Abstract
Botulinum neurotoxin (BoNT) serotype A is the most lethal known toxin and has an occluded structure which prevents direct inhibition of its active site before it enters the cytosol. We combine in situ click target-guided synthesis with synthetic epitope-targeting to exploit the tertiary structure of the BoNT protein as a landscape for assembling a competitive inhibitor. A substrate mimicking peptide macrocycle is used as a direct inhibitor of BoNT. An epitope targeted in situ click screen is utilized to identify a second peptide macrocycle ligand that binds to an epitope that, in the folded BoNT structure, is active site adjacent. A second in situ click screen identifies a molecular bridge between the two macrocycles. The resulting divalent inhibitor exhibits an inhibition constant of 165 pM in vitro against the BoNT/A catalytic chain. The inhibitor is carried into cells by the intact holotoxin, and demonstrates protection and rescue of BoNT intoxication in a human neuron model.
Keywords: target-guided synthesis, botulinum neurotoxin, combinatorial screening, epitope targeting, peptides
Botulinum neurotoxin (BoNT) serotype A is the most lethal known toxin and is produced by some species of the bacterial genus Clostridium. BoNT/A is a chemodenervating zinc-dependent protease that prevent the Ca2+-triggered release of acetylcholine in neuromuscular junctions, by cleaving one of the three SNARE proteins required for synaptic vesicle formation and release[1]. BoNT/A intoxication proceeds with selective binding to neuronal receptors, cell entry through receptor-mediated endocytosis, endosome escape via pH-induced translocation, and, finally, cleavage of its SNAP-25 substrate in the cytosol[2]. BoNT/A is comprised of a receptor-binding heavy chain and disulfide-linked catalytic light chain (LC). This disulfide bond must be intact for the toxin to poison neurons, but must be broken for the LC to act catalytically in the cytosol[3]. A subdomain (the ‘belt’) structurally occludes the intact holotoxin active site (Scheme 1) so that drug-induced inhibition only occurs after belt release, which is promoted by the reduction of the disulfide link by the cytosolic environment[4]. The rapid sequestration of BoNT toxins into motor neurons limits current antibody based therapies, while the occluded active site is undruggable by traditional protease inhibitors[5]. Recently membrane-penetrant small molecule BoNT LC inhibitors have shown promise in vitro, however their reported cytotoxicity indicates significant off-target interactions and have effective doses in the mid to high micromolar range[6]. BoNT is a potentially deadly bioweapon[7], but is also a therapeutic and cosmetic agent, with an accompanying risk of accidental overdosing. Potent and effective inhibitors are needed.
Scheme 1. The development of a biligand inhibitor of BoNT/A.
Inh-1 (orange, Dab(DNP)-R-Lys(N3)-T-Dab-Pra-L-R1) is based on the natural peptide substrate for BoNT LC and existing structural studies of peptidomimetic substrate mimics. Epitope-targeting was used to screen for a secondary ligand targeted to a nearby unstructured epitope (BoNT LC 166–179, green) exposed in the presence of the holotoxin’s occluding belt (red). The ligand selected (L2, red, R2-Pra-NYRWL-Lys(N3)) was selected from a 1.1M member macrocyclic peptide library.
Sharpless’ group first reported on target-guided synthesis by in situ click chemistry[8]. They utilized the active site of an enzyme as a highly selective promoter of the 1,3-dipolar cycloaddition reaction to assemble a divalent inhibitor from two small molecule libraries – one presenting and azide, the other an acetylene. Over the last several years we have applied in situ click to the discovery of peptides targeting a variety of proteins and post-translational modifications[9]. To build a BoNT inhibitor, we sought to further generalize the target-guided synthesis approach by exploiting the tertiary structure of the BoNT LC as a landscape for assembling a potent inhibitor (Scheme 1). We developed a macrocyclic peptide ligand (Inh-1, SI Figure S1–2) that is a substrate mimic for BoNT. Inh-1 binds to the active site with a ~70 nM binding affinity (kD) and similar inhibition constant. We then employ an all synthetic in situ click epitope targeting approach[10] (Scheme 1) to identify a second peptide macrocycle (L2, SI Figure S3–4) that binds to a site a few angstroms away in the folded protein structure from the active site. L2 exhibits a kD ~80 nM, but no inhibitory effects. Finally, we utilize an in situ click screen, promoted by the BoNT LC, to identify a linear peptide that connects the two macrocycles (Scheme 2). The final divalent ligand (Inh-2, SI Figure S5) inhibits the BoNT LC with an IC50 of 165 ± 15 pM. Inh-2 is carried into neuronal cells by BoNT itself, and inhibits the holotoxin in live cells. This technique provides a potentially general route for the development of peripheral and active site binders from naïve libraries for combination through in situ click target-guided synthesis.
Scheme 2. Linker screen for divalent ligand development.
Inh-1 was synthesized with a C-terminal azide and Biotin tag for readout of in situ click screen and used in solution while L2 was synthesized with an N-terminal alkyne and a comprehensive linker library of oligopeptides of length zero to five units on Tentagel resin. An in situ click protein-catalyzed click screen was performed to select for a minimally perturbative correctly oriented linker and resulted in hit sequences Gly-Aib-Leu and Leu-Aib-Gly. A PEG4 linker was also used in preliminary assays as a comparison (See Supplemental Figure S13–14). Inh-2 was the biligand chosen for all future assays.
The active site of the catalytic BoNT/A LC recognizes the 7-residue QRATKML sequence of the SNAP-25 protein. Many inhibitors based on this motif have been reported[5, 11]. One peptidomimetic inhibitor variant resulting from a structure-based search binds BoNT in a tight 310 helical conformation[11b] with low nanomolar inhibition of BoNT LC in vitro (SI Figure S6–7). In an effort to reinforce this conformation in solution and increase chemical and biological stability of this compound, we synthesized a small library of (i, i+3)-click cyclized derivatives[12].
One compound (Inh-1) inhibited BoNT/A LC with a 70 ± 8 nM inhibition constant measured in vitro using a FRET-based substrate cleavage assay (SI Figure S8)[13] and 68 ± 29 nM kD measured by FP and confirmed by sandwich ELISA (Figure 1A and SI Figure S9). However, inhibiting the full BoNT holotoxin with the occluded LC active site is more challenging. Single point ELISA binding data (Figure 1D) shows that the Inh-1 exhibits minimal binding to the holotoxin despite tight binding to the LC. Thus, we sought to expand Inh-1 to include a moiety that binds to a non-occluded region of BoNT LC adjacent to the active site. We targeted an epitope (BoNT LC residues 166–179, SI Figure S10) which is solvent exposed in the occluded conformation of the holotoxin for screening with epitope-catalyzed in situ click.
Figure 1. Characterization of ligand binding and inhibition in vitro.
(A) Inhibition curve (left axis) and fluorescence polarization binding curve (right axis) of Inh-1. Observed IC50 of inhibition is 70 ± 10 nM and observed kD is 68 ± 29 nM by Hill curve fitting. (B) Inhibition curve (left axis) and fluorescence polarization binding curve (right axis) of compound L2. No inhibition is observed, kD observed is 78 ± 8 nM. (C) Inhibition curves comparing compound Inh-1 (pink) to compound Inh-2 (red). Observed IC50 is 60 ± 11 nM and 165 ± 15 pM respectively. (D) Single point ELISA binding data of compounds to 50 nM of the reduced (available active site) and intact (occluded by belt) BoNT/A holotoxins. Error bars indicate standard deviation of replicates.
The epitope was synthesized with an N-terminal azidolysine and a C-terminal biotin and was screened against a 1.1M element library of macrocyclic 5-mer peptides synthesized on Tentagel resin substituted with an N-terminal propargylglycine (Scheme 2) and anti-screened against a scrambled version of the same epitope (SI Figure S10–11). After incubation with the target epitope, beads were stripped, probed and developed with an alkaline phosphatase (AP) anti-biotin mAb. Nine hit beads were sequenced using Edman degradation and found to have significant sequence homology (SI Figure S12). L2 was selected after fluorescence polarization showed a 78 ± 13 nM kD to the BoNT LC and similar binding (kD not quantitated) to the BoNT holotoxin, but no measurable LC inhibition (Figure1B and 1D). Inh-1 and L2 binding sites were verified using competitive ELISA formats with known active site binders and the targeted BoNT epitope respectively (SI Figure S13).
To optimally combine Inh-1 with L2, kinetic in situ target-guided synthesis was used with a combinatorial linker library[14] (Scheme 2) to identify an optimized molecular bridge. To this end, L2 was synthesized on Tentagel resin and appended with an N-terminal linker library that was terminated with a propargylglycine. This appended library was comprehensive from zero to five residues in four non-canonical and alternate chirality amino acids that were selected for structural rigidity. Our rational was to maximize the avidity enhancement achievable when combining two independent binders by prepaying the entropic cost of orientation using a rigid linker of length equal to the root mean square average separation of the two binding sites[15]. Inh-1 was synthesized with a C-terminal azidolysine and a biotin assay handle (SI Figure S14). The BoNT LC was added to promote the target guided synthesis of a bivalent ligand by bringing the reactive azide and alkyne groups in close proximity. After screening, beads were stripped, probed and developed with an AP anti-biotin mAb. Hit beads were sequenced using Edman degradation and resulted in a consensus of two linker sequences: Gly – Aib – Leu and Leu – Aib – Gly (Scheme 2). Inh-2 was selected for all subsequent assays and demonstrated an inhibition constant of 165 ± 15 pM against BoNT LC evaluated in vitro as above. Variants of this ligand were prepared that included a biotin for binding assays, a spontaneously translocating peptide (STP) reported for the delivery of polar cargo to the cytosol, and a PEG4 variant linker[16] (SI Figure S15–17).
We tested the ability of Inh-2 to protect and rescue BoNT intoxication in human neurons (Figures 2 and 3 respectively). iCell neurons are derived from pluripotent stem cells, and have been reported as a model for monitoring SNAP-25 cleavage by BoNT within living cells[17] (SI Figure S18). Cells were plated on poly-D-lysine coated 96-well plates with a laminin matrix and exposed to 2 mouse LD50 units (U) of BoNT pre-incubated with various concentrations of Inh-1 and Inh-2. All cells were lysed after 24 hours and evaluated for SNAP-25 cleavage by western blotting. Cleaved SNAP-25 appeared as a second band under the intact SNAP-25 band (Figure 2A). Western blotting data was quantified by densitometry to extract a dose-response behavior of the ligands in protection against BoNT intoxication. Inh-1 exhibited negligible protective effects at up to 10 µM dosing. This is likely due to the occluded active site being unavailable for binding until BoNT enters the cytosol, and the membrane impermeable nature of Inh-1. Inh-2 is also impermeable to cell membranes, but demonstrated significant protective effect to BoNT intoxication at concentrations as low as 100 nM. This type of protection has been reported for inhibitors of the BoNT LC active site in neuron models, but only at inhibitor concentrations in the mid micromolar range[5]. As a comparison, a sheep antibody acquired from the BEI biodefense repository with known neutralizing effect was evaluated. This antibody protects against BoNT intoxication by interrupting the heavy chain receptor-mediated endocytosis and provides protection into the low to mid nanomolar range.
Figure 2. Ligand inhibition effect in hiPSC-derived neuron system with BoNT holotoxin.
(A) Top shows western blots of neuron lysates for B-tubulin (control) and SNAP (cleaved and uncleaved). The cleaved SNAP product appears as a longer running band below the intact SNAP in the western blot. Assays were performed with varying concentration of compounds Inh-1, Inh-2 along with a known endocytosis-inhibiting polyclonal sheep anti-BoNT heavy chain antibody. Plot below shows percent intact SNAP as a function of ligand concentration. Percent intact was calculated as the ratio of the integrated intact band intensity to the total integrated intensity of the bands. (B) Top shows representative epifluorescence images of live neurons after FM1-43 synaptic vesicle stain loading and after depolarization with and without prior BoNT intoxication, scale bars are 40 µm. Plot below shows average integrated synaptic vesicle stain intensity on a per-cell basis for all conditions with 1 µM added compound where relevant. STP = Spontaneously translocating peptide. Error bars indicate standard deviation of replicates.
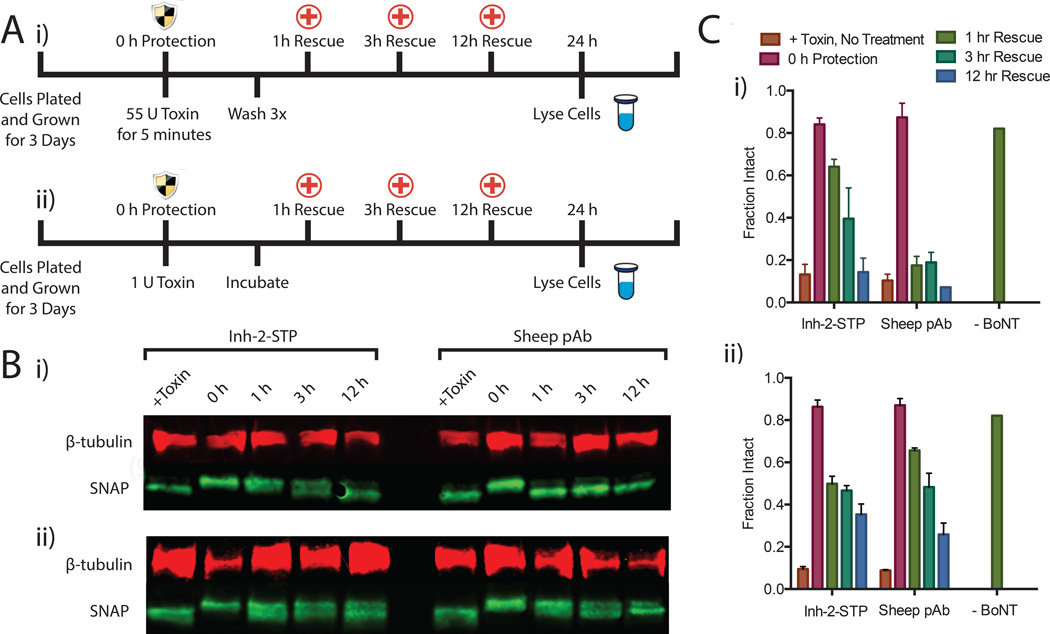
Figure 3. Rescuing hiPSC-derived neurons from BoNT intoxication.
(A) Experimental timeline of 55 U bolus toxin exposure (i) and 24-hr incubation with 2 U toxin (ii). Protection effect was evaluated by preincubation of toxin with 1 µM inhibiting anti-BoNT pAb or Inh-2-STP and rescue effect was evaluated 1hr, 3hrs and 12hrs after exposure to toxin by lysing cells and quantitating SNAP cleavage by western blot and densitometry. In all cases cells were lysed 24 hours after toxin exposure. (B) Representative western blots of neuron lysates for B-tubulin (control) and SNAP (cleaved and uncleaved) after rescue experiments shown in A. The cleaved SNAP product appears as a longer running band below the intact SNAP in the western blot. (C) Percent intact SNAP as a function of exposure time before treatment. Percent intact was calculated as the ratio of the integrated intact band intensity to the total integrated intensity of the bands. Error bars indicate standard deviation of replicates.
After extended maturation, iCell neurons demonstrate functional synaptic vesicle recycling, indicating the development of a presynaptic compartment. Synaptic vesicle recycling was monitored through synaptic vesicle staining with the outer membrane partitioning styryl dye FM1–43 followed by Ca2+/K+ dependent depolarization[18] (SI Figure S19). The synaptic vesicles of BoNT intoxicated neurons can be stained, however Ca2+/K+ depolarization does not induce vesicle exocytosis[19] and subsequent turn-off of FM1–43 fluorescence. Synaptic vesicles appearing as puncta in epifluorescent images of the live neurons were quantified on a per cell basis (SI Figure S20) after exposure to BoNT pre-incubated with 1 µM of inhibitory ligands. Representative cells following depolarization are shown in Figure 2B along with the synaptic vesicle counts for each condition quantified over three wells. As observed in the western blot assay, this functional assay shows negligible protection to BoNT intoxication by Inh-1, while Inh-2 provides full protection. These results are consistent with a Trojan horse-like inhibition mechanism in which the non-inhibiting L2 moiety of Inh-2 binds to the BoNT holotoxin, so that Inh-2 is shuttled into the cell by the holotoxin itself during the endocytosis step of BoNT intoxication. Once the belt is released, the Inh-1 moiety clamps down onto the active site, thus inhibiting the protein. To evaluate the effect of passive delivery to the cytosol, ligands were appended with an STP (SI Figure S21). The Inh-1-STP compound targeting the active site showed a statistically significant increase in protection (p < 0.001) compared to the membrane impermeable Inh-1, though not the complete protection expected at this concentration for the BoNT LC in vitro.
Both the membrane permeable and impermeable Inh-2 variants provided complete functional protection in neurons. We next evaluated the BoNT-independent cellular entry of Inh-2-STP through rescue assays in which BoNT entry and intoxication begin before the addition of the inhibitor. Rescue was evaluated in iCell neurons in a bolus 55U five-minute exposure in cell-stimulating media followed by washing and an extended 1U exposure with no washing. In both experiments 1 µM inhibitory ligand was added to the cells 1, 3 and 12 hours post-exposure. All cells were lysed 24 hours after exposure and SNAP-25 cleavage was evaluated by western blotting (Figure 3B). Two blots per condition were quantified and averaged by densitometry (Fig. 3C). In the bolus exposure, Inh-2-STP exhibits partial rescue from internalized BoNT as late as 3 hours after exposure, with later time points indicating the internalized BoNT had completely cleaved SNAP-25 after 12 hours. This rescue effect is not observed with the Inh-2 compound lacking the STP (SI Figure S22). As expected, the sheep pAb shows no detectable rescue due to its extracellular mechanism of action. In the 1U extended exposure, both the sheep pAb and compound Inh-2-STP showed similar levels of rescue. The process of BoNT endocytosis is slowed significantly at low concentrations and in non-stimulating media, enabling protection through the pAb protective mechanism. The rescue effect of Inh-2-STP in the absence of extracellular BoNT indicates that the STP allowed passive translocation of the Inh-2 cargo across the cell membrane for inhibition of previously internalized BoNT.
We have reported the rational use of the tertiary structure of a protein target as a landscape for the assembly of a highly potent inhibitor. The specific application yielded a ligand, Inh-2 that succeeds as a strong inhibitor in spite of the naturally occluded active site of BoNT holotoxin. Inh-2 effectively acts as a Trojan horse; it is carried into the neuronal cells by the BoNT machinery. Once inside the cytosol, the BoNT LC active site is exposed and Inh-2 performs its inhibitory function. To our knowledge this divalent inhibitory ligand is the most potent in vitro and in cell inhibitor of BoNT/A yet reported and represents the first use of a peripheral binding site to enhance the binding avidity and facilitate BoNT/A holotoxin binding. This basic approach of identifying and combining peripheral binders may have general applicability for developing high affinity, high specificity enzyme inhibitors.
Supplementary Material
Footnotes
This work was supported by the Institute for Collaborative Biotechnologies through grant W911NF-09-0001 from the U.S. Army Research Office and the Defense Advanced Research Projects Agency (DARPA) through the Cooperative Agreement No. HR0011-11-2-0006, and the Jean Perkins Foundation. B.F. is supported by an HHMI International Student Research Fellowship. The following reagents were obtained through the NIH Biodefense and Emerging Infections Research Resources Repository, NIAID, NIH: Polyclonal anti-BoNT/A1 Produced in sheep, NR-9584.
Supporting information for this article is given via a link at the end of the document.
References
- 1.a) Blasi J, Chapman ER, Link E, Binz T, Yamasaki S, Decamilli P, Sudhof TC, Niemann H, Jahn R. Nature. 1993;365:160. doi: 10.1038/365160a0. [DOI] [PubMed] [Google Scholar]; b) Blasi J, Chapman ER, Yamasaki S, Binz T, Niemann H, Jahn R. Embo Journal. 1993;12:4821. doi: 10.1002/j.1460-2075.1993.tb06171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Schiavo G, Benfenati F, Poulain B, Rossetto O, Delaureto PP, Dasgupta BR, Montecucco C. Nature. 1992;359:832. doi: 10.1038/359832a0. [DOI] [PubMed] [Google Scholar]; d) Schiavo G, Rossetto O, Santucci A, Dasgupta BR, Montecucco C. Journal of Biological Chemistry. 1992;267:23479. [PubMed] [Google Scholar]
- 2.Simpson LL. Annu Rev Pharmacol Toxicol. 2004;44:167. doi: 10.1146/annurev.pharmtox.44.101802.121554. [DOI] [PubMed] [Google Scholar]
- 3.Schiavo G, Papini E, Genna G, Montecucco C. Infection and Immunity. 1990;58:4136. doi: 10.1128/iai.58.12.4136-4141.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lacy DB, Tepp W, Cohen AC, DasGupta BR, Stevens RC. Nat Struct Biol. 1998;5:898. doi: 10.1038/2338. [DOI] [PubMed] [Google Scholar]
- 5.Anne C, Turcaud S, Blommaert AGS, Darchen F, Johnson EA, Roques BR. Chembiochem. 2005;6:1375. doi: 10.1002/cbic.200400398. [DOI] [PubMed] [Google Scholar]
- 6.a) Eubanks LM, Hixon MS, Jin W, Hong S, Clancy CM, Tepp WH, Baldwin MR, Malizio CJ, Goodnough MC, Barbieri JT, Johnson EA, Boger DL, Dickerson TJ, Janda KD. Proc Natl Acad Sci U S A. 2007;104:2602. doi: 10.1073/pnas.0611213104. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Capek P, Zhang Y, Barlow DJ, Houseknecht KL, Smith GR, Dickerson TJ. Acs Chemical Neuroscience. 2011;2:288. doi: 10.1021/cn200021q. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Boldt GE, Kennedy JP, Janda KD. Organic Letters. 2006;8:1729. doi: 10.1021/ol0603211. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Burnett JC, Ruthel G, Stegmann CM, Panchal RG, Nguyen TL, Hermone AR, Stafford RG, Lane DJ, Kenny TA, McGrath CF, Wipf P, Stahl AM, Schmidt JJ, Gussio R, Brunger AT, Bavari S. J Biol Chem. 2007;282:5004–5014. doi: 10.1074/jbc.M608166200. [DOI] [PubMed] [Google Scholar]
- 7.a) Burnett JC, Henchal EA, Schmaljohn AL, Bavari S. Nature Reviews Drug Discovery. 2005;4:281. doi: 10.1038/nrd1694. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Clarke SC. British Journal of Biomedical Science. 2005;62:40. doi: 10.1080/09674845.2005.11732685. [DOI] [PubMed] [Google Scholar]; c) USAMRIID. Medical Management of Biological Casualties Handbook. 7th ed. U.S. Government Printing Office; 2011. [Google Scholar]
- 8.Lewis WG, Green LG, Grynszpan F, Radic Z, Carlier PR, Taylor P, Finn MG, Sharpless KB. Angew Chem Int Ed Engl. 2002;41:1053. doi: 10.1002/1521-3773(20020315)41:6<1053::aid-anie1053>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 9.a) Pfeilsticker JA, Umeda A, Farrow B, Hsueh CL, Deyle KM, Kim JT, Lai BT, Heath JR. Plos One. 2013:8. doi: 10.1371/journal.pone.0076224. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Coppock M, Farrow B, Finch AS, Lai B, Maciel J, Heath J, Stratis-Cullum DN. Biopolymers. 2013;100:289. [Google Scholar]; c) Farrow B, Hong SA, Romero EC, Lai B, Coppock MB, Deyle KM, Finch AS, Stratis-Cullum DN, Agnew HD, Yang S, Heath JR. Acs Nano. 2013;7:9452. doi: 10.1021/nn404296k. [DOI] [PubMed] [Google Scholar]; d) Nag A, Das S, Yu MB, Deyle KM, Millward SW, Heath JR. Angewandte Chemie-International Edition. 2013;52:13975. doi: 10.1002/anie.201305882. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Millward SW, Henning RK, Kwong GA, Pitram S, Agnew HD, Deyle KM, Nag A, Hein J, Lee SS, Lim J, Pfeilsticker JA, Sharpless KB, Heath JR. Journal of the American Chemical Society. 2011;133:18280. doi: 10.1021/ja2064389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deyle KM, Farrow B, Hee Y, Work J, Wong M, Lai B, Umeda A, Millward S, Nag A, Das S, Heath JR. Nature Chemistry. 2015 doi: 10.1038/nchem.2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a) Zuniga JE, Hammill JT, Drory O, Nuss JE, Burnett JC, Gussio R, Wipf P, Bavari S, Brunger AT. Plos One. 2010;5 doi: 10.1371/journal.pone.0011378. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zuniga JE, Schmidt JJ, Fenn T, Burnett JC, Arac D, Gussio R, Stafford RG, Badie SS, Bavari S, Brunger AT. Structure. 2008;16:1588. doi: 10.1016/j.str.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobsen O, Maekawa H, Ge NH, Gorbitz CH, Rongved P, Ottersen OP, Amiry-Moghaddam M, Klaveness J. Journal of Organic Chemistry. 2011;76:1228. doi: 10.1021/jo101670a. [DOI] [PubMed] [Google Scholar]
- 13.Caglic D, Bompiani KM, Krutein MC, Capek P, Dickerson TJ. Jove-Journal of Visualized Experiments. 2013 doi: 10.3791/50908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu XD, Manetsch R. Chemical Society Reviews. 2010;39:1316. doi: 10.1039/b904092g. [DOI] [PubMed] [Google Scholar]
- 15.Diestler DJ, Knapp EW. Physical Review Letters. 2008;100 doi: 10.1103/PhysRevLett.100.178101. [DOI] [PubMed] [Google Scholar]
- 16.Marks JR, Placone J, Hristova K, Wimley WC. Journal of the American Chemical Society. 2011;133:8995. doi: 10.1021/ja2017416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whitemarsh RCM, Strathman MJ, Chase LG, Stankewicz C, Tepp WH, Johnson EA, Pellett S. Toxicological Sciences. 2012;126:426. doi: 10.1093/toxsci/kfr354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niedringhaus M, Dumitru R, Mabb AM, Wang Y, Philpot BD, Allbritton NL, Taylor AM. Sci Rep. 2015;5:8353. doi: 10.1038/srep08353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neale EA, Bowers LM, Dunlap V, Williamson LC. Molecular Biology of the Cell. 1995;6:1899. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.