Abstract
Rice tiller angle is an important agronomic trait that contributes to crop production and plays a vital role in high yield breeding. In this study, a recombinant inbred line (RIL) population derived from the cross of a glabrous tropical japonica rice D50 and an indica rice HB277, was used to investigate quantitative trait loci (QTLs) controlling rice tiller angle. Two major QTLs, qTAC8 and qTAC9, were detected. While qTAC9 mapped with a previously identified gene (TAC1), using a BC2F2 population qTAC8 was mapped to a 16.5 cM region between markers RM7049 and RM23175. Position of qTAC8 was narrowed to a 92 kb DNA region by two genetic segregating populations. Finally, one opening reading frame (ORF) was regarded as a candidate gene according to genomic sequencing and qRT-PCR analysis. In addition, a set of four near isogenic lines (NILs) were created to investigate the genetic relationship between those two QTLs, and one line carrying qTAC8 and qTAC9 presented additive effect of tiller angle, suggesting that these QTLs are involved in different genetic pathways. Our results provide a foundation for the cloning of qTAC8 and genetic improvement of the rice plant architecture.
Introduction
Rice (Oryza sativa L.) is one of the most important food crops in China and the world. It is the most effective safeguard for food security and agricultural sustainable development through high-yielding rice breeding. Ideotype breeding strategy is an important approach to increase grain yield potential in rice breeding. Tiller angle, the angle between the main culm and its side tillers [1], is a decisive factor for building ideal plant architecture, whereby neither spread-out rice nor compact type rice is beneficial for grain production [2]. With a spread-out architecture, plants can decrease humidity and escape from some diseases, but they occupy too much space and increase shading and lodging, consequently decreasing photosynthetic efficiency and grain yield per unit area. On the other hand, compact plants prejudice in capturing light and prevention of plant diseases and insect pests, thus appropriate tiller angle is beneficial for improving rice production [3, 4]. Although rice tiller angle has long attracted attention of breeders due to the significant contribution to plant architecture and yield potential, the genetic mechanisms determining its characteristics are not fully understood.
Rice tiller angle has been recognized as a complex quantitative trait, which is not only controlled by genetic factors but also greatly influenced by environmental conditions, such as light intensity, climate, soil, planting density, watering and fertilizing [4]. In the past two decades, a number of QTLs for tiller angle have been identified on chromosomes 1, 2, 5, 7, 8, 9, 11, 12 in various mapping populations of rice. Using an F2 population helped to identify three major genes controlling tiller angle [5]. A major QTL (ta9) on chromosome 9 flanked by RZ228 to RG667 together with other four QTLs (QTa1, QTa2, QTa3, QTa8) were separated in a F2:4 genetic segregating population generated from a cross between Lemont and Teqing [6]. In addition, a doubled haploid population generated from a cross between Zhaiyeqing 8 (loose plant architecture) and Jingxi 7 (compact plant architecture) was used and a total of three controlling tiller angle QTLs named qTA-9a, qTA-9b, qTA-12, that account for 22.7%, 11.9% and 20.9% of the variance, respectively, were detected [7]. Moreover, two major QTLs, qTA8-2 and qTA9-2 were determined in a recombinant inbred line population derived from Xieqingzao B/Miyang 46, and no G×E interaction effect was detected for the additive effect of these two QTLs [1]. Five major QTLs including qTA-9, qTA-2, qTA-7a, qTA-7b and qTA-11 for tiller angle were present in a RIL population from a cross between Asominori and IR24. Then a major QTL, qTA-9, was singled out in a 15 cM region between RFLP markers C609 and C508 by using a CSSLs population [8]. To date, only two major QTLs for tiller angle have been cloned. One is TAC1, a major QTL for tiller angle was isolated by using a large F2 population that derives from the cross between indica rice IR24, and an introgression line IL55. TAC1 harbors three introns in its coding region and a fourth 1.5 kb intron in the 3’ untranslated region, and encodes a 259-amino-acid unknown protein. A mutation in the 3’ splicing site of the fourth intron from ‘AGGA’ to ‘GGGA’ decreases its transcript levels, resulting in compact plant architecture. tac1 has also been extensively utilized in densely planted rice [4]. The second cloned QTL for tiller angle corresponds to PROG1 (PROSTRATE GROWTH 1), a semi-dominant gene encoding a newly identified Cys2-His2 zinc-finger transcription factor located on chromosome 7. PROG1 is predominantly expressed in the axillary meristems and the site of tiller bud formation, and disrupting the prog1 function and inactivate prog1 expression can lead to erect growth, increasing grain number and higher grain yield in cultivated rice [9, 10]. Dong identified three major QTLs (TAC3, DWARF2 (D2) and TAC1) controlling tiller angles by genome-wide association studies. TAC3 encodes a conserved hypothetical protein of 152 amino acids that is preferentially expressed in the tiller base[11].
Previous reports indicate that the tiller angle is not only affected by QTLs but also could be controlled by single genes following the Mendel’s genetics. Thus far, several genes controlling tiller angle have been cloned. LAZY1 (LA1), a novel grass-specific gene, is expressed during gravitropism sensitivity and plays negative role in polar auxin transport (PAT). Loss-of-function of LA1 enhances PAT causing alteration of endogenous IAA distribution in shoots, leading to reduced gravitropism and a tiller-spreading phenotype of rice plants [12, 13]. LPA1 (Loose Plant Architecture 1), identified on chromosome 5, encodes a plant-specific INDETERMINATE DOMAIN protein that regulates tiller angle by controlling the adaxial growth of tiller nodes [3]. PIN-FORMED1 and PIN-FORMED2 are auxin efflux transporters, and suppressing the expression of rice PIN-FORMED1 or enhancing the expression of rice PIN-FORMED2 can alter PAT and increase tiller angles [14, 15].
To further elucidate the genetic control of tiller angle, we used a RIL population that derived from a cross between a japonica and an indica rice cultivars to pinpoint two major QTLs for this trait. qTAC8 and qTAC9 were detected, and qTAC8 was narrowed down to a 92 kb region where one candidate ORF was determined as the one encoding for qTAC8.
Materials and methods
Plant materials
A recombinant inbred line (RIL) population of 190 lines was generated from the cross between D50 and HB277 as described previously by Shao [16]. Briefly, one line (RIL-77) that carried the homozygous target segments RM339-RM210 on chromosome 8 and RM201-RM7306 on chromosome 9 from HB277 (S1A Fig) was chosen from the RILs to backcross with D50. The resultant BC2F1 was selfed to obtain a BC2F2 population containing 178 plants for genotyping and phenotyping, and a progeny (BC2F2:3) was used for phenotyping (S1B Fig). The RILs and BC2F2 population were grown at the experimental site of Fuyang district of Hangzhou in 2012, and the BC2F2:3 plants were grown at the experimental site of Lingshui (Hainan Province, China) and Fuyang district in 2013, respectively. Six plants per row were transplanted with a distance of 18 cm between plants within a row, and 18 cm between rows, and four rows were used to grow each line.
Two large segregating populations BC2F4 and BC2F5 carrying the heterozygous target segment RM339-RM210 on chromosome 8 and the D50 target segment RM201-RM7306 on chromosome 9, were picked for screening recombinant individuals in our target region. As a result, the homozygous recombinants were used for genotyping and phenotyping. One plant from BC2F1 carrying the heterozygous target segment RM339-RM210 and the HB277 target segment RM201-RM7306 was selected to consecutive backcross with D50 and produce BC4F1 and 120 BC4F2 individuals. Finally four NILs designated NIL-qTAC8qtac9, NIL-qtac8qtac9, NIL-qTAC8qtac9 and NIL-qTAC8qTAC9 were developed. This set of four NILs was planted in the experimental site of Lingshui in 2014 following a randomized block design with three repeats. Each line was grown in a six-row plot with 6 plants in each row and spacing of 18 x 18 cm.
PCR and development of molecular markers
DNA was extracted following the protocols described by Murray and Rogers [17, 18]; Each 10 μL PCR reaction system contained 1 μL 10X PCR buffer, 1 μL dNTP (2 mM), 1μL of primer (1 mM), 0.1 μL Taq DNA polymerase (5 U/μL) and 1 μL template DNA. Polymerase chain reaction (PCR) comprised an initial denaturation step (95°C for 3 min), followed by 35 cycles of 95°C for 30 s, 55°C for 30 s and 72°C for 45 s, and ending with an extension step of 72°C for 10 min. PCR products were separated by electrophoresis and silver staining procedure. Simple sequence repeats (SSR) markers were selected covering the target region based on the published linkage map of rice (http://www.gramene.org). InDel (Insertion and Deletion) markers used for fine mapping of qTAC8 were designed based on the reference Nipponbare and 93–11 genomic sequences.
Sequencing and identification of candidate genes
The target gene in the candidate genomic region was predicted using the SIGnAL package (http://signal.salk.edu/). The full-length genomic DNA sequence of the candidate gene was determined by dividing it into several overlapping segments. Sequencing primers were designed according to the sequence of cv. Nipponbare in the target region. The PCR products were sequenced directly. Primer sequences used in this study are listed in S1 Table.
RNA isolation and quantitative real-time PCR
Six plants of each line were pooled for RNA extraction. Total RNA samples from tiller base at four growth stages (seedling, tillering, heading and ripening stages) were extracted using RNAiso Plus (Takara), following the manufacturer’s instructions. The first cDNA strand was synthesized from 3 ug RNA using the First Strand cDNA synthesis Kit-Rever Tra Ace-α (ToYoBo). qRT-PCR analysis was performed on a Roche Light Cycler 480 device using various gene-specific primers. The rice Ubi gene (LOC_Os03g0234200) was chosen as reference gene. Reactions containing SYBR premix Ex TaqII (TaKaRa) were carried out in a final volume of 20 ul. The 2-△△CT method [19, 20] was used to calculate relative levels of transcription. The PCR reaction implied an initial denaturation step (95°C for 4 min), followed by 50 cycles of 95°C for 15 s and 55°C for 30s. Three technical replicates were analyzed for each cDNA sample.
Data analysis
Rice linkage maps were constructed using MAPMAKER/Exp Ver. 3.0, and genetic distances were converted into cM by using the Kosambi function. Composite interval mapping (CIM) analysis of QTL in the RILs and the BC2F2 population was performed with QTL cartographer Ver. 2.5 (statgen.ncsu.edu/qtlcart/WQTLCart.htm). QTLs were called where their LOD value exceeded 2.5. Mean phenotypic values were compared using the Student’s t test. Multiple comparison test and the correlation between genotypes and phenotypes were carried out by the SAS statistical software package.
Results
Primary mapping of tiller angle
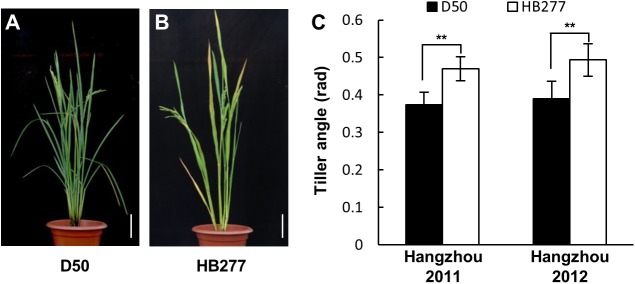
Rice tiller angle serves as an important trait in rice. In this work, a glabrous tropical japonica rice D50 cultivar which exhibits relative compact plant type and an indica rice HB277 cultivar displaying a relative loose architecture were used in this study (Fig 1A and 1B). Then a RIL population was used to detect QTLs for this trait. Measurement of the tiller angle showed a continuous and normal distribution whose variation range was 0.267–1.010 rad (S2 Fig). QTL mapping strategy was conducted and two major QTLs for tiller angle named qTAC8 on chromosome 8 and qTAC9 on chromosome 9 were identified in this RIL population. qTAC8 and qTAC9 were mapped within the region of RM339-RM210 and RM201-RM7306, respectively. qTAC8 could account for almost 33.4% of the variance, while qTAC9 could explain around 17.4% of the variance in tiller angle (Table 1). The positive allele (increasing tiller angle) qTAC9 was inherited from HB277. Unexpectedly, the positive allele qTAC8 derived from D50 which displayed a relative compact plant architecture.
Fig 1. Variation in tiller angle between D50 and HB277.
A and B, the phenotype of D50 and HB277 at heading stage. Scale bar 10 cm. C, mean tiller angle over two years. Error bars represent standard deviations (n = 6); ** indicates significant difference at P≤0.01.
Table 1. Two major tiller angle QTLs detected by analysis of the RIL populations.
| Year/Local | Locus | Chr | Interval | LOD value | Additive | R2 (%) |
|---|---|---|---|---|---|---|
| 2011/Hangzhou | qTAC8 | 8 | RM339-RM210 | 12.87 | 0.125 | 33.4 |
| qTAC9 | 9 | RM201-RM7306 | 9.31 | -0.086 | 17.4 |
qTAC9 is the same gene as TAC1
In accord to reports in the literature, we found that qTAC9 was closely linked to TAC1 flanked by the SSR loci RM201 and RM1026 [4], and a SNP in the 3’ splicing site of the fourth intron of TAC1 was detected after sequencing the TAC1 alleles in D50 and HB277 (S3A Fig). qRT-PCR analysis showed that the expression of TAC1 in HB277 is significant higher than D50 (S3B Fig). These results coincided with the report that a mutation in the 3’ splicing site of the fourth intron from ‘AGGA’ to ‘GGGA’ can decrease the expression of TAC1 and lead to a compact plant. Hence, we can anticipate that qTAC9 and TAC1 may correspond to the same gene.
Characterization of qTAC8
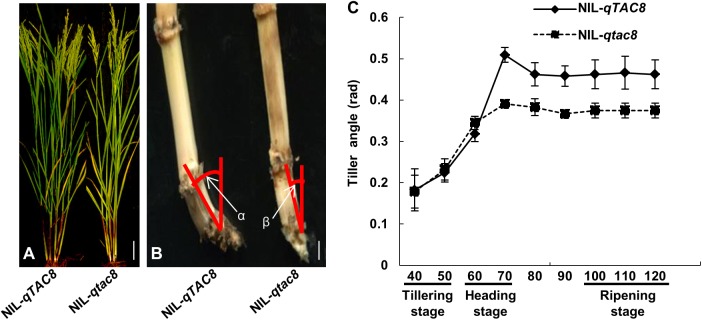
In order to investigate qTAC8, we measured the tiller angle of NIL-qTAC8 and NIL-qtac8. The results showed that the tiller angle of NIL-qTAC8 is significant larger than that in NIL-qtac8 at the ripening stage, and the angle of tiller base in NIL-qTAC8 is significant larger than that in NIL-qtac8 (Fig 2A and 2B). We also found that the two NILs almost exhibited the same tiller angle at tillering stage, but NIL-qTAC8 showed a greater tiller angle than that in NIL-qtac8 since heading stage (Fig 2C). There were no significant differences in tiller numbers, plant height and spikelet fertility between NIL-qTAC8 and NIL-qtac8 (data not shown).
Fig 2. Characterization of NIL-qTAC8 and NIL-qtac8.
A, phenotypes of NIL-qTAC8 and NIL-qtac8 at ripening stage, respectively, Scale bar 10 cm; B, Comparison of the angle of the tiller base between NIL-qTAC8 and NIL-qtac8 at ripening stage, Scale bar 1 cm; C, tiller angle changing during development in NIL-qTAC8 and NIL-qtac8. Error bars represent standard deviations (n = 6). The numbers of X-axis indicate the plant growing days after seed germination.
Fine mapping of qTAC8
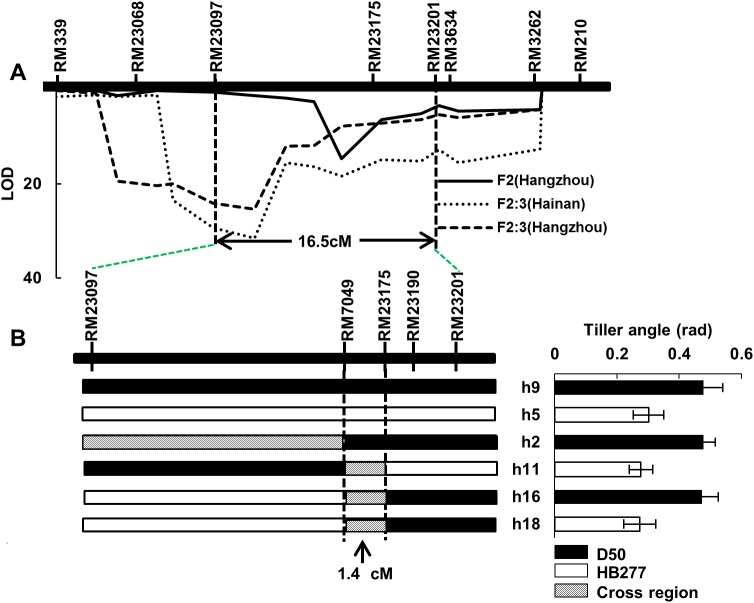
To further investigate the QTL qTAC8, a BC2F2 population containing 178 individuals was established (S1B Fig). The tiller angle of the BC2F2 population showed a normal distribution. According to the progeny test, the BC2F2 individuals could be classified into three subgroups of homozygotes for D50 (DD), for HB277 (HH) and heterozygotes at the targeted region. According to the tiller angle performance in the progeny test, paired t test was used to compare the difference between subgroups. Significant differences occurred between DD and the other two subgroups, HH and DH, suggesting that the DD genotype presented a larger tiller angle (0.462 rad) than HH (0.294 rad), while the tiller angle of the heterozygotes was an intermediate value (S4 Fig). These results showed that qTAC8 is a QTL in D50 that presents a partial dominance and a positive additive effect.
Seven more SSR markers were used to genotype the BC2F2 population and to construct a local linkage group covering 26.2 cM. The phenotype of two populations BC2F2 and BC2F2:3 were investigated in this study and qTAC8 was mapped within a 16.5 cM interval flanked by RM23097-RM23201. qTAC8 could account for 26.3% of the phenotype variance with additive effect of 0.07 rad in BC2F2 population, while in the BC2F2:3, the QTL could explain 47.1% and 51.8% of the variation in Hainan and Hangzhou, respectively (Table 2). In order to test and verify this, four informative homozygous recombinants were identified with four markers within RM23097-RM23201 and grouped into four genotypes according to the positions of recombinant breakpoints and allelic composition. Multiple comparisons between the tiller angle and recombinant individual genotypes, using the two non-recombinant lines as controls (h9 and h5), reflected that qTAC8 was narrowed down to a 1.4 cM interval flanked by RM7049 and RM23175 (Fig 3A and 3B).
Table 2. qTAC8 detected by analysis of the BC2F2 and BC2F2:3 populations.
| Lines | Locus | Chr | Interval | LOD value | Additive | R2 (%) |
|---|---|---|---|---|---|---|
| BC2F2(Hangzhou) | qTAC8 | 8 | RM23097-RM23201 | 14.58 | 0.070 | 26.3 |
| BC2F2:3(Hainan) | qTAC8 | 8 | RM23097-RM23201 | 31.50 | 0.076 | 47.1 |
| BC2F2:3(Hangzhou) | qTAC8 | 8 | RM23097-RM23201 | 25.50 | 0.062 | 51.8 |
Fig 3. Preliminary mapping of qTAC8.
A, qTAC8 was mapped within the SSR loci RM23097 and RM23201 on chromosome 8 by using BC2F2 and BC2F2:3 populations. B, qTAC8 was placed within a 1.4 cM region flanked by RM7049 and RM23175.
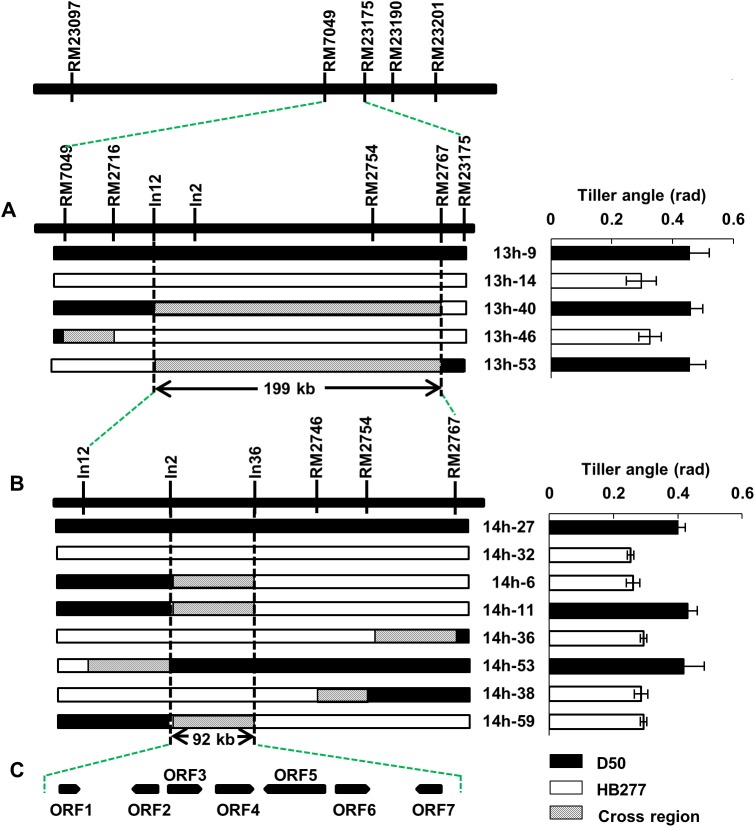
To further fine mapping qTAC8, a segregating population with 2,000 individuals derived from BC2F4 lines that carried a heterozygosis segments at the qTAC8 region, were used to identify the recombinants between RM7049 and RM23175 (Fig 4A). Next, the identified recombinants were analyzed by seven more markers located between RM7049 and RM23175. Multiple comparisons were conducted and qTAC8 was placed in a 199 kb region between In12 and RM2767. To position qTAC8 more precisely, a large segregating population of 6,000 individuals was introduced from BC2F5, and a total of 40 recombinants were identified with the help of two markers In12 and RM2767, and four polymorphism markers within this region were developed to genotype these individuals. Multiple comparisons were also conducted between the genotypes of these homozygous recombinants and the phenotypes of their progeny. Finally, qTAC8 was placed spanning on BAC P0431A03, in a 92 kb region flanked by In2 and In36 (Fig 4B).
Fig 4. Fine mapping of qTAC8.
A, Fine mapping of qTAC8 within a 199 kb region flanked by In2 and RM2767 based on a separation population progenies. B, Fine mapping of qTAC8 within a 92 kb region flanked by In2 and In36. C, Candidate genes for qTAC8 within the region flanked by In12 and In36.
Analysis of candidate genes for qTAC8
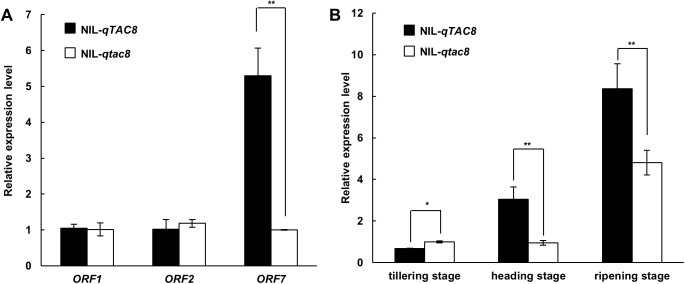
Based on the genome annotated database (http://signal.salk.edu/), the critical 92 kb region contains seven predicted ORFs. They encode a TCP family transcription factor, an ATP-dependent Clp protease adaptor protein, a transposon protein, two retrotransposon proteins, a zinc knuckle family protein and a basic helix-loop-helix protein (Fig 4C and Table 3). First, the transposon protein and retrotransposon proteins were excluded as candidates for qTAC8, leaving ORF1, ORF2, ORF6 and ORF7. Genomic sequencing of these four candidate genes in D50 and HB277 revealed that nucleotide diversity occurred except for ORF2. No products of ORF6 genomic DNA were identified in two parents (data not shown), which might be explained by a deletion of this gene during rice evolution, thus ORF6 was also excluded as candidate gene for qTAC8 (data not shown). Quantitative real-time PCR was used to characterize the transcripts of the three candidate genes left in the tiller base at heading stage of NIL-qTAC8 and NIL-qtac8. No significant difference in expression was observed for ORF1 and ORF2, while RNA expression level of ORF7 in NIL-qTAC8 was significant higher than in NIL-tac8 (Fig 5A). This result indicates that ORF7 is the possible candidate for qTAC8. Next we also found that the expression level of ORF7 was also increased at heading and ripening stages, but not at tillering stage, which coincides with the phenotypes of one NIL pair NIL-qTAC8 and NIL-qtac8 at three developing stages (Fig 2C and Fig 5B). All together, these data suggest that ORF7 is the candidate gene for qTAC8. Its cDNA stretched 1038 bp, comprises two exons, and encodes a 345 amino-acid protein containing a putative bHLH conserved domain. Although we found that seven single nucleotide polymorphisms (SNPs) occurred in ORF7 between the NIL pair, they didn’t cause any variation at the amino acid level. Further genomic sequencing results indicated that several nucleotides difference in the promoter and 3’ UTR region of ORF7 might be responsible for the alteration of gene expression (S5 Fig).
Table 3. The list of candidate genes between In2 and In36.
| ORFs | Gene | Protein function |
|---|---|---|
| ORF1 | Os08g33530 | TCP family transcription factor, putative |
| ORF2 | Os08g33540 | ATP-dependent Clp protease adaptor protein ClpS containg protein, expressed |
| ORF3 | Os08g33550 | transposon protein, putative, Mutator sub-class |
| ORF4 | Os08g33560 | retrotransposon protein, putative, unclassified |
| ORF5 | Os08g33570 | retrotransposon protein, putative, unclassified |
| ORF6 | Os08g33580 | zinc knuckle family protein, putative |
| ORF7 | Os08g33590 | basic helix-loop-helix, putative, expressed |
Fig 5. Relative expression analysis of qTAC8 candidate genes.
A, relative expression analysis of ORF1, ORF2 and ORF7. B, relative expression analysis of ORF7 at different developmental stages between the NIL pair. * indicate significant difference at level P≤0.05 (n = 3); ** indicate significant difference at level P≤0.01 (n = 3).
Expression analysis other tiller angle-related genes
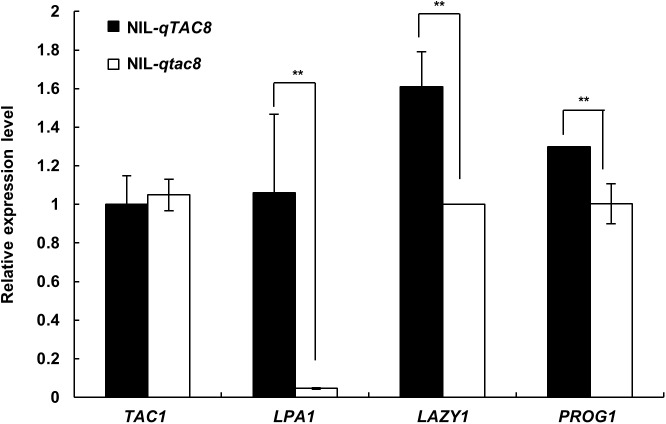
Tiller angle is known to be controlled by TAC1, LPA1, LAZY1 and PROG1. To investigate the expression pattern of those genes in the NILs used in this study, qRT-PCR analysis was conducted. We found that LPA1, LAZY1 and PROG1 were all affected by the positive function of the ORF7 allele, whereas there was no difference in expression of TAC1 between the NIL pair (Fig 6).
Fig 6. Expression analysis of tiller angle-related genes between NIL-qTAC8 and NIL-qtac8 at ripening stage.
Values shown are mean ± SD (n = 3). ** indicate significant difference at level P≤0.01 (n = 3).
Genetic relationship between qTAC8 and qTAC9
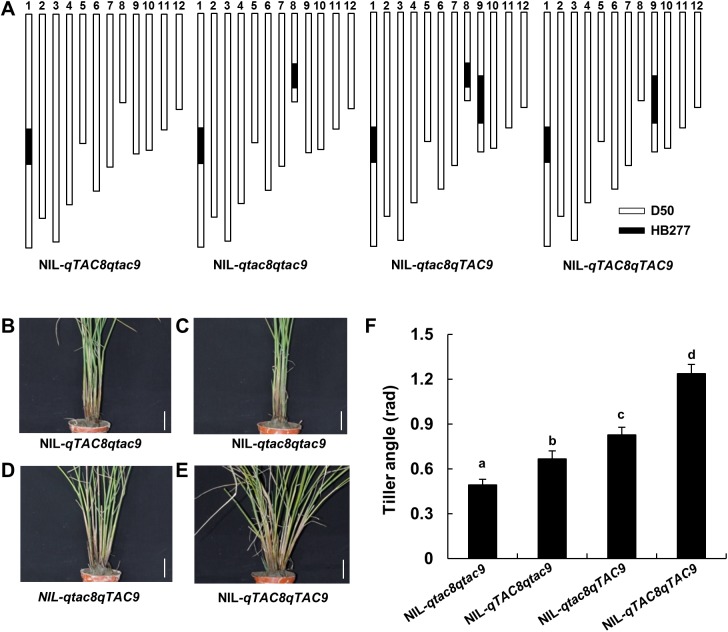
To further study the genetic relationship between qTAC8 and qTAC9, a set of four NILs including NIL-qTAC8qTAC9, NIL-qtac8qTAC9, NIL-qtac8qTAC9 and NIL-qtac8qtac9 was produced to evaluate tiller angle (Fig 7A–7E). The results showed that the tiller angle of NIL-qTAC8qTAC9 was significantly larger than in the other three NILs, with NIL-qtac8qTAC9 and NIL-qtac8/qtac9 presenting the smallest. The tiller angle of NIL-qTAC8qTAC9 displayed an additive effect, indicating that qTAC8 and qTAC9 may be participating in different genetic pathways (Fig 7F).
Fig 7. Genetic relationship between qTAC8 and qTAC9.
A, graphical genotypes of the four NIL pairs, including NIL-qTAC8qtac9, NIL-qtac8qtac9, NIL-qtac8qTAC9 and NIL-qTAC8qTAC9. B-E, phenotypes of NIL-qTAC8qtac9, NIL-qtac8qtac9, NIL-qtac8qTAC9 and NIL-qTAC8qTAC9, respectively. Scale bar 1 cm. F, Tiller angle of four NIL pairs. Error bars represent standard deviations (n = 6). Significant differences revealed by Tukey’s multiple comparison test are indicated by letters above bars (P < 0.05).
Discussion
Plant ideotype has been recognized as an advanced breeding concept and is regarded to be highly associated with high grain yield in rice breeding [21]. Rice traits for plant ideotype include plant height, stem strength, leaf morphology, panicle morphology, tiller angle among other critical traits. Ideal Plant Architecture 1 (IPA1) was a major gene affecting rice productivity. Introduction of the IPA1 semi-dominant gene into the japonica rice Xiushui 11 cultivar resulted in increased seed yield [22]. Tiller angle also plays a central role in rice production formation, and appropriate tiller angle is beneficial for improving rice production [3, 4]. Thus exploration of new genes controlling tiller angle would facilitate strategies to manipulate plant ideotype and increasing rice yield.
In this study, we identified two major QTLs controlling tiller angle on chromosomes 8 and 9 which were named qTAC8 and qTAC9, respectively. Using an F7 RIL population derived from a cross between D50 and HB277 (Table 1), qTAC9 was located between the SSR loci RM201 and RM7306 and the positive allele (increasing tiller angle) at qTAC9 originated from HB277 with a loose plant architecture (Table 1). Previous studies revealed that the target region near qTAC9 was a hot site controlling rice tiller angle on the long arm of chromosome 9 [1, 4, 6, 7], and one QTL named TAC1 was isolated within this interval and cloned using a large F2 population derived from a cross between indica rice IR24 and an introgressiong line IL55 [4]. Sequencing analysis indicated that a SNP (‘AGGA’ to ‘GGGA’) occurred between D50 and HB277, and qRT-PCR analysis revealed that the RNA level of TAC1 was significant reduced in D50 compared to HB277, suggesting that qTAC9 was the same allele of TAC1, a gene that can form a plant with spread-out architecture and is wildly used within indica rice varieties (S3A and S3B Fig). A partially dominant gene qTAC8 originated from D50 with a compact plant architecture and positive allele of the other QTL identified for tiller angle nearby. qTA8-2 which controls rice tiller angle was firstly mapped between R1394 and RZ66 [1], which locates on the same region with qTAC8, suggesting that qTA8-2 may be an allele of qTAC8. However primary QTL mapping, fine mapping, candidate gene prediction and qRT-PCR analysis indicated that ORF7, which encodes a basic helix-loop-helix protein, might be the gene underlying this QTL (Fig 4 and Table 3). Previous studies indicate that the semi-dominant gene PROSTRATE GROWTH 1 which affects plant architecture, also encodes a basic helix-loop-helix transcriptional factor, suggesting that qTAC8 might present a similar function as PROG1 [9, 10].
Analysis of genetic interactions among genes for tiller angle is required to better understand pathways controlling rice tiller angle formation. qTAC8 (the positive allele from D50) and qTAC9 (positive allele from HB277) are two semi-dominant genes for tiller angle, and a double mutant NIL-qTAC8qTAC9 presented additive effect for that trait, suggesting that qTAC8 and qTAC9 are involved in different pathways. This was consistent with the similar expression of TAC1 observed in NIL-qTAC8 and NIL-qtac8 (Fig 6 and Fig 7). Interesting, we found that LAP1, LAZY1 and PROG1 were all down-regulated in NIL-qtac8 at ripening stage, implying that those genes might function in the same pathway with qTAC8 (Fig 6). In addition, qTAC8 encodes a predicted transcription factor, thus, whether qTAC8 acts directly or indirectly on LAP1, LAZY1 and PROG1 remains to be investigated.
Gene expression in plants can be basically of two types, constitutive or with specific expression pattern. Although some genes can be expressed during the whole plant life, it may function during specific growth stages and contribute to a certain phenotype. The genetic control of tiller angle is very complex and phenotypes can change during different developmental stages. The japonica rice ZH11 is a typical rice cultivar which displays large tiller angle at the seedling and tillering stages but usually presents a more compact architecture after the heading stage. The rice mutants lazy1 and prog1 present a large tiller angle during all growing periods, while lpa1 presents a large tiller angle at the seedling stage [3]. In this study, NIL-qTAC8 and NIL-qtac8 exhibit no differences in tiller angle at the tillering stage, but they show increasing TAC8 expression and TAC8 transcript levels are significant different during heading and ripening stages. It would be interesting to uncover the molecular mechanisms of how tiller angle is regulated during different rice development stages.
Supporting information
A, a line derived from the RIL was used to backcross with D50 to obtain backcross populations. The circles indicate the region with QTLs for tiller angle; B, a BC2F1 line derived from RILs used for developing mapping population of qTAC8.
(TIF)
(TIF)
A, Sequencing analysis of TAC1 allele; B, relative expression of the TAC1 allele.
(TIF)
The three genotypes of homozygous D50, HB277 and heterozygote at qtac8 were identified by progeny test.
(TIF)
(TIF)
(DOCX)
Acknowledgments
This work was supported by the National Key Research and Development Program of China (2016YFD0101801), the National S&T Major Project of China (2016ZX08001006) and the National Natural Science Foundation of China (grants 31501285, 31521064).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Key Research and Development Program of China (2016YFD0101801), the National S&T Major Project of China (2016ZX08001006) and the National Natural Science Foundation of China (grants 31501285, 31521064). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Shen SQ, Zhuang JY, Bao JS, Zheng KL, Xia YF, Shu QY. Analysis of QTLs with additive, epistasis and G× E interaction effects of the tillering angle trait in rice. J agric biotech. 2004;13(1):16–20. [Google Scholar]
- 2.Xu YB, McCouch SR, Shen ZT. Transgressive segregation of tiller angle in rice caused by complementary gene action. Crop sci. 1998;38(1):12–9. [Google Scholar]
- 3.Wu X, Tang D, Li M, Wang K, Cheng Z. Loose Plant Architecture1, an INDETERMINATE DOMAIN protein involved in shoot gravitropism, regulates plant architecture in rice. Plant Physiol. 2013;161(1):317–29. Epub 2012/11/06. PubMed Central PMCID: PMCPmc3532263. doi: 10.1104/pp.112.208496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu B, Lin Z, Li H, Li X, Li J, Wang Y, et al. TAC1, a major quantitative trait locus controlling tiller angle in rice. Plant J. 2007;52(5):891–8. Epub 2007/10/03. doi: 10.1111/j.1365-313X.2007.03284.x [DOI] [PubMed] [Google Scholar]
- 5.Xu YB, Shen ZT. Genetic analysis of tiller angles for early season indica rice. Acta Agriculturae Zhejiangensis. 1993;1:000. [Google Scholar]
- 6.Li ZK, Paterson AH, Pinson SR, Stansel JW. RFLP facilitated analysis of tiller and leaf angles in rice (Oryza sativa L.). Euphytica. 1999;109(2):79–84. [Google Scholar]
- 7.Qian Q, He P, Teng S, Zeng DL, Zhu LH. QTLs analysis of tiller angle in rice (Oryza sativa L.). Acta genetica Sinica. 2000;28(1):29–32. [PubMed] [Google Scholar]
- 8.Yu CY, Liu YQ, Jiang L, Wang CM, Zhai HQ, Wan JM. QTLs mapping and genetic analysis of tiller angle in rice (Oryza sativa L.). Acta genetica Sinica. 2005;32(9):948–54. Epub 2005/10/06. [PubMed] [Google Scholar]
- 9.Jin J, Huang W, Gao JP, Yang J, Shi M, Zhu MZ, et al. Genetic control of rice plant architecture under domestication. Nat Genet. 2008;40(11):1365–9. doi: 10.1038/ng.247 [DOI] [PubMed] [Google Scholar]
- 10.Tan L, Li X, Liu F, Sun X, Li C, Zhu Z, et al. Control of a key transition from prostrate to erect growth in rice domestication. Nat Genet. 2008;40(11):1360–4. Epub 2008/09/30. doi: 10.1038/ng.197 [DOI] [PubMed] [Google Scholar]
- 11.Dong H, Zhao H, Xie W, Han Z, Li G, Yao W, et al. A novel tiller angle gene, TAC3, together with TAC1 and D2 largely determine the natural variation of tiller angle in rice cultivars. Plos Genet. 2016;12(11):e1006412 Epub 2016/11/05. PubMed Central PMCID: PMCPmc5096673. doi: 10.1371/journal.pgen.1006412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li P, Wang Y, Qian Q, Fu Z, Wang M, Zeng D, et al. LAZY1 controls rice shoot gravitropism through regulating polar auxin transport. Cell Res. 2007;17(5):402–10. Epub 2007/05/01. doi: 10.1038/cr.2007.38 [DOI] [PubMed] [Google Scholar]
- 13.Yoshihara T, Iino M. Identification of the gravitropism-related rice gene LAZY1 and elucidation of LAZY1-dependent and -independent gravity signaling pathways. Plant Cell Physiol. 2007;48(5):678–88. Epub 2007/04/07. doi: 10.1093/pcp/pcm042 [DOI] [PubMed] [Google Scholar]
- 14.Chen Y, Fan X, Song W, Zhang Y, Xu G. Over-expression of OsPIN2 leads to increased tiller numbers, angle and shorter plant height through suppression of OsLAZY1. Plant Biotechnol J. 2012;10(2):139–49. Epub 2011/07/23. doi: 10.1111/j.1467-7652.2011.00637.x [DOI] [PubMed] [Google Scholar]
- 15.Xu M, Zhu L, Shou H, Wu P. A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice. Plant Cell Physiol. 2005;46(10):1674–81. Epub 2005/08/09. doi: 10.1093/pcp/pci183 [DOI] [PubMed] [Google Scholar]
- 16.Shao G, Tang S, Jiao G, Luo J, Tang A, P. H. Mapping of QTL for cooking quality traits of rice. Chinese J Rice Sci. 2009;1:94–8. [Google Scholar]
- 17.Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8(19):4321–5. Epub 1980/10/10. PubMed Central PMCID: PMCPmc324241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rogers SO, Bendich AJ. Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol Biol. 1985;5(2):69–76. Epub 1985/03/01. doi: 10.1007/BF00020088 [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. Epub 2002/02/16. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 20.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–8. Epub 2008/06/13. [DOI] [PubMed] [Google Scholar]
- 21.Yang SR, Zhang BR, Chen WF, Xu ZJ, Wang JM. Theories and method of rice breeding for maximun yield. Chinese J Rice Sci. 1996;10(2):115–20. [Google Scholar]
- 22.Jiao Y, Wang Y, Xue D, Wang J, Yan M, Liu G, et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet. 2010;42(6):541–4. Epub 2010/05/25. doi: 10.1038/ng.591 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A, a line derived from the RIL was used to backcross with D50 to obtain backcross populations. The circles indicate the region with QTLs for tiller angle; B, a BC2F1 line derived from RILs used for developing mapping population of qTAC8.
(TIF)
(TIF)
A, Sequencing analysis of TAC1 allele; B, relative expression of the TAC1 allele.
(TIF)
The three genotypes of homozygous D50, HB277 and heterozygote at qtac8 were identified by progeny test.
(TIF)
(TIF)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.