Abstract
The Arabidopsis thaliana genome contains five class III homeodomain-leucine zipper genes. We have isolated loss-of-function alleles for each family member for use in genetic analysis. This gene family regulates apical embryo patterning, embryonic shoot meristem formation, organ polarity, vascular development, and meristem function. Genetic analyses revealed a complex pattern of overlapping functions, some of which are not readily inferred by phylogenetic relationships or by gene expression patterns. The PHABULOSA and PHAVOLUTA genes perform overlapping functions with REVOLUTA, whereas the PHABULOSA, PHAVOLUTA, and CORONA/ATHB15 genes perform overlapping functions distinct from REVOLUTA. Furthermore, ATHB8 and CORONA encode functions that are both antagonistic to those of REVOLUTA within certain tissues and overlapping with REVOLUTA in other tissues. Differences in expression patterns explain some of these genetic interactions, whereas other interactions are likely attributable to differences in protein function as indicated by cross-complementation studies.
INTRODUCTION
The innovation of new developmental pathways and the elaboration of existing pathways during evolution are facilitated by gene duplications (Ohno, 1970). Duplicated genes may be preserved either when one paralog gains an advantageous new biochemical function or expression domain or when the two copies each perform a subset of the ancestral gene's functions (or are expressed in subsets of the ancestral expression pattern; Hughes, 1994; Force et al., 1999; Krakauer and Nowak, 1999; Prince and Pickett, 2002; Zhang, 2003). Furthermore, the division of ancestral functions between paralogs may facilitate changes in function because of reduction of the evolutionary constraints imposed by needing to carry out diverse roles. The importance of understanding duplicate gene function and evolution has been underscored by the abundance of duplicated genes in genomes, especially in plants.
Approximately 65% of Arabidopsis thaliana genes are members of gene families (Arabidopsis Genome Initiative, 2000), thus complicating genetic studies. One approach to circumvent problems associated with genetic redundancy is to isolate gain-of-function and dominant-negative alleles (Estelle and Somerville, 1987; Bleecker et al., 1988; Wilson et al., 1996; McConnell and Barton, 1998; Weigel et al., 2000; McConnell et al., 2001; Dievart et al., 2003; Shpak et al., 2003). Reverse genetic analysis has been increasingly productive in studying functional redundancy in Arabidopsis gene families (Hua and Meyerowitz, 1998; Siegfried et al., 1999; Pelaz et al., 2000; Alonso et al., 2003; Franklin et al., 2003a, 2003b; Friml et al., 2003; Pinyopich et al., 2003). These studies have focused either on smaller gene families or a subset of closely related genes within a larger family, but, to our knowledge, a plant gene family with four or more members has not been subjected to comprehensive analysis using loss-of-function mutations. In many cases, studies have focused on only the closest related members of a larger gene family because of the increased effort involved in generating larger-order multiply mutant lines. Although these studies have uncovered genetic redundancy, it is unclear if additional functional overlaps are being overlooked.
REVOLUTA/INTERFASCICULAR FIBERLESS1 (REV) is one of five Arabidopsis Class III homeodomain-leucine zipper (HD-Zip III) proteins. These proteins contain an HD-Zip domain involved in DNA binding and protein dimerization (Sessa et al., 1998), a putative lipid or steroid binding START domain (Ponting and Aravind, 1999), and a conserved C-terminal domain of unknown function. rev loss-of-function mutations cause defects in leaf development and stem cell specification, as well as defects in vascular development and auxin transport (Talbert et al., 1995; Zhong et al., 1997; Zhong and Ye, 1999; Otsuga et al., 2001). athb8 loss-of-function alleles display no detectable phenotypes (Baima et al., 2001), and PHABULOSA (PHB) and PHAVOLUTA (PHV) were recently shown to perform overlapping functions with REV in embryogenesis (Emery et al., 2003). Loss-of-function alleles of CORONA/ATHB15 (CNA) have not yet been described. Gain-of-function alleles caused by mutations in a putative microRNA regulatory target of the PHB and PHV genes result in leaf polarity defects and the formation of meristems in ectopic positions (McConnell and Barton, 1998; McConnell et al., 2001; Tang et al., 2003), and similar mutations in the REV gene result in polarity defects in vascular bundles and leaves (Zhong et al., 1999; Emery et al., 2003; Zhong and Ye, 2004). Ectopic expression of ATHB8 resulted in the overproduction of xylem (Baima et al., 2001).
Given that several of the HD-Zip III genes' roles in development (vascular development, leaf development, and meristem initiation) represent key innovations in land plant evolution (Gifford and Foster, 1989; Graham et al., 2000), we have become interested in the evolution of this gene family. HD-Zip III genes are highly conserved in land plants; >50% of the full-length amino acid sequence is conserved between the moss PpHB10 protein and each of the Arabidopsis HD-Zip III proteins (Sakakibara et al., 2001). Whereas highly conserved, the nonequivalence of HD-Zip III gene function is suggested by the retention of gene pairs from ancient duplication events.
We describe here a comprehensive analysis of HD-Zip III function in Arabidopsis. This includes isolation of loss-of-function alleles for each gene family member, phenotypic analysis of all possible double, triple, quadruple, and quintuple mutants, cross-complementation, and expression analysis for each gene. We have identified a complex combination of overlapping, antagonistic, and divergent gene functions among HD-Zip III gene family members. This thorough analysis of a plant gene family uncovered novel gene functions masked by genetic redundancy but not readily inferred from gene phylogeny. Our results provide key observations for those designing reverse genetic experiments and may lead to insights into the evolution of duplicated gene function.
RESULTS
Loss-of-Function Mutations in the HD-Zip III Genes of Arabidopsis
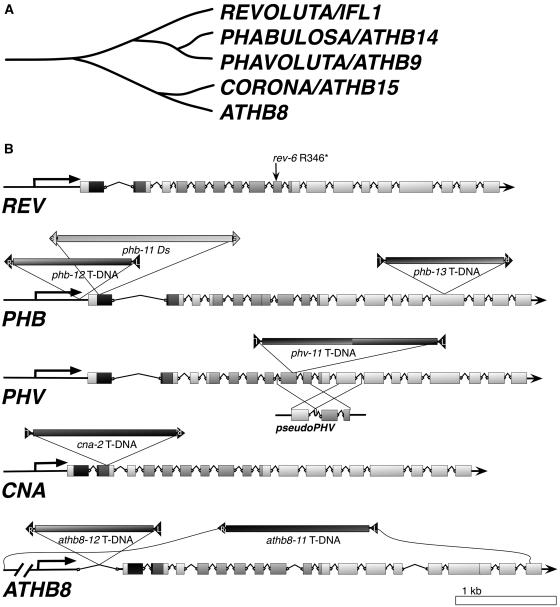
Insertional mutations in the PHB, PHV, CNA, and ATHB8 genes were isolated from Ds insertion and T-DNA insertion collections (Figure 1; see Methods). For each insertion line, border PCR fragments were sequenced to verify the insertion site (see Supplemental Table 2 online), and homozygous isolates were identified. RT-PCR experiments were performed to determine whether the mutated genes were still expressed (see Supplemental Figure 1 online). Based on the nature of the lesions and RT-PCR data, all of the alleles used in this study were strong alleles with the exceptions of phb-12 and possibly athb8-12 (Figure 1; see Supplemental Figure 1 and Table 2 online). Unless otherwise specified, the alleles used for multiply mutant line construction were rev-6, phb-13, phv-11, cna-2, and athb8-11. We have previously shown a strong influence of genetic background on Rev− expressivity (Otsuga et al., 2001). We therefore conducted our analyses in a uniform genetic background. The majority of the mutant alleles we identified were isolated in the Columbia (Col) background, whereas the rev-6 and phb-11 alleles, originally from Landsberg erecta (Ler), were introgressed into the Col background (see Methods). We found that the cna-2 mutant, like phv, phb, and athb8 mutants (Baima et al., 2001; Emery et al., 2003), lacks discernable phenotypes, leaving REV as the only gene family member with identifiable single mutant phenotypes.
Figure 1.
The Arabidopsis HD-Zip III Gene Family.
(A) Phylogram illustrating the relationships between the Arabidopsis HD-Zip III genes.
(B) Diagrams of the Arabidopsis HD-Zip III genes and positions of mutations. The amino acid coding regions are drawn as boxes, and introns are illustrated by bent lines. Horizontal arrows indicate directions of transcription. Exons are shaded to indicate the positions of the homeodomain (dark gray), the Leu zipper (medium gray), and the START domain (light gray); other regions are shown as white boxes. Mutation positions are indicated above the gene diagrams. Where known, orientations of the inserted sequences is indicated by border-sequence labels adjacent to the bars: R, T-DNA right border; L, T-DNA left border; 5, Ds 5′ end; and 3, Ds 3′ end. The phv-11 allele was found to contain left borders at both ends of the inserted fragment but the precise arrangement of T-DNA copies is not known. Also diagrammed is the duplicated fragment of the PHV gene, pseudoPHV, found in the chromosome 5 centromeric region (M.J. Prigge, unpublished data). Regions of sequence similarity are indicated by lines between PHV and pseudoPHV.
REV, PHB, PHV, and CNA Regulate Apical Embryo Patterning
Whereas REV is specifically expressed during most of embryo development (see Supplemental Figure 2 online; Otsuga et al., 2001), no embryonic phenotypes had been identified in rev mutants, leaving the embryonic function of REV unclear. Genetic analysis revealed that REV, PHV, and PHB play key, overlapping roles in two major processes during embryogenesis: the establishment of apical bilateral symmetry and the establishment of the shoot apical meristem (SAM).
The role of REV and PHB in establishing the SAM was apparent from analysis of rev phb plants. These plants usually displayed a shoot meristemless phenotype, characterized by the normal production of all embryonic structures, with the exceptions that the SAM was absent and cotyledons were occasionally absent or display patterning defects (Figures 2B to 2D, Table 1). After a delay, the meristemless rev phb seedlings produced a radially symmetric structure emerging from the region normally occupied by the meristem (Figure 2B). No further postembryonic growth occurred in the rev phb double mutants, most likely because of the previously demonstrated requirement of REV for adventitious shoot formation (Otsuga et al., 2001). By contrast, rev phv double mutants rarely displayed a loss of embryonic SAM formation (Table 1), suggesting PHV plays a lesser role in this process. Whereas the rev phb athb8 triple mutant was similar to the rev phb double mutant, the phv and cna mutations enhanced the embryo patterning defects of the rev phb double mutant phenotype (Table 1; see below).
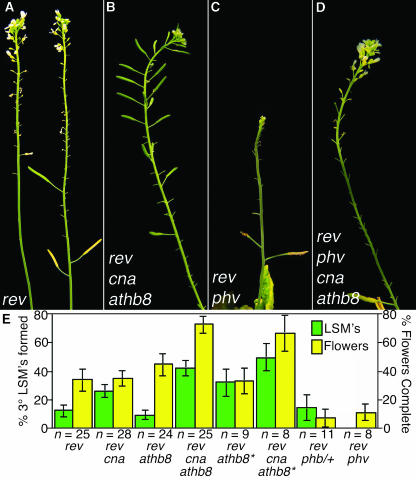
Figure 2.
REV, PHB, PHV, and CNA Regulate Apical Embryo Patterning.
(A) A seedling with a wild-type phenotype derived from a self-fertilized rev-6/+ phb-13 parent.
(B) to (D) Phenotypes of rev-6 phb-13 seedlings. Most rev phb double mutants produce two cotyledons (B), but others produce a single cotyledon (C) or a radially symmetric organ (D). Arrow in (B) indicates the radial symmetric structure emerging from apical region.
(E) to (H) Progeny of rev-6/+ phb-12 phv-11 plants. Approximately three-quarters of the progeny had wild-type heart-stage (E) and torpedo-stage (G) morphologies. The remaining embryos initiated a single radially symmetric cotyledon from the apical end and examples are shown at the heart-stage-equivalent (F) and torpedo-stage-equivalent (H) stages. Arrows indicate the boundary between the cotyledon and hypocotyls domains of embryos.
(I) to (N) Scanning electron micrographs of 10-day-old seedlings. (I) rev-6 phb-13 cna-2. (J) rev-6/+ phb-13 phv-11 cna-2. (K) rev-6 phb-13 phv-11 cna-2. (L) rev-6 phb-13 phv-11 cna-2, upper third of seedling. (M) rev-6 phb-13 phv-11 cna-2, middle third of seedling. (N) rev-6/+ phb-13 cna-2, hypocotyl. Genotypes of plants were inferred by genotyping siblings with similar phenotypes. Scale bars in (I), (J), and (K) = 500 mm. Arrows indicate the boundary between hypocotyl and cotyledon domains.
(O) rev-6 phb-13 phv-11 cna-2 athb8-11/athb8-12 10-d-old seedling. The genotype was determined by PCR.
Table 1.
Segregation of Seedling Phenotypes
| Seedling Phenotypesa
|
||||||
|---|---|---|---|---|---|---|
| Parental Genotype | BG/Mediab | n | Normal | No SAM | Two RSC | One RSC |
| rev | 4XL/Soil | 189 | 99.5 ± 1.0% | 0.5 ± 1.0% | 0 | 0 |
| rev | 5XC/Soil | 149 | 99.3 ± 1.4% | 0.7 ± 1.4% | 0 | 0 |
| rev phb-11/+ | 3XL/Soil | 203 | 79.8 ± 5.6% | 12.8 ± 4.7% | 0 | 7.4 ± 3.7% |
| rev phb-13/+ | 3XL/Soil | 198 | 81.3 ± 5.5% | 10.6 ± 4.4% | 0 | 8.1 ± 3.9% |
| rev/+ phb-13c | 5XC/MS | 935 | 81.7 ± 2.5% | 16.9 ± 2.5% | 0 | 1.4 ± 0.8% |
| rev/+ phb-13c | 5XC/Soil | 476 | 87.6 ± 3.0%d | 12.4 ± 3.0% | 0 | 0 |
| rev/+ phb-13 athb8 | 7XC/Soil | 305 | 80.0 ± 4.6%d | 18.7 ± 4.5% | 0 | 1.3 ± 1.3% |
| rev phv | 3XL/Soil | 58 | 96.6 ± 4.8% | 3.4 ± 4.8% | 0 | 0 |
| rev phv | 5XC/Soil | 104 | 99.0 ± 2.0% | 1.0 ± 2.0% | 0 | 0 |
| rev/+ phb-11 phv | 3XL/MS | 875 | 74.9 ± 2.9% | 2.9 ± 1.1% | 2.6 ± 1.1% | 19.7 ± 2.7% |
| rev/+ phb-11 phv | 2XC/MS | 228 | 78.5 ± 5.4% | 0 | 0 | 21.5 ± 5.4% |
| rev/+ phb-12 phv | 5XC/MS | 1777 | 76.8 ± 2.0% | 0 | 0.2 ± 0.2% | 23.0 ± 2.0% |
| rev/+ phb-13 phv | 5XC/MS | 1056 | 74.9 ± 2.7% | 0.1 ± 0.2% | 0.1 ± 0.2% | 24.9 ± 2.7% |
| rev/+ phb-13 phv athb8 | 7XC/MS | 301 | 73.4 ± 5.1% | 0 | 0.3 ± 0.6% | 26.2 ± 5.1% |
| rev/+ phb-11 cna | 3XL/MS | 395 | 76.5 ± 4.3% | 0.3 ± 0.6% | 7.3 ± 2.6% | 15.9 ± 3.7% |
| rev/+ phb-13 cna | 5XC/MS | 174 | 74.1 ± 6.6% | 0.6 ± 1.2% | 0 | 25.3 ± 6.6% |
| rev/+ phb-13 phv cna | 5XC/MS | 968 | 67.1 ± 3.0% | 9.6 ± 1.9%e | 1.4 ± 0.8%e | 21.8 ± 2.7%e |
| rev/+ phb phv cna athb8/+ | 5XC/MS | 288 | 68.8 ± 5.5% | 10.8 ± 3.7%f | 1.0 ± 1.2% | 19.4 ± 4.7%f |
| rev/+ phb/+ phv cna athb8g | 6XC/MS | 320 | 82.8 ± 4.2% | 11.6 ± 3.6%g | 0.3 ± 0.6%g | 5.3 ± 2.5%g |
Progeny of plants of the given genotype were sown and the percentage in each phenotypic class is reported. Normal, meristem active at least until flowering; No SAM, meristem not formed or terminated before flowering; Two RSC, two radially symmetric cotyledons; One RSC, apical portion consists of a radially symmetric cotyledon. Numbers indicate percent of total ± 2*se of the proportion.
BG, genetic background; numbers indicate the number of times the lines were introgressed into Col (C) or Ler (L). Seeds were sown either on soil or on sterile 0.5× MS media (MS).
Two different batches of rev/+ phb-13 seeds were tested.
Among the Normal seedlings, 66 rev/+ phb-13 (13.9%) and 13 rev/+ phb-13 athb8 (4.3%) plants had narrow-leaf and lateral-meristemless phenotypes and were found to be rev phb double mutants.
Three of three meristemless progeny of the rev/+ phb phv cna plant were found to be rev/+ upon genotyping. One Two RSC and two One RSC seedlings were genotyped and found to be rev/rev.
Three No SAM progeny of the rev/+ phb phv cna athb8-12/+ plant were found to be rev/+ upon genotyping (two of these were ATHB8+ and one was homozygous athb8-12). One Two-RSC seedling was genotyped and found to be rev phb phv cna athb8-12/+. Of five One RSC seedlings genotyped, three were rev phb phv cna athb8-12/+ and two were rev phb phv cna athb8-12.
Two of three No SAM progeny of the rev/+ phb/+ phv cna athb8-11/athb8-12 plant were found to be rev phb/+ phv cna athb8 upon genotyping; the third meristemless plant was rev/+ phb phv cna athb8. Both One RSC seedlings analyzed were found to be quintuple mutants (one was athb8-12 and one was athb8-11/athb8-12).
REV, PHB, PHV, and CNA play overlapping roles in patterning the apical portion of the embryo. Whereas most rev phb double mutants formed two cotyledons (83%; Figure 2B), some formed single or fused cotyledons (8%; Figure 2C) or a single radially symmetric structure occupying the apical portion of the embryo (7%; Figure 2D). The latter phenotypes indicate a profound defect in embryo patterning. A more severe and penetrant loss of bilateral symmetry was observed in rev phb phv triple mutants in which the apical portion of the embryo was replaced by a single radially symmetric structure (Table 1, Figures 2F and 2H).
The CNA gene also plays a role in apical embryo patterning. Mutations in the CNA gene similarly enhanced the apical patterning defect of rev phb embryos such that the triple mutant developed a radially symmetric apical structure similar to that of the rev phb phv triple mutant (Figure 2I, Table 1). The rev phb phv cna quadruple mutant phenotype was similar to that of the rev phb phv and rev phb cna triple mutants (Figure 2K). rev/+ phb phv cna plants displayed an embryonic shoot meristemless phenotype at low penetrance: 93 of 743 nonradially symmetric progeny of a rev/+ phb phv cna plant lacked functional SAM (Figure 2J, Table 1). The genotypes of three such progeny were determined and found to be rev/+ phb phv cna in each case. athb8 mutations did not affect these embryonic phenotypes; the rev phb phv cna athb8 quintuple mutant displayed the same terminal phenotype (Figure 2O, Table 1). Thus, the following mutant combinations resulted in a radially symmetric apical structure phenotype: rev phb phv, rev phb cna, rev phb phv cna, rev phb phv athb8, rev phb cna athb8, and rev phb phv cna athb8 (Table 1).
The morphology and arrangement of cells within the radially symmetric apical structures more closely matched those of cotyledons, and thus they appear to be radially symmetric cotyledons that are fully abaxialized (Figure 2H). This interpretation is consistent with scanning electron micrographs that show a clear boundary separating the upper and lower halves of the structure and also show hypocotyl-like epidermal cell files in the lower one-half (Figures 2I and 2K; compare Figures 2L and 2M to Figure 2N).
All HD-Zip III Genes Regulate Postembryonic Meristem Initiation
The REV, PHB, PHV, CNA, and ATHB8 genes also regulate postembryonic meristem initiation. REV is required for the formation of lateral shoot meristems (LSM) and floral meristems (FM) as well as adventitious shoots (Talbert et al., 1995; Otsuga et al., 2001). rev mutants are characterized by rosette and cauline leaves with barren axils and flowers lacking full meristematic activity, although these phenotypes are variably expressive (Figure 3A; Talbert et al., 1995; Otsuga et al., 2001). Reduction of PHB gene expression in the rev phb/+ lines resulted in a dramatic enhancement of the Rev− phenotype, and these plants produced very few fertile flowers (Figure 3E). The PHV gene appears to play a lesser role in LSM function based on the observation that rev phv double mutants had a FM phenotype similar to that of rev phb/+ plants (Figures 3C and 3E). CNA and ATHB8 play roles antagonistic to REV, PHB, and PHV in the formation of LSM, with CNA and ATHB8 promoting meristem activity as described below.
Figure 3.
phb, phv, cna, and athb8 Modify the Rev− Inflorescence Phenotypes.
(A) Inflorescences of a rev-6 plant.
(B) Inflorescence of a rev-6 cna-2 athb8-11 plant.
(C) Inflorescence of a rev-6 phv-11 plant.
(D) Inflorescence of a rev-6 phv-11 cna-2 athb8-11 plant.
(E) Two phenotypes were compared for each of the indicated genotypes: the percent of cauline leaves on secondary inflorescences with tertiary LSM in their axils (green bars) and the average number of the first 20 flowers that produced carpels (yellow bars). Asterisks indicate PCR-genotyped progeny of the same rev-6/+ cna-2/+ athb8-11 plant. Error bars indicate 2*se of the proportion (LSM) or 2*se of the mean (flowers).
CNA and ATHB8 Antagonize REV Function
The CNA and ATHB8 genes did not appear to share overlapping functions with REV in adult plants because rev cna and rev athb8 double mutants resembled the rev single mutant line (Figure 3E). Unexpectedly, the rev cna athb8 triple mutant produced more LSM and fertile flowers than the rev single mutant (Figures 3B and 3E). This suggests that the CNA and ATHB8 genes antagonize—directly or indirectly—REV gene function in LSM and FM. The flower development defects of rev phv double mutants were also partially suppressed by cna and athb8 mutations. Whereas rev phv flowers were replaced by tiny filamentous structures lacking meristem activity, early-produced flowers of rev phv cna athb8 quadruple mutant were qualitatively different, containing outer-whorl organs (sepals, petals and stamens; Figures 3C and 3D), and later-produced flowers were fertile (data not shown).
REV, PHV, and PHB Regulate Leaf Polarity
Although the roles of the PHB and PHV genes in organ polarity were previously indicated by the dominant mutations that adaxialize lateral organs, further evidence that HD-Zip III genes specify adaxial cell fate came from analysis of loss-of-function mutations. Thirty-four of 100 rev phv plants produced at least one trumpet-shaped leaf. These leaves had adaxial tissue inside the trumpet leaf cone, abaxial tissue surrounding proximal portion of the leaf, and normal adaxial/abaxial polarity distally (Figure 4A). Occasionally, rev phv leaves had ectopic leaf blades emerging from the adaxial surface surrounding patches of abaxial-like tissue (Figure 4B). Such leaves had mirror-image patterns of leaf polarity with abaxial tissue on the top and bottom surfaces and adaxial tissues in between (Figure 4B). The frequency of leaf polarity phenotypes was dominantly enhanced by phb mutations such that nearly all leaves of rev phb/+ phv plants were trumpet-shaped (Figure 4C). Leaf phenotypes for rev phb phv triple mutants could not be assessed because of seedling lethality. Replacement of adaxial tissue with abaxial tissue is consistent with the hypothesized role in specifying adaxial identity by REV, PHB, and PHV (McConnell and Barton, 1998; McConnell et al., 2001; Emery et al., 2003).
Figure 4.
REV, PHB, and PHV Regulate Leaf Abaxial/Adaxial Polarity.
(A) and (B) A partially abaxialized rev-6 phv-11 cauline leaf with trumpet morphology (A) and a partially abaxialized rev-6 phv-11 cauline leaf with a mirror-image duplication (B). Adaxial (Ad) and abaxial (Ab) surfaces are indicated by the arrows.
(C) A rev-6 phb-11/+ phv-11 plant with multiple trumpet leaves (arrows).
PHB, PHV, and CNA Have Overlapping Functions Independent of REV
Because the two most recent persisting duplication events gave rise to the PHB/PHV and CNA/ATHB8 gene pairs, we characterized the phb phv and cna athb8 double mutants. Both lines were found to be indistinguishable from wild-type plants. Interestingly, a novel phenotype appeared in the phb phv cna triple mutant; these plants had extra cotyledons, enlarged SAM, fasciated stems, flowers with extra organs, and reduced fertility (Figure 5B). Flowers of phb-12 phv cna triple mutants contained 4.15 ± 0.16 sepals, 4.10 ± 0.13 petals, 6.55 ± 0.39 stamens, and 3.73 ± 0.10 carpels (mean ± twice the se [2*se] of the mean; wild-type flowers contain 4 sepals, 4 petals, 5–6 stamens, and 2 carpels). In addition, phb phv cna flowers developed additional whorls of organs within the whorl 4 gynoecium (Figure 5C). The inflorescences of phb cna double mutants exhibited reduced internode lengths and occasionally developed flowers containing extra carpels (Figure 5E). The reduced fertility of phb phv cna flowers was likely attributable to defects in ovule development; many ovules of phb phv cna plants had short integuments (data not shown). Plants with all of the other combinations of the phb, phv, cna, and athb8 alleles (except phb phv cna athb8; see below) were similar to wild-type plants with respect to these phenotypes.
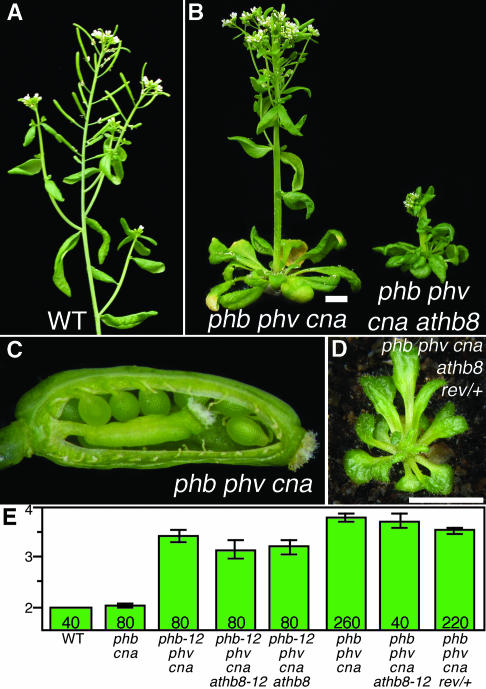
Figure 5.
Defects in Meristem Regulation and Plant Size.
(A) Inflorescence of a wild-type (er-2) plant.
(B) Similar aged phb-13 phv-11 cna-2 (left) and phb-13 phv-11 cna-2 athb8-12 (right) plants. Scale bar = 5 mm.
(C) Dissected silique of a phb-13 phv-11 cna-2 plant showing fifth whorl gynoecium and unfertilized ovules.
(D) Phenotype of a rev-6/+ phb-13 phv-11 cna-2 athb8-12 plant. Scale bar = 5 mm.
(E) Bars indicate the average numbers of carpels in flowers 1 through 20 for plant lines of the indicated genotypes. Error bars indicate ± 2*se of the mean.
The role of the PHB, PHV, and CNA genes in meristem regulation is independent of REV and ATHB8. Evidence that the meristem regulation defect is independent of REV comes from analysis of rev/+ phb phv cna plants. If REV was involved in the process, the reduction of REV activity would be expected to enhance the phenotype when, in fact, a slight suppression was observed (Figure 5E). Similarly, the SAM and FM defects of phb phv cna athb8 quadruple mutant plants were similar to or slightly suppressed relative to phb phv cna triple mutants (Figure 5E).
All five HD-Zip III genes affect plant stature. The phb phv cna athb8 quadruple mutant plants were significantly smaller (in stem height and leaf length) than the phb phv cna triple mutant lines, indicating that these genes have redundant roles in plant stature (Figure 5B; rosette diameters at flowering: phb phv cna, 29 ± 4 mm; phb phv cna athb8, 21 ± 2 mm; mean ± 2*se of the mean). To assess whether REV affects plant size, we identified a rev/+ phb phv cna athb8-12 seedling among the progeny of a rev/+ phb phv cna athb8-12/+ plant by PCR (Figure 5D). The plant was extremely dwarfed, having a rosette diameter of only 11 mm, and did not flower during 9 weeks of growth. The rev/+ phb phv cna athb8-12 genotype was underrepresented because of both meristem-defective phenotypes (above) and poor survival on soil (see Methods). Such sensitivity of plant stature to REV levels was not seen when comparing phb phv cna and rev/+ phb phv cna plants, indicating that plant stature is redundantly regulated by each of the HD-Zip III genes.
Roles of HD-Zip III Genes in Vascular Development
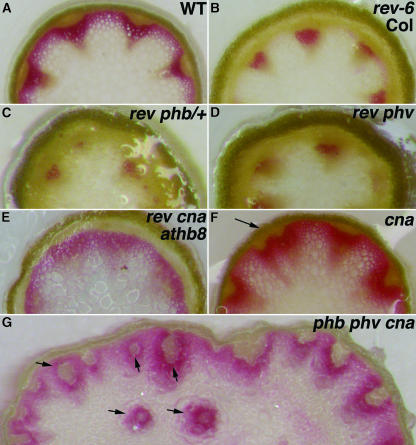
The HD-Zip III gene family is thought to play a role in vascular development based on rev loss-of-function and gain-of-function mutations, ectopic ATHB8 expression, and gene expression analyses in Zinnia (Baima et al., 1995, 2001; Zhong et al., 1997, 1999; Zhong and Ye, 1999, 2004; Ohashi-Ito et al., 2002; Ye, 2002; Emery et al., 2003; Ohashi-Ito and Fukuda, 2003). As with the ifl1 alleles of REV, the rev-6 mutation affected interfascicular fiber differentiation, but the effects were variable in different genetic backgrounds. In a Col background, the interfascicular fibers are absent (Figure 6B), yet the fibers were largely unaffected in the Ler background (data not shown). Mutations in the PHB and PHV genes enhanced the vascular defects of the rev-6 mutant, similar to genetic interactions observed in other aspects of inflorescence development (Figures 6C and 6D). By contrast, the Rev− vascular defects were partially suppressed in the rev cna athb8 triple mutant (Figure 6E). This result suggests that the antagonistic interactions of the REV, CNA, and ATHB8 genes described above might be indirectly attributable to suppression of the vascular defect of rev mutants by the cna and athb8 mutations.
Figure 6.
HD-Zip III Genes Regulate Vascular Patterning in Inflorescence Stems.
(A) to (H) Cross sections of inflorescence stems from ∼1 cm above the rosettes were stained with phloroglucinol (red) to detect lignified tissues.
(A) Wild type (er-2).
(B) rev-6 (Col).
(C) rev-6 phb-13/+.
(D) rev-6 phv-11.
(E) rev-6 cna-2 athb8-11.
(F) cna-2. Note irregularly spaced vascular bundles (arrow).
(G) phb-13 phv-11 cna-2. Note internal bundles (arrows).
Arrows indicate aberrant vascular bundles.
Slight perturbations in vascular development were seen in the cna single mutant stems, and more pronounced defects were seen in the phb phv cna triple mutant stems. The vascular bundles in cna single mutants were frequently less well distributed around the stem periphery (Figure 6F). The phb phv cna triple mutant produced bundles internally away from the stem periphery (Figure 6G). It is unclear whether the internalized vascular bundles reflect a specific patterning defect or are merely a secondary consequence of the larger meristem where the bundles are initiated.
HD-Zip III Proteins Are Not All Equivalent in Function
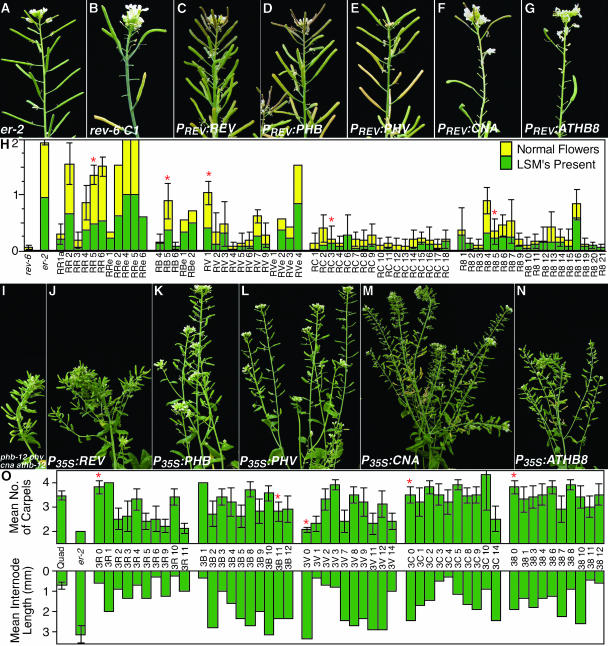
To test whether the proteins encoded by each of the five HD-Zip III genes are functionally equivalent, the corresponding cDNA sequences were fused to the REV (PREV) and cauliflower mosaic virus 35S (P35S) regulatory sequences and introduced into the rev-6 mutant and the phb-12 phv cna athb8-12 quadruple mutant, respectively. As previously reported, the PREV:REV transgene complemented the rev-6 phenotype (Figure 7C; Zhong and Ye, 1999). Complete rescue of Rev− phenotype was not observed for any other cDNA, although expression of the PHB and PHV cDNAs resulted in significant suppression of the mutant phenotype (Figures 7D, 7E, and 7H). Significantly fewer transformants were recovered after introduction of the PREV:PHB construct, and approximately one-half of the transformants exhibited a meristemless phenotype like that seen with the rev phb double mutant suggesting cosuppression of the endogenous PHB gene. The few nonseedling-lethal PREV:PHB lines that were recovered displayed phenotypes ranging from strongly suppressed to that of the rev-6 mutant (Figures 7D and 7H). The PREV:CNA and PREV:ATHB8 constructs did not obviously suppress the rev-6 mutant phenotype, although quantitative measurements revealed a weak partial suppression of the Rev− phenotype (Figures 7F to 7H). It is unclear whether this suppression is attributable to CNA and ATHB8 expression or to cosuppression of the endogenous loci, as we observed that loss-of-function mutations in these genes also suppress the Rev− phenotype (see above).
Figure 7.
Complementation of Mutant Phenotypes by HD-Zip III Gene Expression.
Each HD-Zip III gene was expressed in the rev-6 ([B] to [H]), and phb-12 phv cna athb8-12, ([I] to [O]), mutant backgrounds, and the abilities to suppress mutant phenotypes were assessed.
(A) Wild-type inflorescence.
(B) Inflorescence of a rev-6 “C1” plant.
(C) Inflorescence of RR5a, a T2 rev-6 “C1” plant with the PREV:REV transgene.
(D) Inflorescence of RB5a, a T2 rev-6 “C1” plant with the PREV:PHB transgene.
(E) Inflorescence of RV1a, a T2 rev-6 “C1” plant with the PREV:PHV transgene.
(F) Inflorescence of RC3a, a T2 rev-6 “C1” plant with the PREV:CNA transgene.
(G) Inflorescence of R85a, a T2 rev-6 “C1” plant with the PREV:ATHB8 transgene.
(H) A stacked bar graph illustrating the severity of two Rev− phenotypes in wild type (er-2), the rev-6 C1 line, and transgenic rev-6 C1 plant lines carrying a PREV:cDNA transgene. The names of the lines indicate which transgene is present: RR, PREV:REV, RB, PREV:PHB, RV, PREV:PHV, RC, PREV:CNA, R8, PREV:ATHB8. Each sample indicates an independent transgenic line. The lower (green) bar indicates the average proportion of cauline leaves on secondary inflorescences that bore tertiary inflorescences. The upper (yellow) bar indicates the average proportion of flowers 11 through 20 on primary inflorescences that developed inner-whorl organs. Nine Basta-resistant individual T3 or T2 plants from each line were scored for both traits except RV 3′ (3 plants), RV 6 (8), RC 13 (3), R8 10 (2), and those denoted RRe, RBe, and RVe where measurements from the T1 plants are shown. The error bars indicate 2*se of the mean. The red stars (*) indicate the plant lines shown in panels (C) through (G).
(I) Inflorescence of a phb-12 phv-11 cna-2 athb8-12 plant.
(J) Inflorescence of the 3R 0 T1 plant (P35S:REV; phb-12 phv-11 cna-2 athb8-12).
(K) Inflorescence of the 3B 11 T1 plant (P35S:PHB; phb-12 phv-11 cna-2 athb8-12).
(L) Inflorescence of the 3V 0 T1 plant (P35S:PHV; phb-12 phv-11 cna-2 athb8-12).
(M) Inflorescence of the 3C 0 T1 plant (P35S:CNA; phb-12 phv-11 cna-2 athb8-12).
(N) Inflorescence of the 38 0 T1 plant (P35S:ATHB8; phb-12 phv-11 cna-2 athb8-12).
(O) Graphs illustrating the average number of carpels in flowers 11 to 20 on the primary inflorescence (top graph) and the average internode length between flowers 11 to 20 on the primary inflorescence of the T1 plants. For comparisons, the averages for the untransformed quadruple mutant (quad) and wild type (er-2) are provided. The line names indicate the transgene present: 3R, P35S:REV; 3B, P35S:PHB; 3V, P35S:PHV; 3C, P35S:CNA; and 38, P35S:ATHB8. The error bars, where provided, indicate 2*se of the mean.
To assess complementation of phb-12 phv cna athb8-12, four phenotypes were monitored: fasciation of the primary inflorescence, average numbers of carpels per flower, average internode lengths between flowers, and rosette diameter. The P35S:PHB and P35S:PHV transgenes most efficiently suppressed each of the quadruple mutant phenotypes, although the suppression was variable (Figures 7K, 7L, and 7O; data not shown). The P35S:CNA construct did not complement the quadruple mutant fasciation phenotype and rarely suppressed the carpel number phenotype, but frequently complemented the internode length and rosette diameter defects (Figures 7M and 7O; data not shown). Consistent with the genetic results showing ATHB8 does not have functional overlap with PHB, PHV, and CNA in regulating meristem size, the P35S:ATHB8 transgene did not complement the quadruple mutant fasciation phenotype, but partially suppressed the internode length and rosette diameter phenotypes (Figures 7N and 7O; data not shown). The P35S:REV transgene had the least effect on stem fasciation, internode length, and rosette diameter; but the greatest effect on carpel number (Figures 7J and 7O; data not shown). Plants with strong suppression of the carpel phenotype, however, frequently also had barren leaf axils, suggesting that the apparent suppression may be attributable to silencing the endogenous REV locus rather than true complementation (data not shown).
DISCUSSION
HD-Zip III Genes Play Overlapping and Divergent Roles in Arabidopsis Development
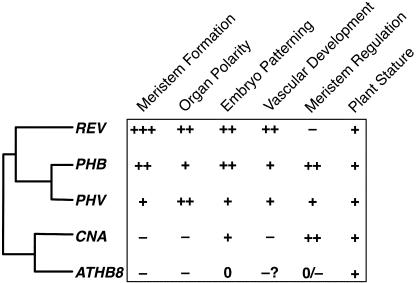
Analysis of loss-of-function alleles of the HD-Zip III family members revealed roles for these genes in embryo patterning, meristem initiation, organ polarity, meristem regulation, and vascular development, with different subsets of the genes being involved in each process (Figure 8). In addition to being differentially expressed, the protein-coding sequences have evolved distinct functions and are not fully interchangeable.
Figure 8.
HD-Zip III Genes Play Overlapping, Antagonistic, and Distinct Roles in Development.
The phylogram shows the phylogenetic relationships of Arabidopsis HD-Zip III genes, and the chart shows the functional relationships. The + symbols indicate the relative strength of defects seen in single and multiply mutant lines lacking the genes. The − symbol indicates that the gene antagonizes the roles of other genes with respect to this function. A 0 indicates that no role was observed. Meristem Formation, development of LSM and FM; Organ Polarity, specification of adaxial polarity during leaf development; Embryo Patterning, patterning the apical end of the embryo; Vascular Development, formation of lignified cells in inflorescence stems; Meristem Regulation, control of SAM and FM size; Plant Stature, role in growth to normal size.
The resulting picture that emerged from our analyses is a complex set of overlapping, distinct, and antagonistic functions. Importantly, several of the relationships among the HD-Zip III family were not readily inferable from phylogenetic relationships (Figure 8). The REV gene is the only member of the gene family whose loss is apparent in single-mutant plants. REV plays roles in apical embryo patterning, all embryonic and postembryonic SAM and FM initiation, lateral organ patterning, vascular development, and plant stature. The roles of the PHB and PHV genes overlap more than any other pair; this is consistent with their phylogenetic distinction of having duplicated relatively recently within the eudicot lineage. PHB and PHV play significant roles in postembryonic SAM and FM initiation, lateral organ patterning (all in conjunction with REV), apical embryo patterning (with REV and CNA), and meristem size regulation (with CNA, but not with REV or ATHB8). Mutations in the CNA and ATHB8 genes were found to partially suppress the rev and rev phv mutant phenotypes.
Some of the differences in HD-Zip gene function can be explained by differences in expression patterns. All of the HD-Zip III genes except ATHB8 play roles in patterning the apical portion of embryos (Figure 2; Emery et al., 2003), and ATHB8 is the only HD-Zip III gene not expressed in the apical half of globular embryos (see Supplemental Figure 2 online; McConnell et al., 2001; Emery et al., 2003). Later in embryogenesis, PHV expression is restricted to adaxial cotyledon tissue whereas REV and PHB are also expressed in the developing SAM (see Supplemental Figure 2 online; McConnell et al., 2001; Otsuga et al., 2001; Emery et al., 2003). This likely explains the meristemless phenotype seen in rev phb double mutants but rarely in rev phv double mutants. Interestingly, CNA is expressed in a pattern similar to that of REV and PHB in later stages of embryogenesis (see Supplemental Figure 2 online), but rev cna double mutants do not show enhanced embryonic meristem phenotypes. Consistent with their overlapping roles in vascular development, organ polarity, and meristem initiation during inflorescence development, we found that the REV and PHB are both expressed in developing vascular tissues, adaxial cells of floral organs, FM, and at presumptive FM initiation sites, and PHV is expressed in the same tissues with the exception that it is only weakly expressed in some vascular tissues (see Supplemental Figure 2 online). The CNA gene is expressed at relatively high levels, particularly in vascular tissue, but is also detectable in developing LSM and FM and in pith cells, whereas the ATHB8 gene expression is restricted to vascular development (see Supplemental Figure 2 online; Baima et al., 1995; Kang and Dengler, 2002; Ohashi-Ito and Fukuda, 2003).
Based upon ATHB8 gene expression patterns and overexpression phenotypes, it has been suggested that ATHB8 plays a major role in vascular development (Baima et al., 1995, 2001; Kang and Dengler, 2002), although we saw little evidence to support this contention. The smaller plant stature phenotype of phb phv cna athb8 and rev/+ phb phv cna athb8 plants could result from a reduction in vascular system function, although our superficial analysis of phloroglucinol staining of lignified cells did not reveal evidence of this. It remains possible that (1) the vascular system function—but not lignification—is altered, or (2) that vascular tissue development is redundantly regulated by all of the HD-Zip III genes, but defects eluded genetic analysis because of lethality of the quintuple mutant.
Many of the functions we identified for HD-Zip III genes had previously been indicated by studies of selected subsets of the HD-Zip III genes (Talbert et al., 1995; Zhong et al., 1997; McConnell and Barton, 1998; Zhong and Ye, 1999; McConnell et al., 2001; Otsuga et al., 2001; Emery et al., 2003), but the regulation of meristem size by PHB, PHV, and CNA and the antagonism between the REV and the ATHB8 and CNA genes in inflorescence development were surprising. The phb phv cna triple mutant phenotypes are very similar to those of clavata (clv) mutants (Clark et al., 1993, 1995; Dievart et al., 2003). The relative severity of the increase in organ number in each whorl was nearly identical to that caused by intermediate-strength clv alleles and quite different from other floral organ number mutants such as fas1, pan, ult, or se (Leyser and Furner, 1992; Running and Meyerowitz, 1996; Fletcher, 2001; Prigge and Wagner, 2001). Similarly, the appearance of the “fifth-whorl” is also a hallmark of clv mutants as is stem fasciation (Clark et al., 1993, 1995; Shannon and Meeks-Wagner, 1993; Dievart et al., 2003). A dominant-negative mutation in CNA has been identified as an enhancer of the Clv− phenotype in that clv cna-1 SAM are greatly enlarged and later exhibit profound defects in meristem function (Pogany et al., 1998; K.A. Green, M.J. Prigge, R.B. Katzman, and S.E. Clark, unpublished data). The striking similarities between the phenotypes of clv and phb phv cna plants suggest that HD-Zip III genes and the CLV pathway regulate meristem function in a similar manner, and further dissection may lead to new insights into how stem cell maintenance and organ formation are balanced.
Several lines of evidence indicate a mutual antagonism between the REV and the CNA and ATHB8 genes during postembryonic development. We observed suppression of the Rev− phenotype by the cna and athb8 mutations and suppression of the phb phv cna triple mutant phenotype by a rev allele. Furthermore, it has recently been reported that the avb1 mutation—a dominant, gain-of-function rev allele immune to repression by microRNA—results in stem fasciation similar to the phb phv cna mutants (Zhong and Ye, 2004). Because of a combination of conserved and divergent biochemical properties, it is possible that binding of CNA and ATHB8 proteins to specific DNA elements or cofactors may compete with the binding of REV (and PHV or PHB) protein(s). This interpretation is seemingly at odds with the apparent functional overlap between the PHB, PHV, and CNA genes in meristem regulation, but it remains possible that the synergistic triple mutant phenotype does not necessarily reflect functional redundancy. These genetic interactions may result from the combination of distinct defects conditioned by the losses of PHB/PHV and of CNA.
Pleiotropy Can Complicate the Identification of Primary and Secondary Defects
Because members of the HD-Zip III gene family are expressed in diverse tissues and mutations in these genes cause pleiotropic effects on development, the genes may have evolved diverse functions in angiosperm development. We cannot at present rule out that some or all of the pleiotropic phenotypes may result from defects in a single process such as vascular development or auxin signaling; however, the expression of each of these genes in each of the affected tissues indicates that this is unlikely.
Despite extensive research into the auxin-signaling pathway, the full extent of its role in plant development is not well understood. Auxin signaling has been implicated in many of the processes affected by the HD-Zip III mutations, and it has been shown that treatment with a polar auxin transport inhibitor can mimic the effects of rev mutations (Zhong and Ye, 2001). Furthermore, expression of ATHB8 has been shown to be regulated by auxin and to be dependent upon the activity of members of the auxin response factor family of transcription factors, although there is no evidence of direct regulation of the other HD-Zip III genes by auxin signaling (Baima et al., 1995; Sawa et al., 2002; Mattsson et al., 2003; Zhao et al., 2003). Even if the HD-Zip III genes are not directly involved in auxin signaling, it is possible that the two pathways were linked initially in plant evolution and were recruited together as the pathways were incorporated into patterning new tissues during evolution.
Ancestral Function of HD-Zip III Genes
The HD-Zip III genes are involved in the development of several structures thought of as important innovations in plant evolution (Gifford and Foster, 1989; Graham et al., 2000). Because this gene family diverged in the tracheophyte lineage, the evolution of the functions of this gene family may be tied to important evolutionary adaptations in the seed plant lineage. To study the incorporation of the genes' functions into new processes, it is important to understand the ancestral function of HD-Zip III genes. Given the high conservation of HD-Zip III gene sequences from moss to seed plants (Sakakibara et al., 2001), the ancestral function of these genes is not likely related to organ polarity, shoot branching, meristem size regulation, or vascular development. The last common ancestor of bryophytes and vascular plants was likely similar to present-day bryophytes in having dichotomously branching shoots with a single apical cell (rather than a meristem) and lacking vascular tissues (but may have had specialized water-conducting cells and single-cell thick leaf-like appendages; Gifford and Foster, 1989; Niklas, 1997; Graham et al., 2000; Ligrone et al., 2000). Despite not having many of the anatomical features regulated by the HD-Zip III genes in Arabidopsis, bryophytes do have auxin signaling pathways (Cooke et al., 2002; Sakakibara et al., 2003). This is consistent with the hypothesis that these genes were initially involved in auxin signaling and evolved with the auxin-signaling pathway as it was incorporated into developmental processes during seed plant evolution.
It is also possible that ancestral HD-Zip III genes regulated tissue polarity. Our results corroborate previous results implicating REV, PHB, and PHV in specifying polarity of stems and lateral organs (McConnell and Barton, 1998; McConnell et al., 2001; Emery et al., 2003). Furthermore, the defects of rev phb cna embryos and the vascular defects of phb phv cna stems suggest that CNA may also be involved in specifying polarity. Given the telome theory that lateral organs of euphyllophytes evolved from branched shoot systems, pathways specifying stem polarity and organ polarity in these plants may share common ancestry with a pathway that specified central versus peripheral polarity in ancestral plants (Bower, 1935; Gifford and Foster, 1989). This hypothesis may be tested by analyzing the HD-Zip III gene families in plants such as the lycophyte, Selaginella, and the moss, Physcomitrella patens. Expression analysis may provide support given that the hypothesis predicts that the genes in these organisms would be expressed in cells centrally located in the shoot stems, but such a result would not rule out a primary role in other, more basic, signaling pathways, such as auxin signaling. Functional analysis in these plants would provide much stronger evidence.
Insights into Apical Patterning of Plant Embryos
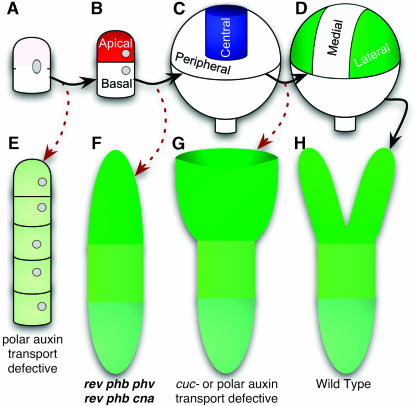
Unique among embryo patterning mutant phenotypes, the apical portion of both rev phb phv and rev phb cna embryos is cleanly replaced with a single, radially symmetric cotyledon-like organ whereas the embryonic hypocotyl and root develop normally (Figure 9F). Similar phenotypes have been reported for the rev ago1 double mutant (Kidner and Martienssen, 2004). In contrast with our results, Emery and coworkers found that the rev phb phv triple mutant frequently displayed weaker phenotypes (Emery et al., 2003). The discrepancy might be explained by differences in allele strengths. In addition to these multiply mutant lines lacking REV, a superficially similar morphology is seen occasionally in topless mutants (Long et al., 2002). Embryos lacking bilateral symmetry have been observed previously in, for example, pin1 stm double mutant and pin1 cuc1 cuc2 triple-mutant lines (Aida et al., 2002) as well as in embryos treated with polar auxin transport inhibitors (Schiavone and Cooke, 1987; Liu et al., 1993). These radially symmetric embryos differ from rev phb phv (and rev phb cna) embryos in that radially symmetric cotyledon envelopes the persisting apical pole resulting in an embryo shaped like a wine glass (Figure 9G). This distinction suggests that the HD-Zip III genes may be involved in differentiating between central and peripheral cells (Figure 9C). In the absence of HD-Zip III function, all apical cells have a peripheral identity, giving rise to a fully abaxialized cotyledon (Figure 9F). In pin1 stm and pin1 cuc1 cuc2, central cells would be differentiated from peripheral cells, but later subdivision of the peripheral domain would fail to occur (Figures 9D and 9G). Such a central versus peripheral distinction in the globular embryo had previously been hypothesized to exist based on gene expression data (Long and Barton, 1998), but genetic evidence was lacking. It will be interesting to see whether marker gene expression analyses corroborate this interpretation of the HD-Zip III gene family function in embryo patterning.
Figure 9.
Apical Patterning of the Globular Embryo.
(A) to (D) Illustrations of patterning steps in wild-type embryogenesis. (A), Zygote; (B), Specification of apical-basal polarity by an asymmetric cell division; (C), Specification of the central versus peripheral domains; and (D), Specification of medial versus lateral (cotyledon-forming) domains.
(E) to (G) Illustrations depicting mutants defective in certain patterning steps. (E) In pin1 pin3 pin4 pin7 quadruple mutants, the zygote divides symmetrically and frequently fails to form an embryo proper (Friml et al., 2003). (F) In the absence of the central domain specification by HD-Zip III genes, the apical portion of the embryo lacks central tissue and forms a single, radially symmetric cotyledon. (G) When the medial domain is not distinguished from the lateral domains—such as when polar auxin transport is lost or when the CUC genes are mutated—a single cup-shaped cotyledon is formed (Schiavone and Cooke, 1987; Liu et al., 1993; Aida et al., 2002).
(H) Wild-type mature embryo.
Solid, black arrows indicate progression through the patterning steps; and the red, dashed arrows indicate the phenotypic consequences of patterning defects.
METHODS
Plant Growth and Analysis
Plants were grown in 10-cm pots with 1 to 2 cm topsoil overlying a 1:1:1 mixture of topsoil:coarse vermiculite:perlite and ∼1 g of Osmocote fertilizer. The plants were grown at 20°C under cool-white fluorescent lights with a 4-h night break. For aseptic plant growth, seeds were surface sterilized and spread on Petri dishes containing 0.5× MS media.
The rev-6 allele was described previously (Otsuga et al., 2001). The phb-11 allele was previously identified in the CSHL/IMA Ds gene trap collection (Parinov et al., 1999), and the phb-12 (SALK 023802), phb-13 (SALK 021684), phv-11 (SALK JP91_0F10L.47.75), and athb8-12 (SALK 114415) alleles were identified using the SIGnAL T-DNAexpress service (Alonso et al., 2003). T-DNA enhancer trap lines were screened by PCR using pools of genomic DNA supplied from the Arabidopsis Biological Resource Center; cna-2 (SP4380) and athb8-11 (SP2460) were thus identified (McKinney et al., 1995; Krysan et al., 1996; Campisi et al., 1999). To confirm the positions of insertions, the T-DNA left border-flanking DNA was sequenced for each T-DNA allele after PCR amplification. The phv-11 allele has a complex T-DNA insertion with left border sequences on both sides; sequences flanking both left borders were determined. The T-DNA right border-flanking DNA was sequenced for the athb8-11 and athb8-12 alleles, but T-DNA right border-flanking DNA could not be amplified for the phb-12, phb-13, and cna-2 alleles. DNA sequences flanking both sides of the Ds insertion were determined for the phb-11 allele. Primers used for genotyping and sequencing the alleles are reported in Supplemental Table 1 online, and junction sequences are reported in Supplemental Table 2 online.
All lines also carried the er-2 allele (also known as er-106) to facilitate growth of large numbers of plant lines in our plant growth facilities. The background of the er-2 allele was confirmed to be Col using several PCR-based markers from loci spanning the genome (data not shown). All the multiply mutant lines were identified in segregating populations by PCR-based genotyping. The rev-6 allele was followed using a cleaved amplified polymorphic sequence marker, and the insertion alleles were followed using combinations of two gene-specific primers and an insertion-specific primer (see Supplemental Table 1 online). Because rev-6 was the only frequently used allele not originally isolated in the Col background, care was taken to eliminate Ler background modifiers both before crossing to other mutants and during the multiple-mutant construction. The original rev-6 ‘C1’ isolate (Col/Ler mix with an intermediate phenotype) was crossed to Col-1 then to er-2 (Col-1) twice with the second and third crosses using (PCR-determined) rev-6/+ F1 individuals. After the third cross, all F2 rev progeny had a uniformly strong phenotype and were used for subsequent crosses. In most cases, more than one plant was found for each multiply mutant genotype of interest. The only genotypes for which only one individual was identified were those without a discernible phenotype (e.g., phb-13 phv-11 athb8-11). Because rev-6 had the largest effect on development, multiply mutant lines involving rev-6 were generally identified in progeny of plants found by PCR to be heterozygous for rev-6 and homozygous for the other mutations. These plants rarely displayed any abnormal phenotypes that could bias plant selection. The reciprocal combinations were additionally looked at for many of the genotypes, but were usually less desirable for establishing seed stock lines because of low fertility. The rev/+ phb phv cna athb8-12 genotype was underrepresented among the soil-sown progeny of a rev/+ phb phv cna athb8-12/+ plant because of a high mortality rate on soil. Less skewed segregation ratios were seen when germinated on sterile media (Table 1). Initially, 16 nonseedling-lethal progeny were genotyped by PCR. (Ten of these were selected for smaller rosette sizes because we surmised that rev/+ phb phv cna athb8-12 would likely resemble phb phv cna athb8 plants.) Four of the 16 were homozygous athb8-12, but these four were also homozygous REV+. The rev/+ phb phv cna athb8 plant discussed in the Results section was subsequently identified by searching for plants with more severe phenotypes.
Shortly after flowers first became visible, rosette diameters were measured as the maximum distance from leaf-tip to leaf-tip. Hand-sectioned stems, 1 to 2 cm above the rosette, were stained in a solution of 82% ethanol, 14% hydrochloric acid, and 0.09% (w:v) phloroglucinol for approximately one-half h and photographed immediately (Ruzin, 1999).
Photographs were captured using a Nikon Coolpix 995 digital camera directly or attached to a Zeiss Stemi SV 11 dissecting microscope, a Nikon Optiphot-2 microscope, or a Zeiss Axioskop microscope with differential interference contrast optics. Images were imported and manipulated using Apple iPhoto 2.0 and Adobe Photoshop 7.0.1 software.
Cross-Complementation
Each HD-Zip III cDNA was amplified by RT-PCR using first-strand cDNA derived from mRNA from Col-1 shoot apices using primers that added either a BamHI or MluI sites just 5′ from the start codons and either BstBI or SacI sites 3′ of the stop codon. PREV was amplified by PCR using the MUP24 P1 artificial chromosome clone as a template and contained the region from −2748 (EcoRI site) to −6 (followed by an engineered BamHI site; numbered relative to the REV start codon). Pfu DNA polymerase or a mixture of Pfu and Taq DNA polymerases was used to minimize PCR-induced mutations, and error-free subcloned PCR products were identified by sequencing. The P35S promoter fragment was excised from pLhG4 supplied by Ian Moore (Oxford; Moore et al., 1998). The promoters and cDNAs were inserted into pCB321 or pMP535, derivatives of pCB302 containing the Agrobacterium nos terminator sequence or the pea rbcS (E9) polyadenylation sequence, respectively (Xiang et al., 1999). The constructs were introduced into either the rev-6 C1 line or phb-12 phv-11 cna-2 athb8-12 quadruple mutants by in planta transformation using the AGL1 Agrobacterium strain (Clough and Bent, 1998).
Accession numbers for the genomic sequences containing HD-ZIP III genes are the following: REV, AB005246; PHB, AC003096; PHV, AC009917; CNA, AC006216; ATHB8, AL049915.
Supplementary Material
Acknowledgments
We thank the Arabidopsis Biological Resource Center for providing seeds and the pooled T-DNA genomic DNA stocks, and we thank Venkatesan Sundaresan (Institute of Molecular Agrobiology and University of California, Davis) for providing the phb-11 allele. Kirsten Green provided many helpful discussions, and Sang-Kee Song provided advice on microscopy. We also thank Jianzhi Zhang and Yin-Long Qiu for comments on the manuscript. This project was supported by National Science Foundation Grant IBN-0131492 to S.E.C. and National Institutes of Health/National Research Service Postdoctoral Fellowship GM20900 to M.J.P.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Steven E. Clark (clarks@umich.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.026161.
References
- Aida, M., Vernoux, T., Furutani, M., Traas, J., and Tasaka, M. (2002). Roles of PIN-FORMED1 and MONOPTEROS in pattern formation of the apical region of the Arabidopsis embryo. Development 129, 3965–3974. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Baima, S., Nobili, F., Sessa, G., Lucchetti, S., Ruberti, I., and Morelli, G. (1995). The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development 121, 4171–4182. [DOI] [PubMed] [Google Scholar]
- Baima, S., Possenti, M., Matteucci, A., Wisman, E., Altamura, M.M., Ruberti, I., and Morelli, G. (2001). The Arabidopsis ATHB-8 HD-zip protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol. 126, 643–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleecker, A.B., Estelle, M.A., Somerville, C., and Kende, H. (1988). Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science 241, 1086–1089. [DOI] [PubMed] [Google Scholar]
- Bower, F.O. (1935). Primitive Land Plants, Also Known as the Archegoniatae. (London: Macmillan).
- Campisi, L., Yang, Y., Yi, Y., Heilig, E., Herman, B., Cassista, A.J., Allen, D.W., Xiang, H., and Jack, T. (1999). Generation of enhancer trap lines in Arabidopsis and characterization of expression patterns in the inflorescence. Plant J. 17, 699–707. [DOI] [PubMed] [Google Scholar]
- Clark, S.E., Running, M.P., and Meyerowitz, E.M. (1993). CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119, 397–418. [DOI] [PubMed] [Google Scholar]
- Clark, S.E., Running, M.P., and Meyerowitz, E.M. (1995). CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121, 2057–2067. [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–744. [DOI] [PubMed] [Google Scholar]
- Cooke, T.J., Poli, D., Sztein, A.E., and Cohen, J.D. (2002). Evolutionary patterns in auxin action. Plant Mol. Biol. 49, 319–338. [PubMed] [Google Scholar]
- Dievart, A., Dalal, M., Tax, F.E., Lacey, A.D., Huttly, A., Li, J., and Clark, S.E. (2003). CLAVATA1 dominant-negative alleles reveal functional overlap between multiple receptor kinases that regulate meristem and organ development. Plant Cell 15, 1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery, J.F., Floyd, S.K., Alvarez, J., Eshed, Y., Hawker, N.P., Izhaki, A., Baum, S.F., and Bowman, J.L. (2003). Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13, 1768–1774. [DOI] [PubMed] [Google Scholar]
- Estelle, M.A., and Somerville, C.R. (1987). Auxin-resistant mutants of Arabidopsis with an altered morphology. Mol. Gen. Genet. 206, 200–206. [Google Scholar]
- Fletcher, J.C. (2001). The ULTRAPETALA gene controls shoot and floral meristem size in Arabidopsis. Development 128, 1323–1333. [DOI] [PubMed] [Google Scholar]
- Force, A., Lynch, M., Pickett, F.B., Amores, A., Yan, Y.L., and Postlethwait, J. (1999). Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin, K.A., Davis, S.J., Stoddart, W.M., Vierstra, R.D., and Whitelam, G.C. (2003. a). Mutant analyses define multiple roles for phytochrome C in Arabidopsis photomorphogenesis. Plant Cell 15, 1981–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin, K.A., Praekelt, U., Stoddart, W.M., Billingham, O.E., Halliday, K.J., and Whitelam, G.C. (2003. b). Phytochromes B, D, and E act redundantly to control multiple physiological responses in Arabidopsis. Plant Physiol. 131, 1340–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml, J., Vieten, A., Sauer, M., Weijers, D., Schwarz, H., Hamann, T., Offringa, R., and Jurgens, G. (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426, 147–153. [DOI] [PubMed] [Google Scholar]
- Gifford, E.M., and Foster, A.S. (1989). Morphology and Evolution of Vascular Plants. (New York: W.H. Freeman and Co.).
- Graham, L.E., Cook, M.E., and Busse, J.S. (2000). The origin of plants: Body plan changes contributing to a major evolutionary radiation. Proc. Natl. Acad. Sci. USA 97, 4535–4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua, J., and Meyerowitz, E.M. (1998). Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94, 261–271. [DOI] [PubMed] [Google Scholar]
- Hughes, A.L. (1994). The evolution of functionally novel proteins after gene duplication. Proc. R. Soc. Lond. B. Biol. Sci. 256, 119–124. [DOI] [PubMed] [Google Scholar]
- Kang, J., and Dengler, N. (2002). Cell cycling frequency and expression of the homeobox gene ATHB-8 during leaf vein development in Arabidopsis. Planta 216, 212–219. [DOI] [PubMed] [Google Scholar]
- Kidner, C.A., and Martienssen, R.A. (2004). Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 428, 81–84. [DOI] [PubMed] [Google Scholar]
- Krakauer, D.C., and Nowak, M.A. (1999). Evolutionary preservation of redundant duplicated genes. Semin. Cell Dev. Biol. 10, 555–559. [DOI] [PubMed] [Google Scholar]
- Krysan, P.J., Young, J.C., Tax, F., and Sussman, M.R. (1996). Identification of transferred DNA insertions within Arabidopsis genes involved in signal transduction and ion transport. Proc. Natl. Acad. Sci. USA 93, 8145–8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser, H.M.O., and Furner, I.J. (1992). Characterisation of three shoot apical meristem mutants of Arabidopsis thaliana. Development 116, 397–403. [Google Scholar]
- Ligrone, R., Ducket, J.G., and Renzaglia, K.S. (2000). Conducting tissues and phyletic relationships of bryophytes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355, 795–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C., Xu, Z., and Chua, N.H. (1993). Auxin polar transport is essential for the establishment of bilateral symmetry during early plant embryogenesis. Plant Cell 5, 621–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, J.A., and Barton, M.K. (1998). The development of apical embryonic pattern in Arabidopsis. Development 125, 3027–3035. [DOI] [PubMed] [Google Scholar]
- Long, J.A., Woody, S., Poethig, S., Meyerowitz, E.M., and Barton, M.K. (2002). Transformation of shoots into roots in Arabidopsis embryos mutant at the TOPLESS locus. Development 129, 2797–2806. [DOI] [PubMed] [Google Scholar]
- Mattsson, J., Ckurshumova, W., and Berleth, T. (2003). Auxin signaling in Arabidopsis leaf vascular development. Plant Physiol. 131, 1327–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell, J.R., and Barton, M.K. (1998). Leaf polarity and meristem formation in Arabidopsis. Development 125, 2935–2942. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R., Emery, J., Eshed, Y., Bao, N., Bowman, J., and Barton, M.K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411, 709–713. [DOI] [PubMed] [Google Scholar]
- McKinney, E.C., Aali, N., Traut, A., Feldmann, K.A., Belostotsky, D.A., McDowell, J.M., and Meagher, R.B. (1995). Sequence-based identification of T-DNA insertion mutations in Arabidopsis: Actin mutants act2–1 and act4–1. Plant J. 8, 613–622. [DOI] [PubMed] [Google Scholar]
- Moore, I., Galweiler, L., Grosskopf, D., Schell, J., and Palme, K. (1998). A transcription activation system for regulated gene expression in transgenic plants. Proc. Natl. Acad. Sci. USA 95, 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niklas, K.J. (1997). The Evolutionary Biology of Plants. (Chicago: University of Chicago Press).
- Ohashi-Ito, K., Demura, T., and Fukuda, H. (2002). Promotion of transcript accumulation of novel Zinnia immature xylem-specific HD-Zip III homeobox genes by brassinosteroids. Plant Cell Physiol. 43, 1146–1153. [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito, K., and Fukuda, H. (2003). HD-Zip III homeobox genes that include a novel member, ZeHB-13 (Zinnia)/ATHB-15 (Arabidopsis), are involved in procambium and xylem cell differentiation. Plant Cell Physiol. 44, 1350–1358. [DOI] [PubMed] [Google Scholar]
- Ohno, S. (1970). Evolution by gene duplication. (Berlin, New York: Springer-Verlag).
- Otsuga, D., Deguzman, B., Prigge, M., Drews, G., and Clark, S. (2001). REVOLUTA regulates meristem initiation at lateral positions. Plant J. 25, 223–236. [DOI] [PubMed] [Google Scholar]
- Parinov, S., Sevugan, M., Ye, D., Yang, W.C., Kumaran, M., and Sundaresan, V. (1999). Analysis of flanking sequences from dissociation insertion lines: A database for reverse genetics in Arabidopsis. Plant Cell 11, 2263–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaz, S., Ditta, G.S., Baumann, E., Wisman, E., and Yanofsky, M.F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405, 200–203. [DOI] [PubMed] [Google Scholar]
- Pinyopich, A., Ditta, G.S., Savidge, B., Liljegren, S.J., Baumann, E., Wisman, E., and Yanofsky, M.F. (2003). Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424, 85–88. [DOI] [PubMed] [Google Scholar]
- Pogany, J.A., Simon, E.J., Katzman, R.B., De Guzman, B.M., Yu, L.P., Trotochaud, A.E., and Clark, S.E. (1998). Identifying novel regulators of shoot meristem development. J. Plant Res. 111, 307–313. [Google Scholar]
- Ponting, C.P., and Aravind, L. (1999). START: A lipid-binding domain in StAR, HD-ZIP and signalling proteins. Trends Biochem. Sci. 24, 130–132. [DOI] [PubMed] [Google Scholar]
- Prigge, M.J., and Wagner, D.R. (2001). The Arabidopsis SERRATE gene encodes a zinc-finger protein required for normal shoot development. Plant Cell 13, 1263–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince, V.E., and Pickett, F.B. (2002). Splitting pairs: The diverging fates of duplicated genes. Nat. Rev. Genet. 3, 827–837. [DOI] [PubMed] [Google Scholar]
- Running, M.P., and Meyerowitz, E.M. (1996). Mutations in the PERIANTHIA gene of Arabidopsis specifically alter floral organ number and initiation pattern. Development 122, 1261–1269. [DOI] [PubMed] [Google Scholar]
- Ruzin, S.E. (1999). Plant Microtechnique and Microscopy. (New York: Oxford University Press).
- Sakakibara, K., Nishiyama, T., Kato, M., and Hasebe, M. (2001). Isolation of homeodomain-leucine zipper genes from the moss Physcomitrella patens and the evolution of homeodomain-leucine zipper genes in land plants. Mol. Biol. Evol. 18, 491–502. [DOI] [PubMed] [Google Scholar]
- Sakakibara, K., Nishiyama, T., Sumikawa, N., Kofuji, R., Murata, T., and Hasebe, M. (2003). Involvement of auxin and a homeodomain-leucine zipper I gene in rhizoid development of the moss Physcomitrella patens. Development 130, 4835–4846. [DOI] [PubMed] [Google Scholar]
- Sawa, S., Ohgishi, M., Goda, H., Higuchi, K., Shimada, Y., Yoshida, S., and Koshiba, T. (2002). The HAT2 gene, a member of the HD-Zip gene family, isolated as an auxin inducible gene by DNA microarray screening, affects auxin response in Arabidopsis. Plant J. 32, 1011–1022. [DOI] [PubMed] [Google Scholar]
- Schiavone, F.M., and Cooke, T.J. (1987). Unusual patterns of somatic embryogenesis in the domesticated carrot: Developmental effects of exogenous auxins and auxin transport inhibitors. Cell Differ. 21, 53–62. [DOI] [PubMed] [Google Scholar]
- Sessa, G., Steindler, C., Morelli, G., and Ruberti, I. (1998). The Arabidopsis Athb-8, -9 and -14 genes are members of a small gene family coding for highly related HD-ZIP proteins. Plant Mol. Biol. 38, 609–622. [DOI] [PubMed] [Google Scholar]
- Shannon, S., and Meeks-Wagner, D.R. (1993). Genetic interactions that regulate inflorescence development in Arabidopsis. Plant Cell 5, 639–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak, E.D., Lakeman, M.B., and Torii, K.U. (2003). Dominant-negative receptor uncovers redundancy in the Arabidopsis ERECTA Leucine-rich repeat receptor-like kinase signaling pathway that regulates organ shape. Plant Cell 15, 1095–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried, K.R., Eshed, Y., Baum, S.F., Otsuga, D., Drews, G.N., and Bowman, J.L. (1999). Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126, 4117–4128. [DOI] [PubMed] [Google Scholar]
- Talbert, P.B., Adler, H.T., Parks, D.W., and Comai, L. (1995). The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development 121, 2723–2735. [DOI] [PubMed] [Google Scholar]
- Tang, G., Reinhart, B.J., Bartel, D.P., and Zamore, P.D. (2003). A biochemical framework for RNA silencing in plants. Genes Dev. 17, 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel, D., et al. (2000). Activation tagging in Arabidopsis. Plant Physiol. 122, 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, K., Long, D., Swinburne, J., and Coupland, G. (1996). A Dissociation insertion causes a semidominant mutation that increases expression of TINY, an Arabidopsis gene related to APETALA2. Plant Cell 8, 659–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, C., Han, P., Lutziger, I., Wang, K., and Oliver, D.J. (1999). A mini binary vector series for plant transformation. Plant Mol. Biol. 40, 711–717. [DOI] [PubMed] [Google Scholar]
- Ye, Z.H. (2002). Vascular tissue differentiation and pattern formation in plants. Annu. Rev. Plant Biol. 53, 183–202. [DOI] [PubMed] [Google Scholar]
- Zhang, J. (2003). Evolution by gene duplication: An update. Trends Ecol. Evol. 18, 292–298. [Google Scholar]
- Zhao, Y., Dai, X., Blackwell, H.E., Schreiber, S.L., and Chory, J. (2003). SIR1, an upstream component in auxin signaling identified by chemical genetics. Science 301, 1107–1110. [DOI] [PubMed] [Google Scholar]
- Zhong, R., and Ye, Z.H. (1999). IFL1, a gene regulating interfascicular fiber differentiation in Arabidopsis, encodes a homeodomain-leucine zipper protein. Plant Cell 11, 2139–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., and Ye, Z.H. (2001). Alteration of auxin polar transport in the Arabidopsis ifl1 mutants. Plant Physiol. 126, 549–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., and Ye, Z.H. (2004). amphivasal vascular bundle 1, a gain-of-function mutation of the IFL1/REV gene, is associated with alterations in the polarity of leaves, stems and carpels. Plant Cell Physiol. 45, 369–385. [DOI] [PubMed] [Google Scholar]
- Zhong, R., Taylor, J.J., and Ye, Z.H. (1997). Disruption of interfascicular fiber differentiation in an Arabidopsis mutant. Plant Cell 9, 2159–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., Taylor, J.J., and Ye, Z.H. (1999). Transformation of the collateral vascular bundles into amphivasal vascular bundles in an Arabidopsis mutant. Plant Physiol. 120, 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.