observed.
Fig. 4.
Differentially expressed genes between wild-type and rtcs embryos at 25, 30, and 35 d after pollination (dap) involved in cell proliferation (A) and plant hormone metabolism and signaling (B). Values are fold-changes between wild-type and rtcs. Green boxes indicate preferential expression in wild-type and red boxes indicate preferential expression in rtcs. Blank boxes indicate genes that did not meet the FDR<5% and |log2Fc|≥1 criteria. Gene IDs in bold indicate that these genes contain at least one LDB motif within 1 kb upstream of the ATG start codon.
Differential expression of genes involved in hormone metabolism and hormone signaling
Phytohormones are essential for the regulation of plant development. Therefore, the functional category ‘hormone metabolism’ enriched among differentially expressed genes at 25 and 30 dap was investigated in more detail. In total, 73 genes differentially expressed between rtcs and wild-type embryos are involved in hormone metabolism, hormone signaling, or hormone response (Fig. 4B; see Supplementary Table S7). Among these, increased or decreased expression of 29 genes related to auxin was observed in rtcs embryos. All but one of these genes were related to auxin signaling (5) or response (23). For instance, nine genes belonged to the early auxin-responsive SMALL AUXIN UP RNA (SAUR) gene family. Interestingly, 13 of 14 genes involved in ethylene biosynthesis (6) and signaling (8) were expressed at 2–6-fold higher levels in rtcs embryos. Furthermore, expression of five brassinosteroid (BR) -related genes were elevated in the wild-type embryos, including a BR-activated transcription factor gene (similar to OsBZR1) and four BR biosynthesis genes. Moreover, 13 genes related to abscisic acid (ABA) biosynthesis (6), signaling (1), and response (6) were differentially expressed. Finally, differential expression of six genes involved in the biosynthesis or signaling of gibberelin (GA) and six genes related to jasmonate (JA) was observed.
Identification of putative RTCS target genes
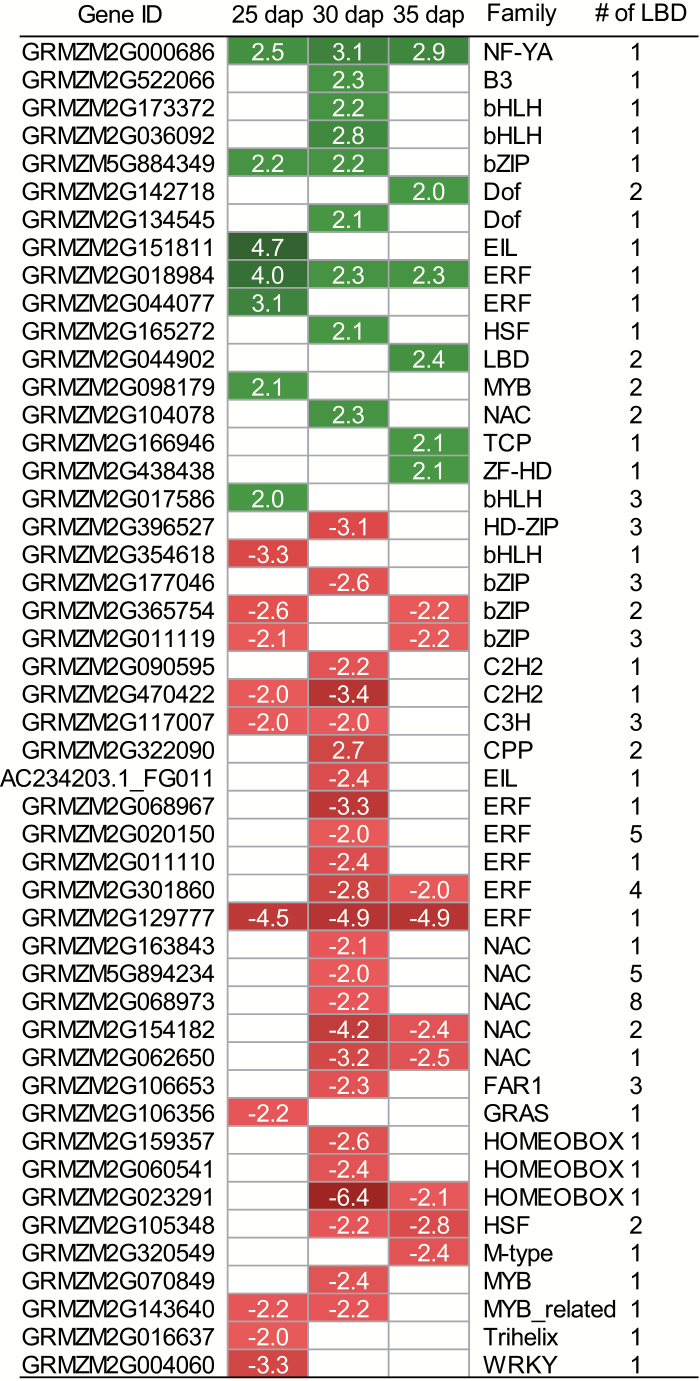
LBD transcription factors such as rtcs can bind to LBD promoter motifs of target genes. We therefore specifically examined the abundance of LBD motifs 1 kb upstream of the ATG start codon among 131 differentially expressed genes encoding for transcription factors. This analysis revealed that 48 of 131 (37%) genes contained at least one LBD motif (Fig. 5). Among those, 17 were expressed at higher levels in wild-type embryos while 31 were preferentially expressed in rtcs. These putative RTCS target genes mainly belonged to the ERF, NAC, bHLH, bZIP, and HOMEOBOX families of transcription factors. Moreover, the abundance of LBD motifs was examined among differentially expressed genes involved in hormone function. In total, 31 of 73 (43%) genes contained at least one LBD motif (Fig. 4B). Among those, 12 were preferentially expressed in wild-type and 19 in rtcs embryos. Finally, at least one LBD motif was found in four of 18 genes associated with cell cycle and division, including preferential expression of two in wild-type embryos and two in rtcs embryos (Fig. 4A).
Fig. 5.
Differentially expressed genes encoding transcription factors between wild-type and rtcs embryos at 25, 30, and 35 d after pollination (dap) containing at least one LBD motif. Values in boxes are fold-changes between wild-type and rtcs embryos. Green and red boxes indicate preferential expression of genes in wild-type and rtcs embryos, respectively. Blank boxes indicate genes that did not meet the FDR<5% and |log2Fc|≥1 criteria.
Evolutionary origin of differentially expressed genes
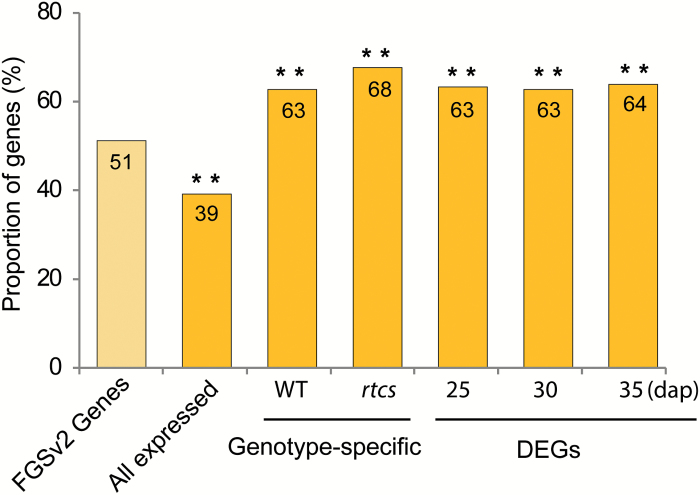
Non-syntenic genes emerged after the last whole-genome duplication of maize by individual duplications of syntenic genes. To survey their evolutionary origin, differentially expressed genes were classified into syntenic and non-syntenic genes. The maize filtered gene set (FGSv2) consists of 51% (20291/39656) non-syntenic genes, and 49% (19356/39656) syntenic genes (Schnable et al., 2011). Among all expressed genes in this study, 39% (10117/25822) were of non-syntenic origin (Fig. 6; see Supplementary Table S8). Hence, non-syntenic genes were significantly under-represented among all expressed genes relative to all genes of the maize genome. This suggests that the evolutionary older syntenic genes are expressed more frequently in maize embryos. By contrast, among genes active in only one genotype (see Fig. 3C), 63% (627/999) of wild-type-specific and 68% (616/911) of rtcs-specific genes were non-syntenic (Fig. 6; Supplementary Table S8). Similarly, non-snyntenic genes were significantly over-represented among genes differentially expressed between wild-type and rtcs embryos (see Fig. 3B) at 25 dap (63%; 984/1556), 30 dap (64%; 1516/2418), and 35 dap (64%; 803/1260) (Fig. 6; Supplementary Table S8). Thus, significant over-representation of the evolutionary younger non-syntenic genes among genes displaying genotype-specific and differential expression reflects a putative involvement of these genes in the RTCS-dependent molecular networks related to seminal root formation.
Fig. 6.
Proportion of syntenic and non-syntenic genes among differential and genotype-specific genes. Significant differences to the proportion of non-syntenic genes in FGSv2 are indicated by red asterisks (χ 2 test with Yates’ correction). **, P<0.001; FGSv2, Maize B73 filtered Gene Set Version 5b.60; DEGs, differentially expressed genes.
Discussion
RTCS substantially regulates the transcriptomic landscape of maize embryos during seminal root development
Seminal root primordia in maize are formed during embryogenesis in a multi-step process during which cells inside the scutellar node divide (Hochholdinger, 2009). The timing of seminal root initiation depends on the genotype and the growth conditions, and can be observed between 22 and 40 d after pollination (dap) (Sass, 1977; Erdelska and Vidovencova, 1993). In the present study, formation of seminal root primordia was studied in developing homozygous wild-type and rtcs embryos using microscopic sections. As described in the Methods section, wild-type and rtcs embryos were genetically closely related and highly isogenic. Histological analyses revealed that at 25 dap no seminal root primordia were initiated in wild-type embryos, while early primordia were visible at 30 dap and fully developed primordia were formed at 35 dap under our growth conditions (Fig. 1A). The gene rtcs is a major regulator of seminal root initiation as illustrated by the rtcs mutant, which does not form any seminal root primordia during embryogenesis (Hetz et al., 1996). Consistently, seminal root primordia were not detected in rtcs mutant embryos at any stage between 25 and 35 dap (Fig. 1A). Otherwise, mutant rtcs embryos were histologically indistinguishable from their wild-type siblings (Fig. 1B).
These three developmental stages were subsequently selected for transcriptome profiling of rtcs versus wild-type embryos by RNA-Seq. Hierarchical clustering revealed that the transcriptomes of wild-type and rtcs embryos were more distinct at all three developmental stages than the transcriptomes representing different stages of either genotype (Fig. 2B), suggesting a substantial RTCS-dependent transcriptomic regulation during the formation of seminal root primordia. Similarly, RTCS-dependent transcriptome regulation has been observed during formation of crown root primordia in coleoptilar nodes of maize (Muthreich et al., 2013).
RTCS-dependent control of cell cycle regulators during formation of seminal root primordia
Formation of lateral organ primordia is driven by the coordinated division of cells and the specification of boundaries of different tissues (Aida and Tasaka, 2006). Hence, formation of lateral root primordia is controlled by the subtle regulation of cell cycle genes in maize pericycle cells (Yu et al., 2015). In this study, functional annotation of differentially expressed genes revealed that RTCS regulates the expression of genes involved in cell cycle control at different stages of formation of seminal root primordia (Fig. 4A). Most of these genes were differentially expressed at 30 dap, suggesting active cell division at this stage. Consistently, seminal root primordia were observed in wild-type embryos from 30 dap onwards (Fig. 1A). Differential regulation of these cell cycle regulators in wild-type versus rtcs embryos suggests a role of these genes in the coordination of cell cycle progression in the formation of seminal root primordia. Among these, two genes (GRMZM2G031204 homologous to AtFKBP15 and GRMZM2G162388 homologous to AtCYP2) displaying consistently higher expression levels in wild-type embryos are members of the peptidyl-prolyl isomerase (PPIase) gene family essential for regulation of mitosis and cell growth (Lu et al., 2002). Moreover, the regulator of chromosome condensation 1 (RCC1) protein is a critical regulator of the cell cycle and is required for chromatin-induced formation of the mitotic spindle (Carazo-Salas et al., 1999). Accordingly, several genes encoding the RCC1 protein family were differentially expressed.
RTCS orchestrates phytohormone-dependent transcriptional networks during embryogenesis
The rtcs gene encodes an LBD transcription factor that is a central regulator of auxin signaling (Taramino et al., 2007; Majer et al., 2012). LBD proteins are essential in lateral organ initiation and patterning, and several members have been demonstrated to be involved in controlling different aspects of root development. For instance, LBD16, LBD18, LBD29, and LBD33 cooperatively regulate lateral root initiation and emergence in Arabidopsis (Okushima et al., 2007; Lee et al., 2009; Feng et al., 2012), RL1 is instrumental in lateral and shoot-borne root formation in rice (Inukai et al., 2005), and RTCS and its paralog RTCL regulate shoot-borne root initiation and elongation in maize (Xu et al., 2015). Differentially expressed genes associated with hormone functions between rtcs and wild-type embryos provide an opportunity to study RTCS-dependent regulation of auxin signaling and its interplay with other phytohormones during embryogenesis. In the present study, 29 auxin-related genes (mainly associated with auxin signaling) were differentially expressed (Fig. 4B). Among those, nine early auxin-responsive SMALL AUXIN UP RNA (SAUR) genes, which have been suggested to play a role in maize root development (Chen et al., 2014), were identified. Moreover, ZmIAA27 was expressed consistently higher in rtcs embryos than in wild-type embryos at all three developmental stages (Fig. 4B). ZmIAA27 is a key component of auxin signaling modules regulating primordia formation of maize tassels and ears (Galli et al., 2015).
Crosstalk between ethylene and auxin has been demonstrated for lateral root initiation in Arabidopsis (Ivanchenko et al., 2008). Moreover, ethylene synthesis involved in stress responses inhibits organ growth (Achard et al., 2006). Elevated expression of a number of ethylene biosynthesis genes was observed in rtcs embryos in the present study, including two genes encoding ACC oxidases (GRMZM2G013448: AtACO1, and GRMZM2G052422: AtACO4; Fig. 4B). Ethylene interacts with abscisic acid (ABA) signaling during seed development (Finkelstein et al., 2008). In the present study, a maize homolog (GRMZM2G149940) of the Arabidopsis ABA signaling gene ABI3 was expressed at significantly higher levels in wild-type embryos at all three stages (Fig. 4B). In Arabidopsis ABI3 is involved in lateral root development (Brady et al., 2003). The balance between ABA and gibberelic acid (GA) plays an important role in seed development (Finkelstein et al., 2008). Moreover, GA has been demonstrated to control root meristem size (Ubeda-Tomás et al., 2009). In the present study, a GA biosynthesis gene (GRMZM2G164090) homologous to OsGASR6 was expressed 100-fold higher in rtcs than in wild-type embryos at 25 dap (Fig. 4B). OsGASR6 is highly expressed in rice embryos and is suggested to be a key regulator of GA biosynthesis during seed development (Xue et al., 2012). Jasmonic acid (JA) promotes formation of lateral root primordia by an auxin-dependent mechanism (Raya-González et al., 2012). Moreover, it has been demonstrated that the LBD protein OsIG1 positively regulates the JA biosynthesis gene EG1 (Zhang et al., 2015). In this study, a JA biosynthesis gene, similar to Arabidopsis AtAOS, was expressed 5–11-fold higher in wild-type embryos (Fig. 4B). Brassinosteroid (BR) homeostasis and signaling are necessary for proper organ boundary formation (Gendron et al., 2012). Expression of five BR-related genes was up-regulated in wild-type embryos (Fig. 4B). Moreover, a feedback regulation between the Arabidopsis LBD gene LOB and BR-related genes has been demonstrated to limit growth in organ boundaries (Bell et al., 2012).
Taken together, differential expression of diverse hormone-related genes between rtcs and wild-type embryos suggests that the RTCS-dependent transcriptional regulation of genes associated with different phytohormones coordinates the complex interplay of molecular networks involved in the formation of seminal root primordia during embryogenesis.
Potential RTCS target genes during embryogenesis
To date, little is known about direct RTCS target genes in seminal root initiation. It has been demonstrated that RTCS is able to regulate the expression of target genes by binding to LBD promoter motifs (Majer et al., 2012). An in silico promoter analysis of genes differentially expressed between wild-type and rtcs during embryogenensis revealed that 37% (48/131) of transcription factor genes (Fig. 5) and 43% (31/73) of hormone-related genes contain at least one LBD motif (Figs 4A and 5). Among all differentially expressed genes, only 28% (970/3526) contained at least one LBD motif. Regulatory genes are typically expressed at low levels and are thus difficult to measure in classical microarray studies. As a consequence, in a comparative microarray study of rtcs and wild-type coleoptilar nodes, a significantly lower proportion (12%) of putative RTCS target genes was detected (Muthreich et al., 2013) compared with 28% in the present study. In Arabidopsis, it was demonstrated that the DNA-binding affinity of AtLOB to downstream target genes is regulated by interaction with bHLH048 (Husbands et al., 2007). Consistently, several members of the bHLH family were differentially expressed in this presnt study (Fig. 5), suggesting that proteins encoded by these genes could interact with RTCS and control their regulation of downstream genes.
The present study thus provides initial clues on putative RTCS target genes as a basis for further genetic investigations.
Evolutionary origin of RTCS-regulated genes during embryogenesis
Embryonic seminal roots are formed in maize, whereas they are missing in its close relative sorghum (Sorghum bicolor) (Singh et al., 2010). Modern maize was domesticated ~9000 years ago from its ancestor teosinte in the Mexican highlands, and after initial diversification it spread to the lowlands and thus into a completely different ecosystem (Matsuoka et al., 2002). As a consequence of this change, it can be hypothesized that ancient farmers inadvertently selected for genotypes with root architectures that performed better under lowland conditions where, for instance, phosphorus is less available (Lynch and Brown, 2008). One root trait that is associated with the domestication of teosinte into maize is the number of seminal roots. It has been demonstrated that the majority of teosinte accessions (62%) do not form seminal roots (Burton et al., 2013). While the remaining teosinte accessions form a maximum of three seminal roots, maize landraces form up to 11 (Burton et al., 2013) and modern maize varieties up to 13 seminal roots (reviewed in Hochholdinger, 2009).
Analyses of mutants have demonstrated that activity of the genes rtcs (Hetz et al., 1996) and rum1 (Woll et al., 2005) is necessary, albeit not sufficient, to form seminal roots. This is illustrated by the fact that both rtcs and rum1 (Taramino et al., 2007; von Behrens et al., 2011) are syntenic between maize and sorghum, which does not form seminal roots (Woodhouse et al., 2010; Schnable et al., 2011). In the present study, non-syntenic genes were significantly over-represented among differentially expressed genes (Fig. 6). This suggests that genes acting downstream of rtcs have mainly been recruited from non-syntenic genes that emerged by individual gene duplications and therefore have no homologs in sorghum. These genes might not have had crucial functions in maize development before their association with RTCS-dependent functions. This is in line with observations that non-syntenic genes might play an essential role in adaptation to environmental challenges (Schnable, 2015). Moreover, the anatomical organization of seminal roots suggests that they are better equipped for efficient soil resource absorption compared with more anciently formed primary and shoot-borne crown roots (Tai et al., 2016).
Conclusions
Taken together, this study provides first insights into the molecular framework underlying seminal root initiation in maize. Our data suggest that during seminal root evolution the syntenic gene rtcs has recruited evolutionary younger non-syntenic genes particularly associated with auxin signaling, transcription factor action, and cell proliferation by transcriptional regulation. Interaction of these genes with RTCS probably resulted in the specification of seminal root primordia and as a consequence contributed to the agronomic success of modern maize.
Supplementary data
Supplementary data are available at JXB online.
Table S1. RNA-Seq output and mapping results.
Table S2. Stage and genotype-specific gene activity patterns.
Table S3. Transcription factors among genotype-specific genes.
Table S4. Genes assigned to eight K-means clusters in wild-type and rtcs embryos.
Table S5. The 3526 genes differentially expressed between wild-type and rtcs embryos.
Table S6. Assignment of differentially expressed genes to MapMan categories.
Table S7. Genes related to hormone metabolism and cell proliferation differentially expressed between wild-type and rtcs embryos.
Table S8. Non-syntenic genotype-specific and differentially expressed genes.
Supplementary Material
Acknowledgements
We thank Stefan Hey and Jutta Baldauf (University of Bonn) for stimulating discussions of these results. HT was supported by a China Scholarship Council fellowship (number: 201206350004). Root research in the laboratory of FH is supported by the Deutsche Forschungsgemeinschaft (DFG) and the University of Bonn.
References
- Abdel-Ghani AH, Kumar B, Pace J, Jansen C, Gonzalez-Portilla PJ, Reyes-Matamoros J, San Martin JP, Lee M, Lübberstedt T. 2015. Association analysis of genes involved in maize (Zea mays L.) root development with seedling and agronomic traits under contrasting nitrogen levels. Plant Molecular Biology 88, 133–147. [DOI] [PubMed] [Google Scholar]
- Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd NP. 2006. Integration of plant responses to environmentally activated phytohormonal signals. Science 311, 91–94. [DOI] [PubMed] [Google Scholar]
- Aida M, Tasaka M. 2006. Morphogenesis and patterning at the organ boundaries in the higher plant shoot apex. Plant Molecular Biology 60, 915–928. [DOI] [PubMed] [Google Scholar]
- Bell EM, Lin W-C, Husbands AY, Yu L, Jaganatha V, Jablonska B, Mangeon A, Neff MM, Girke T, Springer PS. 2012. Arabidopsis lateral organ boundaries negatively regulates brassinosteroid accumulation to limit growth in organ boundaries. Proceedings of the National Academy of Sciences, USA 109, 21146–21151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SM, Sarkar SF, Bonetta D, McCourt P. 2003. The ABSCISIC ACID INSENSITIVE 3 (ABI3) gene is modulated by farnesylation and is involved in auxin signaling and lateral root development in Arabidopsis. The Plant Journal 34, 67–75. [DOI] [PubMed] [Google Scholar]
- Burton AL, Brown KM, Lynch JP. 2013. Phenotypic diversity of root anatomical and architectural traits in Zea species. Crop Science 53, 1042–1055. [Google Scholar]
- Carazo-Salas RE, Guarguaglini G, Gruss OJ, Segref A, Karsenti E, Mattaj IW. 1999. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature 400, 178–181. [DOI] [PubMed] [Google Scholar]
- Chen Y, Hao X, Cao J. 2014. Small auxin upregulated RNA (SAUR) gene family in maize: Identification, evolution, and its phylogenetic comparison with Arabidopsis, rice, and sorghum. Journal of Integrative Plant Biology 56, 133–150. [DOI] [PubMed] [Google Scholar]
- Erdelska O, Vidovencova Z. 1993. Development of adventitious seminal root primordia of maize during embryogenesis. Biologia Plantarum 48, 85–88. [Google Scholar]
- Feng Z, Zhu J, Du X, Cui X. 2012. Effects of three auxin-inducible LBD members on lateral root formation in Arabidopsis thaliana . Planta 236, 1227–1237. [DOI] [PubMed] [Google Scholar]
- Finkelstein R, Reeves W, Ariizumi T, Steber C. 2008. Molecular aspects of seed dormancy. Annual Review of Plant Biology 59, 387–415. [DOI] [PubMed] [Google Scholar]
- Galli M, Liu Q, Moss BL, Malcomber S, Li W, Gaines C, Federici S, Roshkovan J, Meeley R, Nemhauser JL. 2015. Auxin signaling modules regulate maize inflorescence architecture. Proceedings of the National Academy of Sciences, USA 112, 13372–13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron JM, Liu J-S, Fan M, Bai M-Y, Wenkel S, Springer PS, Barton MK, Wang Z-Y. 2012. Brassinosteroids regulate organ boundary formation in the shoot apical meristem of Arabidopsis. Proceedings of the National Academy of Sciences, USA 109, 21152–21157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkes CV, DeAngelis KM, Firestone MK. 2007. Root interactions with soil microbial communities and processes. In: Cardon ZG, Whitbeck JL, eds. The rhizosphere, an ecological perspective. Amsterdam: Elsevier, 1–29. [Google Scholar]
- Hetz W, Hochholdinger F, Schwall M, Feix G. 1996. Isolation and characterization of rtcs, a maize mutant deficient in the formation of nodal roots. The Plant Journal 10, 845–857. [Google Scholar]
- Hochholdinger F. 2009. The maize root system: morphology, anatomy, and genetics. In: Bennetzen J, Hake S, eds. Handbook of maize: its biology, New York: Springer, 145–160. [Google Scholar]
- Hochholdinger F, Park WJ, Sauer M, Woll K. 2004. From weeds to crops: genetic analysis of root development in cereals. Trends in Plant Science 9, 42–48. [DOI] [PubMed] [Google Scholar]
- Hund A, Fracheboud Y, Soldati A, Frascaroli E, Salvi S, Stamp P. 2004. QTL controlling root and shoot traits of maize seedlings under cold stress. Theoretical and Applied Genetics 109, 618–629. [DOI] [PubMed] [Google Scholar]
- Husbands A, Bell EM, Shuai B, Smith HM, Springer PS. 2007. LATERAL ORGAN BOUNDARIES defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Research 35, 6663–6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inukai Y, Sakamoto T, Ueguchi-Tanaka M, Shibata Y, Gomi K, Umemura I, Hasegawa Y, Ashikari M, Kitano H, Matsuoka M. 2005. Crown rootless 1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. The Plant Cell 17, 1387–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanchenko MG, Muday GK, Dubrovsky JG. 2008. Ethylene–auxin interactions regulate lateral root initiation and emergence in Arabidopsis thaliana . The Plant Journal 55, 335–347. [DOI] [PubMed] [Google Scholar]
- Jin J, Zhang H, Kong L, Gao G, Luo J. 2013. PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Research 42, 1182–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HW, Kim NY, Lee DJ, Kim J. 2009. LBD18/ASL20 regulates lateral root formation in combination with LBD16/ASL18 downstream of ARF7 and ARF19 in Arabidopsis. Plant Physiology 151, 1377–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Helariutta Y, Specht CD, Jung J, Sims L, Bruce WB, Diehn S, Benfey PN. 2000. Molecular analysis of the SCARECROW gene in maize reveals a common basis for radial patterning in diverse meristems. The Plant Cell 12, 1307–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu KP, Liou YC, Xiao Zhen Zhou XZ. 2002. Pinning down proline-directed phosphorylation signaling. Trends in Cell Biology 12, 164–172. [DOI] [PubMed] [Google Scholar]
- Lynch JP. 2011. Root phenes for enhanced soil exploration and phosphorus acquisition: tools for future crops. Plant Physiology 156, 1041–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JP. 2013. Steep, cheap and deep: an ideotype to optimize water and N acquisition by maize root systems. Annals of Botany 112, 347–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JP, Brown KM. 2008. Root strategies for phosphorus acquisition. In: White PJ, Hammond JP, eds. The ecophysiology of plant–phosphorus interactions, The Netherlands: Springer, 83–116. [Google Scholar]
- Majer C, Hochholdinger F. 2011. Defining the boundaries: structure and function of LOB domain proteins. Trends in Plant Science 16, 47–52. [DOI] [PubMed] [Google Scholar]
- Majer C, Xu C, Berendzen KW, Hochholdinger F. 2012. Molecular interactions of ROOTLESS CONCERNING CROWN AND SEMINAL ROOTS, a LOB domain protein regulating shoot-borne root initiation in maize (Zea mays L.). Philosophical Transactions of the Royal Society of London B: Biological Sciences 367, 1542–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka Y, Vigouroux Y, Goodman MM, Sanchez J, Buckler E, Doebley J. 2002. A single domestication for maize shown by multilocus microsatellite genotyping. Proceedings of the National Academy of Sciences, USA 99, 6080–6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthreich N, Majer C, Beatty M, Paschold A, Schützenmeister A, Fu Y, Malik WA, Schnable PS, Piepho H-P, Sakai H. 2013. Comparative transcriptome profiling of maize coleoptilar nodes during shoot-borne root initiation. Plant Physiology 163, 419–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. 2007. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. The Plant Cell 19, 118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B, De Rybel B, Casimiro I, Benková E, Swarup R, Laplaze L, Beeckman T, Bennett MJ. 2009. Arabidopsis lateral root development: an emerging story. Trends in Plant Science 14, 399–408. [DOI] [PubMed] [Google Scholar]
- Raya-González J, Pelagio-Flores R, López-Bucio J. 2012. The jasmonate receptor COI1 plays a role in jasmonate-induced lateral root formation and lateral root positioning in Arabidopsis thaliana . Journal of Plant Physiology 169, 1348–1358. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A. 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biology 11, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rue H, Martino S, Chopin N. 2009. Approximate Bayesian inference for latent Gaussian models by using integrated nested Laplace approximations. Journal of the Royal Statistical Society: Series B, Statistical Methodology 71, 319–392. [Google Scholar]
- Salvi S, Giuliani S, Ricciolini C, Carraro N, Maccaferri M, Presterl T, Ouzunova M, Tuberosa R. 2016. Two major quantitative trait loci controlling the number of seminal roots in maize co-map with the root developmental genes rtcs and rum1 . Journal of Experimental Botany 67, 1149–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanguineti M, Giuliani M, Govi G, Tuberosa R, Landi P. 1998. Root and shoot traits of maize inbred lines grown in the field and in hydroponic culture and their relationships with root lodging. Maydica 43, 211–216. [Google Scholar]
- Sass J. 1977. Morphology. In: Sprague GF, ed. Corn and corn improvement. Madison, WI: American Society of Agronomy Publishers, 89–110. [Google Scholar]
- Schnable JC. 2015. Genome evolution in maize: from genomes back to genes. Annual Review of Plant Biology 66, 329–343. [DOI] [PubMed] [Google Scholar]
- Schnable JC, Springer NM, Freeling M. 2011. Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proceedings of the National Academy of Sciences, USA 108, 4069–4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai B, Reynaga-Peña CG, Springer PS. 2002. The Lateral Organ Boundaries gene defines a novel, plant-specific gene family. Plant Physiology 129, 747–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V, van Oosterom EJ, Jordan DR, Messina CD, Cooper M, Hammer GL. 2010. Morphological and architectural development of root systems in sorghum and maize. Plant and Soil 333, 287–299. [Google Scholar]
- Tai H, Lu X, Opitz N, Marcon C, Paschold A, Lithio A, Nettleton D, Hochholdinger F. 2016. Transcriptomic and anatomical complexity of primary, seminal, and crown roots highlight root type-specific functional diversity in maize (Zea mays L.). Journal of Experimental Botany 67, 1123–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taramino G, Sauer M, Stauffer J, Multani D, Niu X, Sakai H, Hochholdinger F. 2007. The RTCS gene in maize (Zea mays L.) encodes a LOB domain protein that is required for post-embryonic shoot-borne and embryonic seminal root initiation. The Plant Journal 50, 649–659. [DOI] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M. 2004. MapMan: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. The Plant Journal 37, 914–939. [DOI] [PubMed] [Google Scholar]
- Tuberosa R, Salvi S, Sanguinetti MC, Landi P, Maccaferri M, Conti S. 2002. Mapping QTLs regulating morpho-physiological traits and yield: Case studies, shortcomings and perspectives in drought-stressed maize. Annals of Botany 89, 941–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda-Tomás S, Federici F, Casimiro I, Beemster GT, Bhalerao R, Swarup R, Doerner P, Haseloff J, Bennett MJ. 2009. Gibberellin signaling in the endodermis controls Arabidopsis root meristem size. Current Biology 19, 1194–1199. [DOI] [PubMed] [Google Scholar]
- Van De Wiel MA, Leday GG, Pardo L, Rue H, Van Der Vaart AW, Van Wieringen WN. 2012. Bayesian analysis of RNA sequencing data by estimating multiple shrinkage priors. Biostatistics 10, 1–16. [DOI] [PubMed] [Google Scholar]
- Villordon AQ, Ginzberg I, Firon N. 2014. Root architecture and root and tuber crop productivity. Trends in Plant Science 19, 419–425. [DOI] [PubMed] [Google Scholar]
- von Behrens I, Komatsu M, Zhang Y, Berendzen KW, Niu X, Sakai H, Taramino G, Hochholdinger F. 2011. Rootless with undetectable meristem 1 encodes a monocot-specific AUX/IAA protein that controls embryonic seminal and post-embryonic lateral root initiation in maize. The Plant Journal 66, 341–353. [DOI] [PubMed] [Google Scholar]
- Woll K, Borsuk LA, Stransky H, Nettleton D, Schnable PS, Hochholdinger F. 2005. Isolation, characterization, and pericycle-specific transcriptome analyses of the novel maize lateral and seminal root initiation mutant rum1 . Plant Physiology 139, 1255–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhouse MR, Schnable JC, Pedersen BS, Lyons E, Lisch D, Subramaniam S, Freeling M. 2010. Following tetraploidy in maize, a short deletion mechanism removed genes preferentially from one of the two homeologs. PLoS Biology 8, e1000409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Tai H, Saleem M, et al. 2015. Cooperative action of the paralogous maize lateral organ boundaries (LOB) domain proteins RTCS and RTCL in shoot borne root formation. New Phytologist 207, 1123–1133. [DOI] [PubMed] [Google Scholar]
- Xue LJ, Zhang JJ, Xue HW. 2012. Genome-wide analysis of the complex transcriptional networks of rice developing seeds. PloS ONE 7, e31081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P, Eggert K, von Wirén N, Li C, Hochholdinger F. 2015. Cell-type specific gene expression analyses by RNA-Seq reveal local high nitrate triggered lateral root initiation in shoot-borne roots of maize by modulating auxin-related cell cycle-regulation. Plant Physiology 169, 690–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P, Gutjahr C, Li C, Hochholdinger F. 2016. Genetic control of lateral root formation in cereals. Trends in Plant Science 21, 951–961. [DOI] [PubMed] [Google Scholar]
- Zhang J, Tang W, Huang Y, Niu X, Zhao Y, Han Y, Liu Y. 2015. Down-regulation of a LBD-like gene, OsIG 1, leads to occurrence of unusual double ovules and developmental abnormalities of various floral organs and megagametophyte in rice. Journal of Experimental Botany 66, 99–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Mickelson SM, Kaeppler SM, Lynch JP. 2006. Detection of quantitative trait loci for seminal root traits in maize (Zea mays L.) seedlings grown under differential phosphorus levels. Theoretical and Applied Genetics 113, 1–10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.