Plants are remarkable with regard to their capability to go beyond diploidy and exist as stable polyploids. It is estimated that ∼50 to 70% of flowering plants are polyploids (Masterson, 1994; Otto and Whitton, 2000). Theory predicts that gene duplicates degenerate and disappear, but the extensive retention of paralogs in polyploids or paleopolyploids runs counter to this prediction. What is the fate of gene duplicates in polyploids? Multiple evolutionary fates for paralogs have been explored from different perspectives (Lynch and Conery, 2000; Wolfe, 2001). Here, this question is revisited. Specifically, several scenarios will be explored based on the notion that point mutations or deletions of the paralogs can be deleterious if their products are involved in protein–protein or protein–DNA complexes. This perspective is not exclusive but rather complementary to the ones developed in previous studies.
The extent of paralog survival in plants is so important that protein diversity in this kingdom has been hypothesized to be generated primarily through gene duplication rather than by alternative splicing, as has been proposed for vertebrates (Hodgkin, 2000; Yu et al., 2002). However, one cannot overlook that much of vertebrate diversity comes from the fishes, major lineages of which appear to be the result of genome duplication events. While still controversial, it would not be unreasonable to say that data are suggestive of a role for polyploidy in the majority of vertebrate diversity (Makalowski, 2001). Thus, while the focus of this review is the fate of paralogs in plants, the ideas developed are applicable to other organisms.
Force et al. (1999) proposed that random mutation and drift can alter the expression patterns of paralogous genes and segregate their regulation so that both (together) cover the full range of functions of the ancestral gene. Accordingly, both copies would be needed. For instance, each paralogous gene might have a specific domain of expression. This process is known as subfunctionalization. Once subfunctional mutations arise and become fixed, selection will maintain both paralogous copies. Acquisition of a new function by one of the paralogs, neofunctionalization (a rare event at least in small populations), can also explain evolutionary retention (Ohno, 1970; Force et al., 1999).
Another mechanism that can lead to the retention of paralogs, not exclusive of subfunctionalization, is a requirement for the preservation of stoichiometry within complexes or pathways. Gene products that participate together in a macromolecular complex or pathway are thought to require a dosage balance for proper function (Veitia, 2002, 2004). The importance of gene dosage balance is obvious in organisms that alternate between haploid and diploid phases, especially haplodiplobiontics, such as plants, which undergo development in both phases. During the haploid gametophyte phase, deletion of genes required for viability cannot be tolerated, and the balance among interacting components is achieved with one gene copy of each locus. In the diploid state, a heterozygous deletion of a gene can be deleterious to diploid development if it leads to a stoichiometric imbalance among components of a complex (Veitia, 2002, 2003a).
General theories of dominance have stated that one of the advantages of diploidy is that one allele can mask deleterious mutations in the other allele at a particular locus (Orr and Otto, 1994). At dosage sensitive genes, however, one active allele in a diploid cannot mask a complete lack of activity of the other. Accordingly, a recent genomic survey has shown that deletion of one allele of 21% of the yeast genes analyzed leads to defective growth (i.e., these genes are haplo-insufficient; Kondrashov and Koonin, 2004). In humans, the effects of dosage sensitivity might be more dramatic because haplo-insufficient mutations responsible for disease seem to outnumber the recessive ones (Kondrashov and Koonin, 2004). Further evidence for the importance of dosage balance has been found in yeast, which is able to undergo vegetative growth in both haploid and diploid conditions. Dosage-sensitive genes (whose underexpression or overexpression induces a growth defect) are more likely to encode proteins involved in macromolecular complexes than genes with low dosage sensitivity (Papp et al., 2003). Birchler et al. (2001) also presented evidence suggesting that the stoichiometric relationships among the components of regulatory complexes affect target gene expression.
DOSAGE EFFECTS AND THE SURVIVAL OF PARALOGS
The contribution of polyploids to evolution, either with multisomic inheritance or as diploidized forms, presumably would be minor if most duplicated copies were to be silenced (nonfunctionalized). Walsh (1995) estimated that ∼99% of duplicate genes would become pseudogenes under certain conditions of population size and mutation rates. The analysis of ancient polyploids shows that a much larger fraction of functional paralogs is actually retained. In the paleopolyploid yeast Saccharomyces cerevisiae, ∼12% of paralogs have retained functionality for 100 million years (Kellis et al., 2004). The fraction of retained paralogs rises to 72% in maize, through 11 million years of evolution (Ahn and Tanksley, 1993). However, their functionality is yet to be assessed. As discussed below, Arabidopsis thaliana also provides many examples of functional paralogs retained over several cycles of polyploidization.
Gene duplicates generated by genome duplication are likely to be retained if they undergo selection for absolute gene product dosage (Otto and Whitton, 2000). This simple idea entails more complexity than might be suspected at first sight. Thus, before proceeding with the discussion, we must consider that on the one hand, in plant ploidy series the mRNA level of most genes is roughly proportional to ploidy (Guo et al., 1996). On the other hand, when similar cell types are compared, cell volume tends to be proportional to DNA content (Cavalier-Smith, 1982; Gregory, 2001). For instance, pollen grain volume and stomatal cell size correlate with ploidy (Bennett, 1972; Masterson, 1994). Even subtle changes in genomic DNA content (i.e., by adding or substracting small amounts of DNA) have an impact on cell volume (Gregory, 2001). Thus, when correlations between gene expression, cell volume, and ploidy are considered together, an increase in gene dosage after autopolyploidization is not expected to be paralleled by an increase in the concentrations of gene products. An increase in the absolute gene product concentration can only be accomplished by a reduction in cell volume if expression rates of the paralogous alleles do not change and if there is some degree of saturation in protein degradation. A reduction in cell volume could be achieved by the deletion of DNA. A number of other characteristics of the three-dimensional cell will be altered with an increase in ploidy. For example, the surface-to-volume ratio of the plasma membrane to the cytosol, or of the nuclear envelope to the nucleoplasm and cytoplasm, might be altered. Because these effects have been less well studied, the discussion will focus on gene products in suspension in the cytosol or the nucleoplasm.
Absolute increases in the quantity of functional gene products are straightforward when the proteins act as monomers poorly connected within the cellular interaction network, for which increased dosage can be achieved through segmental DNA duplication. However, for dosage-sensitive genes, increases in the expression level of one component can lead to an alteration in the amount of functional complex. In such cases, a coordinated increase of dosage of the interacting gene products must be achieved, possibly through a reduction in cellular volume after genome duplication. The survival of one paralog will depend on the retention of paralogs encoding the other stoichiometric interactors (see below).
In yeast, homozygous deletions of many genes are associated with little or no apparent phenotypic effects. At least one-quarter of those gene deletions that have no growth phenotype are expected to be compensated for by duplicate genes. Gu et al. (2003) found a higher probability of phenotypic compensation for a gene with two recognizable copies than for singletons, and higher sequence similarities between duplicates correlated with higher frequencies of compensation. Interestingly, when the more highly expressed paralog was deleted, there was a higher probability of a severe fitness effect. This observation points to the existence of absolute or relative dosage requirements for many duplicated genes in yeast.
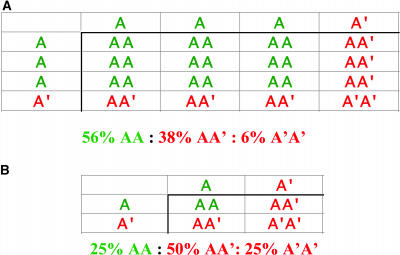
In addition, it has been argued that paralogous genes encoding multidomain proteins may be retained in polyploids because mutations in such genes can produce dominant negative phenotypes (Gibson and Spring, 1998). Alterations that fully obliterate a gene, such as mutations in the promoter or full deletions, are rarer than mutations leading to negative dominance. Although Gibson and Spring (1998) focused on genes encoding multidomain proteins, the key point regarding negative dominance is that new variants can be incorporated into macromolecular complexes, which they inactivate. For example, within the context of the dimer AA, consider that a mutation A′ renders AA′ and A′A′ inactive. In a diploid, only 25% of the complex would remain active. Whereas additional alleles in a tetraploid can buffer deleterious effects, one mutant allele results in only 56% of active AA being produced (Figure 1). Such a decrease in protein activity might be associated (i.e., at the borderline) with a fitness defect. A similar drop in active complex would be experienced by the trimer A-B-A if A-B-A′ is inactive. Moreover, if selection also acts during the gametophyte phase (which has a reduced ploidy), an allele with negative fitness consequences in a diploid heterozygote (i.e., AA′) can be swept by selection if fitness depends critically on the concentration of AA or ABA. In these cases, the amount of active AA or ABA will be as low as 25% of the normal resulting in a greater than twofold decrease in activity of the complex and a concomitant increase in the efficiency of negative selection. It is worth noting that dominant negative effects can also arise in the context of transcription complexes. For example, a truncating mutation can remove the transactivation domain of a transcription factor and leave the DNA binding domain intact. This truncated factor will behave as a competitive inhibitor of transcription, and its effect can be even stronger than that of a deletion.
Figure 1.
Dominant Negative Effects and Retention of Paralogs.
(A) Punnett square showing all possible combinations of dimers produced from three normal A alleles and one mutant allele A′ that renders the dimer inactive. The presence of only one mutant allele out of four leads to 56% of normal dimer AA.
(B) Passage through a reduced ploidy phase is even worse because only 25% of dimers will be active. This fraction corresponds, in genetic terms, to the proportion of homozygotes AA, which are favored by selection. Removal of the deleterious A′ allele from the population can be extremely effective during this phase if fitness depends tightly on AA. Similar results are obtained for the trimer A-B-A.
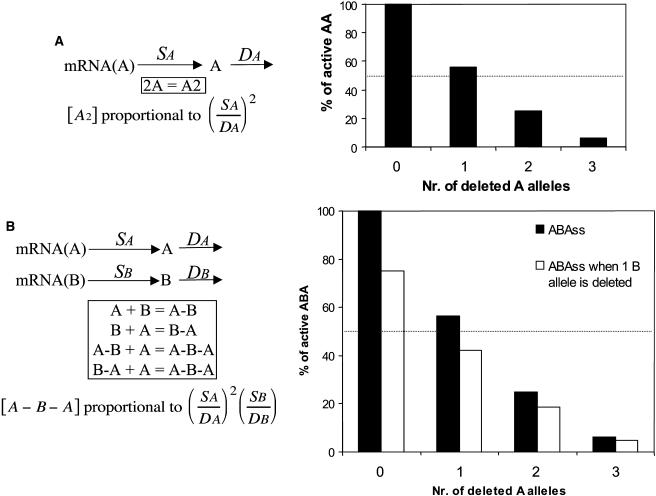
Gene dosage effects also provide a selective force tending to preserve paralogs. Indeed, a dosage alteration may lead to biochemical and potential fitness effects that are comparable to the negative dominance described above in the context of macromolecular complexes. For the dimer AA, if the rate of production of A is proportional to the number of gene copies and degradation is proportional to monomer concentration, after deleting one allele in a tetraploid the steady state concentration of AA drops to as low as 56% of normal (Figure 2A). For the trimer A-B-A, where B is a bridge between the two A subunits, the precise effect of a stoichiometric imbalance between A and B depends critically on the route that assembly follows. For example, if all steps are reversible and only the monomers are degradable, then a deletion of one A allele leads to 56% of ABA (Figure 2B). Absence of one allele of B in the tetraploid leads to 75% of ABA, but a missing allele of both A and B would lead to only 42% of ABA. Thus, deletion of one copy of B constrains survival of all four alleles of A. Similar epistatic interactions might also play a role in the case of dominant negative mutations described earlier. Dosage effects may also appear in other complexes, such as A-B-C. If we allow the existence of intermediates AB and BC (i.e., random assembly) under the same assumptions as above, then the deletion of any component separately translates into a proportional diminution of ABC. However, deletion of one allele (out of four) of two components translates into a more drastic diminution of ABC (56% of normal), and, of course, deletions in a diploid state would have more dramatic consequences. This example shows again how a deletion of one allele at any locus can constrain the survival of all other remaining interacting alleles. While the selective forces acting on negative dominance or dosage imbalances may have a low impact on organisms with high ploidy levels (i.e., an octoploid), a passage through a reduced ploidy phase will enhance their stringency. Consistent with the ideas outlined above, many retained paralogs in the recent allotetraploid Xenopus laevis are involved in protein–protein interactions (Hughes and Hughes, 1993). Not surprisingly, yeast genes encoding interacting subunits tend to have the same number of duplicates (Papp et al., 2003; Yang et al., 2003).
Figure 2.
Dosage Effects and Retention of Paralogs.
(A) In the context of the homodimer AA, consider that the synthesis of A results from the translation of the mRNA(A), present at a stable concentration, then A undergoes reversible dimerization or is degraded with a rate proportional to the concentration of monomers. According to this scheme, the concentration of AA in the steady state (ss) is proportional to the square of the ratio of specific rates of synthesis of A (Sa) and its degradation (Da; derivation in Veitia, 2004). Deletion of one allele out of four leads to 56% of normal AA. The graph shows the effects of deleting one, two, or three alleles expressed as the percentage of active AA with respect to the normal tetraploid system (the dotted line represents 50% of concentration of dimer). In a diploid phase, obliteration of one copy of A leads to 25% AA. Results are different if, for instance, AA becomes the degradable entity, as in a fast irreversible dimerization. When AA is degraded with a rate proportional to its concentration, a change in monomer input will have a proportional impact on the concentration of the dimer.
(B) What happens to the trimer A-B-A in the steady state? Under similar assumptions as above, the yield of ABA depends again on the square of Sa. The black bars in the graph show the steady state amounts of ABA for different doses of the wild-type A allele (for the maximum/tetraploid dose of B). The gray bars show how deleting one allele of B increases dosage sensitivity of A. Again, the results are slightly different if the dimers or trimer are allowed to be degraded, and less dramatic effects are obtained when ABA is the only degradable entity.
REGULATORY DOSAGE UPSET IN SUCCESSFUL ALLOPOLYPLOIDS AND THE FATE OF PARALOGS
There is increasing evidence for nonequivalent contributions of the two parents to gene expression in allopolyploids and hybrids. A classical example is nucleolar dominance, in which one parental set of rRNA genes is silenced in an interspecific hybrid or allopolyploid (Pikaard, 2000). For instance, in A. suecica, an allotetraploid of A. thaliana and Cardaminopsis arenosa, there is silencing of the rRNA genes contributed by the A. thaliana subgenome (Chen et al., 1998). A similar phenomenon has been observed for many genes in other polyploids. For example, a recent analysis by Adams et al. (2003) has shown that expression levels of paralogous genes in synthetic allopolyploids simulating the ancestor of the most widely cultivated cotton, Gossypium hirsutum, were not additive. That is, the parental subgenomes did not contribute equally to the transcriptome of the polyploid. Indeed, expression of each paralog varied among organs and was suggestive of differential developmental regulation. Interestingly, the expression patterns in the synthetic polyploids can recapitulate those found in the naturally occurring cotton (Adams et al., 2003). This suggests that certain patterns of expression appear just after a polyploidization event and persist over time. Classical epigenetic mechanisms, such as altered DNA methylation or chromatin structure, have been proposed to explain this phenomenon (Kellogg, 2003). Heritable changes, such as DNA methylation, deletions, and chromosomal rearrangements, have been documented in other cases of allopolyploidy (Chen et al., 1998; Ozkan et al., 2001; Pires et al., 2004). Alternatively and not exclusively, sudden changes in transcription can be attributable to a regulatory mismatch between the effectors and the target genes contributed by the merging genomes. This mismatch can arise from the divergence of the relevant regulatory mechanisms in the parents and/or from global effects of the genome on gene regulation.
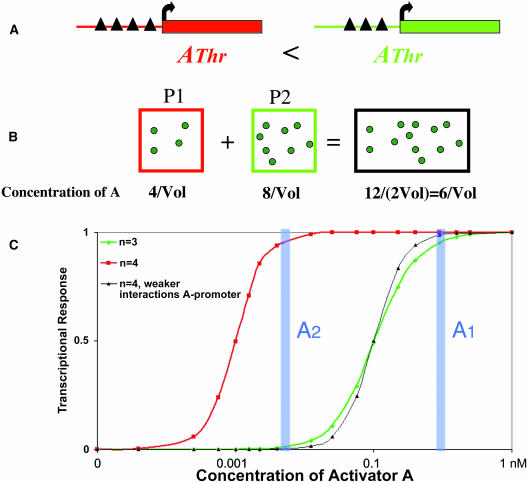
How can a regulatory mismatch arise? During allopolyploidization or hybridization between divergent species, there is a merger of two genomes that usually differ in DNA content (i.e., by a factor of two in Adams et al., 2003). By virtue of the rough proportionality between DNA content and nuclear and cellular volumes, this merger can lead to new cellular and nuclear volumes that are suboptimal for the expression of the parental subgenomes. As a mechanistic example, let us consider what happens to transcription from a promoter containing multiple (three to four) binding sites for the transactivator A (Figure 3A). Protein–DNA interactions are assumed to be cooperative, which enhances specific recognition of the promoter (Veitia, 2003b). Such a simple device works like a switch, leading to a transcriptional response that is sigmoidal with respect to the concentration of the transactivator A. In this system, the concentration of A must remain above the threshold AThr to maintain full transcription. As a general rule, the lower the number of similar binding sites, the higher the concentration of A required to keep the switch on (details in Veitia, 2003b). In the parental individuals, the threshold concentrations of A are successfully attained because the responding promoters have the right settings (i.e., through coevolution of the promoter and the transactivator). This is so even if evolutionary divergence led the parents to generate different threshold settings and different transcriptional responses. For instance, a point mutation increasing the rate of production of A followed by a mutation abolishing one binding site in the promoter yields a functional switch triggered by a higher concentration of A. After polyploidization or hybridization, the concentration of A can be different from, and not necessarily at the midpoint, of the parental values. If the concentration of A is not high enough, the promoter having the lowest threshold will be activated while the other will remain silent.
Figure 3.
Potential Impact of Allopolyploidization or Hybridization on Transcription.
(A) Effect of an allopolyploidization event involving the two parental cells P1 and P2 on transcription from homologous promoters responding to the transactivator A. The black triangles represent the binding sites for A.
(B) In the simplest case, the parental cells have similar volumes (Vol) and similar DNA contents but might contain different amounts of A and promoters with different thresholds (AThr). After the merger, let us suppose that the volume doubles and that the regulation of the expression of A is unaltered. It is easy to see that, in this particular case, the expected concentration of A is intermediate. For instance, this concentration can be suitable to trigger expression from the promoter coming from P1 but not from P2.
(C) Transcriptional responses from promoters with three or four binding sites allowing cooperative interactions with the transactivator A. The solid curves are redrawn from Figure 5 of Veitia (2003b), with the same parameters. Decreasing the number of similar binding sites (n) shifts the position of the threshold to higher concentrations of A. In the case of a newly formed allopolyploid, if the resulting concentration of A is higher than the thresholds of both types of promoters (case A1), transcription is expected from both. If, on the contrary, the concentration of A is not high enough (i.e., A2), the red promoter will be on, while the green one will be off. Of course, cases of incomplete silencing are expected depending on the actual A concentration with respect to AThr. The thin black sigmoid represents a promoter with four binding sites that produces a response similar to that with three binding sites. It can be shown that there is an infinity of ways to obtain such a result, simply by changing the strength of the interaction between A and its binding sites, as long as cooperativity is maintained. This can explain the outcome of compensatory mutations.
At another extreme, the transcriptional responses of the parents can be very similar, while the underlying system's promoter-activator can be different. Indeed, similar responses are obtained with mutations decreasing (or increasing) the number of binding sites compensated by increasing (or decreasing) the affinity of A for DNA (Figures 3B and 3C). Accordingly, an allopolyploid or hybrid can carry a mixture of A molecules that have different affinities for DNA in a volume that is different from that of the parents. Several outcomes are possible, but as a rule the promoter with a higher number of binding sites is more likely to be triggered by preferential interaction with A molecules having the highest affinity for DNA. In these simple cases involving only one activator, the promoter having the lowest AThr, whatever its parental origin, is more likely to remain active after the genomes merge. Although this does not involve any mark on the chromatin, it provides a mechanism allowing transmission of regulatory patterns. In the case of nucleolar dominance, chromatin modifications seem to be involved in epigenetic transmission (Chen et al., 1998). In this case, the factor A might attract a histone acetylase to promote transcription of the dominant genotype, or it might somehow induce repression of the silent genotype. The results of Adams et al. (2003) show that the relative expression of many paralogous gene pairs is dependent on tissue, organ, and/or developmental stage. This complexity could easily stem from the interaction of many loci, inhibitors, and activators that have an impact on the same promoter (Birchler et al., 2001). Indeed, this must also be the case for nucleolar dominance because a model relying on a promoter responding to a single factor cannot account for all the findings described by Chen et al. (1998), but this analysis is beyond the scope of this essay.
Sudden alterations in gene expression patterns in a newly formed allopolyploid are not likely to be adaptive or optimal. However, the evolutionary persistence of a similar pattern of expression in a polyploid, as noted by Adams et al. (2003), is suggestive of some underlying selective advantage or constraint. The emergence of a new pattern of expression provides a further explanation for the retention of the paralogs, even if the initial event was a regulatory mismatch. Genes involved in the same pathway or macromolecular complex are often coregulated, responding basically to the same regulators. If a regulatory mismatch arises after allopolyploidization, it is expected that genes from each paralogous pathway or complex will undergo a concerted change in their regulation (e.g., interacting genes from one genome might have lower AThr). To work properly, each of these (now) differentially regulated paralogous pathways or complexes will need to retain a copy of each component, irrespective of their dosage sensitivity. Thus, an alteration in the mechanism of coregulation can provide a simple explanation for both the coordinated divergence of duplicated pathways or complexes and paralog retention. This effect would be partly due to releasing one paralogous system from the original source of selective pressure. Any interference between duplicated pathways can be avoided if they are expressed in different tissues or at different developmental stages (i.e., neo/subfunctionalized). This process could be thought of as an enhancer of diploidization and may contribute to the return of polyploids to diploidy.
A. thaliana, a modern diploid, has undergone at least three detectable rounds of polyploidy (Bowers et al., 2003). Recently, Blanc and Wolfe (2004) have assessed patterns of functional divergence of paralogs formed during the most recent polyploidy event. They found overrepresentation of retained paralogs encoding proteins involved in signal transduction and transcription, whereas paralogs of housekeeping genes have been preferentially lost. Another genomic analysis in the same plant has shown that the same classes of genes have a higher probability of being retained from one round of genome duplication to another (Seoighe and Gehring, 2004). According to Blanc and Wolfe (2004) more than 50% of the paralogous pairs in A. thaliana display significantly different patterns of expression. Similar results have been obtained for gene duplicates in mouse and human (Huminiecki and Wolfe, 2004), suggesting that this might be a more general phenomenon. Moreover, in the context of A. thaliana, genes whose expression profiles were significantly correlated, which are likely to interact, diverged from the corresponding paralogs in the same direction (Blanc and Wolfe, 2004). The aforementioned results contrast, in quantitative terms, with those of Haberer et al. (2004) who found more coexpressed paralogs than divergently expressed ones in A. thaliana. Yet, this study also found that large fractions of duplicated gene pairs exhibit expression divergence. They detected overall similarity between promoters of duplicated genes, which suggests that the process of transcriptional neofunctionalization and subfunctionalization is restricted to a fraction of cis-regulatory elements or that we are currently unable to match expression differences and subtle sequence divergence.
Macromolecular assembly and signaling through a network are complex processes, and similar outputs can be obtained with different allelic combinations at different loci. In such systems, compensatory mutations provide stability in the responses while allowing the existence of high levels of heterozygosity in the population. As a side product, great phenotypic and genotypic differences can be obtained when selection favors extreme phenotypes. The components of such systems can codiverge in levels or patterns of expression, if they coevolve to minimize the deleterious effects of dosage imbalances. Examples bearing the potential signature of this process have been documented. For instance, Lemos et al. (2004) have studied how the number of interactions a protein has within a network constrains genetic variation of gene expression in yeast and fruitfly populations. They found that gene expression variation was negatively correlated with the number of protein–protein interactions. Moreover, the expression levels of interactors were positively correlated across strains, and the extent of variation in expression among genes encoding interactors was smaller than that of random pairs of genes. This might result from the maintenance of balance between interactors within complexes or among complexes of the same pathway. It is therefore conceivable that directional selection might lead to the concerted retention of specific sets of allelic variants at interacting loci involved in two paralogous pathways so as to resolve the pathways. That is, before selection the paralogous pathways respond to similar signals in basically the same way. After selection, the pathways will respond to different signals in different ways and avoid cross talk. This can be compared with a selection-driven process of neo/subfunctionalization, not of a single gene but of a whole complex/pathway.
CONCLUSIONS
Beyond the simple case of components poorly connected within the cellular interaction network, when selection occurs on one element of a pathway or complex involved in a balanced relationship with other elements, coevolution with its partners is expected. This may explain retention (or erasure) of modules or entire pathways or complexes through rounds of polyploidization. All in all, the ultimate fate of one paralog must be considered from the perspective of the selective advantage provided (or not) by the whole.
Acknowledgments
I wish to thank the editors R. Jorgensen, J. Birchler, B. Gaut, and especially N. Eckardt for their helpful editorial and scientific advice on many occasions. I have appreciated the comments and suggestions of one anonymous referee. I am particularly indebted to B. Dilkes and L. Comai, who were also referees of this essay, for sharing with me a lot of insights on several occasions that helped me to improve the manuscript.
References
- Adams, K.L., Cronn, R., Percifield, R., and Wendel, J.F. (2003). Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc. Natl. Acad. Sci. USA 100, 4649–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn, S., and Tanksley, S.D. (1993). Comparative linkage maps of the rice and maize genomes. Proc. Natl. Acad. Sci. USA 90, 7980–7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, M.D. (1972). Nuclear DNA content and minimum generation time in herbaceous plants. Proc. R. Soc. Lond. B Biol. Sci. 181, 109–135. [DOI] [PubMed] [Google Scholar]
- Birchler, J.A., Bhadra, U., Bhadra, M.P., and Auger, D.L. (2001). Dosage-dependent gene regulation in multicellular eukaryotes: Implications for dosage compensation, aneuploid syndromes, and quantitative traits. Dev. Biol. 234, 275–288. [DOI] [PubMed] [Google Scholar]
- Blanc, G., and Wolfe, K.H. (2004). Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16, 1679–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers, J.E., Chapman, B.A., Rong, J., and Paterson, A.H. (2003). Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422, 433–438. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith, T. (1982). Skeletal DNA and the evolution of genome size. Annu. Rev. Biophys. Bioeng. 11, 273–302. [DOI] [PubMed] [Google Scholar]
- Chen, Z.J., Comai, L., and Pikaard, C.S. (1998). Gene dosage and stochastic effects determine the severity and direction of uniparental ribosomal RNA gene silencing (nucleolar dominance) in Arabidopsis allopolyploids. Proc. Natl. Acad. Sci. USA 95, 14891–14896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force, A., Lynch, M., Pickett, F.B., Amores, A., Yan, Y.L., and Postlethwait, J. (1999). Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, T.J., and Spring, J. (1998). Genetic redundancy in vertebrates: Polyploidy and persistance of genes encoding multidomain proteins. Trends Genet. 14, 46–49. [DOI] [PubMed] [Google Scholar]
- Gregory, T.R. (2001). Coincidence, coevolution, or causation? DNA content, cell size, and the C-value enigma. Biol. Rev. Camb. Philos. Soc. 76, 65–101.11325054 [Google Scholar]
- Gu, Z., Steinmetz, L.M., Gu, X., Scharfe, C., Davis, R.W., and Li, W.H. (2003). Role of duplicate genes in genetic robustness against null mutations. Nature 421, 63–66. [DOI] [PubMed] [Google Scholar]
- Guo, M., Davis, D., and Birchler, J.A. (1996). Dosage effects on gene expression in a maize ploidy series. Genetics 142, 1349–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberer, G., Hindemitt, T., Meyers, B.C., and Mayer, K.F. (2004). Transcriptional similarities, dissimilarities, and conservation of cis-elements in duplicated genes of Arabidopsis. Plant Physiol. 136, 3009–3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin, J. (2000). A view of mount Drosophila. Nature 404, 442–443. [DOI] [PubMed] [Google Scholar]
- Hughes, M.K., and Hughes, A.L. (1993). Evolution of duplicate genes in a tetraploid animal, Xenopus laevis. Mol. Biol. Evol. 10, 1360–1369. [DOI] [PubMed] [Google Scholar]
- Huminiecki, L., and Wolfe, K.H. (2004). Divergence of spatial gene expression profiles following species-specific gene duplications in human and mouse. Genome Res. 14, 1870–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis, M., Birren, B.W., and Lander, E.S. (2004). Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428, 617–624. [DOI] [PubMed] [Google Scholar]
- Kellogg, E.A. (2003). What happens to genes in duplicated genomes. Proc. Natl. Acad. Sci. USA 100, 4369–4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov, F.A., and Koonin, E.V. (2004). A common framework for understanding the origin of genetic dominance and evolutionary fates of gene duplications. Trends Genet. 20, 287–290. [DOI] [PubMed] [Google Scholar]
- Lemos, B., Meiklejohn, C.D., and Hartl, D.L. (2004). Regulatory evolution across the protein interaction network. Nat. Genet. 36, 1059–1060. [DOI] [PubMed] [Google Scholar]
- Lynch, M., and Conery, J.S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155. [DOI] [PubMed] [Google Scholar]
- Makalowski, W. (2001). Are we polyploids? A brief history of one hypothesis. Genome Res. 11, 667–670. [DOI] [PubMed] [Google Scholar]
- Masterson, J. (1994). Stomatal size in fossil plants: Evidence for polyploidy in majority of angiosperms. Science 264, 421–423. [DOI] [PubMed] [Google Scholar]
- Ohno, S. (1970). Evolution by Gene Duplication. (London: George Allen and Unwin).
- Otto, S.P., and Whitton, J. (2000). Polyploid incidence and evolution. Annu. Rev. Genet. 34, 401–437. [DOI] [PubMed] [Google Scholar]
- Orr, H.A., and Otto, S.P. (1994). Does diploidy increase the rate of adaptation? Genetics 136, 1475–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkan, H., Levy, A.A., and Feldman, M. (2001). Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops-Triticum) group. Plant Cell 13, 1735–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp, B., Pal, C., and Hurst, L.D. (2003). Dosage sensitivity and the evolution of gene families in yeast. Nature 424, 194–197. [DOI] [PubMed] [Google Scholar]
- Pikaard, C.S. (2000). The epigenetics of nucleolar dominance. Trends Genet. 16, 495–500. [DOI] [PubMed] [Google Scholar]
- Pires, J.C., Zhao, J.W., Schranz, M.E., Leon, E.J., Quijada, P.A., Lukens, L.N., and Osborn, T.C. (2004). Flowering time divergence and genomic rearrangements in resynthesized Brassica polyploids (Brassicaceae). Biol. J. Linnean Soc. Lond. 82, 675–688. [Google Scholar]
- Seoighe, C., and Gehring, C. (2004). Genome duplication led to highly selective expansion of the Arabidopsis thaliana proteome. Trends Genet. 20, 461–464. [DOI] [PubMed] [Google Scholar]
- Veitia, R.A. (2002). Exploring the etiology of haploinsufficiency. Bioessays 24, 175–184. [DOI] [PubMed] [Google Scholar]
- Veitia, R.A. (2003. a). Nonlinear effects in macromolecular assembly and dosage sensitivity. J. Theor. Biol. 220, 19–25. [DOI] [PubMed] [Google Scholar]
- Veitia, R.A. (2003. b). A sigmoidal transcriptional response: Cooperativity, synergy and dosage effects. Biol. Rev. Camb. Philos. Soc. 78, 149–170. [DOI] [PubMed] [Google Scholar]
- Veitia, R.A. (2004). Gene dosage balance in cellular circuits and pathways: Implications for dominance and gene duplicability. Genetics 168, 569–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, J.B. (1995). How often do duplicated genes evolve new functions? Genetics 139, 421–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, K.H. (2001). Yesterday's polyploids and the mystery of diploidization. Nat. Rev. Genet. 2, 333–341. [DOI] [PubMed] [Google Scholar]
- Yang, J., Lusk, R., and Li, W.H. (2003). Organismal complexity, protein complexity, and gene duplicability. Proc. Natl. Acad. Sci. USA 100, 15661–15665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296, 79–92. [DOI] [PubMed] [Google Scholar]