Abstract
Adenosine-to-inosine (A-to-I) editing is a highly prevalent posttranscriptional modification of RNA, mediated by ADAR (adenosine deaminase acting on RNA) enzymes. In addition to RNA editing, additional functions have been proposed for ADAR1. To determine the specific role of RNA editing by ADAR1, we generated mice with an editing-deficient knock-in mutation (Adar1E861A, where E861A denotes Glu861→Ala861). Adar1E861A/E861A embryos died at ~E13.5 (embryonic day 13.5), with activated interferon and double-stranded RNA (dsRNA)–sensing pathways. Genome-wide analysis of the in vivo substrates of ADAR1 identified clustered hyperediting within long dsRNA stem loops within 3′ untranslated regions of endogenous transcripts. Finally, embryonic death and phenotypes of Adar1E861A/E861A were rescued by concurrent deletion of the cytosolic sensor of dsRNA, MDA5. A-to-I editing of endogenous dsRNA is the essential function of ADAR1, preventing the activation of the cytosolic dsRNA response by endogenous transcripts.
Adenosine-to-inosine (A-to-I) editing is the most prevalent form of RNA base modification in mammals. Hundreds of thousands of A-to-I editing events have been reported in the human transcriptome (1–3). A-to-I editing is catalyzed by ADAR (adenosine deaminase acting on RNA) enzymes, which deaminate genomically encoded A-to-I in double-stranded RNA (dsRNA). A-to-I editing predominantly occurs in noncoding, repetitive elements such as inverted Alus and short interspersed nuclear elements (SINEs) (3, 4). There are three mammalian ADAR proteins: ADAR1, -2, and -3. ADAR1 is widely expressed during embryonic and postnatal development and is present as a predominantly nuclear, constitutive ADAR1p110 isoform expressed in all tissues and an additional interferon (IFN)–inducible ADAR1p150 isoform that is found in both the nucleus and the cytoplasm (5). Adar1−/− (null for both isoforms) and Adar1p150−/− mice die in utero at E11.5 (embryonic day 11.5) to E12.5, due to failed erythropoiesis and fetal liver (FL) disintegration (6–8).
Only ADAR1 and ADAR2 demonstrate editing activity in vitro. The AMPA receptor GluA2 pre-mRNA is edited by ADAR2 in the brain, converting a glutamine residue to an arginine in a functionally critical position (9). Adar2−/− mice die from seizures and were rescued by the genomic substitution of the single edited adenosine for guanosine, mimicking editing of the transcript (10). In contrast to this elegant single-substrate paradigm that describes the Adar2−/− phenotype, no such editing site(s) described to date have resolved the physiological requirement for ADAR1.
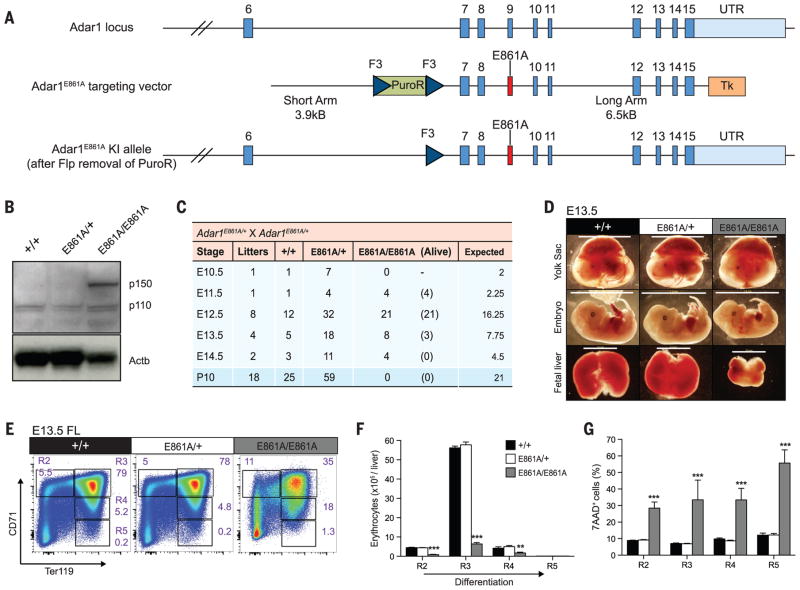
To directly determine the contribution of A-to-I editing to ADAR1’s biological function, we generated a constitutive knock-in of an editing-deficient ADAR1 allele in mice (Adar1E861A, where E861A denotes Glu861→Ala861) (Fig. 1A and fig. S1A), homologous to the human ADAR1E912A allele, which is catalytically inactive in vitro (11) (also see supplementary materials and methods). In E12.5 whole embryos, ADAR1p110 protein was detected in Adar1E861A/E861A samples at comparable levels to Adar1+/+ and Adar1E861A/+ controls (Fig. 1B). Additionally, Adar1E861A/E861A embryos had elevated expression of ADAR1p150 (Fig. 1B).
Fig. 1. Adar1E861A/E861A embryos die in utero.
(A) Schematic of Adar1E861A knock-in allele. (B) ADAR1 protein expression in whole E12.5 embryos of the indicated genotypes. (C) Survival data at the indicated stages. (D) Images of viable E13.5 yolk sacs, embryos, and FL. Scale: yolk sac and embryo, 1 cm; FL, 2 mm. Representative (E) fluorescence-activated cell sorting (FACS) profiles, (F) cell numbers, and (G) frequency of 7AAD+ FL erythroblasts at E13.5. Results are mean ± SEM (+/+, n = 5 embryos; E861A/+, n = 18; E861A/E861A, n = 3). **P < 0.005 and ***P < 0.0005 compared with Adar1+/+. R2 to R5 denote erythroblast populations.
Adar1E861A/+ heterozygous mice were normal, were present at the expected Mendelian ratio, and had no discernible phenotype. No viable Adar1E861A/E861A pups were born from Adar1E861A/+ intercrosses and were found to be dying at ~E13.5 (Fig. 1C). Adar1E861A/E861A yolk sacs were pale, and embryos appeared developmentally delayed compared with controls (Fig. 1D), despite being normal at E12.5 (fig. S1, D and E). At E13.5, Adar1E861A/E861A FL was smaller, with eightfold fewer viable cells (Fig. 1D and fig. S1E). We observed a failure in erythropoiesis with a severe loss of erythroblast populations (Fig. 1, E and F) and increased cell death (Fig. 1G), also apparent in other hematopoietic populations (fig. S2). Consistent with our previous analysis of the ADAR1 conditional allele (12), adult hematopoietic stem cells (HSCs) expressing only the catalytically inactive ADAR1 could not be maintained in vivo and were lost over time (fig. S3). The A-to-I editing activity of ADAR1 is essential for embryonic development and the maintenance of hematopoiesis in vivo.
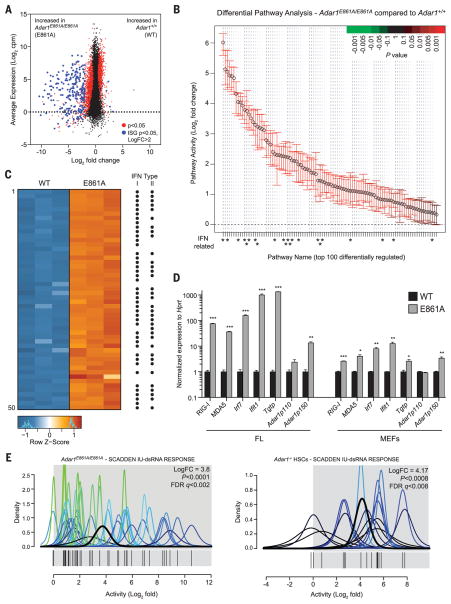
We performed transcriptional profiling on three independent littermate Adar1+/+ and Adar1E861A/E861A FLs. An absence of ADAR1 editing resulted in the up-regulation of 383 transcripts (log2FC > 2), 258 of which are IFN-stimulated genes (ISGs) (Fig. 2A and table S1). Pathway analysis revealed a profound enrichment of signatures activated after IFN treatment or viral infection (Fig. 2B and table S1). Of the 50 most differentially expressed genes, 44 were up-regulated by either type I IFN, type II IFN, or both (Fig. 2C and table S1). The derepression of ISGs was confirmed by quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) (Fig. 2D). Furthermore, we demonstrated that the transcriptional response in Adar1E861A/E861A FLs closely parallels that of complete ADAR1 deficiency (12) (fig. S4A).
Fig. 2. Absence of editing transcriptionally phenocopies loss of ADAR1.
(A) MA plot comparing gene expression in WT and E861A E12.5 FL. Red dots, differentially expressed genes; blue dots, differentially expressed ISGs. (B) QuSAGE analysis of the top 100 differential pathway signatures ranked by fold enrichment and P value. (C) Heat map of the 50 most differentially expressed genes. Black dots indicate known ISGs. (D) qRT-PCR of ISGs in E861A compared with controls in FL and MEFs. Results are mean ± SEM (n = 3). *P < 0.05, **P < 0.005, and ***P < 0.0005 compared with Adar1+/+. (E) IU-dsRNA response gene set in E861A compared with WT samples (left) and Adar1−/− HSCs compared with controls (right).
Synthetic dsRNA sequences based on endogenous RNA containing adenosine but not inosine stimulate ISGs and apoptosis in vitro by binding to MDA5 and RIG-I (13). We defined a gene set associated with the response to IU-dsRNA (inosine-uracil–paired dsRNA) in human cells (13). Gene signatures of both Adar1E861A/E861A FL and Adar1−/− HSCs were highly enriched for the IU-dsRNA response (Fig. 2E), demonstrating a species-conserved response to dsRNA that is constrained by the presence of inosine residues. Therefore, the deamination of adenosine in dsRNA by ADAR1 is necessary for suppression of the IFN response under homeostatic conditions. Furthermore, it suggested that the absence of A-to-I substitutions in endogenous dsRNA may initiate this response.
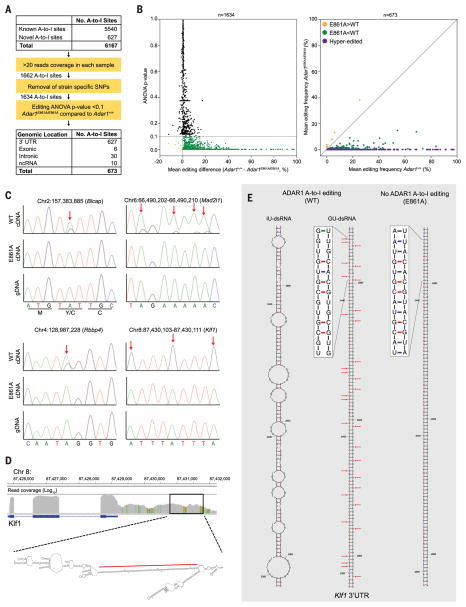
To define ADAR1-specific editing events in vivo, we analyzed A-to-I (G) mismatches within the RNA sequencing data. Across all samples, 6167 A-to-I editing sites were identified: 5540 known (14) and 627 previously undiscovered (Fig. 3A). Strain-specific single-nucleotide polymorphisms were excluded, and only A-to-I mismatch frequencies that differed significantly between genotypes were considered (Fig. 3B). Using these criteria, 673 A-to-I editing sites were defined (Fig. 3A). Of these, 666 sites had reduced editing in mutants, including hyperedited loci, which had no detectable editing (Fig. 3B and tables S2 and S3). This confirmed that the Adar1E861A allele is catalytically inactive and the majority of FL A-to-I editing is ADAR1 dependent. We validated 281 of the identified A-to-I sites on independent Adar1E861A/E861A and Adar1+/+ FL samples (15). All tested sites were confirmed as differentially edited in both FL and mouse embryonic fibroblasts (MEFs) (table S4). Sanger sequencing validated ADAR1-specific BlcapY2C, Mad2l1, Rbbp4, and Klf1 editing sites from independent FL samples (Fig. 3C).
Fig. 3. Defining the ADAR1 FL editome.
(A) Summary of A-to-I editing site analysis in FL. SNPs, single-nucleotide polymorphisms; ANOVA, analysis of variance; ncRNA, noncoding RNA. (B) Mean editing difference for sites with >20 reads (left, n = 1634). Mean editing frequency of differentially edited sites between E861A and controls (right, n = 673). (C) Genomic DNA (gDNA) (bottom) and complementary DNA (cDNA) (top and middle) Sanger sequencing validation of editing sites. Red arrows highlight edited adenosine. (D) Integrative Genomics Viewer image of Klf1 in WT E12.5 FL and predicted secondary structure of 3′UTR. The red line denotes the region depicted in (E). (E) Predicted secondary structure ofa 212–base pairdsRNA stem loop from the 3′UTRof Klf1, with inosine (IU-dsRNA, left) and guanosine (GU-dsRNA, middle) in place of adenosine at the edited sites. Right, predicted secondary structure with no A-to-I editing.
ADAR1 is required for erythropoiesis (Fig. 1, E to G). Interestingly, 40% (264 of 666) of the ADAR1-specific editing sites were within hyperedited clusters of only three genes—Klf1, Oip5, and Optn—which have peak or restricted expression in the erythroid lineage (table S3). Seventy, 61, and 133 ADAR1-specific A-to-I hyperedited loci were within long 3′ untranslated regions (3′UTRs) of Klf1, Oip5, and Optn, respectively. An example of hyperediting in Klf1 is depicted in Fig. 3D. An absence of ADAR1 editing of these loci provides a possible link to the FL failure in Adar1E861A/E861A embryos.
Modeling of the predicted secondary structure of the hyperedited 3′UTRs showed that in the absence of editing there was the potential for long perfect dsRNA segments to be formed through duplexing of repetitive regions (Fig. 3E and fig. S7, A to C). The thermodynamics of inosine base pairing is not defined. Therefore, we modeled secondary structures for hyperedited substrates in two ways: (i) by predicting secondary structures of Klf1 (Fig. 3E, left), Optn, and Oip5 (fig. S7) 3′UTRs with inosine in place of adenosine, assuming no base pairing, or (ii) by replacing adenosine with guanosine (Fig. 3E, middle), assuming wobble base paring. In both cases, the predicted dsRNA structures have higher free-energy states in the presence of A-to-I or A-to-G substitutions (fig. S7, A to F). Therefore, extensive I-U mismatches would be predicted to destabilize perfect dsRNA stem loops within hyperedited 3′UTRs (Fig. 3E and fig. S7, A to F). Thus, we hypothesized that ADAR1 editing remodeled the secondary structure of endogenous RNA to abrogate the formation of long matched dsRNAs.
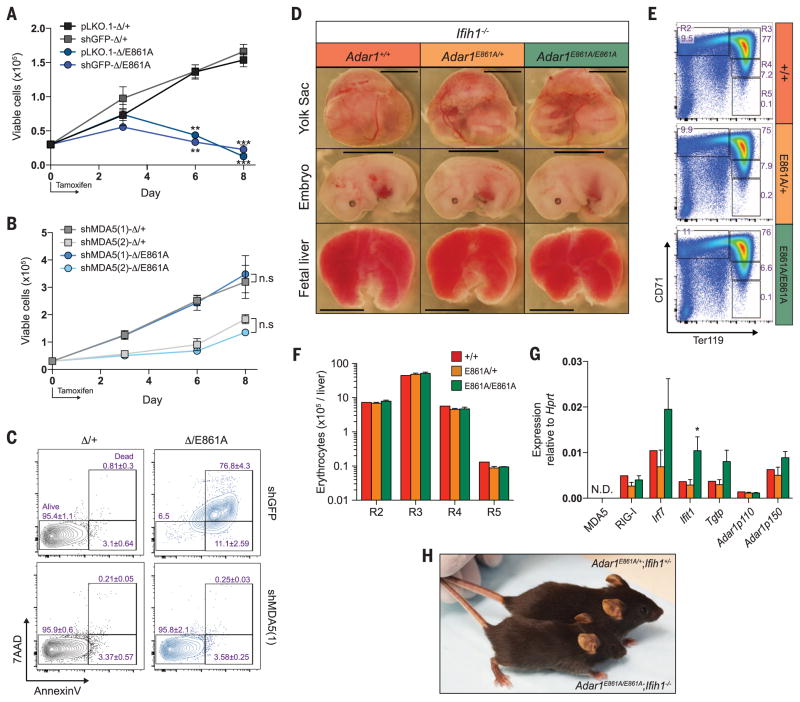
ThetranscriptionalsignaturesintheAdar1E861A/E861A FL resembled that of RIG-I and MDA5 activation (13). We postulated that in the absence of ADAR1 editing, endogenous dsRNAs—such as the 3′UTRs of Klf1, Oip5, and Optn—could be bound by MDA5 and/or RIG-I and could activate the cellular dsRNA response. Short hairpin RNA (shRNA) knockdown of MDA5 rescued proliferation and viability (Fig. 4, A to C) and suppressed ISG induction in hematopoietic stem and progenitor cells expressing only the Adar1E861A allele to control levels, using two independent MDA5 shRNAs (fig. S8).
Fig. 4. Loss of MDA5 rescues Adar1E861A/E861Aviability.
(A) LKS+ cells isolated from Rosa26CreERT2 Adar1fl/+ (Δ/+) and Rosa26CreERT2 Adar1fl/E861A (Δ/E861A) infected with pLKO.1 empty vector, shGFP, or (B) two independent shMDA5 [shMDA5(1) and shMDA5(2)] were cultured for 8 days. Results are mean ± SEM (n = 3). **P < 0.005 and ***P < 0.0005 compared with Δ/+. n.s, not significant. (C) Analysis of apoptosis on day 8. (D) Images of viable E13.5 yolk sac, embryo, and FL of the indicated genotype (all Ifih1−/−). Scale: yolk sac and embryo, 5 mm; FL, 1.6 mm. Representative (E) FACS profiles and (F) numbers of FL erythrocytes at E13.5. (G) E13.5 FL qRT-PCR of ISGs. Results are mean ± SEM (+/+, n = 2; E861A/+, n = 8; E861A/E861A, n = 4). *P < 0.05 compared with Adar1+/+Ifih1−/− controls. (H) Photo of an Adar1E861A/E861A Ifih1−/− mouse and an Adar1E861A/+Ifih1+/− littermate at 26 days of age.
Based on the in vitro rescue, we crossed Adar1E861A/E861A to MDA5−/− (Ifih1−/−) mice. Initially we assessed embryos at E13.5, a time point when Adar1E861A/E861A embryos are no longer viable (Fig. 1C). Adar1E861A/E861AIfih1−/− double mutant yolk sac, embryo, and FL were comparable to controls and were present at the expected ratio (Fig. 4D and fig. S9A). Erythropoiesis was completely rescued, and FL ISGs were equivalent to those of control littermates (Fig. 4, E to G). Most surprisingly, viable Adar1E861A/E861AIfih1−/− mutants that survived past weaning have been identified (fig. S9B). The viable double mutants are outwardly healthy, albeit slightly smaller than littermate controls, and have not demonstrated additional phenotypes to date (Fig. 4H). The rescue of both developmental and adult viability demonstrates that MDA5 is the primary sensor of endogenous dsRNA in the absence of ADAR1 editing.
Unlike ADAR2, the primary role of ADAR1 has not been clearly defined. Although it was assumed that RNA editing was its central function, additional RNA editing–independent roles have been proposed. The murine phenotypes and transcriptional consequences of complete ADAR1 deficiency and the specific loss of A-to-I editing are markedly similar, demonstrating that RNA editing is the primary and physiologically most important function of ADAR1. Adar1E861A/E861A embryos die ~1 to 1.5 days later than Adar1−/− embryos, suggesting limited contributions from nonediting functions of ADAR1. We believe the difference in survival is accounted for by the catalytically inactive ADAR1 retaining the ability to sequester immunogenic dsRNA (16). ADAR1 sequestration is not sufficient, however, to completely prevent MDA5 recognition of unedited dsRNA and subsequent signaling. Therefore, the extended survival of the editing-deficient animals reflects a delay rather than a fundamental difference in the presentation of the same phenotype.
The data from the Adar1E861A mutants are consistent with the type I interferonopathies of Aicardi-Goutières syndrome (AGS) patients bearing ADAR1 mutations (17, 18). Editing of endogenous transcripts in ADAR1-mutant AGS patients is probably reduced, leading to retention of paired endogenous dsRNA that can be sensed by MDA5. Consistent with this, gain-of-function mutations of MDA5 have been identified in non–ADAR1-mutant AGS patients (18). AGS MDA5 mutations caused higher-affinity binding to self-dsRNA, resulting in the inappropriate detection of self-dsRNA (18). Thus, the physiological function of ADAR1 is probably conserved in mammals.
Approximately half of the mammalian genome is composed of noncoding retrotransposons such as SINEs and Alus (19, 20), which typically form dsRNA duplexes. Retrotransposons are subjected to extensive A-to-I RNA editing (3, 4). The location of repetitive elements may determine their immunogenicity. Retrotransposons located within introns do not persist in the cytosol and therefore cannot activate MDA5. Repetitive elements in 3′UTRs, though rare (4), can be retained and form duplexes, harboring the potential for recognition by MDA5. We propose that hyperediting of self-dsRNA by ADAR1 (such as the 3′UTRs of Klf1, Optn, and Oip5) generates multiple I-U mismatches that act to prevent MDA5 oligomerization (21). In the absence of ADAR1 editing, long dsRNA stem loops can form that activate MDA5 (fig. S10). However, we cannot rule out an alternate possibility that edited substrates preferentially bind MDA5 to prevent its activation by other (unidentified and nonedited) dsRNA transcripts (13).
Concurrent ablation of MAVS, the downstream adaptor of MDA5 and RIG-I, rescues ADAR1-null mice to birth (22), demonstrating a role for ADAR1 in the suppression of the RLR pathway. Our study specifically identifies the critical cytosolic sensor upstream of MAVS and demonstrates that the Adar1E861A/E861A phenotype and, by extension, Adar1−/− can be ascribed to the lack of editing of multiple substrates, resulting in the inappropriate activation of MDA5. We speculate that these unedited transcripts are sensed as nonself by MDA5 and activate innate immune signaling. ADAR1’s primary physiological function is to edit endogenous dsRNA to prevent sensing of endogenous dsRNA as nonself by MDA5.
Supplementary Material
Acknowledgments
We thank V. Sankaran, S. Orkin, L. Purton, and J. Heierhorst for discussion and SVH BioResources Centre for animal care. Data sets described in the paper are deposited in Gene Expression Omnibus (accession number GSE58917). This work was supported by the Leukaemia Foundation (C.R.W.), a Leukaemia Foundation Ph.D. scholarship (B.J.L), National Health and Medical Research Council (NHMRC) Project Grant 1021216, NHMRC Career Development Award 559016 (C.R.W.), a German Academic Exchange Service Postdoctoral Fellowship (R.P.), a Stanford University Dean’s Fellowship (R.P.), Stanford Genome Training Program (NIH grant T32 HG000044) and Stanford Graduate Fellowship (G.R.), NIH grant R01GM102484 (J.B.L.), the Ellison Medical Foundation (J.B.L.), and the Stanford University Department of Genetics (J.B.L.). This work was also supported in part by the Victorian State Government Operational Infrastructure Support Scheme (to St. Vincent’s Institute of Medical Research). C. R.W. was the Leukaemia Foundation Phillip Desbrow Senior Research Fellow. J.C.H. is an employee of Taconic Biosciences. Taconic Biosciences had no role in the preparation and content of this Report. All other authors declare no conflicts of interest. Author contributions: M.H., P.H.S., and J.C.H. generated the knock-in mouse line; B.J.L., R.P., A.M.C., G.R., J.C.H., J.B.L., and C.R.W. performed experiments and analyzed and interpreted data; M.H., P.H.S., and J.C.H. provided intellectual input and conceptual advice; and B.J.L. and C.R.W. wrote the manuscript.
Footnotes
REFERENCES AND NOTES
- 1.Danecek P, et al. Genome Biol. 2012;13:R26. [Google Scholar]
- 2.Li JB, et al. Science. 2009;324:1210–1213. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- 3.Ramaswami G, et al. Nat Methods. 2012;9:579–581. doi: 10.1038/nmeth.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neeman Y, Levanon EY, Jantsch MF, Eisenberg E. RNA. 2006;12:1802–1809. doi: 10.1261/rna.165106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patterson JB, Samuel CE. Mol Cell Biol. 1995;15:5376–5388. doi: 10.1128/mcb.15.10.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartner JC, et al. J Biol Chem. 2004;279:4894–4902. doi: 10.1074/jbc.M311347200. [DOI] [PubMed] [Google Scholar]
- 7.Wang Q, et al. J Biol Chem. 2004;279:4952–4961. doi: 10.1074/jbc.M310162200. [DOI] [PubMed] [Google Scholar]
- 8.Ward SV, et al. Proc Natl Acad Sci USA. 2011;108:331–336. doi: 10.1073/pnas.1017241108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Higuchi M, et al. Cell. 1993;75:1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- 10.Higuchi M, et al. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 11.Lai F, Drakas R, Nishikura K. J Biol Chem. 1995;270:17098–17105. doi: 10.1074/jbc.270.29.17098. [DOI] [PubMed] [Google Scholar]
- 12.Hartner JC, Walkley CR, Lu J, Orkin SH. Nat Immunol. 2009;10:109–115. doi: 10.1038/ni.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vitali P, Scadden AD. Nat Struct Mol Biol. 2010;17:1043–1050. doi: 10.1038/nsmb.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramaswami G, Li JB. Nucleic Acids Res. 2014;42:D109–D113. doi: 10.1093/nar/gkt996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang R, et al. Nat Methods. 2014;11:51–54. doi: 10.1038/nmeth.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Washburn MC, et al. Cell Reports. 2014;6:599–607. doi: 10.1016/j.celrep.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rice GI, et al. Nat Genet. 2012;44:1243–1248. doi: 10.1038/ng.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rice GI, et al. Nat Genet. 2014;46:503–509. doi: 10.1038/ng.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lander ES, et al. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 20.Mouse Genome Sequencing Consortium. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 21.Wu B, et al. Cell. 2013;152:276–289. doi: 10.1016/j.cell.2012.11.048. [DOI] [PubMed] [Google Scholar]
- 22.Mannion NM, et al. Cell Reports. 2014;9:1482–1494. doi: 10.1016/j.celrep.2014.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.