Abstract
Bovine paratuberculosis (PTB) is a chronic enteric inflammatory disease of ruminants caused by Mycobacterium avium subsp. paratuberculosis (MAP) that causes large economic losses in the dairy industry. Spread of PTB is mainly provoked by a long subclinical stage during which MAP is shed into the environment with feces; accordingly, detection of subclinical animals is very important to its control. However, current diagnostic methods are not suitable for detection of subclinical animals. Therefore, the current study was conducted to develop a diagnostic method for analysis of the expression of genes of prognostic potential biomarker candidates in the whole blood of cattle naturally infected with MAP. Real-time PCR with nine potential biomarker candidates was developed for the diagnosis of MAP subclinical infection. Animals were divided into four groups based on fecal MAP PCR and serum ELISA. Eight genes (Timp1, Hp, Serpine1, Tfrc, Mmp9, Defb1, Defb10, and S100a8) were up-regulated in MAP-infected cattle (p <0.05). Moreover, ROC analysis revealed that eight genes (Timp1, Hp, Serpine1, Tfrc, Mmp9, Defb1, Defb10, and S100a8) showed fair diagnostic performance (AUC≥0.8). Four biomarkers (Timp1, S100a8, Defb1, and Defb10) showed the highest diagnostic accuracy in the PCR positive and ELISA negative group (PN group) and three biomarkers (Tfrc, Hp, and Serpine1) showed the highest diagnostic accuracy in the PCR negative and ELISA positive group (NP group). Moreover, three biomarkers (S100a8, Hp, and Defb10) were considered the most reliable for the PCR positive and ELISA positive group (PP group). Taken together, our data suggest that real-time PCR based on eight biomarkers (Timp1, Hp, Serpine1, Tfrc, Mmp9, Defb1, Defb10, and S100a8) might be useful for diagnosis of JD, including subclinical stage cases.
Introduction
Johne’s disease (JD) is a chronic inflammatory disease of the gastrointestinal tract of ruminants with granulomatous lesions that is caused by Mycobacterium avium subsp. paratuberculosis (MAP) (Whitlock et al., 1996). Johne’s disease can be divided into four stages depending on the clinical signs and MAP shedding levels including the silent, subclinical, clinical, and advanced clinical stage [1]. In the silent stage, the infected animals do not show any clinical sign or excrete MAP into the environment [2]. During the subclinical stage, animals still do not have clinical symptoms; however, they shed low numbers of MAP into environment, which can be circulated in the herd and infect other animals [2]. After subclinical stage, animals enter clinical stage and start to show clinical signs such as gradual weight loss, diarrhea, and decreased milk production [2]. Finally, animals become cachectic and lethargic in advanced clinical stage [2]. Accordingly, it is very important to remove animals in the subclinical stage to control the disease. However, current diagnostic methods are insufficient for diagnosis of subclinical stages of disease [3]. Although fecal culture has been considered a gold standard for the diagnosis of MAP [4], this method is time-consuming and shows low sensitivity, especially in subclinical stages of the disease [5, 6]. PCR allows rapid detection of MAP in clinical samples such as feces, milk and blood [7]; however, PCR-based methods are also limited in their usefulness for diagnosis of subclinical stages of disease because of the low sensitivity [8] and low specificity caused by genetic similarities with other mycobacteria [9, 10]. Although ELISA has been used for detection of antibodies to MAP in clinical samples such as serum and milk, this method is also inadequate for diagnosis of fecal shedders in the subclinical stage, especially in 1–2 year old cattle [11]. Therefore, new diagnostic tools have been requested to detect MAP-infected animals at early stage of infection.
Biomarkers, which are considered indicators of specific pathogenic conditions or therapeutic responses to treatment [12], are commonly used as diagnostic tools for various diseases [13–16]. Recently, host biomarkers discovered using transcriptomics, metabolomics, and proteomics have been proposed as alternative diagnostic methods for paratuberculosis [17–20]. Biomarkers indicating early stages of MAP-infection were proposed by analyzing gene expression profiles of blood in cattle with experimental MAP infection [17, 18]. A metabolic profiling in cattle with experimental infection of MAP revealed that four metabolites (iso-butyrate, branched chain amino acids, leucine, and isoleucine) were increased in serum of the MAP-infected cattle while citrate was decreased [19]. Moreover, six proteins (transferrin, gelsolin isoforms α & β, complement subcomponent C1r, complement component C3, amine oxidase-copper containing 3, and coagulation factor II) were proposed as biomarkers after they were found to increase by at least 2-fold in MAP-infected cattle, as were two proteins (coagulation factor XIII-B polypeptide, and fibrinogen γ chain and its precursor) that were reduced by nearly two-fold in MAP-infected cattle [20]. Our previous studies also proposed several biomarkers that were up-regulated in MAP infected macrophages, mice, and cattle [21–23]. Transcriptional profiles of MAP-infected macrophage RAW 264.7 cells and a mouse model suggested five and three genes as prognostic biomarkers, respectively [21, 22]. β-defensins were also suggested as prognostic biomarkers in subclinical animals of MAP-naturally infected cattle [23]. However, application of those biomarkers for diagnosis of JD has yet to be investigated. Therefore, we developed a real-time PCR method using the biomarkers for diagnosis of bovine paratuberculosis by measuring the gene expression level of several biomarkers in whole blood.
Materials and methods
Experimental design and animals
About 300 Holstein cattle were raised on the national farm in Cheonan city which located in mid-west region of the South Korea. The cattle were regularly tested for absence of JD two times per year using fecal PCR and serum ELISA. A total of three to eight year old fourty-four cows were selected for further analysis after detection of MAP-specific antibodies using a commercial ELISA kit (IDEXX Laboratories, Inc., Westbrook, ME, USA) and MAP in the feces by PCR [24]. The detection was performed four times with a 6-month interval to enable accurate classification of infection status. The animals were divided into the following groups based on the results of PCR and ELISA: NN, ELISA and PCR negative; PN, ELISA negative and PCR positive; NP, ELISA positive and PCR negative; PP, ELISA positive and PCR positive. All animal procedures were approved by the National Institute of Animal Science (2013–046). Detailed characteristics of study subjects are shown in Table 1.
Table 1. Characteristics of study subjects.
| Number of subjects | All (n = 44) | NN group (n = 11) | PN group (n = 12) | NP group (n = 14) | PP group (n = 7) | |
|---|---|---|---|---|---|---|
| Heifers, n (%) | 44 (100) | 11 (100) | 12 (100) | 14 (100) | 7 (100) | |
| Median age (Years) | 6 (4 to 9) | 4 (4 to 7) | 6 (4 to 9) | 6.5 (4 to 8) | 6 (5 to 8) | |
| Serum ELISA | Positive, n (%) | 21 (47.7) | 0 (0) | 0 (0) | 14 (100) | 7 (100) |
| Negative, n (%) | 23 (52.3) | 11 (100) | 12 (100) | 0 (0) | 0 | |
| Fecal PCR | Positive, n (%) | 19 (43.2) | 0 (0) | 12 (100) | 0 (0) | 7 (100) |
| Negative, n (%) | 25 (56.8) | 11 (100) | 0 (0) | 14 (100) | 0 (0) | |
Selection of biomarker candidates
Nine genes that were significantly up-regulated in MAP infected macrophages, mice, and cattle were selected for use as diagnostic biomarkers based on our previous studies (Table 2) [21–23, 25]. All datasets used in selection of the biomarkers are available at Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo website) under accession number GSE62836, http://dx.doi.org/10.4014/jmb.1302.02021, and http://dx.doi.org/10.4014/jmb.1408.08059.
Table 2. Mean fold change of selected biomarker genes between infected animals and non-infected animals.
| Accession No. | Gene symbol | Gene name | Location | Mean fold change (log2 value) | |||||
|---|---|---|---|---|---|---|---|---|---|
| PN vs. NN | P value | NP vs. NN | P value | PP vs. NN | P value | ||||
| NM_003234.2 | Tfrc | Transferrin receptor (p90, CD71) | Plasma membrane | 1.6 | 0.0005 | 1.3 | 0.0021 | 1.9 | 0.0004 |
| NM_174744 | Mmp9 | Matrix metallopeptidase 9 | Extracellular space | 2.9 | 0.008 | 2.4 | 0.0342 | 3.6 | 0.0052 |
| NM_002964.4 | S100a8 | S100 calcium binding protein A8 | Cytoplasm | 1.6 | 0.0039 | 0.9 | 0.146 | 1.7 | 0.0069 |
| NM_002965.3 | S100a9 | S100 calcium binding protein A9 | Cytoplasm | 0.4 | 0.6228 | 0.6 | 0.2596 | 1.1 | 0.0548 |
| NM_174137 | Serpine1 | Serpin peptidase inhibitor | Extracellular space | 1.9 | 0.0041 | 1.6 | 0.0183 | 2.6 | 0.0009 |
| NM_005143.3 | Hp | Haptoglobin | Extracellular space | 2.3 | 0.0031 | 3.2 | <0.0001 | 3.3 | 0.0003 |
| NM_174471.3 | Timp1 | Tissue inhibitor of metallopeptidase 1 | Extracellular space | 1.7 | <0.0001 | 1.4 | 0.0002 | 0.5 | 0.415 |
| NM_001324544.1 | Defb1 | Defensin beta 1 | Extracellular space | 5.2 | 0.0009 | 3.3 | 0.039 | 1.8 | 0.5842 |
| NM_001115084.1 | Defb10 | Defensin beta 10 | Extracellular space | 2.3 | 0.0017 | 1.6 | 0.0313 | 1.6 | 0.1009 |
Extraction of total RNA from blood
Peripheral blood was collected from the tail vein of cattle using a BD Vacutainer® Plus Plastic K2EDTA Tubes. A total of 125 μl of whole blood was then mixed with 125 μl of RNase-free water and 750 μl of Trizol LS reagent (Ambion) and incubated at room temperature for 5 min. Next, 200 μl of chloroform was added to the mixture and it was centrifuged at 13,523 g and 4°C for 15 min. The supernatant was subsequently transferred to an RNAeasy column (Qiagen, Hilden, Germany) and centrifuged at 8,500 g for 15 sec. After washing, RNA was eluted in 30 μl of RNase-free water and immediately stored at -80°C until use.
Optimization of primer and probe concentrations
The optimal concentration of primer and probe concentration was determined with an identical cDNA template for each biomarker gene. Three concentrations (0.5μM, 0.75μM, 1μM) of both forward and reverse primers with a constant probe concentration were tested. The combination showing the highest fluorescence value was tested at three different concentrations of the probe (0.1μM, 0.2μM, 0.3μM). For further experiment, primer and probe concentration that showing the highest fluorescence value was selected.
Real-time PCR
Total RNA was employed to prepare cDNA with random primers using a QuantiTect® Reverse Transcription Kit (Qiagen Inc., Valencia, CA, USA) according to the manufacturer’s instructions. The expression of nine biomarker genes was measured by quantitative real time RT-PCR, which was conducted using a Rotor-Gene multiplex PCR kit (Qiagen Inc). In brief, total of 18μl reaction mixture was prepared consists of 10μl Master mix, RNase-free water, 0.5μM forward and reverse primers, and 0.1μM probe for each of the biomarker genes. After that, 2μl of cDNA template was added to a final volume of 20μl. The specificity of the primers and probes for each biomarker genes was confirmed by homology search (https://www.ncbi.nlm.nih.gov/tools/primer-blast) and agarose gel electrophoresis. The primers and probe used in this study are shown in Table 3. Sensitivity of real-time PCR reactions was confirmed by real-time PCR reaction using the known copy numbers calculated from purified PCR products which serially diluted from 109 to 102 copies of the templates. The real-time PCR was conducted for 45 cycles and CT values were obtained. Negative control was included with no template. Real-time PCR was conducted by subjecting the samples to 95°C for 10 min, followed by 50 cycles of 95°C for 15 s and 60°C for 45 s. The expression level was determined by the 2-ΔΔCt method using the housekeeping gene, β-actin, as a reference.
Table 3. Oligonucleotide sequence of primer and probe used for real-time PCR in this study.
| Target gene | Primer sequence(5’ to 3’) | PCR product size(base pair) | Reference | |
|---|---|---|---|---|
| β- actin | F | GCAAGCAGGAGTACGATGAG | 134 | In this study |
| R | GCCATGCCAATCTCATCTCG | |||
| Probe | FAM-TTCTAGGCGGACTGTTAGCTGCGTTACAC-BHQ1 | |||
| Mmp9 | F | CCCGGATCAAGGATACAGCC | 177 | [25] |
| R | GGGCGAGGACCATACAGATG | |||
| Probe | HEX-AGTTTGGCCACGCGCTGGGCTTAGAT-BHQ1 | |||
| Serpine1 | F | CTGCGAAATTCAGGATGCGG | 191 | [25] |
| R | GGGTGAGAAAACCACGTTGC | |||
| Probe | FAM-AGACTTTGGAGTGAAGGTGTTTCAGCAGG-BHQ1 | |||
| Timp1 | F | TCTGCAACTCCGATGTCGTC | 125 | In this study |
| R | CCTCAAGGCACTGAACCCTT | |||
| Probe | HEX-GTTCGTGGGGACCGCAGAAGTCAATG-BHQ1 | |||
| Hp | F | CCAAGTACCAGGACGACACC | 131 | In this study |
| R | ACCATACTCAGCCACAGCAC | |||
| Probe | FAM-ACGACAAGGAAGACGACACCTGGTATGC-BHQ1 | |||
| S100a8 | F | ATTTTGGGGAGACCTGGTGG | 124 | [25] |
| R | ACGGCGTGGTAATTCCCTTT | |||
| Probe | FAM-TAACTCCCTGATTGACGTCTACCACAAG-BHQ1 | |||
| S100a9 | F | AGGCTACGGGAAGGGCAG | 134 | [25] |
| R | GCTGGCCTCCTGATTAGTGG | |||
| Probe | HEX-ATGGAGGTCACGGCCACAGCCAC-BHQ1 | |||
| Tfrc | F | CAAAGTTTCTGCCAGCCCAC | 188 | [25] |
| R | AACAGAAAGAGACCGCTGGG | |||
| Probe | HEX-TATCGGGACAGCAACTGGATCAGCAAAG-BHQ1 | |||
| Defb1 | F | CGAATGGAGGCATCTGTTTG | 110 | In this study |
| R | CTTCGCCTTCTTTTACCACGA | |||
| Probe | FAM-TGCCCTGGACACATGATACAGATTGGCA-BHQ1 | |||
| Defb10 | F | ATCTAAGCTGCTGGGGGAAT | 97 | In this study |
| R | CATTTTACTCGGGGCGCTAA | |||
| Probe | HEX-GTTTGCTTAACAGGTGCCCTGGAC-BHQ1 | |||
Statistical analysis
Data are reported as the means ± the standard error of the mean (S.E.M.) of three independent experiments. Statistical significance was determined by ANOVA (p ≤ 0.05) with Dunnett’s post hoc test using the GraphPad Prism software version 7.00 (GraphPad Software, Inc., La Jolla, CA, USA). Receiver operator characteristics (ROC) curve analysis was conducted using the statistical package for social science (SPSS) software version 21.0 (SPSS Inc., Chicago, IL, USA) and the MedCalc Statistical Software version 13.3.3 (MedCalc Software, Ostend, Belgium). Higher AUC scores were considered to show better discriminatory powers as follows: excellent discriminatory power, AUC ≥0.9; good discriminatory power, 0.8≤AUC<0.9; fair discriminatory power, 0.7≤AUC<0.8; poor discriminatory power, AUC<0.7 [26]. The optimal cutoff values were calculated for each ROC curve while maximizing the Youden Index. Sensitivity and specificity were calculated based on cut-off value which showed highest AUC value in the ROC curve for each biomarker gene. A p <0.05 was considered to indicate statistical significance.
Results
Specificity of probe and primers
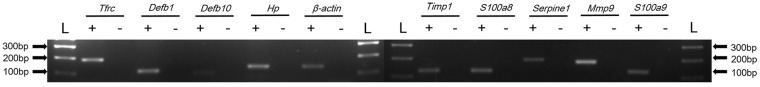
Specificity of primers and probes were confirmed by homology search. Also, to confirm the specificity for each biomarker genes, RT-PCR and agarose gel electrophoresis was performed. Single PCR band were confirmed for each biomarker gene and the β-actin gene and non-specific PCR product was not observed confirmed in the negative control with no cDNA sample (Fig 1).
Fig 1. Gel electrophoresis of PCR products of biomarkers genes and β-actin gene.
The biomarker genes and β-actin gene expression from bovine whole blood cDNA were confirmed by RT-PCR. A single PCR product was observed with expected size for each biomarker and β-actin gene. No band was observed in the PCR products of negative control without template DNA sample. In the figure (L) indicates 100bp DNA size marker, (+) indicates PCR product with template cDNA, (-) indicates PCR product without cDNA.
Sensitivity of real-time PCR reactions
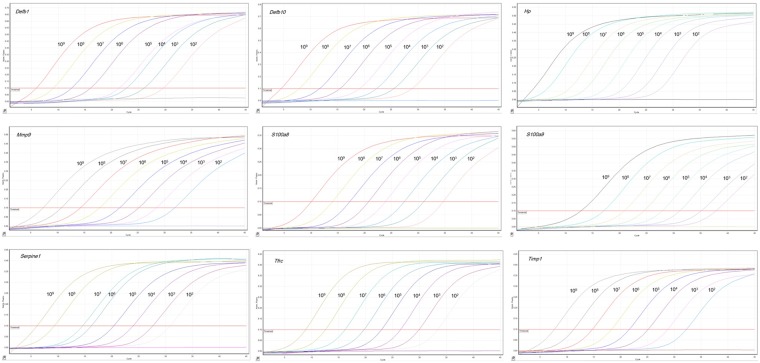
Real-time PCR for the each biomarker gene was performed using the specific primers, probes and the purified PCR products. Amplification plots were presented for biomarker genes with increased template copy numbers from 102 to 109. Amplification plot shows that fluorescence increase with increased template copy numbers (Fig 2). Also, real-time PCR was highly sensitive to detect low level of gene expression of biomarker genes (about 102 copies of the template cDNA) and negative control sample with no template DNA showed no increasing of fluorescence (Fig 2).
Fig 2. Amplification plots of the biomarker genes in the real-time PCR.
Real-time PCR was conducted with PCR product that serially diluted 10-fold from 109 to 102 copy numbers. The emission of fluorescence was measured at each cycle numbers and negative control sample with no template DNA showed no increasing of fluorescence.
Optimization of primer and probe concentrations
The optimal concentration of primer and probe concentration was determined by conducting real-time PCR with three primer and probe concentrations. The combination of forward and reverse primer at 0.5μM for biomarker genes and β-actin gene revealed highest florescence and lowest CT value. With this primer concentration, 0.1μM of probe showed highest florescence and lowest CT value. Combination of 0.5μM forward and reverse primers and 0.1μM probe concentration was used in further analysis.
Gene expression level of biomarkers in MAP infected cattle
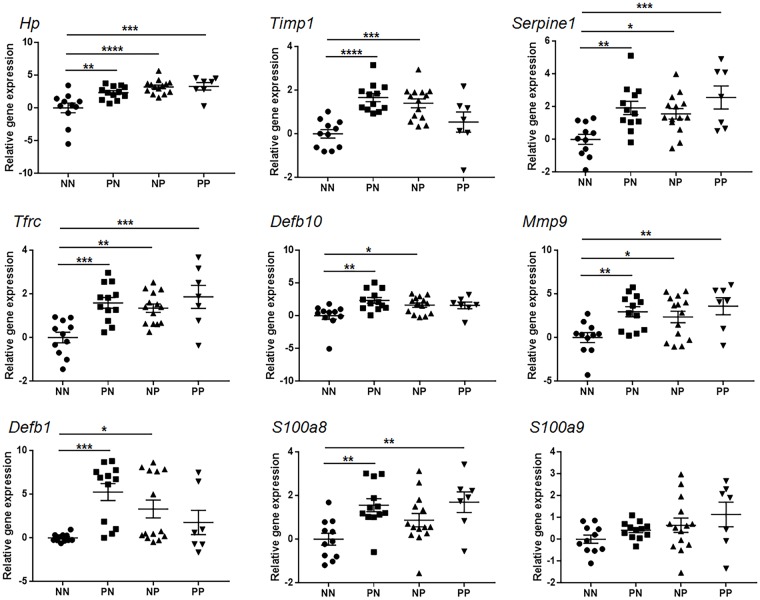
Experimental animals were divided into four groups based on the results of fecal PCR and serum ELISA conducted three times with a 6 month interval (Table 1). When compared with the non-infected NN group, expression of eight genes (S100a8, Defb1, Defb10, Mmp9, Timp1, Hp, Serpine1, and Tfrc) showed higher expression in the PN group (p<0.05), while higher expression of seven other genes (Timp1, Hp, Serpine1, Tfrc, Defb1, Defb10, and Mmp9) was observed in the NP group (p <0.05). Moreover, in the PP group, five genes (S100a8, Mmp9, Hp, Serpine1, and Tfrc) showed significantly higher expression in the PP group (p <0.05). Four genes (Tfrc, Hp, Serpine1, and Mmp9) were up-regulated in all infected groups, while three genes (Timp1, Defb1, and Defb10) were up-regulated in the PN group and the NP group, and S100a8 was up-regulated in the PN group and the PP group (Fig 3). The mean fold changes of each biomarker are shown in Table 2.
Fig 3. Gene expression level of biomarkers in MAP-infected cattle.
The gene expression level of biomarker genes in cattle infected with Mycobacterium avium subspecies paratuberculosis compared to non-infected cattle. The data are shown as scatter plots with each dot representing a single animal. (*, p<0.05 **, p<0.01 ***, p<0.001 ****, p<0.0001).
Discrimination between infected and non-infected animals
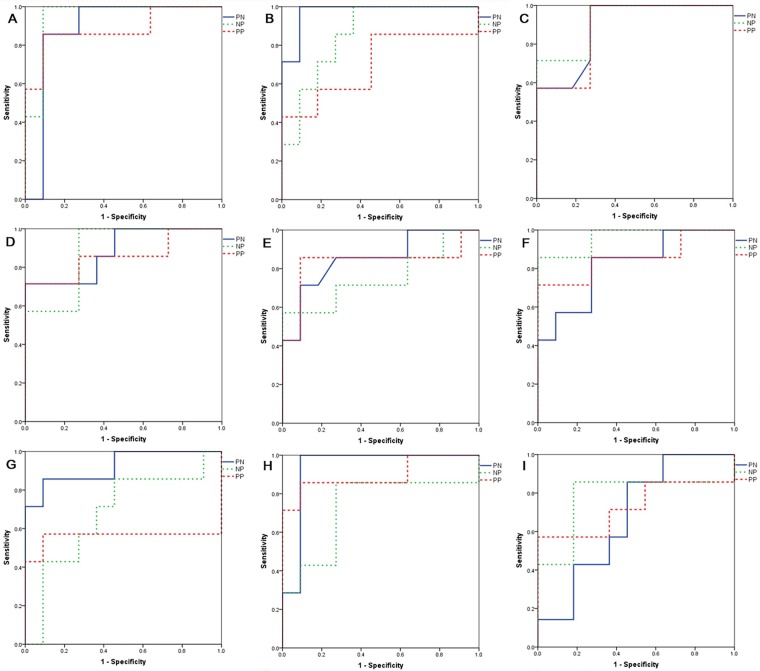
The AUC score of biomarkers was calculated during ROC analysis. In the PN group, the AUC scores of eight genes (Timp1, Defb1, Tfrc, Defb10, S100a8, Serpine1, Mmp9, and Hp) were ≥0.8. In the NP group, four genes (Hp, Timp1, Tfrc, and Serpine1) had AUC scores ≥0.8, while six genes (S100a8, Hp, Serpine1, Tfrc, Mmp9, and Defb10) in the PP group had AUC scores ≥0.8 (Fig 4). When the diagnostic accuracies of individual biomarkers were calculated by ROC curve analysis, the most accurate biomarker in the PN group was Timp1, with an AUC value of 0.985, while the most accurate biomarker in the NP group was Hp, with an AUC value of 0.942. Additionally, the most accurate biomarker in the PP group was S100a8, with an AUC value of 0.896. Similarly, in the PN group, Timp1 showed the most accurate diagnostic performance, with a sensitivity of 100% and a specificity of 90.9%. In the NP group, Hp showed the most accurate diagnostic performance, with a sensitivity of 92.9% and a specificity of 90.9%. Moreover, S100a8 showed the most accurate diagnostic performance in the PP group, with a sensitivity of 85.7% and a specificity of 90%. Other details pertaining to the diagnostic performance of biomarkers are shown in Table 4.
Fig 4. Discriminatory ability of biomarkers between infected animals and control animals.
Receiver operator characteristics curves of biomarker genes in cattle infected with Mycobacterium avium subspecies paratuberculosis compared to non-infected cattle. (A) Hp; (B) Timp1; (C) Serpine1; (D) TFRC; (E) Defb10; (F) Mmp9; (G) Defb1; (H) S100a8; (I) S100a9
Table 4. Diagnostic performance of biomarkers for diagnosis of JD.
| Biomarker | AUC | P value | Cut-off (fold change) | Sensitivity (%) | Specificity (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PN vs. NN | NP vs. NN | PP vs. NN | PN vs. NN | NP vs. NN | PP vs. NN | PN vs. NN | NP vs. NN | PP vs. NN | PN vs. NN | NP vs. NN | PP vs. NN | PN vs. NN | NP vs. NN | PP vs. NN | |
| Tfrc | 0.909 | 0.89 | 0.857 | <0.0001 | <0.0001 | 0.0012 | >0.94 | >0.47 | >0.94 | 75 | 92.9 | 71.4 | 100 | 72.7 | 100 |
| Mmp9 | 0.871 | 0.76 | 0.857 | <0.0001 | 0.0102 | 0.0012 | >0.455 | >2.73 | >2.73 | 91.7 | 57.1 | 71.4 | 72.7 | 100 | 100 |
| S100a8 | 0.894 | 0.721 | 0.896 | <0.0001 | 0.045 | <0.0001 | >0.829 | >-0.095 | >0.829 | 91.7 | 92.9 | 85.7 | 90.9 | 54.5 | 90 |
| S100a9 | 0.693 | 0.669 | 0.727 | 0.11 | 0.1315 | 0.1365 | >-0.07 | >0.511 | >0.856 | 91.7 | 57.1 | 57.1 | 54.5 | 81.8 | 100 |
| Serpine1 | 0.875 | 0.864 | 0.883 | <0.0001 | <0.0001 | <0.0001 | >0.463 | >1.022 | >0.372 | 91.7 | 78.6 | 100 | 72.7 | 100 | 72.7 |
| Hp | 0.864 | 0.942 | 0.883 | <0.0001 | <0.0001 | 0.0001 | >1.806 | >1.806 | >1.806 | 75 | 92.9 | 85.7 | 90.9 | 90.9 | 90.9 |
| Timp1 | 0.985 | 0.929 | 0.701 | <0.0001 | <0.0001 | 0.1815 | >0.682 | >0.682 | >1.022 | 100 | 78.6 | 42.9 | 90.9 | 90.9 | 100 |
| Defb1 | 0.955 | 0.786 | 0.558 | <0.0001 | 0.0024 | 0.7681 | >0.951 | >0.325 | >0.325 | 83.3 | 64.3 | 57.1 | 100 | 90.9 | 90.9 |
| Defb10 | 0.905 | 0.773 | 0.831 | <0.0001 | 0.0052 | 0.0118 | >1.154 | >1.815 | >1.154 | 83.3 | 57.1 | 85.7 | 90.9 | 100 | 90.9 |
Discussion
Early diagnosis of JD is the most important requirement to eradicate it from MAP-infected herds. However, current diagnostic methods are not sufficient for the diagnosis of subclinical stage animals that are actively dispersing MAP into the environment via fecal shedding [2]. Recently, several studies have attempted to diagnose subclinical stages of JD by analyzing host-pathogen interactions, including gene expression, miRNA, protein, and metabolites to MAP infection [17–20, 27]. Some of the studies have been conducted to identify prognostic biomarkers of JD by understanding host response to infection during the progression of JD [21, 28–31]. However, no attempt has been made to apply biomarkers as diagnostic tools. Therefore, the present study was conducted to diagnose MAP infection using a real-time PCR method based on potential prognostic biomarkers.
In the present study, several biomarkers showed good discriminatory ability (AUC≥0.8) between MAP-infected cattle and non-infected cattle. Three genes (Hp, Serpine1, and Tfrc) showed good discriminatory ability (AUC≥0.8) in fecal PCR-positive and/or serum ELISA-positive groups (PN, NP, PP). Acute phase proteins are blood proteins that respond to infection and inflammation and have been used as diagnostic and prognostic biomarkers in veterinary medicine [32]. Hp is the major acute phase protein of cattle that responds to infection [33, 34]. Moreover, Hp is known to exert anti-inflammatory activity by down-regulating neutrophil activity via inhibition of both lipoxygenase and cycloxygenase [35] and to inhibit bacterial growth by interfering with iron acquisition by the host cell [36]. Moreover, Hp inhibits phagocytosis and intracellular killing of pathogens [37]. This anti-inflammatory response induced by Hp might reduce the harmful aspects of inflammation that could be destructive to the host itself. In that regard, up-regulation of Hp in MAP-infected animals might be a host response to early infection of MAP. Hp showed highest diagnostic accuracy for the NP group and whole infected animals, with AUC values of 0.942 and 0.901, respectively.
The initial response to MAP infection is dominant cell-mediated immunity, which is characterized by increasing interferon gamma release [38]. Serpine1 is known to be an essential element of the fibrinolytic system that is related to blood coagulation [39]. Serpine1 also acts as an inflammatory mediator by increasing the level of interferon gamma in blood to eliminate the pathogen in the early phase of an infectious disease [40, 41]. Therefore, increasing gene expression levels of Serpine1 might be related to interferon gamma release due to MAP infection. In addition, expression of MAP0403 in MAP was increased in infected macrophages and MAC-T cells in recent study [42]. MAP0403 is kind of serine protease which served as a key element of the stress response network in intraphagosomal survival of MAP [42]. Up-regulation of Serpine1 might be a counter response to intraphagosomal survival of MAP in host cells. The diagnostic accuracy of Serpine1 was good (AUC≥0.8) in all infected animals (PN, NP, PP group).
Iron is an important nutrient in innate immune response to bacterial pathogen [43]. Tfrc, which is one of the key elements of iron metabolism, transfers iron to cells from transferrin protein [43]. Tfrc is known to down-regulated in response to intracellular pathogen infection; however, its expression was significantly increased in all infected animals in the present study. This phenomenon might be related to the alternative iron acquisition system of MAP, which acts in a host-independent manner using mycobactin [44]; however, further studies are needed to confirm this.
Mmp9 is a matrix metalloproteinase related to leukocyte migration to infection sites and tissue destruction if it is secreted in excess amounts [45]. The level of Mmp9 was regulated by Timp1, which inhibits the activity of MMP9 [45]. Mmp9 and Timp1 are known to be up-regulated in tuberculosis infection and have therefore been proposed as biomarkers for diagnosis of tuberculosis [46]. The simultaneous up-regulation of Mmp9 and Timp1 in infected animals might be caused by inflammatory conditions due to the early stages of MAP infection. Two genes (Mmp9 and Timp1) showed good discriminatory ability (AUC≥0.8) in the PN group.
β-defensins exhibit antimicrobial functions, providing first protection against pathogens while playing an immune-modulation role [47]. Moreover, β-defensins interplay between innate and adaptive immune responses by down-regulating pro-inflammatory cytokines [48]. In the present study, Defb1 and Defb10 were significantly up-regulated in both the PN group and the NP group. Moreover, Defb1 and Defb10 showed excellent discriminatory ability (AUC≥0.9) in the PN group.
S100a8 and S100a9 are members of a calcium-binding cytosolic protein family that are located in the cytoplasm [49]. S100a8 and S100a9 form a heterodimer known as calprotectin that induces an inflammatory response via activation of TLR4 signaling [50]. Moreover, calprotectin is known to induce leukocyte migration in the early phase of bacterial infection [51]. In previous studies, serum S100A8/A9 have been proposed as prognostic biomarkers for disease progression and therapeutic response in inflammatory bowel diseases (IBD)[52, 53]. In the present study, S100a8 showed good discriminatory ability (AUC≥0.8) in the PN and PP groups. However, gene expression of S100a9 was not significant in all infected animals. Generally, S100a8 and S100a9 exist as heterodimers, but they also exist as homodimers [54]. The inconsistent gene expression levels between S100a8 and S100a9 might be related to the presence of the homodimer form.
An ideal biomarker for diagnosis of JD should be able to discriminate between infected and non-infected animals with high sensitivity and specificity. Our data showed that the response of eight biomarkers (Hp, Timp1, Mmp9, Serpine1, Tfrc, S100a8, Defb1, and Defb10) significantly discriminated MAP-infected and non-infected animals. Moreover, eight biomarkers (Hp, Timp1, Mmp9, Serpine1, Tfrc, S100a8, Defb1, and Defb10) showed good accuracy (AUC≥0.7) for diagnosis of subclinical animals. Additionally, four genes (Timp1, S100a8, Defb1, and Defb10) showed sensitivity over 80% and specificity over 90%. It is generally very difficult to detect subclinical stages of JD using currently available diagnostic methods such as bacterial culture, fecal PCR and serum ELISA [3]. Fecal PCR is a reliable method for diagnosis of MAP infection; however, intermittent shedding of MAP into feces because of immunological changes during the progress of disease can interfere with accurate diagnosis [55]. Moreover, although serum ELISA is a simple, fast and cost-effective method for diagnosis of JD, it is known to have low sensitivity for MAP-infected animals that do not show clinical signs [56]. However, our real-time PCR method based on biomarkers showed relatively precise diagnostic results. In that regard, combination of eight biomarker genes (Hp, Timp1, Mmp9, Serpine1, Tfrc, S100a8, Defb1, and Defb10) might be used for diagnosis of JD, including in subclinical stage animals.
In conclusion, a real-time PCR method was developed based on eight biomarkers that can be used as a new diagnostic tool for JD with good diagnostic performance. Moreover, this real-time PCR based on biomarkers might be used for diagnosis of JD, especially in subclinical stage animals that cannot be detected by current diagnostic methods. Although our developed diagnostic method might be applied to field test, this method will be more concreted if possible limitations in our study such as the low number of samples and sampling times would be addressed in future studies by including large scale field investigations.
Acknowledgments
We are grateful to all our research participants, and staff of the National Instiute of Animal Science, who contributed to this study.
Data Availability
All datasets used in selection of the biomarkers are available at Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geowebsite) under accession number GSE62836, http://dx.doi.org/10.4014/jmb.1302.02021, and http://dx.doi.org/10.4014/jmb.1408.08059.
Funding Statement
This work was supported by the Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ00897001) to HSY, Rural Development Administration, National Research Foundation (No. 2014R1A2A2A01007291) to HSY, BK21 PLUS to HSY and the Research Institute for Veterinary Sciences, Seoul National University, Republic of Korea to HSY. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Whitlock RH, Buergelt C. Preclinical and clinical manifestations of paratuberculosis (including pathology). Vet Clin North Am Food Anim Pract. 1996;12: 345–3 [DOI] [PubMed] [Google Scholar]
- 2.Tiwari A, VanLeeuwen JA, McKenna SL, Keefe GP, Barkema HW. Johne's disease in Canada Part I: clinical symptoms, pathophysiology, diagnosis, and prevalence in dairy herds. Can Vet J. 2006;47: 874–882. [PMC free article] [PubMed] [Google Scholar]
- 3.Mortier RA, Barkema HW, De Buck J. Susceptibility to and diagnosis of Mycobacterium avium subspecies paratuberculosis infection in dairy calves: A review. Prev Vet Med. 2015;121(3–4): 189–98. 10.1016/j.prevetmed.2015.08.011 [DOI] [PubMed] [Google Scholar]
- 4.Whittington RJ, Whittington AM, Waldron A, Begg DJ, de Silva K, Purdie AC, et al. Development and validation of a liquid medium (M7H9C) for routine culture of Mycobacterium avium subsp. paratuberculosis to replace modified Bactec 12B medium. J Clin Microbiol. 2013;51(12): 3993–4000 10.1128/JCM.01373-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bögli-Stuber K, Kohler C, Seitert G, Glanemann B, Antognoli MC, Salman MD, et al. Detection of Mycobacterium avium subspecies paratuberculosis in swiss dairy cattle by real-time PCR and culture: a comparison of the two assays. J Appl Microbiol. 2005;99: 587–597. 10.1111/j.1365-2672.2005.02645.x [DOI] [PubMed] [Google Scholar]
- 6.Sockett DC, Carr DJ, Collins MT. Evaluation of conventional and radiometric fecal culture and a commercial DNA probe for diagnosis of Mycobacterium paratuberculosis infections in cattle. Can J Vet Res. 1992;56(2): 148–53. [PMC free article] [PubMed] [Google Scholar]
- 7.Sevilla IA, Garrido JM, Molina E, Geijo MV, Elguezabal N, Vázquez P, et al. Development and evaluation of a novel multicopy-element-targeting triplex PCR for detection of Mycobacterium avium subsp. paratuberculosis in feces. Appl Environ Microbiol. 2014;80(12): 3757–68. 10.1128/AEM.01026-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wells SJ, Collins MT, Faaberg KS, Wees C, Tavornpanich S, Petrini KR, et al. Evaluation of a rapid fecal PCR test for detection of Mycobacterium avium subsp. paratuberculosis in dairy cattle. Clin Vaccine Immunol. 2006;13(10): 1125–30. 10.1128/CVI.00236-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cousins DV, Whittington R, Marsh I, Masters A, Evans RJ, Kluver P. Mycobacteria distenct from Mycobacterium avium subsp. paratuberculosis isolated from the faeces of ruminants possess IS900-like sequences detectable IS900 polymerase chain reaction: implications for diagnosis. Mol Cell Probes. 1999;13(6): 431–42. 10.1006/mcpr.1999.0275 [DOI] [PubMed] [Google Scholar]
- 10.Englund S, Bölske G, Johansson KE, An IS900-like sequence found in a Mycobacterium sp. other than Mycobacterium avium subsp. paratuberculosis. FEMS Microbiol Lett. 2002;209: 267–271. [DOI] [PubMed] [Google Scholar]
- 11.Al Hajri SM, Alluwaimi AM. ELISA and PCR for evaluation of subclinical paratuberculosis in the Saudi dairy herds. Vet Microbiol. 2007;121: 384–385. 10.1016/j.vetmic.2007.01.025 [DOI] [PubMed] [Google Scholar]
- 12.Strimbu K, Tavel JA. What are biomarkers? Curr Opin HIV AIDS. 2010;5(6): 463–6. 10.1097/COH.0b013e32833ed177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dehnad A, Ravindran R, Subbian S, Khan IH. Development of immune-biomarkers of pulmonary tuberculosis in a rabbit model. Tuberculosis (Edinb). 2016;101: 1–7. [DOI] [PubMed] [Google Scholar]
- 14.Goyal N, Kashyap B, Singh NP, Kaur IR. Neopterin and oxidative stress markers in the diagnosis of extrapulmonary tuberculosis. Biomarkers. 2016;8: 1–6. [DOI] [PubMed] [Google Scholar]
- 15.Walker SJ, Beavers DP, Fortunato J, Krigsman A. A Putative Blood-Based Biomarker for Autism Spectrum Disorder-Associated Ileocolitis. Sci Rep. 2016;6: 35820 10.1038/srep35820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waters WR, Maggioli MF, Palmer MV, Thacker TC, McGill JL, Vordermeier HM, et al. Interleukin-17A as a Biomarker for Bovine Tuberculosis. Clin Vaccine Immunol. 2015;23: 168–80. 10.1128/CVI.00637-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.David J, Barkema HW, Mortier R, Ghosh S, le Guan L, De Buck J. Gene expression profiling and putative biomarkers of calves 3 months after infection with Mycobacterium avium subspecies paratuberculosis. Vet Immunol Immunopathol. 2014; 160: 107–117. 10.1016/j.vetimm.2014.04.006 [DOI] [PubMed] [Google Scholar]
- 18.David J, Barkema HW, le Guan L, De Buck J. Gene-expression profiling of calves 6 and 9 months after inoculation with Mycobacterium avium subspecies paratuberculosis. Vet Res. 2014;45: 96 10.1186/s13567-014-0096-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Buck J, Shaykhutdinov R, Barkema HW, Vogel HJ. Metabolomic profiling in cattle experimentally infected with Mycobacterium avium subsp. paratuberculosis. PLoS One. 2014;9(11): e111872 10.1371/journal.pone.0111872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.You Q, Verschoor CP, Pant SD, Macri J, Kirby GM, Karrow NA. Proteomic analysis of plasma from Holstein cows testing positive for Mycobacterium avium subsp. paratuberculosis (MAP). Vet Immunol Immunopathol. 2012;148(3–4): 243–51. 10.1016/j.vetimm.2012.05.002 [DOI] [PubMed] [Google Scholar]
- 21.Cha SB, Yoo A, Park HT, Sung KY, Shin MK, Yoo HS. Analysis of transcriptional profiles to discover biomarker candidates in Mycobacterium avium subsp. paratuberculosis-infected macrophages, RAW 264.7. J Microbiol Biotechnol. 2013;23: 1167–1175. [DOI] [PubMed] [Google Scholar]
- 22.Shin MK, Park HT, Shin SW, Jung M, Im YB, Park HE, et al. Whole-blood gene-expression profiles of cows infected with Mycobacterium avium subsp. paratuberculosis reveal changes in immune response and lipid metabolism. J Microbiol Biotechnol. 2015;25: 255–267. [DOI] [PubMed] [Google Scholar]
- 23.Shin MK, Park H, Shin SW, Jung M, Lee SH, Kim DY, et al. Host Transcriptional Profiles and Immunopathologic Response following Mycobacterium avium subsp. paratuberculosis Infection in Mice. PLoS One. 2015;10(10): e0138770 10.1371/journal.pone.0138770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park HT, Shin MK, Park HE, Cho YI, Yoo HS. PCR-based detection of Mycobacterium avium subsp. paratuberculosis infection in cattle in South Korea using fecal samples. J Vet Med Sci. 2016;78(9): 1537–1540. 10.1292/jvms.15-0271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park HE, Shin MK, Park HT, Jung M, Cho YI, Yoo HS. Gene expression profiles of putative biomarker candidates in Mycobacterium avium subsp. paratuberculosis-infected cattle. Pathog Dis. 2016;74(4): ftw022 10.1093/femspd/ftw022 [DOI] [PubMed] [Google Scholar]
- 26.Muller MP, Tomlinson G, Marrie TJ, Tang P, McGeer A, Low DE, et al. Can routine laboratory tests discriminate between severe acute respiratory syndrome and other causes of community-acquired pneumonia? Clin Infect Dis. 2005;40(8): 1079–86. 10.1086/428577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malvisi M, Palazzo F, Morandi N, Lazzari B, Williams JL, Pagnacco G, et al. Responses of Bovine Innate Immunity to Mycobacterium avium subsp. paratuberculosis Infection Revealed by Changes in Gene Expression and Levels of MicroRNA. PLoS One. 2016;11(10): e0164461 10.1371/journal.pone.0164461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plain KM, de Silva K, Earl J, Begg DJ, Purdie AC, Whittington RJ. Indoleamine 2,3-dioxygenase, tryptophan catabolism, and Mycobacterium avium subsp. paratuberculosis: a model for chronic mycobacterial infections. Infect Immun. 2011; 79(9): 3821–32. 10.1128/IAI.05204-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seth M, Lamont EA, Janagama HK, Widdel A, Vulchanova L, Stabel JR, et al. Biomarker discovery in subclinical mycobacterial infections of cattle. PLoS One. 2009;4(5): e5478 10.1371/journal.pone.0005478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verschoor CP, Pant SD, You Q, Kelton DF, Karrow NA. Gene expression profiling of PBMCs from Holstein and Jersey cows sub-clinically infected with Mycobacterium avium ssp. paratuberculosis. Vet Immunol Immunopathol. 2010;137: 1–11 10.1016/j.vetimm.2010.03.026 [DOI] [PubMed] [Google Scholar]
- 31.Wang X, Wang H, Aodon-geril, Shu Y, Momotani Y, Nagata R, et al. Decreased expression of matrix metalloproteinase-9 and increased expression of tissue inhibitors of matrix metalloproteinase-1 in paratuberculosis-infected cattle in the ELISA-negative subclinical stage. Anim Biotechnol. 2011;22(1): 44–9. 10.1080/10495398.2010.536096 [DOI] [PubMed] [Google Scholar]
- 32.Eckersall PD, Bell R. Acute phase proteins: Biomarkers of infection and inflammation in veterinary medicine. Vet J. 2010;185(1): 23–7. 10.1016/j.tvjl.2010.04.009 [DOI] [PubMed] [Google Scholar]
- 33.Seppä-Lassila L, Orro T, Lassen B, Lasonen R, Autio T, Pelkonen S, et al. Intestinal pathogens, diarrhoea and acute phase proteins in naturally infected dairy calves. Comp Immunol Microbiol Infect Dis. 2015;41: 10–6. 10.1016/j.cimid.2015.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El-Deeb WM, Elmoslemany AM. Acute phase proteins as biomarkers of urinary tract infection in dairy cows: diagnostic and prognostic accuracy. Jpn J Vet Res. 2016; 64(1): 57–66. [PubMed] [Google Scholar]
- 35.Saeed SA, Ahmad N, Ahmed S. Dual inhibition of cyclooxygenase and lipoxygenase by human haptoglobin: its polymorphism and relation to hemoglobin binding. Biochem Biophys Res Commun. 2007;353(4): 915–20. 10.1016/j.bbrc.2006.12.092 [DOI] [PubMed] [Google Scholar]
- 36.Eaton JW, Brandt P, Mahoney JR, Lee JT Jr. Haptoglobin: a natural bacteriostat. Science. 1982;215(4533): 691–3. [DOI] [PubMed] [Google Scholar]
- 37.Rossbacher J, Wagner L, Pasternack MS. Inhibitory effect of haptoglobin on granulocyte chemotaxis, phagocytosis and bactericidal activity. Scand J Immunol 1999;50(4): 399–404. [DOI] [PubMed] [Google Scholar]
- 38.Stabel JR. Transitions in immune responses to Mycobacterium paratuberculosis. Vet Microbiol. 2000;77(3–4): 465–73. [DOI] [PubMed] [Google Scholar]
- 39.Furie B, Furie BC. The molecular basis of blood coagulation. Cell. 1988;53(4): 505–18. [DOI] [PubMed] [Google Scholar]
- 40.Lim JH, Woo CH, Li JD. Critical role of type 1 plasminogen activator inhibitor (PAI-1) in early host defense against nontypeable Haemophilus influenzae (NTHi) infection. Biochem Biophys Res Commun. 2011;414(1): 67–72. 10.1016/j.bbrc.2011.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Z, Zhao Q, Han Y, Zhang D, Zhang L, Luo D. PAI-1 and IFN-γ in the regulation of innate immune homeostasis during sublethal yersiniosis. Blood Cells Mol Dis. 2013; 50(3): 196–201. 10.1016/j.bcmd.2012.11.005 [DOI] [PubMed] [Google Scholar]
- 42.Kugadas A, Lamont EA, Bannantine JP, Shoyama FM, Brenner E, Janagama HK, et al. A Mycobacterium avium subsp. paratuberculosis Predicted Serine Protease Is Associated with Acid Stress and Intraphagosomal Survival. Front Cell Infect Microbiol. 2016;6: 85 10.3389/fcimb.2016.00085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson EE, Wessling-Resnick M. Iron metabolism and the innate immune response to infection. Microbes Infect. 2012;14(3): 207–16. 10.1016/j.micinf.2011.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang J, Moolji J, Dufort A, Staffa A, Domenech P, Reed MB, et al. Iron Acquisition in Mycobacterium avium subsp. paratuberculosis. J Bacteriol. 2015;198(5): 857–66. 10.1128/JB.00922-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goetzl EJ, Banda MJ, Leppert D. Matrix metalloproteinases in immunity. J Immunol. 1996;156(1): 1–4. [PubMed] [Google Scholar]
- 46.Chen Y, Wang J, Ge P, Cao D, Miao B, Robertson I, et al. Tissue inhibitor of metalloproteinases 1, a novel biomarker of tuberculosis. Mol Med Rep. 2017;15(1): 483–487. 10.3892/mmr.2016.5998 [DOI] [PubMed] [Google Scholar]
- 47.Meade KG, Cormican P, Narciandi F, Lloyd A, O'Farrelly C. Bovine β-defensin gene family: opportunities to improve animal health? Physiol Genomics. 2014;46(1): 17–28. 10.1152/physiolgenomics.00085.2013 [DOI] [PubMed] [Google Scholar]
- 48.Allaker RP. Host defence peptides-a bridge between the innate and adaptive immune responses. Trans R Soc Trop Med Hyg. 2008;102(1): 3–4. 10.1016/j.trstmh.2007.07.005 [DOI] [PubMed] [Google Scholar]
- 49.Schiopu A, Cotoi OS. S100A8 and S100A9: DAMPs at the crossroads between innate immunity, traditional risk factors, and cardiovascular disease. Mediators Inflamm. 2013;2013: 828354 10.1155/2013/828354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vogl T, Tenbrock K, Ludwig S, Leukert N, Ehrhardt C, van Zoelen MA, et al. Mrp8 and Mrp14 are endogenous activators of Toll-like receptor 4, promoting lethal, endotoxin-induced shock. Nat Med. 2007; 13(9): 1042–9. 10.1038/nm1638 [DOI] [PubMed] [Google Scholar]
- 51.Achouiti A, Vogl T, Urban CF, Röhm M, Hommes TJ, van Zoelen MA, et al. Myeloid-related protein-14 contributes to protective immunity in gram-negative pneumonia derived sepsis. PLoS Pathog. 2012;8(10): e1002987 10.1371/journal.ppat.1002987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leach ST, Yang Z, Messina I, Song C, Geczy CL, Cunningham AM, et al. Serum and mucosal S100 proteins, calprotectin (S100A8/S100A9) and S100A12, are elevated at diagnosis in children with inflammatory bowel disease. Scand J Gastroenterol. 2007; 42(11): 1321–31. 10.1080/00365520701416709 [DOI] [PubMed] [Google Scholar]
- 53.Cayatte C, Joyce-Shaikh B, Vega F, Boniface K, Grein J, Murphy E, et al. Biomarkers of Therapeutic Response in the IL-23 Pathway in Inflammatory Bowel Disease. Clin Transl Gastroenterol. 2012;3: e10 10.1038/ctg.2012.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vogl T, Ludwig S, Goebeler M, Strey A, Thorey IS, Reichelt R, et al. MRP8 and MRP14 control microtubule reorganization during transendothelial migration of phagocytes. Blood. 2004;104: 4260–8. 10.1182/blood-2004-02-0446 [DOI] [PubMed] [Google Scholar]
- 55.Mitchell RM, Schukken Y, Koets A, Weber M, Bakker D, Stabel J, et al. Differences in intermittent and continuous fecal shedding patterns between natural and experimental Mycobacterium avium subspecies paratuberculosis infections in cattle. Vet Res. 2015;19: 46–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nielsen SS, Toft N. Ante mortem diagnosis of paratuberculosis: a review of accuracies of ELISA, interferon-gamma assay and faecal culture techniques. Vet Microbiol. 2008; 129: 217–35. 10.1016/j.vetmic.2007.12.011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All datasets used in selection of the biomarkers are available at Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geowebsite) under accession number GSE62836, http://dx.doi.org/10.4014/jmb.1302.02021, and http://dx.doi.org/10.4014/jmb.1408.08059.