Abstract
Renal cell carcinoma (RCC) is one of the most malignant tumors in human. Here, we found that odd-skipped related transcription factor 1 (OSR1) was downregulated in 769-P and 786-O cells due to promoter CpG methylation. OSR1 expression could be restored by pharmacological demethylation treatment in silenced cell lines. Knockdown of OSR1 in two normal expressed cell lines- A498 and ACHN promoted cell invasion and cellular proliferation. RNA-Sequencing analysis showed that expression profile of genes involved in multiple cancer-related pathways was changed when OSR1 was downregulated. By quantitative real-time PCR, we confirmed that depletion of OSR1 repressed the expression of several tumor suppresor genes involved in p53 pathway, such as p53, p21, p27, p57 and RB in A498 and ACHN. Moreover, knockdown of OSR1 suppressed the transcriptional activity of p53. Of note, OSR1 depletion also led to increased expression of a few oncogenic genes. We further evaluated the clinical significance of OSR1 in primary human RCC specimens by immunohistochemical staining and found that OSR1 expression was downregulated in primary RCC and negatively correlated with histological grade. Thus, our data indicate that OSR1 is a novel tumor suppressor gene in RCC. Downregulation of OSR1 might represent a potentially prognostic marker and therapeutic target for RCC.
Keywords: OSR1, methylation, invasion, proliferation, RCC
INTRODUCTION
Renal cell carcinoma (RCC) is one of the most malignant tumors, which caused more than 140,000 deaths per year [1]. In 2013, more than 350,000 people were diagnosed with RCC worldwide. Smoking tobacco, hypertension and obesity are considered as risk factors for RCC [1]. Despite the development of therapeutic regimens [2], the prognosis of patients with RCC is still poor, mainly due to delayed diagnosis and a relatively high incidence of metastasis. Thus, there is an urgent need for identification of novel diagnosis and therapeutic targets for RCC. However, the molecular mechanism underlying the tumorigenesis of RCC remains elusive.
It is well known that abnormal genetic and epigenetic pattern will lead to tumorigenesis [3]. Currently, it is well accepted that epigenetic alterations even precede genetic changes during tumorigenesis [3]. An increasing number of epigenetic silenced tumor suppressor genes (TSGs) were identified in multiple cancers [4–8], which exert antitumor effects but silenced by promoter methylation in tumor specific manner.
OSR1 gene, located on human chromosome 2p24.1, contains three C2H2 zinc finger domains. OSR1 is reported to be involved in embryonic heart and urogenital formation. It also plays key roles in the development of the metanephric kidney and regulates formation and differentiation of kidney precursor cells. Furthermore, OSR1 is considered as a negative feedback regulator of nodal-induced endoderm development [9–12]. Of note, recent studies have shown tumor specific silencing of OSR1 by promoter methylation in gastric and lung cancer [13, 14]. Whereas overexpression of OSR1 significantly inhibited cell growth, arrested cell cycle, and induced apoptosis in the gastric cancer cell lines AGS, MKN28, and MGC803, knockdown of OSR1 led to enhancement of cell proliferation and inhibition of apoptosis in the normal gastric epithelial cell line GES1[13], indicating that OSR1 is a functional tumor suppressor in gastric cancer.
In this study, we found that OSR1 expression was frequently silenced in some of the RCC cells, and the expression silencing could be restored by 5-Aza-2′-deoxycytidine (DEC) treatment. Its downregulation was caused by promoter methylation as validated by quantitative methylation-specific PCR (qMSP). Knockdown of OSR1 in normal expressed cancer cell lines elevated invasion ability and cellular proliferation. RNA-Sequencing of RCC cell lines following OSR1 depletion has identified hundreds of potential target genes of OSR1, which are involved in DNA replication, cell cycle, mismatch repair, p53 and Wnt pathway. A few of downregulated TSGs (p53, p21, p27, p57 and RB) and upregulated oncogenes (MYC, FRA1, MET, HMGA1 and PIK3CA) were further confirmed by real-time PCR. In addition, knockdown of OSR1 repressed the transcriptional activity of p53. We further evaluated the clinical significance of OSR1 in primary human RCC specimens by immunohistochemical staining and found that OSR1 expression was downregulated in primary RCC and negatively correlated with histological grade. Thus, our data indicate that OSR1 functions as a novel TSG in RCC but is frequently epigenetically silenced in this cancer. Downregulation of OSR1 might represent a potentially prognostic marker and therapeutic target for RCC.
RESULTS
Expression profile of OSR1 in RCC cells
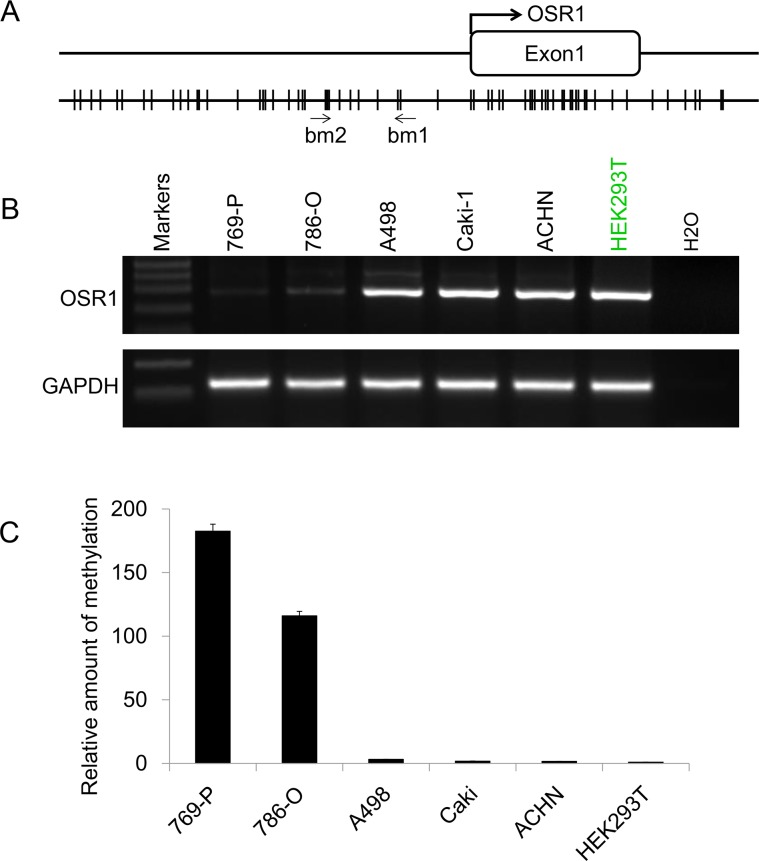
Promoter sequence analysis of the OSR1 gene revealed a typical CpG island spanning the proximal promoter and exon 1 regions (Figure 1A). We then checked the expression profile of OSR1 in five RCC cell lines and one immortalized human renal epithelial cell line HEK293T by semi-quantitative RT-PCR. We found that OSR1 was expressed in A498, Caki-1, ACHN and HEK293T, but downregulated in 769-P and 786-O (Figure 1B). This tumor specific silenced pattern suggested that OSR1 was potential silenced by promoter methylation in a tumor specific manner.
Figure 1.
A. Schematic structure of OSR1 promoter CGI. The transcription start site is indicated by a curved arrow. qMSP primers are indicated. Bm1 and bm2 designate primers designed according to the sequence of the bottom strain. B. The expression profile of OSR1 in a series of RCC cell lines. GAPDH was used as an internal control. C. qMSP results of OSR1 promoter in RCC cell lines.
Downregulation of OSR1 was caused by promoter methylation
To determine whether methylation of OSR1 results in its downregulation in specific RCC cell lines, the methylation status of OSR1 promoter was examined by qMSP with primers OSR1bm1 and OSR1bm2 (Figure 1A). ACTB was used as internal control to monitor the DNA quantity and quality. We found that promoter of OSR1 was methylated in 769-P and 786-O, where expression of OSR1 was downregulated, but not in cell lines of ACHN, A498, Caki-1 and HEK293T, where OSR1 was normal expressed (Figure 1C). Our data suggested that promoter methylation of OSR1 led to its downregulation in RCC.
Pharmacological demethylation restored OSR1 expression in RCC cell lines
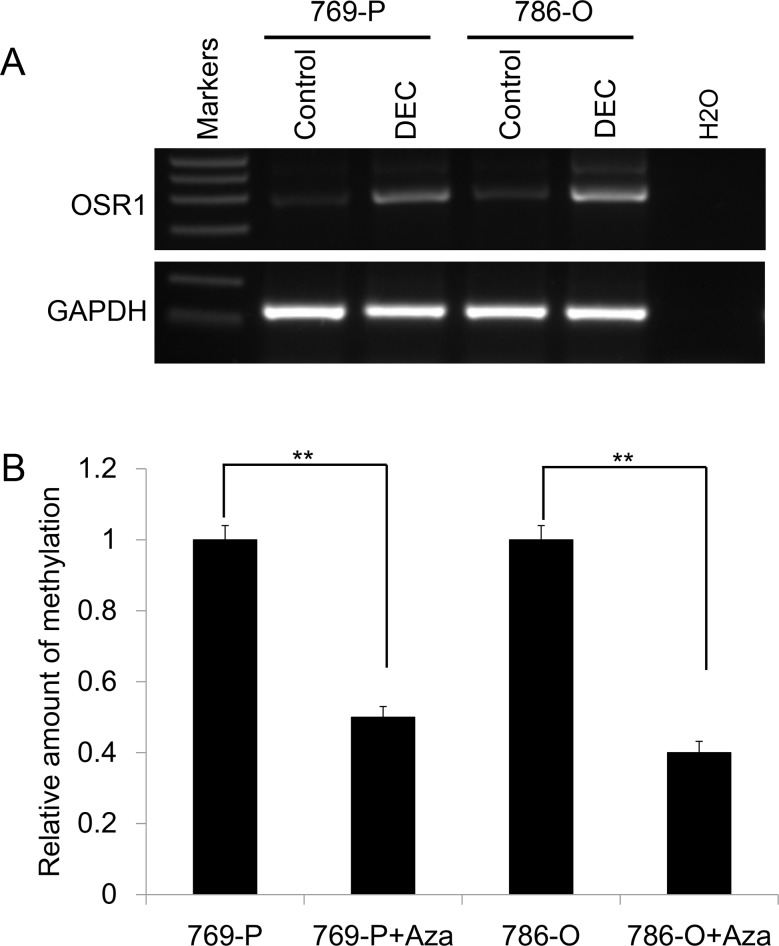
To further validate our hypothesis that downregulation of OSR1 was directly mediated by promoter methylation, 769-P and 786-O cells with methylated and downregulated OSR1 were treated with DNA methytransferase inhibitor DEC. Pharmacological demethylation treatment with DEC resulted in the upregulation of OSR1 expression (Figure 2A) accompanied by a decrease in the methylated alleles of OSR1 (Figure 2B) in 769-P and 786-O cells. These results indicated that downregulation of OSR1 was directly caused by promoter methylation in RCC cells.
Figure 2.
A. Pharmacological demethylation with DEC restored the expression of OSR1 in silenced cells. B. qMSP results of OSR1 promoter in pharmacological demethylated cells and untreated cells.
Loss of OSR1 promoted cell invasion in RCC
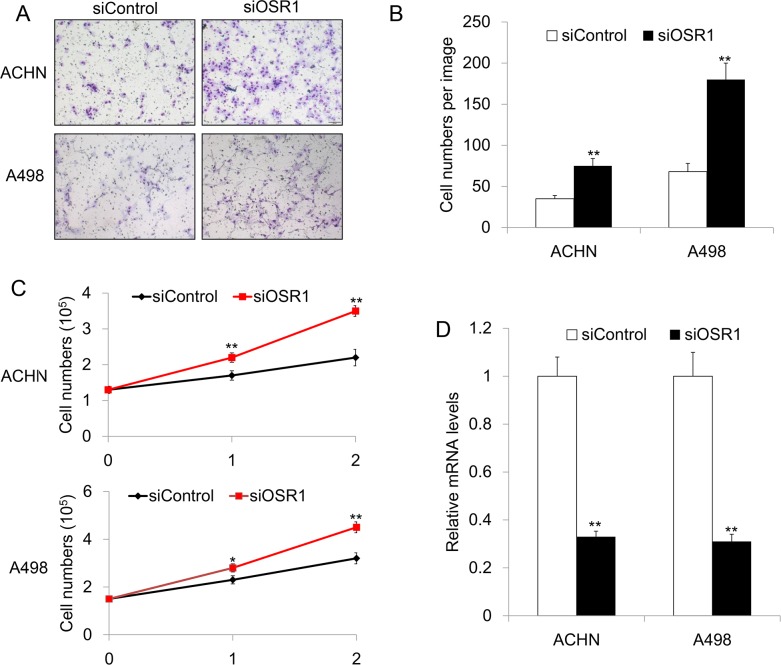
Previous study showed that OSR1 is a functional tumor suppressor in gastric cancer [13]. The expression profile of OSR1 in RCC indicated that OSR1 might also have tumor suppressor function in renal cancer. In order to investigate the role of OSR1 in RCC, siRNA knockdown of OSR1 was performed in RCC cell lines of A498 and ACHN that show normal OSR1 expression. We examed the role of OSR1 in renal cancer cell invasion by transwell invasion assays. The number of siOSR1-transfected A498 or ACHN cells observed on the filter was significantly increased compared with the number of siControl-transfected cells (P<0.01). Our data revealed that knockdown of OSR1 in A498 and ACHN increased RCC cell invasive ability in vitro (Figure 3A&3B), suggesting that OSR1 is a negative regulator of cell invasion in RCC.
Figure 3.
A. Representative invasion image of OSR1 in siControl-transfected or siOSR1-transfected ACHN and A498 cells. B. Quantitative analysis of invasive cell numbers in siControl-transfected or siOSR1-transfected ACHN and A498 cells. **, P<0.01. C. Growth curve of ACHN and A498 cells without or with OSR1 silencing, respectively. **, P<0.01. D. Knockdown efficacy of OSR1 in ACHN an A498 cells. **, P<0.01.
Loss of OSR1 enhanced cellular proliferation in RCC
We further test proliferation rate in OSR1 knockdown cells. Firstly, we seed the cells at appropriate density in six-well plate. After 16 hours, cells were transfected with siControl or siOSR1, respectively. Cell numbers were counted at 0 h, 24 h and 48 h after transfection. Interestingly, we found that loss of OSR1 lead to higher proliferation rate in both ACHN and A498 cells (Figure 3C&3D), indicating that OSR1 could inhibit cell proliferation in RCC cells.
OSR1 regulated multiple genes expression
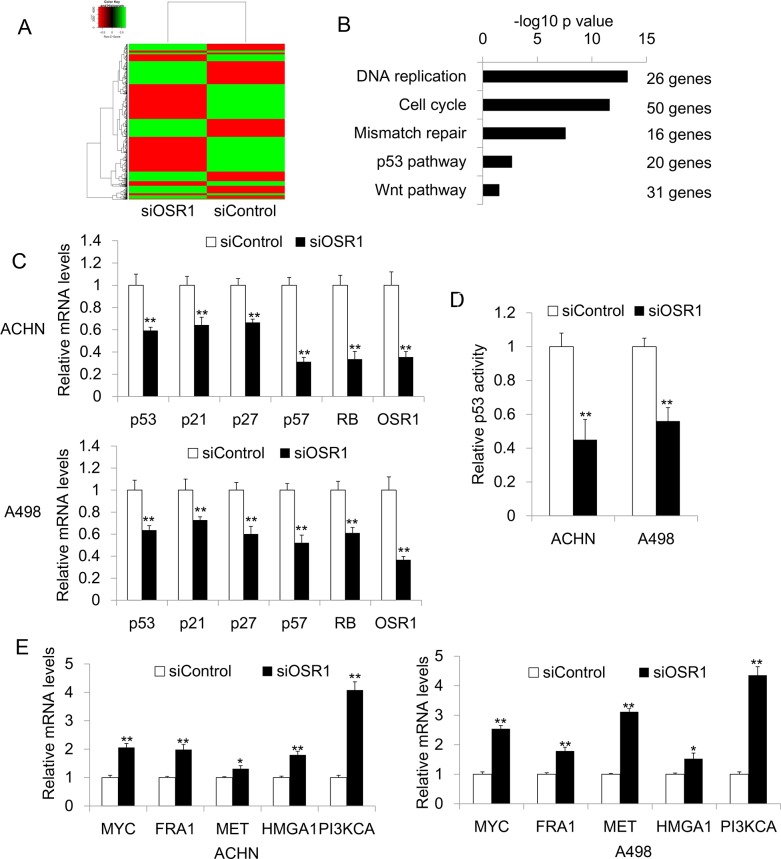
Our functional study suggested that OSR1 is a functional tumor suppressor in RCC. To explore the underlying mechanism by which OSR1 exerts tumor suppressive function in RCC, we performed RNA-Sequencing analysis to identify genes that were differentially expressed in OSR1 knockdown ACHN cells and control ACHN cells. Genes with 2 fold changes were considered as significant (Figure 4A). Firstly, we analyzed the candidate genes by Go analysis. We found that most of the downstream genes are involved in DNA replication, cell cycle, mismatch repair, p53 and Wnt pathway (Figure 4B). The involvement of those cancer related pathway indicated that OSR1 has functional role in tumorigenesis.
Figure 4.
A. Heatmap for RNA sequencing results from OSR1 knockdown ACHN cells and control ACHN cells. B. Go analysis for RNA sequencing results. C. Confirmation of downregulated genes in ACHN and A498 cells by quantitative real-time PCR. **, P<0.01. D. P53 luciferase assay in siControl-transfected or siOSR1-transfected ACHN and A498 cells. **, P<0.01. E. Validation of upregulated genes by real-time PCR in ACHN an A498 cells. *, P<0.05; **, p<0.01.
We further confirmed the expression of potential OSR1 target gene by quantitative real-time PCR. In both ACHN and A498 cells, knockdown of OSR1 inhibited the tumor suppressor genes, including p53, p21, p27, p57 and RB gene expression (Figure 4C). Moreover, OSR1 knockdown clearly suppressed p53 promoter activity in ACHN and A498 cells (Figure 4D). In addition, we found that loss of OSR1 increased the mRNA levels of several oncogenes including MYC, FRA1, MET, HMGA1, and PIK3CA (Figure 4E). Our data suggested that OSR1 acted as a TSG through regulating multiple TSGs and oncogene expression.
OSR1 was downregulated in primary RCC and correlated with histological grade
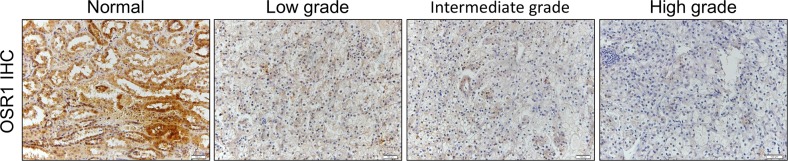
Our study suggested that OSR1 is a silenced tumor suppressor in specific RCC cell lines due to promoter methylation. We further investigate the clinical significance of OSR1 in primary human RCC specimens by immunohistochemical staining. We found that OSR1 was downregulated in 82.7% (62/75) primary RCC tissues (Table 1, Figure 5). Its expression was significantly lower in primary RCC tissues compared to that in normal tissues (P<0.0001, Table 1). Of note, OSR1 expression was negatively correlated with histological grade (P=0.002). However, no correlation was found between OSR1 expression and age, gender, and clinical stage. Our data suggested that downregulation of OSR1 might represent a potentially prognostic marker for RCC.
Table 1. Relationship between Clinicopathological Variables and OSR1 Expression Level in RCC Patients.
| Classification | Number | Low expression, n(%) | High expression, n(%) | P |
|---|---|---|---|---|
| Tissues | ||||
| Normal | 75 | 20(26.7) | 55(73.3) | <0.0001* |
| RCC | 75 | 62(82.7) | 13(17.3) | |
| Age (year) | ||||
| <60 | 29 | 23(79.3) | 6(20.7) | 0.542 |
| ≥60 | 46 | 39(84.8) | 7(15.2) | |
| Gender | ||||
| male | 50 | 40(80.0) | 10(20.0) | 0.524 |
| female | 25 | 22(88.0) | 3(12.0) | |
| Clinical stage | ||||
| I~II | 52 | 44(84.6) | 8(15.4) | 0.503 |
| III~IV | 23 | 18(78.3) | 5(21.7) | |
| Histologic grade | ||||
| poorly differentiated | 28 | 26 (92.8) | 2(7.1) | 0.002* |
| moderate differentiated | 22 | 20 (90.9) | 2(9.1) | |
| well differentiated | 25 | 16(64) | 9(36) |
Low expression including no(−) and weak(+) staining, high expression including moderate (++) and strong (+++) staining.
Figure 5.
A. Representative IHC image of OSR1 in primary RCC samples. OSR1 was downregulated in patient samples and negatively correlated with histological grade of primary RCC.
DISCUSSION
In this study, we identified OSR1 as a novel TSG in RCC. We found that OSR1 was downregulated by promoter methylation in RCC cells. Inhibition of OSR1 promoted cell invasion and proliferation. Expression profile of genes involved in multiple cancer-related pathways was changed when OSR1 was downregulated. A few of representative downregulated TSGs (p53, p21, p27, p57 and Rb) and upregulated oncogenes (Myc, Fra1, MET, HMGA1, STAT2, PIK3CA and L1CAM) were further confirmed by real-time PCR. We also found that OSR1 was downregulated in primary RCC and correlated with histological grade. Thus, our present study indicated that OSR1 is a novel TSG in RCC but is frequently silenced by promoter methylation in this cancer. Downregulation of OSR1 might represent a potentially prognostic marker and therapeutic target for RCC.
Development of RCC from a normal cell is a complex and multi-step process with multiple oncogenes, TSGs and signaling transduction pathways involved in this process [1–3, 15–20]. Increasing numbers of promoter methylated TSGs identified in RCC [21–23] contribute to elucidating the molecular mechanisms of RCC tumorigenesis. Here, we identified OSR1 as a novel TSG in RCC. OSR1 contains three C2H2 zinc finger domain. Previous studies suggested that OSR1 was involved in embryonic heart and urogenital formation, development of the metanephric kidney, negative feedback regulator of nodal-induced endoderm development [9–12]. OSR1 expression is also regulated by Runx2 and Ikzf1, which are known as master-gene of osteogenesis and hematopoiesis, respectively [24]. But the function of OSR1 in cancer is largely unknown. Previous study of OSR1 in gastric cancer suggested that OSR1 is a functional tumor suppressor in gastric cancer. It is frequently silenced by promoter methylation in gastric cell line and primary tumor samples [13]. The present study demonstrated for the first time that OSR1 is a novel TSG in RCC, which is downregulated by promoter methylation. Remarkly, OSR1 depletion promoted renal cancer cell invasion and proliferation at least partially through p53 pathway and other important cellular regulators.
The p53 pathway can regulate the basic cellular activity such as proliferation, apoptosis, cell cycle and cellular senescence [25]. Upon a stress signal, activated p53 will bind to p53-responsive DNA sequence elements in the genome. It increases p21 for cell cycle arrest which results in proliferation inhibition. P27 is involved in cell cycle progression and acts as a tumor suppressor to control both tissue expansion and cell proliferation [26]. p57(KIP2) regulates several hallmarks of cancer, including cell invasion, metastasis, apoptosis, and angiogenesis [27]. Tumor suppressor RB contributes to a diversity of cellular functions, including cell proliferation, differentiation, cell death, and genome stability [28]. Interestingly, genes involved in p53 pathway, such as p21, p27, p57, and RB, were significantly downregulated in OSR1 knockdown RCC cells. Besides, we further confirmed the effect of OSR1 on p53 pathway by luciferase assay. We found that knockdown of OSR1 significantly downregulates p53 activity in ACHN and A498 cell lines, which further confirmed our finding. In consistent with our finding, previous study of OSR1 in gastric cancer also found that OSR1 upregulates p53 in gastric cancer. The role and the regulation of p53 were also reported and summarized in many studies [23, 29–33]. TSGs Mir-22 and ATS/TMS1 could regulate p53 activity in RCC cells [30, 32]. The p53 also regulates several TSGs in RCC [23, 34]. All these suggested that OSR1 functions as a critical TSG in RCC in part through regulation of p53 signaling pathway.
Besides its effect on tumor suppressor genes, we also found that repression of OSR1 led to increased expression of a few of oncogenes. We confirmed the upregulation of several oncogenes by real-time PCR, including MYC, FRA1, MET, HMGA1 and PIK3CA. MYC is correlated with cell growth, proliferation and apoptosis [35]. FRA1 is a component of AP-1 transcription factor complex, which could promote the cell ability of invasion and migration [36]. MET has been implicated in a variety of cellular processes, including cell proliferation, survival, migration, motility and invasion [37]. HMGA1 upregulates cellular proliferation and invasion in multiple cancers [38]. PIK3CA has been shown to be important for tumor cell survival, adhesion, motility and proliferation [39]. The real-time PCR results could explain how OSR1 downregulates invasion and proliferation in RCC. Our future work will focus on how OSR1 regulates those genes' expression.
We further investigate the clinical significance of OSR1 in primary human RCC specimens and found that OSR1 was downregulated in primary RCC tissues. Importantly, OSR1 expression was negatively correlated with histological grade, indicating a potential role of OSR1 as a prognostic marker for RCC.
In summary, we found that OSR1 was downregulated in RCC cells by promoter methylation. OSR1 can function as a tumor suppressor via inhibition of invasion and proliferation in RCC cells, possibly via upregulating tumor suppressor genes and downregulating oncogenes. Downregulation of OSR1 was observed in primary RCC and its downregulation was correlated with histological grade, making it a potentially prognostic marker and therapeutic target for RCC.
MATERIALS AND METHODS
Cell culture and transfection
A series of RCC cell lines (769-P, 786-O, A498, Caki-1, and ACHN) and an immortalized human embryonic kidney cell line - HEK293T were used for this study. Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin was used for cell culture. Cells were cultured in DMEM in a humidified chamber maintained at 37°C and 5% CO2. OSR1-short interfering RNA (siOSR1) and control siRNA (siControl) were purchased from Santa Cruz Biotechnology (Dallas, TX, USA). Transfection was carried out according to the manufacturer's instruction using RNAiMAX transfection reagent (Invitrogen, Eugene, OR, USA).
Pharmacological demethylation with DEC
The method of DEC treatment was described before [4, 6]. In brief, ACHN cells or A498 cells (1 × 105/mL) were allowed to grow overnight in 10 cm cell culture dishes. The cell culture medium was replaced with fresh medium containing 50 μmol/L DEC for every 24 h, for three consecutive days. Then ACHN or A498 cells were harvested for RNA extraction.
RNA extraction, semi-quantitative reverse transcription PCR (RT-PCR) and real-time PCR
RNA was extracted with Trizol reagent according to manufacturer's protocol (Invitrogen, Eugene, OR, USA). The cDNA was synthesized using Random hexamers and SuperScript-III (Invitrogen, Eugene, OR, USA). Semi-quantitative RT-PCR was performed for 32 cycles with AccuPrime™ DNA polymerases, according to manufacturer's protocol (Invitrogen, Eugene, OR, USA). All primers used are listed in Supplementary Table 1. Quantitative real-time PCR was carried out with the Applied Biosystems 7300 real-time systems using real-time PCR Master Mix (SYBR Green). Each experiment was conducted in triplicate in three independent experiments.
DNA bisulfite treatment and quantitative methylation-specific PCR (qMSP)
The bisulfite treatment was carried out with the EpiTect Bisulfite kit (QIAGEN) by following the manufacturer's instructions. In brief, around 2 ug DNA was used for each reaction and mixed with 85 μL bisulfite mix and 35 μL DNA protect buffer. Bisulfite conversion was performed on a thermocycler followed the manufacturer's instructions. After that, the bisulfite-treated DNA was recovered by EpiTect spin column and used for qMSP. The qMSP was performed with SYBR Green master mix. To correct for differences in both quality and quantity between samples, ACTB was used as an internal control.
Growth curve
ACHN or A498 cells were seeded in six-well plates at an appropriate density. After 16 hours, cells were transfected with siControl or siOSR1 through RNAiMax. The cell numbers were counted at 0 h, 24 h, and 48 h after transfection. Each experiment was conducted in triplicate in three independent experiments.
Invasion assay
Cell invasion assay was performed using 24-well culture plates (Millipore, Billerica, MA) with inserts of 8-μm pore membranes pre-coated with Matrigel (BD Bioscience, San Jose, CA). Briefly, ACHN or A498 cells were transfected with siControl or siOSR1 through RNAiMax at an appropriate density in six-well plates. Twenty-four hours after transfection, cells were trypsinized and transferred to the upper Matrigel chamber in 100 μL of serum-free medium supplementing with 1 × 105 cells. The lower chamber was supplemented with medium containing 10% FBS. The invaded cells were stained with 0.1% crystal violet according to manufacturer's protocol (Fisher scientific, Atlanta, GA, USA) after 48 h. Cell numbers per microscopic image field were counted to compare the invasion ability between siControl and siOSR1. Each experiment was conducted in triplicate in three independent experiments.
RNA sequencing
RNA was purified with an RNeasy Mini kit (QIAGEN). The RNA-Sequencing library preparation was performed according to the manual of manufacturers (KAPA biosystems). Sequencing reactions were performed with the Illumina HiSeq platform. RNA-seq reads were mapped to the human genome (hg19) using Burrows-Wheeler Aligner (bwa) [40]. We then marked the duplicate reads using picard [41]. The HTseq tool [42] was used to calculate the reads count for each gene. Finally, we used the Reads Per Kilobase per Million mapped reads (rpkm) command in edgeR [43] package to calculate the rpkm of each gene.
Luciferase assay
To exam the effect of OSR1 on p53 pathway, ACHN or A498 was transfected with p53-luc (Stratagene) and pRL-TK by lipofectamine 2000 (Invitrogen). After 6 hours, medium was changed and cells were transfected with siControl or siOSR1 with RNAiMAX (Invitrogen). After 48 hours, cells were harvested and the activity of p53 was analyzed by Promega dual luciferase reporter assay system. All the experiments were performed in triplicates in three independent experiments.
Patient samples and immunohistochemistry (IHC)
Primary tumor tissues and adjacent normal kidney tissues from 75 different cases of RCC patients were collected in Shenzhen People's Hospital with patients' permission. IHC was performed on 4-μm sections of formalin-fixed, paraffin-embedded human RCC tissues. Sections were deparaffinized, rehydrated and subjected to heat induced antigen retrieval. After incubation with blocking solution, sections were incubated with anti-OSR1 antibody (Abcam) for 1 h, biotinylated secondary antibody for 30 min, and then with streptavidin horseradish peroxidase for another 10 min. Sections were developed with 3,3′-diaminobenzidine chromogen and further stained with hematoxylin. An H-score was assigned to each tissue based on the product of staining intensity ((−), nostaining; (+), weak; (++), moderate; and (+++), strong) and percentage of stained cells (0-0%, 1-1% to 30%, 2-31% to 70%, and 3-71% to 100%). Chi-squared and Fisher's exact test were performed to analyze the association between OSR1 expression and clinicopathological characteristics.
Statistical analysis
Data are presented as mean ± standard deviation. Statistical assessments were carried out using Student's t test. P < 0.05 was considered statistically significant.
SUPPLEMENTARY TABLE
Acknowledgments
This work was supported by the Shenzhen Commission of Science and Innovation program (No JCYJ20150403101028172), National Natural Science Foundation of China (81670760, 81301783), Shenzhen Dedicated Funding for Strategic Development of Emerging Industry (JCYJ20140418091413584) and funding from Guangdong Provincial Department of Science and Technology (2014A020212370).
Footnotes
CONFLICTS OF INTEREST
No potential conflicts of interest were disclosed.
REFERENCES
- 1.Capitanio U, Montorsi F. Renal cancer. Lancet. 2016;387:894–906. doi: 10.1016/S0140-6736(15)00046-X. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 3.Coleman WB, Tsongalis GJ. Molecular mechanisms of human carcinogenesis. Exs. 2006:321–349. doi: 10.1007/3-7643-7378-4_14. [DOI] [PubMed] [Google Scholar]
- 4.Cheng Y, Liang P, Geng H, Wang Z, Li L, Cheng SH, Ying J, Su X, Ng KM, Ng MH, Mok TS, Chan AT, Tao Q. A novel 19q13 nucleolar zinc finger protein suppresses tumor cell growth through inhibiting ribosome biogenesis and inducing apoptosis but is frequently silenced in multiple carcinomas. Molecular cancer research. 2012;10:925–936. doi: 10.1158/1541-7786.MCR-11-0594. [DOI] [PubMed] [Google Scholar]
- 5.Wang S, Cheng Y, Du W, Lu L, Zhou L, Wang H, Kang W, Li X, Tao Q, Sung JJ, Yu J. Zinc-finger protein 545 is a novel tumour suppressor that acts by inhibiting ribosomal RNA transcription in gastric cancer. Gut. 2013;62:833–841. doi: 10.1136/gutjnl-2011-301776. [DOI] [PubMed] [Google Scholar]
- 6.Cheng Y, Geng H, Cheng SH, Liang P, Bai Y, Li J, Srivastava G, Ng MH, Fukagawa T, Wu X, Chan AT, Tao Q. KRAB zinc finger protein ZNF382 is a proapoptotic tumor suppressor that represses multiple oncogenes and is commonly silenced in multiple carcinomas. Cancer research. 2010;70:6516–6526. doi: 10.1158/0008-5472.CAN-09-4566. [DOI] [PubMed] [Google Scholar]
- 7.Fu L, Dong SS, Xie YW, Tai LS, Chen L, Kong KL, Man K, Xie D, Li Y, Cheng Y, Tao Q, Guan XY. Down-regulation of tyrosine aminotransferase at a frequently deleted region 16q22 contributes to the pathogenesis of hepatocellular carcinoma. Hepatology. 2010;51:1624–1634. doi: 10.1002/hep.23540. [DOI] [PubMed] [Google Scholar]
- 8.Shu XS, Li L, Ji M, Cheng Y, Ying J, Fan Y, Zhong L, Liu X, Tsao SW, Chan AT, Tao Q. FEZF2, a novel 3p14 tumor suppressor gene, represses oncogene EZH2 and MDM2 expression and is frequently methylated in nasopharyngeal carcinoma. Carcinogenesis. 2013;34:1984–1993. doi: 10.1093/carcin/bgt165. [DOI] [PubMed] [Google Scholar]
- 9.Wang Q, Lan Y, Cho ES, Maltby KM, Jiang R. Odd-skipped related 1 (Odd 1) is an essential regulator of heart and urogenital development. Developmental biology. 2005;288:582–594. doi: 10.1016/j.ydbio.2005.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.James RG, Kamei CN, Wang Q, Jiang R, Schultheiss TM. Odd-skipped related 1 is required for development of the metanephric kidney and regulates formation and differentiation of kidney precursor cells. Development. 2006;133:2995–3004. doi: 10.1242/dev.02442. [DOI] [PubMed] [Google Scholar]
- 11.Tena JJ, Neto A, de la Calle-Mustienes E, Bras-Pereira C, Casares F, Gomez-Skarmeta JL. Odd-skipped genes encode repressors that control kidney development. Developmental biology. 2007;301:518–531. doi: 10.1016/j.ydbio.2006.08.063. [DOI] [PubMed] [Google Scholar]
- 12.Terashima AV, Mudumana SP, Drummond IA. Odd skipped related 1 is a negative feedback regulator of nodal-induced endoderm development. Developmental dynamics. 2014;243:1571–1580. doi: 10.1002/dvdy.24191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Otani K, Dong Y, Li X, Lu J, Zhang N, Xu L, Go MY, Ng EK, Arakawa T, Chan FK, Sung JJ, Yu J. Odd-skipped related 1 is a novel tumour suppressor gene and a potential prognostic biomarker in gastric cancer. The Journal of pathology. 2014;234:302–315. doi: 10.1002/path.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rauch TA, Wang Z, Wu X, Kernstine KH, Riggs AD, Pfeifer GP. DNA methylation biomarkers for lung cancer. Tumour biology. 2012;33:287–296. doi: 10.1007/s13277-011-0282-2. [DOI] [PubMed] [Google Scholar]
- 15.Cheng Y, Tu Y, Liang P. Promoter Methylated Tumor Suppressor Genes in Glioma. Cancer Translational Medicine. 2015;1:123–130. [Google Scholar]
- 16.Sun S, Xu A, Yang G, Cheng Y. Galanin is a novel epigenetic silenced functional tumor suppressor in renal cell carcinoma. Cancer Translational Medicine. 2015;1:183–187. [Google Scholar]
- 17.Chen D, Dai C, Jiang Y. Histone H2A and H2B Deubiquitinase in Developmental Disease and Cancer. Cancer Translational Medicine. 2015;1:170–175. [Google Scholar]
- 18.Zhun F, Liang Y, Chen D, Li Y. Melanoma Antigen Gene Family in the Cancer Immunotherapy. Cancer Translational Medicine. 2016;2:85–89. [Google Scholar]
- 19.Peng L, Hu Y, Chen D, Jiao S, Sun S. Ubiquitin specific peptidase 21 regulates interleukin-8 expression, stem-cell like property of human renal cell carcinoma. Oncotarget. 2016;7:42007–42016. doi: 10.18632/oncotarget.9751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang Y, Zhu F, Zhang H, Chen D, Zhang X, Gao Q, Li Y. Conditional ablation of TGF-beta signaling inhibits tumor progression and invasion in an induced mouse bladder cancer model. Scientific reports. 2016;6:29479. doi: 10.1038/srep29479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Q, Zhang L, Li L, Wang Z, Ying J, Fan Y, Xu B, Wang L, Liu Q, Chen G, Tao Q, Jin J. Interferon regulatory factor 8 functions as a tumor suppressor in renal cell carcinoma and its promoter methylation is associated with patient poor prognosis. Cancer letters. 2014;354:227–234. doi: 10.1016/j.canlet.2014.07.040. [DOI] [PubMed] [Google Scholar]
- 22.Xu X, Wu J, Li S, Hu Z, Xu X, Zhu Y, Liang Z, Wang X, Lin Y, Mao Y, Chen H, Luo J, Liu B, et al. Downregulation of microRNA-182-5p contributes to renal cell carcinoma proliferation via activating the AKT/FOXO3a signaling pathway. Molecular cancer. 2014;13:109. doi: 10.1186/1476-4598-13-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin J, Deng Z, Tanikawa C, Shuin T, Miki T, Matsuda K, Nakamura Y. Downregulation of the tumor suppressor HSPB7, involved in the p53 pathway, in renal cell carcinoma by hypermethylation. International journal of oncology. 2014;44:1490–1498. doi: 10.3892/ijo.2014.2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamauchi M, Kawai S, Kato T, Ooshima T, Amano A. Odd-skipped related 1 gene expression is regulated by Runx2 and Ikzf1 transcription factors. Gene. 2008;426:81–90. doi: 10.1016/j.gene.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 25.Levine AJ, Hu W, Feng Z. The P53 pathway: what questions remain to be explored? Cell death and differentiation. 2006;13:1027–1036. doi: 10.1038/sj.cdd.4401910. [DOI] [PubMed] [Google Scholar]
- 26.Wander SA, Zhao D, Slingerland JM. p27: a barometer of signaling deregulation and potential predictor of response to targeted therapies. Clinical cancer research. 2011;17:12–18. doi: 10.1158/1078-0432.CCR-10-0752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kavanagh E, Joseph B. The hallmarks of CDKN1C (p57, KIP2) in cancer. Biochimica et biophysica acta. 2011;1816:50–56. doi: 10.1016/j.bbcan.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 28.Gordon GM, Du W. Conserved RB functions in development and tumor suppression. Protein & cell. 2011;2:864–878. doi: 10.1007/s13238-011-1117-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noon AP, Vlatkovic N, Polanski R, Maguire M, Shawki H, Parsons K, Boyd MT. p53 and MDM2 in renal cell carcinoma: biomarkers for disease progression and future therapeutic targets? Cancer. 2010;116:780–790. doi: 10.1002/cncr.24841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang S, Zhang D, Yi C, Wang Y, Wang H, Wang J. MicroRNA-22 functions as a tumor suppressor by targeting SIRT1 in renal cell carcinoma. Oncology reports. 2016;35:559–567. doi: 10.3892/or.2015.4333. [DOI] [PubMed] [Google Scholar]
- 31.Liu Q, Jin J, Ying J, Cui Y, Sun M, Zhang L, Fan Y, Xu B, Zhang Q. Epigenetic inactivation of the candidate tumor suppressor gene ASC/TMS1 in human renal cell carcinoma and its role as a potential therapeutic target. Oncotarget. 2015;6:22706–22723. doi: 10.18632/oncotarget.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ku BM, Kim DS, Kim KH, Yoo BC, Kim SH, Gong YD, Kim SY. Transglutaminase 2 inhibition found to induce p53 mediated apoptosis in renal cell carcinoma. FASEB journal. 2013;27:3487–3495. doi: 10.1096/fj.12-224220. [DOI] [PubMed] [Google Scholar]
- 33.Caratozzolo MF, Valletti A, Gigante M, Aiello I, Mastropasqua F, Marzano F, Ditonno P, Carrieri G, Simonnet H, D'Erchia AM, Ranieri E, Pesole G, Sbisa E, et al. TRIM8 anti-proliferative action against chemo-resistant renal cell carcinoma. Oncotarget. 2014;5:7446–7457. doi: 10.18632/oncotarget.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma JJ, Liao CG, Jiang X, Zhao HD, Yao LB, Bao TY. NDRG2 suppresses the proliferation of clear cell renal cell carcinoma cell A-498. Journal of experimental & clinical cancer research. 2010;29:103. doi: 10.1186/1756-9966-29-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cai Q, Medeiros LJ, Xu X, Young KH. MYC-driven aggressive B-cell lymphomas: biology, entity, differential diagnosis and clinical management. Oncotarget. 2015;6:38591–38616. doi: 10.18632/oncotarget.5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Milde-Langosch K. The Fos family of transcription factors and their role in tumourigenesis. European journal of cancer. 2005;41:2449–2461. doi: 10.1016/j.ejca.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Du Z, Zhang M. Biomarker development in MET-targeted therapy. Oncotarget. 2016;7:37370–37389. doi: 10.18632/oncotarget.8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Di Cello F, Shin J, Harbom K, Brayton C. Knockdown of HMGA1 inhibits human breast cancer cell growth and metastasis in immunodeficient mice. Biochemical and biophysical research communications. 2013;434:70–74. doi: 10.1016/j.bbrc.2013.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lai K, Killingsworth MC, Lee CS. Gene of the month: PIK3CA. Journal of clinical pathology. 2015;68:253–257. doi: 10.1136/jclinpath-2015-202885. [DOI] [PubMed] [Google Scholar]
- 40.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. http://broadinstitute.github.io/picard.
- 42.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.