Abstract
Affinity purification coupled to mass spectrometry (AP-MS) is the method of choice to analyze protein-protein interactions, but common protocols frequently recover only the most stable interactions and tend to result in low bait yield for membrane proteins. Here, we present a novel, deep interactome sequencing approach called CoPIT (for Co-interacting Protein Identification Technology), which allows comprehensive identification and analysis of membrane protein interactomes and their dynamics. CoPIT integrates experimental and computational methods for a co-immunoprecipitation (Co-IP)-based workflow from preparing the sample for mass spectrometric analysis to generating protein-protein interaction networks. The protocol addresses several limitations of current methods for protein interaction analysis. The approach particularly improves the results for membrane protein interactomes, which proved to be difficult to identify and analyze. CoPIT was used successfully to identify the interactome of the Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) and to demonstrate its validity and performance. The experimental step achieved up to 100 fold higher bait yield than previous methods by optimizing lysis, elution, sample-clean up and detection of interacting proteins with a well established, highly sensitive mass spectrometric detection method called Multidimensional protein identification technology (MudPIT). Here, we provide evidence that CoPIT is applicable to other types of proteins as well, and can be successfully used as a general Co-IP method. The protocol describes all steps from considerations for experimental design, Co-IP, preparation of the sample for mass spectrometric analysis as well as data analysis steps including discrimination of background proteins from true interactors, comparison of samples, and visualization of interaction networks. While the experimental part can be performed in less than three days, data analysis may take up to a few weeks.
Introduction
The proper functioning of an organism is orchestrated by highly complex protein networks, which are dynamically regulated in time and space. The combination of affinity-purification and mass spectrometry (AP-MS) has become an important method to advance the discovery and functional characterization of such networks by facilitating the analysis of protein-protein interactions 1,2. Discovering protein interaction networks is still challenging though because a comprehensive analysis requires high yields of the “bait” protein for robust co-purification of its interactors in order to differentiate true interactors from non-specific background and to allow comparisons between different samples based on the relative quantification of protein-protein interactions.
The generally lower abundance, hydrophobic nature, and partial protection of protease cleavage sites by lipid layers exacerbate experimental and analytical challenges for membrane proteins. Because of these technical challenges, the interactome of many membrane proteins has remained unknown or is only poorly characterized. Yet, there is a particular need for elucidating the interactomes of membrane proteins because membrane proteins make up about a third of the human genome, include many physiological important proteins such as ion channels, transporters and receptors, and represent the majority of the “druggable genome” 3-6. In general, shotgun proteomic methods have improved the identification of membrane proteins and topology mapping, but these methods are not compatible with the identification of protein interaction partners 7-13. To study the interactome of membrane proteins, the bait is typically tagged with an epitope. While epitope tagging strategies can provide efficient enrichment because of the availability of high affinity reagents and antibodies, epitope tagging increases the potential for mis-sorting or mis-folding of membrane proteins during their complex biogenesis 14-16 and may render AP-MS data potentially less informative 17,18. Once mass spectrometric data is acquired and proteins in the sample are identified, the data needs to be analyzed to rank interactors according to confidence, construct interactomes and finally visualize interactome networks and changes thereof. Several recently published methods including SAINT 19,20, CompPASS 21 and MiST 22 analyze AP-MS experiments performed with epitope tagged proteins, but do not provide an integrated solution from experiment to network for immunoprecipitations of non-tagged proteins or require prior knowledge such as typical background in the immunoprecipitations or expected interactors.
To facilitate the identification and analysis of membrane protein interactomes we developed a novel, highly sensitive “deep proteomic interactome profiling approach”, called CoPIT (for Co-interacting Protein Identification Technology) 23,24. CoPIT is an experimental and computational framework that allows the comprehensive characterization of endogenous membrane as well as non-membrane protein interactomes with three individual workflows as illustrated in Figure 1. The first workflow is an optimized experimental protocol for co-immunoprecipitation which provides enhanced sensitivity and efficiency by addressing several issues associated with enrichment of protein interactomes in general and with membrane protein interactomes in particular. Difficulties are (a) highly efficient recovery of membrane proteins from cell lysates while preserving interactions, (b) insolubility and/or aggregation of membrane proteins during IP and subsequent elution, (c) removal of lipids and other contaminants that may cause signal suppression during electrospray ionization, and (d) compatibility of Co-IP procedure with in-solution digestion, chromatographic separation and mass spectrometric detection of interactors. The second workflow includes novel data analysis algorithms that allow to discriminate highly confident from less confident interactors in comparison to control experiments for label-free data as well as to determine interactome changes between experimental conditions using spectral counting. Finally, interacting proteins can be graphed in a specialized network visualization tool (Radial Topology Viewer) that maps interactors for example according to statistical significance of the interactions, whereby the arithmetic distance to the bait in the center can reflect a dynamic variable of the user's choice like confidence in or change of protein-protein interaction. Additional relational information on protein-protein interactions gathered from other databases can be included to enable a contextual interpretation of the interactome.
Figure 1.

Schematic outline and workflow of CoPIT. The three individual workflows performed in CoPIT are shown. Experimental Protocol: Experimental protocols were optimized for high bait and interactor recovery by adjusting cell lysis-, Co-IP and elution conditions. Data analysis: Resulting Raw-data are searched with ProLuCID and uniformly filtered to a peptide FDR of ≤ 0.5 %. Signal-to-noise discrimination is carried out by including different negative controls, such as experiments from CFTR null cells, and significantly regulated interactions are identified using a counting statistics. Network comparison: Protein IDs and results are stored in a relational database, which contains also further ontological information. The Radial Topology Viewer visualizes ranking, functional annotation and connectivity of interactors in the resulting networks as well as differences between conditions.
The CoPIT method was developed to determine the interactome of the Cystic Fibrosis Transmembrane Conductance Regulator (CFTR), an ion channel with twelve transmembrane segments that belongs to the family ABC transporter proteins 25-27, and to compare it against the interactome of the most common CFTR mutation, an in-frame deletion of phenylalanine 508, that causes Cystic Fibrosis (CF) in 70 - 90% of all CF patients 28. CFTR can be considered a touchstone for methods that aim at determining membrane protein interactomes, because of its low abundance (≤ 100 molecules per cell for the HBE41o- cell line), above average protein size (168 kDa), rapid turn over in the cell, and short protein half-life 29,30, – all of which contribute to difficulties associated with membrane protein analysis.
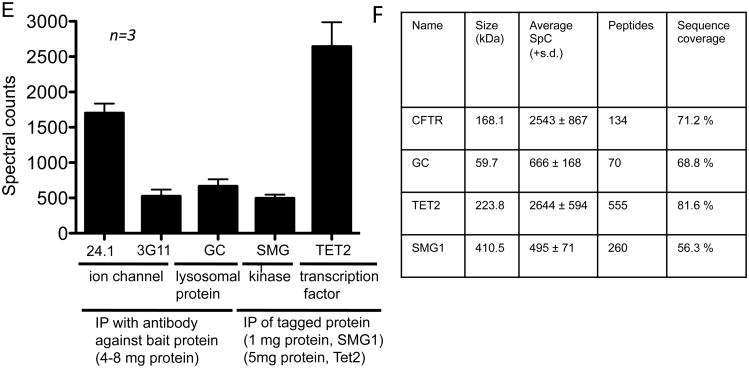
In summary, CoPIT enables the comprehensive characterization of a membrane protein interactome for low-abundant membrane proteins such as CFTR as long as an antibody with good immunoprecipitation capabilities is available, – even if only low amounts of protein and sample are provided. These characteristics make CoPIT ideal for comparative interactome analysis of neuronal receptors and membrane proteins in different brain regions or cell types (e.g. the synaptic interactome) for example. In addition, the capability and applicability of CoPIT to other types of proteins is illustrated with IPs for the peripheral membrane protein glucocerebrosidase (GC), the soluble protein kinase SMG1 as well as the transcription factor Tet2 (Figure 2). It is important to bear in mind that the success of any immunoprecipitation is dependent on a high quality antibody-antigen interaction, and quantitative immunoprecipitation of the antigen (the bait protein) has to be established before attempting CoPIT experiments.
Figure 2.

High sensitivity of CoPIT. A. Western blot depicting improved recovery of ΔF508 CFTR from CFBE41o- cells with TNI in comparison with different lysis buffers. A,B,C indicate the different CFTR glycoforms. B. Western blot showing enhanced recovery of ΔF508 CFTR from beads after Co-IP with detergent and heat aided low pH elution in comparison to other directly mass spectrometric compatible elution methods. Lane Wang et al. 2006: Elution conditions as described in Wang et al., 2006 41. Gly. Glycine C. Enhanced sensitivity of the CFTR Co-IP and chromatography is reflected by enhanced spectral counts for CFTR itself and well-established interactors like HSP70 and HSP90. E. Recovery of baits from different protein classes with CoPIT. Data are shown as spectral counts for the bait in independent biological triplicates. Error bars indicate standard deviation. The starting amount is given below as number of cells that were lysed for CoPIT. F. Table showing the performance of CoPIT for proteins of different classes and molecular weights giving the average of recovered spectral counts per experiment, and sequence coverage.
Figure 2 A-C is taken from Pankow, Bamberger et al. 2015 24.
Experimental Design
The sensitivity of CoPIT depends on the inclusion of proper controls. CoPIT does not rely on pre-established lists of background or contaminant proteins because the determination of what is background in an actual experiment greatly varies with the choice of the bait, cellular or organismal model system. Instead, CoPIT relies on an unbiased statistical approach that makes use of the high recovery of bait and interactors and a set of specific control experiments (c), which determine the level of background within the CoPIT procedure (overview in Figure 3). Appropriate controls are “mock-IPs” that are performed with beads devoid of any antibody in order to determine the background caused by non-specific binding to the beads. Further antibodies might bind several antigens in a cell in addition to the antigen of interest. Samples that are devoid of the bait protein should be used in an additional set of control experiments in order identify contaminant proteins. Ideally, an isogenic cell line or tissue that does not express the bait protein is available or, alternatively, can be generated with current gene editing tools. In case the protein is essential to cell viability, RNA interference might be an option to at least transiently down-regulate the antigen of interest by at least more than 10-fold. We do not recommend to use Co-IPs with antibodies against proteins “unrelated” to the intended bait as controls. The reason is that the interactomes of two different proteins are compared rather than a bait-unspecific background subtracted. Thus the results are difficult to interpret with regards to being a control experiment and this interpretation may result in an unwanted bias in the interactome as proteins that interact with the bait and the “unrelated” protein are removed during subsequent data analysis.
Figure 3.

Determination of a specific interactome. A. The schematic shows the experimental details for background reduction. B. Schematic of the experiment design and computational procedure. Boxes indicate distinct steps in the procedure. C. Frequency distribution Nrp ec of all rp ec determined for the experimental condition wt CFTR to control condition. Individual points (black dots) indicate the individual νrp ec values. The two-term Gaussian fit is shown in grey. The individual Gaussian describing the distribution of non-specific binding is colored in brown, whereas the Gaussian describing the enrichment for weak specific interactors is indicated in light green. The black arrow marks the rp ec determined for CFTR, the bait protein. D. Example P -values for well known CFTR interactors (light green) and proteins commonly identified as background in Co-IP experiments (light brown). Threshold for a high-confidence ΔF508-CFTR interactor was calculated at ≥ 0.92. E. Proportional Venn Diagram showing the overlap between the determined wt CFTR and ΔF508 CFTR core-interactomes. 62 proteins were found bound only to wt CFTR, whereas 208 proteins were detected only in ΔF508 CFTR Co-IPs. 368 proteins were detected in both wt and ΔF508 CFTR Co-IPs. Data represent independent biological replicates (wt CFTR, n=7; ΔF508, n=8.).
Figure 3 C is taken from Pankow, Bamberger et al. 2015 24.
Due to the high sensitivity of the mass spectrometry instrumentation, efficient removal of non-peptide contaminants prior to mass spectrometry is further required for highest sensitivity and reproducibility of the analysis. Contaminating lipids are a particular problem for membrane protein analysis, because they impede both the tryptic digest of proteins as well as mass spectrometric detection of peptides. High levels of residual lipids might contribute to reduced peptide ionization during electrospray ionization orincrease the complexity of mass spectra which in turn can result in suppression of low abundant peptides. Therefore, CoPIT takes several precautions to reduce background at different steps during the procedure. First, samples are pre-cleared using non-antibody-coupled beads to reduce the background of proteins that bind non-specifically to beads. Second, antibody-coupled beads are saturated with antibody during the coupling procedure to maximize antigen retrieval and minimize non-specific binding of contaminant proteins and, third, excess lipids as well as the detergent and salts are removed by precipitation with a mixture of methanol and chloroform prior to proteolytic digest of the sample (Figure 3A).
While mass spectrometry of proteins is a very sensitive detection method, it is heavily dependent on the quality of the immunoprecipitation itself and cannot correct for sub-optimal performance. Thus it is recommended that the suitability of an antibody for Co-IP is tested first in a small scale experiment followed by western blotting. Not all antibodies are suitable for Co-IP, and success of the method is greatly dependent on the affinity and specificity of the antibody, which needs to be determined empirically for each new bait. Finding the best antibody for an immunoprecipitation of interest can be a substantial effort: Sometimes, it can be quicker and more successful to test a different antibody than trying to further modify experimental conditions in hopes of improving the performance of a relatively poor immunoprecipitation. In general, we greatly prefer to use monoclonal antibodies for CoPIT because large amounts of antibody can be generated in hybridoma cell lines at reasonable cost to ensure saturation of beads with the antibody. Saturation of the beads to maximum capacity helps to prevent non-specific binding of proteins and concentrates protein complexes of interest in a small volume. In general, binding capacity of the Sepharose beads ranges from 5 mg/ml to 15 mg/ml (mg antibody per ml packed beads), and antibodies should be coupled to the beads at this ratio. It is recommended to check the amount of antibody loaded to beads and the crosslinking of antibody to protein-G with SDS gel electrophoresis as pointed out in the protocol.
Another critical variable is the starting amount of the sample. Main parameters that influence how much starting material is required and thereby determine the success of a CoPIT experiment are: the abundance of the bait protein per cell, the relative contribution of the cell type of interest in a tissue preparation and the affinity of the antibody for the bait. Therefore it is good practice to carefully consider cell and tissue preparation and enrichment techniques prior to sample lysis for immunoprecipitation. We suggest starting with 1 × 108 cells for low abundant bait proteins like CFTR. Samples can be prepared with much success from much lower numbers of cells depending on above parameters and to a small degree on the experience of the person performing the experiment.
Materials
Reagents:
Monoclonal Antibody (preferred) or polyclonal antibody
Protein G or Protein A Sepharose 4 Fast Flow beads (Cat No. 17-0618-01, GE Healthcare)
Sodium Borate 10-Hydrate, Crystal (Cat No. 3568-01, JT Baker)
Ethanolamine (Cat No. 02400-250ML, Sigma, Aldrich) CAUTION: corrosive. Proper protective equipment should be worn.
Dimethylpimelimidate (DMP) (Cat No. 21667, Pierce) CAUTION: toxic. Dust mask and eye-shield should be worn when handling powder.
Dulbecco's Phosphate buffered Saline (DPBS) without CaCl2, and MgCl2 (Cat No. 14190-144, Gibco, Invitrogen)
Ultra-Pure Tris (Cat No. EC-406, National Diagnostics)
Igepal CA-630 (Cat No. 18896-50ML, Sigma-Aldrich)
Ultra-pure EDTA (Cat No. 3579, Gibco)
Sodium Chloride (Cat No. S7653-5KG, Sigma-Aldrich)
PhosStop (Cat No. 04906845001, Roche)
Complete ULTRA, EDTA-free Protease Inhibitor cocktail (Cat No. 05892953001, Roche)
Non-stick surface microcentrifuge tubes (e.g. Cat No. 20170-650, VWR)
Glycine, electrophoretic grade (Cat. No. 100191, Roche)
Methanol, HPLC grade (Cat. No. 9093-03, J.T.Baker). CAUTION: Vapor and liquid are toxic. Wear proper protective clothing and handle in a fume hood.
Chloroform, 99.8+%, ACS reagent, stabilized with ethanol (Cat. No. 423555000, Acros Organics). CAUTION: toxic if inhaled, handle in a fume hood.
Rapigest SF Surfactant (Cat. No. 186001861, Waters)
TCEP (Cat. No. 77720, Thermo)
Iodoacetamide (Cat. No. 90034, Thermo)
Sequencing grade recombinant trypsin (Cat. No. V5111, Promega)
Formic acid (Cat. No. A117, Fisher Scientific)
Acetonitrile, HPLC grade, ≥ 99.8% purity (Cat. No. LC015-2.5, Honeywell)
Ammonium acetate, HPLC grade (Cat. No. A2149, Spectrum)
Kasil No. 1624 potassium silicate solution (PQ Corporation)
Kasil No.1 potassium silicate solution (PQ Corporation)
Fused Silica (Cat. No. 1068150047, i.d. 100 μm, Polymicro Technologies)
Fused Silica (Cat. No. 160-2250-5, i.d. 250 μm, Agilent Technologies)
Aqua C18 resin, 3 μm and 5 μm particle size (Cat. No 00B-4299-B0, Phenomenex)
Strong cation exchange resin (e.g. Cat. No. 4621-0507, Hichrom Limited)
Cells (see REAGENT SETUP) or Tissue
Equipment
High-pressure column and sample loader (pressure bomb)
HPLC (e.g. HP 1200; Agilent Technologies) with post-HPLC split for nanoflow
P-2000 microcapillary laser puller (Sutter Instrument Co.)
Mass spectrometer (LTQ Velos Pro Orbitrap or similar, Thermo)
Water bath sonicator (55kHz, 80 kHz, Branson)
Cooled Microcentrifuge
Thermomixer (e.g. Eppendorf Thermomixer R)
Insulin injection syringe (BD bioscience)
Rotator
Cell scraper
Vacuum concentrator (e.g. Speedvac, Thermo)
Software
Xcalibur software (Thermo Electron Corporation)
RawExtract or RawConverter
SEQUEST or ProLuCID
DTASelect
Census
Matlab
Cytoscape
Genemania
Access to Local computer cluster or cloud computing
Reagent setup
Cell Culture
CoPIT was tested on HBE41o- or CFBE41o- cells as well as Hek293T, HeLa, and HACAT cells, and primary human keratinocytes expanded in cell culture. Cells should be cultivated according to the needs of the specific cell line and harvested at a density that is maximal but does not compromise homogeneity of the cell phenotype (state of differentiation). For CoPIT experiments with CFTR isogenic human bronchial epithelial cell lines carrying the ΔF508 CFTR mutation (CFBE41o-), or the wt CFTR allele (HBE41o-) and the isogenic CFTR null cell line were kindly provided by Dr. J. Clancy (University of Alabama, Birmingham, AL). Cells were cultured in Advanced-MEM (GIBCO, Carlsbad, CA) supplemented with 1 % Penicillin/Streptomycin (GIBCO), 10% fetal bovine serum (GIBCO) and 2 mM L-Glutamine (GIBCO) at 37 °C, 5% CO2. Cells were harvested at confluence.
<CRITICAL> All subsequent procedures should be carried out immediately after harvesting cells or tissues. In case cells, tissues or lysates thereof were frozen, CoPIT will work, but recovery of interactors, in particular of weak or transient interactions, will be significantly reduced.
Equipment setup
Fritted micro-capillary column (MudPIT microcapillary column): This column should be prepared as described elsewhere 31-33. <CRITICAL> To avoid cross contaminations between the different samples, each sample is loaded onto a single, freshly prepared MudPIT microcapillary column 13,34. The number of MudPIT columns needed is identical to the number of replicates as well as experimental conditions and controls.
Mass spectrometer setup
The procedure has been successfully carried out using LTQ, LTQ Orbitrap, LTQ Velos and Velos Pro Orbitrap as well as Orbitrap Elite mass spectrometers (all from Thermo Fisher). The MudPIT approach 13,33 to run samples is recommended for greatest yield of bait and interactors. Briefly, purified peptides are re-suspended in 50 μl of buffer A (95% H2O, 5.0% MeCN, 0.1% formic acid) and pressure loaded onto the back-end of a preparative MudPIT microcapillary column consisting of fused silica (i.d. 250 μm) packed in-house with 2.5 cm of 5 μm Aqua C18 resin and 2.5 cm of strong cation exchange resin. The preparative column should be connected by a small union body to an analytical reversed-phase column (115 mm fused silica i.d. 100 μm) packed with 3 μm Aqua C18 resin.
In our laboratory, samples were analyzed by nano-ESI-LC/LC-MS/MS on an LTQ-Orbitrap XL, LTQ or Orbitrap Elite by placing the triphasic MudPIT column inline with an Agilent quaternary HPLC pump and separating the peptides in multiple dimensions with a modified 6-step gradient containing 0%, 20%, 40%, 60%, and 100% of Buffer C (500 mM ammonium acetate, 5% MeCN, 0.1% formic acid) over 12 h or a 10-step gradient (0% to 90% Buffer C) over 20 h as described previously 13. Each full scan mass spectrum (400-2000 m/z) was followed by 6 (LTQ, LTQ-Orbitrap XL) or 20 (Orbitrap Elite, Velos or Velos Pro) data-dependent MS/MS scans at 35% normalized collisional energy and an ion count threshold of 1000 (LTQ-Orbitrap XL, Elite or Velos) or 500 counts (LTQ). Dynamic exclusion was used with an exclusion list of 500, repeat time of 60 s and asymmetric exclusion window of -0.51 and +1.5 Da.
Software
RawExtract, Sequest, ProLuCID, DTASelect2 and Census may be obtained from the Yates laboratory (www.fields.scripps.edu). Detailed instructions on how to download and use additional software specifically designed for analysis and visualization of CoPIT experiments (CoPITgenerator, RadialTopologyViewer) are available at proteomicswiki.com: http://proteomicswiki.com/wiki/index.php/CoPITgenerator and http://proteomicswiki.com/wiki/index.php/RadialTopologyViewer.
Procedure
Coupling of antibodies to Sepharose beads
Timing: ∼4.5 h
<CRITICAL> 100 mM Sodium Borate, pH 9.0, and 200 mM Ethanolamine, pH 8.0, solutions should be preparedly freshly. pH must be adjusted exactly, otherwise the crosslinking reaction may be inefficient. Avoid any contamination of these buffers with primary amines from sources such as pH meter devices.
<CRITICAL> To check for crosslinking efficiency, take small sample aliquots of antibody, beads and after steps 1.2, 1.5 (the supernatant), and the final step and run an SDS-Page gel (10%) and stain with Coomassie brilliant blue G-250.
Aliquot the appropriate amount of Sepharose beads into a microcentrifuge tube and wash four times with 10 volumes of DPBS. Spin at 500 × g, 3 min, RT.
-
Remove excess PBS (). Add the appropriate amount of antibody to the beads. Save a small aliquot corresponding to 1 μg of antibody for SDS-Page gel control (“input”). Gently mix antibody and beads for 2 h at RT on a rotator to allow binding of the antibody to the beads.
<CRITICAL STEP> Never let Sepharose beads become dry.
Centrifuge the binding reaction for 3 min, 500 × g, RT. Wash twice with 10 bead volumes of 100 mM Sodium Borate, pH 9.0. Save a small aliquot corresponding to 1 μg of antibody bound to beads for SDS-Page gel control (“bound”).
-
Re-suspend the beads in 10 bead volumes of 100 mM Sodium Borate, pH 9.0. Start the crosslinking reaction by adding DMP powder to a final concentration of 20 mM.
Critical STEP Add the appropriate amount of DMP powder needed for crosslinking directly to the solution containing the beads after re-suspending the beads in the sodium borate solution. DMF quickly hydrolyzes in water and cannot be stored in solution for prolonged times.
Mix for 30 min (±5 min) at room temperature on a rotator. Critical: Do not exceed this time to avoid over-crosslinking.
Centrifuge for 3 min, 500 × g, RT. Remove supernatant. Wash with 10 bead volumes of 200 mM Ethanolamine, pH 8.0 to stop the reaction. Save a small aliquot corresponding to 1 μg of antibody bound to beads for SDS-Page gel (“crosslinked”).
Centrifuge for 3 min, 500 × g, RT. Remove supernatant. Add 10 bead volumes of 200 mM Ethanolamine, pH 8.0. Incubate for 2 hours at RT with gentle mixing.
-
Wash five times with 10 bead volumes of 1 × DPBS to eliminate Ethanolamine interfering with experiments. Save a small aliquot corresponding to 1 μg of antibody bound to beads for Western blot (“final”).
PAUSE POINT. Beads can be prepared in advance and stored for up to 1 yr at 4 °C.
9. Optional: Perform an SDS-Page gel (10%) separation with the aliquots as indicated in the protocol and with a protein standard series in order to determine how much antibody was used in the initial binding to the beads, how much antibody was bound to the beads and how well the antibody was crosslinked to the beads. Optimal crosslinking of antibody to the beads is revealed by detection of the light chain only.
Co-immunoprecipitation
Timing: 1.5 days
<CRITICAL> All steps should be performed as quickly as possible, on ice and with pre-chilled buffers. Centrifugation steps should be performed at 4°C. Beads dry out extremely fast, do not let them become dry for any period of time.
10. Rinse cells with at least 10 volumes (v/v) of 1 × DPBS. Remove excess 1 × DPBS by decanting or by vacuum suction and tapping on a paper towel.
11. Immediately add 1.5 ml of ice-cold TNI lysis buffer per 15 cm cell culture dish (TNI: 0.5% Igepal CA-630, 50 mM Tris [pH 7.5], 250 mM NaCl, 1 mM EDTA and 1× Complete ULTRA EDTA-free Protease Inhibitor cocktail, 1× PhosStop).
12. Incubate for 20 min on ice on an orbital shaker.
13. Scrape off cell lysate with a large cell scraper and transfer to a 1.5 ml microcentrifuge tube.
14. Sonicate the cell lysate for 3 min in a water bath sonicator operating at 55 kHz. Critical: Do not use a probe sonicator, this will result in loss of interactors.
15. Centrifuge for 30 min. (18,000 × g, 4 °C).
16. Preclear the cell lysate with an appropriate amount of Protein G Sepharose 4 Fast Flow beads (100 μl of 50% slurry in lysis buffer) for 2 h at 4 °C with head-over-head rotation. The volume of preclear beads is the same as the volume of antibody coupled beads. For volumes larger than 1.4 ml, pool the cell lysates and transfer into a 15 ml tube.
17. Centrifuge for 3 min, 500 × g, 4°C.
18. Transfer supernatant to a new microcentrifuge tube containing the
19. appropriate amount of antibody-coupled sepharose slurry (e.g. 100 μl of 50% antibody slurry, equaling 200 μg of antibody for starting amount of ≈10 mg). Incubate over night with head-over-head rotation at 4 °C.
20. Centrifuge the binding reaction for 3 min at 500 × g, 4 °C.
21. Remove the supernatant, transfer the beads to a new tube and wash the beads three times with 20 – 100 bead volumes of TNI lysis buffer.
-
22. Centrifuge for 3 min at 500 × g, 4 °C, carefully remove supernatant, and wash beads two times with TN lysis buffer containing no Igepal CA-630.
23. Carefully remove all of the supernatant with an insulin syringe.
PAUSE POINT: beads can be stored at -80°C for up to two weeks.
24. Optional: Freeze beads for >1 h at −80 °C to increase yield in the following steps.
25. Elute proteins twice with at least four to 10 bead volumes of elution buffer (0.2 M glycine, pH 2.3/ 0.5% Igepal CA-630) for 20 min, 37 °C, with shaking.
-
26. Combine the eluates and transfer to a new microcentrifuge tube.
Critical STEP: Make sure to get no beads into the eluate, they might clog microcapillary columns and produce background signal later in the mass spectrometer.
27. Neutralize eluate with 1/10 vol (v/v) freshly prepared 1 M NH4CO3 .
28. Add 4 vol (v/v) Methanol to the eluate and vortex.
-
29. Add 1 vol (v/v) Chloroform and vortex well for 0.5 - 1 min.
CAUTION: Methanol/Chloroform extraction should be carried out in a fume hood, as chloroform and Methanol vapors are toxic.
30. Centrifuge for 10 min at 18,000 × g.
31. Remove the supernatant without disturbing the pellet. Note: The pellet maybe very tiny or hardly visible at all.
32. Wash pellet with 3 vol (v/v) Methanol.
33. Centrifuge for 10 min at 18,000 × g, RT.
34. Remove the supernatant without disturbing the pellet.
PAUSE POINT: the pellet can be stored at -80°C for up to four weeks.
Digestion of eluted proteins
Timing: ∼15 h
35. Re-solubilize the methanol / chloroform precipitated proteins (pellet) in 100 mM Tris, pH 8.5, 0.2% Rapigest and sonicate for 1 h in a water bath sonicator (Branson).
36. Reduce cysteine disulfide bonds with TCEP [5 mM final concentration] for 20 min.
37. Alkylate reduced cysteine residues with 10 mM Iodoacetamide [finalconcentration] or Chloroacetamide for 30 min. The reaction should beshielded from light.
38. Digest proteins with recombinant trypsin (30:1 ratio protein:trypsin) over night at 37°C with shaking (e.g. in an Eppendorf Thermomixer).
39. Inactivate Rapigest by adding formic acid to 9% final concentration and incubate for at least 1 h, 37°C, with shaking.
-
40. Reduce samples to near dryness in vacuo using a vacuum concentrator (approximately 45 min).
PAUSE POINT: Samples can be stored at -80°C for up to four weeks.
41. Resolubilize sample in a small amount of buffer A, load onto a preparative MudPIT column and perform a MudPIT run as described in 32,33.
Data analysis
Timing: ∼2 d to several weeks
<CRITICAL> The following steps can be performed individually or performed in an integrated software solution such as IP2 (Integrated Proteomics Applications, Inc., San Diego).
42. Extract raw files into text file format.
43. Search the MS/MS spectra for matching peptide sequences using an appropriate protein database that is well annotated. It is advised to include a reversed or scrambled database of the same size to allow for an estimation of the false discovery rate.
-
44. Combine Search engine results from MS/MS spectra for all biological replicates before assigning peptides to proteins and filtering the results, for example by using DTASelect 2.1. We strongly recommend to conservatively filter and adjust the false positive rate to a peptide false positive rate of less than 0.5%, and a protein false positive rate of less than 1.0%. Experiments with insufficient recovery of bait (for example, bait protein not within the top ten most identified proteins or less than 50 SpC as a guideline) may be discarded.
Critical STEP: Ambiguities in protein identification caused by peptides matching to multiple protein isoforms should be resolved, because the resulting redundancy is problematic for statistical analysis as it skews the analysis towards proteins with many isoform entries. The current CoPIT approach is based on a “gene centric” strategy. Therefore all protein isoforms are assigned to their respective genes by converting the protein ID to Entrez Gene Symbols. This is important when isoform-exhaustive databases are used. For the global analysis of the interactome, CoPIT currently retains the highest intensity value obtained from the different isoforms identified.
Determine which protein interactors are specific
<CRITICAL> To distinguish specific protein interactors from non-specifically binding proteins, perform the following steps in Matlab (Mathworks) or similar software. Alternatively, run CoPIT.jar, which performs the steps up to step 4.10, and is available at http://proteomicswiki.com/wiki/index.php/CoPITgenerator. All steps to use the software are documented on that website.
45. Determine an experiment-to-control ratio rpec for a protein p based on the sum of the intensities from all measurements for the experiment ( ) over the control ( ). The unit for intensity I is spectral counts, SpC. The ratio is expressed on a log10 scale , where n is the number of independent measurements of experimental condition e or of control condition c. The number of biological replicates n should be similar for both, experimental condition e and control condition c.
<CRITICAL STEP> The identification of proteins enriched in bait-specific immunoprecipitations is based on two approximations: First, proteins binding non-specifically and non-selectively to beads or antibody are detected in both, experimental and control measurements. As shown before 35,36, the frequency distribution for non-enriched proteins can be assumed to follow a normal distribution, which is centered close to rpec=1.0 as all non-specific proteins are expected to be detected with equal intensities in both control and experimental condition. Second, bait-interacting proteins can be grouped as high and low abundant interactors. Interactors of high abundance are measured with high intensity values and in large excess over control values (rpec ≫1), clearly indicating enrichment. Less abundant interactors may represent a weak or transient interaction or an interaction with the bait that happens only in a very specific subcellular location. These interactors are typically measured at lower levels over control values (rPec >1). In order to delineate a specificity of interaction, CoPIT approximates that the distribution of specific interactors follows a normal distribution independent of the normal distribution of proteins binding non-specifically to beads or antibodies only. The center of this second normal distribution significantly depends on the yield of the immunoprecipitations as higher yields result in larger signal intensities and thus in lager contribution of the specific binding intensity to the ratio rpec.
46. Determine the frequency distribution of all ratios rpec: bin the frequency distribution with 25 to 30 bins (in intervals of 0.1 to 0.2) depending on the sample measured and generate a histogram covering a range of binned ratios ρ (ρmin to ρmax, typically −2 to +4).
-
47. Fit a function to the histogram. In case of the CFTR interactome a Gaussian with two terms was fitted to the histogram with a goodness of fit 0.90 ≤ R2 ≤ 0.98 depending on the experimental condition:
<CRITICAL STEP> If appropriate for the respective frequency distribution obtained, CoPIT carries out a Gaussian fitting comprising two terms, of which the first describes an approximation of the distribution of ratios measured for non-specific binding of proteins, e.g. background signal bg, whereas the second term describes an approximation of the distribution of ratios for proteins with specific binding affinity to the bait sp.
48. Calculate Confidence Values P for each protein measured, taking the measurement errors into account according to: where σbg, μbg, σsp and μsp are derived from the respective terms of the Gaussian fit. In case the protein has not been detected in the control condition can be set manually to a value different than 0, for example to 0.1 SpC.
-
49. Calculate ratios rp for individual proteins from the sum of all intensities per protein and experiment condition e1 and e2 and normalize to the sum of all bait intensities (SpC) according to:
-
50. Calculate errors according to random error of measurement:
The P-value cutoff can be adjusted according to the ratio of bait in the different experimental conditions:
Network representation and data presentation
51. Obtain interactions between the identified interactors with the GeneMANIA 2.2 Plugin in Cytoscape 2.8.2 37,38 and export connectivity information to a .txt or .csv file to load into Radial Topology Viewer. A step-by-step instruction how to obtain networks from GeneMania and Cytoscape is available in Morris et al., 2014 39. Detailed instruction on how to load and visualize data in Radial Topology Viewer are provided at http://proteomicswiki.com/wiki/index.php/RadialTopologyViewer. Radial Topology Viewer particularly enables a user to define the length of each individual edge in a network (e.g. according to strength of interaction), and allows users to group proteins, e.g. based on annotation information or other relational information. This information should be provided in a .txt or .csv file.
Troubleshooting
Troubleshooting guidelines can be found in Table 1.
Table 1. Troubleshooting.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 1.1-1.8 | No antibody or only small portion is coupled to beads. | pH of sodium borate solution is incorrect. | Make sure all sodium borate is dissolved before use, and pH is adjusted to the correct value. |
| DMP is degraded. | Light and moisture rapidly inactivate DMP. Buy and use small amounts that are stored dry (with desiccant). Allow DMP to come to room temperature before opening to prevent condensation. | ||
| Incorrect sepharose beads | Check the binding strength of the antibody to the particular type of sepharose, like protein A or protein G coupled sepharose. | ||
| 2.9 | Recovery of bait is low. | Antibody does not bind bait specifically. | Make sure that the antibody recognizes the bait protein specifically by including positive and negative controls, e.g. overexpress the bait protein in a cell system and if available compare signal with a knockout strain or a cell or tissue that does not express the bait protein. |
| 2.9 | Antibody does not recognize native epitope. | Even though an antibody may work well for western blotting, it may not recognize the epitope in non-denatured protein. Try a different antibody for the bait. | |
| 2.1-2.23 | Too little antibody coupled beads were used or too many beads were lost during procedure. | Use an insulin syringe to remove solutions during washing of the Co-IP. Check that sufficient quantities of antibody have been coupled to the beads and enough beads were used for the Co-IP procedure. | |
| 2.1-2.23 | pH of solutions was incorrect. | All solutions need to have the correct pH, in particular the lysis buffer and elution buffer. It is critical that detergent is added to the elution buffer for elution of membrane proteins. | |
| 2.1 | Not enough starting material. | Increase the starting material amount. “The more the merrier” still holds true for Co-IP experiments if low abundant proteins are used as bait. Confirm presence of the protein in the starting material. | |
| 3 | Liquid chromatography or mass spectrometer problem | Ensure that chromatographic separation is good and the mass spectrometer is calibrated correctly. Run a test sample if necessary. | |
| 3 | Wrong protein database used for search | Download the correct protein database for the species that was used in the experiment, it should contain the bait protein of interest. | |
| 2.9 | Many background proteins were identified, but little of the bait. | Antibody is not suitable for IP. | If the antibody binds to many other proteins in addition to the bait, it may not be suitable for Co-IP. Try a different antibody if available or affinity purification of an antiserum. |
| 2.10-2.12 | Insufficient washing of the IP. | Carry out all five wash steps and remove washing solution carefully. If necessary an additional wash step with lysis buffer containing no detergent can be included. Washing helps to increase signal to noise ratio. | |
| 2.7 | Pre-clearing was not sufficient. | Pre-clear the cell lysate before incubation with antibody-coupled beads. Remove insoluble material by centrifugation and be careful to remove it completely. | |
| 4 | CoPIT cannot distinguish background from specific interactors. | Too much background was present in the IPs or bait was not enriched enough. | Optimize experimental procedure. |
Anticipated results
CoPIT identifies and quantifies the interaction between a bait and interacting proteins in three steps that comprise experimentation, data analysis and comparative analysis. Although CoPIT was developed for facilitating analysis of membrane protein interactomes, it is applicable to other protein classes as well, such as transcription factors, which are typically underrepresented in proteomics experiments due to low abundance and binding to DNA.
In particular, we tested applicability of CoPIT to the transcription factor Tet2, to the peripheral membrane protein human glucoceramidase (GC) as well as to soluble proteins such as the kinase SMG1. IPs on these different protein classes led to good recovery of the bait, with an average of 2,644 SpCs for the transcription factor Tet2, 495 SpCs for SMG1 and 666 SpCs for GC per run (all acquired on an LTQ Orbitrap XL) and showed high reproducibility of CoPIT with small standard deviations in the recovery of the bait (Figure 2E). It should be noted, that the yield of an IP varies depending on the suitability of the antibody. For instance, recovery of CFTR could be further increased by using a different anti-CFTR antibody (monoclonal antibody 24-1, R&D Systems), which resulted in up to 3,441 spectral counts (316 peptides) from a single IP and MudPIT run, with 5 mg of protein from a whole cell lysate as starting amount and using an LTQ Orbitrap Elite.
A successful experiment should be further reflected in high sequence coverage of the bait and its interactors: For example, up to 60% CFTR could be covered in a single experiment, and up to 71% with additional non-tryptic digest. High sequence coverage in a single run could also be achieved for the other baits, with 68.8% for GC, 56.3% for SMG1, which is an exceptionally large kinase of 410 kD, and 81.6%, for Tet2 (Figure S1A). In addition, a successful experiment will also increase recovery and sequence coverage for interacting proteins (Figure S1B). If neither high spectral counts for the bait nor high sequence coverage are observed, each step of the experimental protocol should be carefully assessed for mistakes, and the suitability of the antibody for the IP should be confirmed. Usually the bait is within the ten proteins recovered with the highest spectral counts in an experiment. Additionally, using MudPIT instead of a single reverse phase approach will increase the recovery of peptides, in particular of hydrophobic ones (Figure S2).
The success of the second workflow of CoPIT critically builds on the quality of the results obtained in the first step. To achieve sufficient discriminative power to differentiate specific from non-specific interactors, a broad coverage of the interactome as well as of background proteins is required, and thus it heavily depends on a robust performance of the liquid chromatography as well as best detection sensitivity of the mass spectrometric set-up. Usually, best results are achieved using a MudPIT experimental setup rather than a single reverse phase set up because MudPIT greatly reduces ion suppression that is caused by co-eluting peptide ions.
The distribution of the relative number of interactors to background shown for CFTR CoPIT experiments in Figure 3C. The left ascending flank of the distribution reflects the ascending slope of non-specific binding proteins assuming a normal distribution that is centered close to 0. The left-most descending flank of the observed distribution of proteins represents weak, specific interactors that are also present in the background interactome and that are assumed to follow a second normal distribution. Strong interactors show up at the outmost right tail of the descending flank of tis second normal distribution. In addition to a good P-value (typically P ≥ 0.9), the interacting protein should be detected in at least two independent biological replicates of the same experimental condition to be considered an interactor.
We tested this approach by comparing the scores obtained for typical, putative background proteins like Keratins and antibody sequences and for established CFTR interactors like HSP90 and HSP70, which are also often detected in the general background and thus challenge any method used to discriminate interactor from background. Results show that background protein candidates were eliminated based on low confidence scores whereas known CFTR interactors including Hsp70 and Hsp90 were identified with high specificity (Figure 3D). This procedure allows to determine high-confidence interactors for each experimental condition which constitute a respective core interactome. In addition, an extended interactome can be defined, which additionally contains all medium confidence interactors that are enriched, but fail to pass the stringent criteria set for high-confidence interactors.
The third workflow in CoPIT is tailored to comparing and visualizing the results obtained. The comparison of individual experimental conditions is based on a counting statistics and associated errors because the way a mass spectrometer acquires spectra in data dependent mode can be described as a counting experiment. The comparison of interactomes also relies on having a large number of interactors identified. This contrasts with data analysis frameworks for co-immunoprecipitations tailored to experiments that identify only a small number of highly specific interactors across a range of baits.
A challenge in proteomic analysis of Co-IPs can be a high variability in absolute spectrum counts between individual experiments. This variability has been attributed in part to the limitations of data dependent acquisition by the mass spectrometer and in part to variation between biological experiments. In CoPIT individual immunoprecipitation experiments are therefore normalized to differences in the amount of whole cell lysate (e.g. starting material) and differences in the abundance of the bait protein to compensate for changes in expression of the bait protein upon, for example, a specific treatment. In addition, P-value cut offs for interactors should and can be adjusted according to the ratio of bait protein between two experimental conditions. For example, interactors of ΔF508 CFTR required 2-fold higher rp ec than wt CFTR (P ≥ 0.93), because about twice as much CFTR was recovered from ΔF508 CFTR immunoprecipitations than from wt CFTR immunoprecipitations.
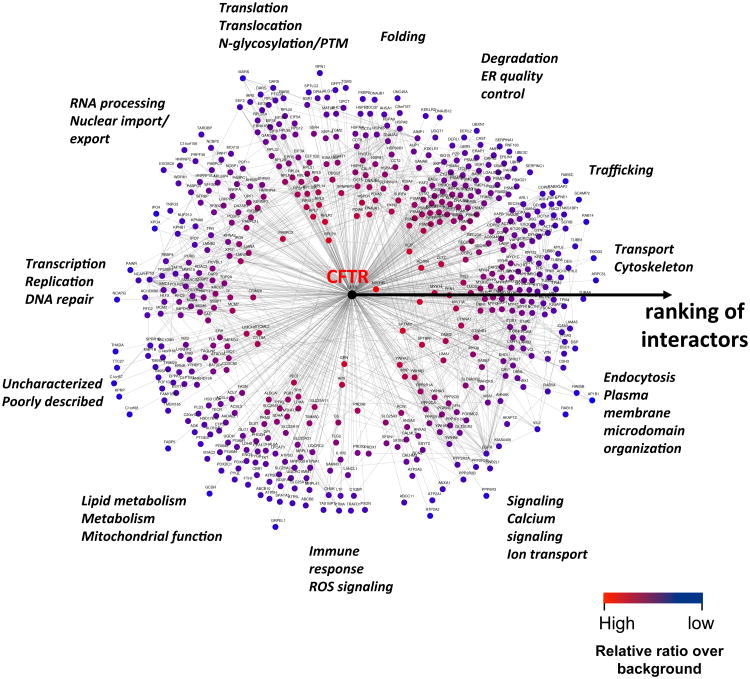
While flexible tools for network-representation are readily available (such as Cytoscape and Osprey), CoPIT uses a specialized network viewer named Radial Topology Viewer, which was based on Medusa 40. The quantitative relationship of the interacting protein with the bait is reflected in the length of individual edges creating an “interactome-radar”, which can help to quickly distinguish strong and weak interactions based on their distance to the bait, while grouping by ontological information allows easy identification of cellular processes a particular interactor or group of interactors is involved in (Figure 4).
Figure 4.
Schematic depicting a Radial Topology Viewer map of the ΔF508 CFTR interactome. Distance to CFTR in the center and node color as well as the color gradient of the circle around the interactome reflect the ranking of identified interactors. Interactors are further grouped into categories according to ontological information (for example “folding”).
Interpretation of Results
While the complexity of a specific protein interactome certainly depends on the protein of interest, a large but specific interactome may be rationalized by an estimation of the degrees of interaction. Based on the CFTR interactome, an estimated 20 to 25 1st degree interactors directly bind to CFTR at different stages of its life cycle. These interacting proteins reflect essentially different subcellular compartments to which ΔF508 CFTR localization can be tethered according to the specific steps or specific time points during its biogenesis and can be visualized by grouping interactors according to subcellular function using the Radial Topology Viewer. Accordingly, the connectivity between nodes is highest within the individual functional groups for the CFTR interactome map. Thus, in each specific subcellular compartment or time point ΔF508 CFTR appears to interact with a few primary interactors followed by secondary interacting proteins. Given that the average shortest distance from one protein to the next protein is 4.1 in the human proteome and the average number of interacting proteins ranges from 1.8 to 3.9 (depending on the study), 80 to 100 proteins are expected as 2nd degree interactors of the initial 20 to 25 1st degree interactors of CFTR. With CoPIT, we provide evidence for 3rd degree interactors, which we expect to be 320 to 400 proteins. Thus the total number of proteins that direct and indirectly interact with CFTR can be estimated to be roughly 420 to 525 proteins, which is in the range of the interactome size that was determined experimentally with CoPIT. This estimation also explains the larger size of the ΔF508 CFTR interactome relative to the wt CFTR interactome: Difficulties in translation, insertion into the ER, folding, enhanced degradation and altered trafficking of ΔF508 CFTR all contribute to an increased and more diverse interactome for ΔF508 CFTR.
Supplementary Material
Figure S1. A. Sequence coverage of SMG1, GC and Tet2 indicating identified peptides (green). B. Sequence coverage of Calnexin indicating identified peptides (green), transmembrane domain (orange) and cleaved signal peptide.
Figure S2. High salt concentrations are necessary for elution of hydrophobic CFTR peptides from SCX. The graph shows the distribution of Kyte-Dolittle (KD) values indicating hydrophobicity of identified ΔF508 CFTR peptides over the different MudPIT steps and corresponding salt concentrations. Error bars indicate mean with s.e.m.
Acknowledgments
We would like to thank Dr. Daniel Cociorva and Dr. Tao Xu for suggestions and comments on data analysis strategies, Yun Lei Tan from Dr. Jeffrey Kelly's laboratory for performing the GC-IPs and Dr. Nancy Huang from Dr. Anjana Rao's laboratory for performing the Tet2- IPs. This work is supported by NIH grants 5R01HL079442-08 (J.R.Y.), P01AG031097 (J.R.Y.), P41 RR011823 (J.R.Y), DK075295 (J.W.K), HHSN268201000035C (J.R.Y), and CFF mass spectrometry fellowship BALCH050X6 (S.P. and J.R.Y).
Footnotes
Author contributions: S.P. and C.B. developed experimental methods and performed experiments, data analysis and mass spectrometric measurements. C.B., A.B. and S.P. developed the statistical analysis. D.C. and C.B. developed the Radial Topology Viewer. J.R.Y, S.P. and C.B. designed the study and wrote the manuscript.
Author information: The authors declare no competing financial interests.
Literature
- 1.Gingras AC, Gstaiger M, Raught B, Aebersold R. Analysis of protein complexes using mass spectrometry. Nat Rev Mol Cell Biol. 2007;8:645–654. doi: 10.1038/nrm2208. [DOI] [PubMed] [Google Scholar]
- 2.Cravatt BF, Simon GM, Yates JR., 3rd The biological impact of mass-spectrometry-based proteomics. Nature. 2007;450:991–1000. doi: 10.1038/nature06525. [DOI] [PubMed] [Google Scholar]
- 3.Russ AP, Lampel S. The druggable genome: an update. Drug discovery today. 2005;10:1607–1610. doi: 10.1016/S1359-6446(05)03666-4. [DOI] [PubMed] [Google Scholar]
- 4.Hopkins AL, Groom CR. The druggable genome. Nature reviews. Drug discovery. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 5.Wallin E, von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 1998;7:1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Almen MS, Nordstrom KJ, Fredriksson R, Schioth HB. Mapping the human membrane proteome: a majority of the human membrane proteins can be classified according to function and evolutionary origin. BMC biology. 2009;7:50. doi: 10.1186/1741-7007-7-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blonder J, et al. Enrichment of integral membrane proteins for proteomic analysis using liquid chromatography-tandem mass spectrometry. J Proteome Res. 2002;1:351–360. doi: 10.1021/pr0255248. [DOI] [PubMed] [Google Scholar]
- 8.Howell KE, Palade GE. Hepatic Golgi fractions resolved into membrane and content subfractions. J Cell Biol. 1982;92:822–832. doi: 10.1083/jcb.92.3.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu CC, MacCoss MJ, Howell KE, Yates JR., 3rd A method for the comprehensive proteomic analysis of membrane proteins. Nat Biotechnol. 2003;21:532–538. doi: 10.1038/nbt819. [DOI] [PubMed] [Google Scholar]
- 10.Santoni V, Kieffer S, Desclaux D, Masson F, Rabilloud T. Membrane proteomics: use of additive main effects with multiplicative interaction model to classify plasma membrane proteins according to their solubility and electrophoretic properties. Electrophoresis. 2000;21:3329–3344. doi: 10.1002/1522-2683(20001001)21:16<3329::AID-ELPS3329>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 11.Mitra SK, Gantt JA, Ruby JF, Clouse SD, Goshe MB. Membrane proteomic analysis of Arabidopsis thaliana using alternative solubilization techniques. J Proteome Res. 2007;6:1933–1950. doi: 10.1021/pr060525b. [DOI] [PubMed] [Google Scholar]
- 12.Macher BA, Yen TY. Proteins at membrane surfaces-a review of approaches. MolBiosyst. 2007;3:705–713. doi: 10.1039/b708581h. [DOI] [PubMed] [Google Scholar]
- 13.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 14.Palade G. Intracellular aspects of the process of protein synthesis. Science. 1975;189:347–358. doi: 10.1126/science.1096303. [DOI] [PubMed] [Google Scholar]
- 15.Wickner W, Schekman R. Protein translocation across biological membranes. Science. 2005;310:1452–1456. doi: 10.1126/science.1113752. [DOI] [PubMed] [Google Scholar]
- 16.Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- 17.Moore I, Murphy A. Validating the location of fluorescent protein fusions in the endomembrane system. Plant Cell. 2009;21:1632–1636. doi: 10.1105/tpc.109.068668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sastry MS, Zhou W, Baneyx F. Integrity of N- and C-termini is important for E. coli Hsp31 chaperone activity. Protein Sci. 2009;18:1439–1447. doi: 10.1002/pro.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi H, et al. SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat Methods. 2011;8:70–73. doi: 10.1038/nmeth.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Breitkreutz A, et al. A global protein kinase and phosphatase interaction network in yeast. Science. 2010;328:1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jager S, et al. Global landscape of HIV-human protein complexes. Nature. 2012;481:365–370. doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pankow S, Bamberger C, Calzolari D, Bamberger A, Yates JR. Characterization of membrane protein interactomes by Co-interacting Protein Identification Technology (CoPIT) Protocol Exchange. 2015 doi: 10.1038/nprot.2016.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pankow S, et al. F508 CFTR interactome remodelling promotes rescue of cystic fibrosis. Nature. 2015;528:510–516. doi: 10.1038/nature15729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zielenski J, et al. Genomic DNA sequence of the cystic fibrosis transmembrane conductance regulator (CFTR) gene. Genomics. 1991;10:214–228. doi: 10.1016/0888-7543(91)90503-7. [DOI] [PubMed] [Google Scholar]
- 26.Rommens JM, et al. Identification of the cystic fibrosis gene: chromosome walking and jumping. Science. 1989;245:1059–1065. doi: 10.1126/science.2772657. [DOI] [PubMed] [Google Scholar]
- 27.Sheppard DN, Welsh MJ. Structure and function of the CFTR chloride channel. Physiol Rev. 1999;79:S23–45. doi: 10.1152/physrev.1999.79.1.S23. [DOI] [PubMed] [Google Scholar]
- 28.Collins FS. Cystic fibrosis: molecular biology and therapeutic implications. Science. 1992;256:774–779. doi: 10.1126/science.1375392. [DOI] [PubMed] [Google Scholar]
- 29.Jensen TJ, et al. Multiple proteolytic systems, including the proteasome, contribute to CFTR processing. Cell. 1995;83:129–135. doi: 10.1016/0092-8674(95)90241-4. [DOI] [PubMed] [Google Scholar]
- 30.Lukacs GL, et al. The delta F508 mutation decreases the stability of cystic fibrosis transmembrane conductance regulator in the plasma membrane. Determination of functional half-lives on transfected cells. J Biol Chem. 1993;268:21592–21598. [PubMed] [Google Scholar]
- 31.Abelin JG, et al. Complementary IMAC enrichment methods for HLA-associated phosphopeptide identification by mass spectrometry. Nat Protoc. 2015;10:1308–1318. doi: 10.1038/nprot.2015.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schieltz DM, Washburn MP, Hays LG. Analysis of Complex Protein Mixtures Using Nano-LC Coupled to MS/MS. CSH protocol. 2006:2006. doi: 10.1101/pdb.prot4553. [DOI] [PubMed] [Google Scholar]
- 33.Wolters DA, Washburn MP, Yates JR., 3rd An automated multidimensional protein identification technology for shotgun proteomics. Anal Chem. 2001;73:5683–5690. doi: 10.1021/ac010617e. [DOI] [PubMed] [Google Scholar]
- 34.Schieltz DM, Washburn MP. Analysis of Complex Protein Mixtures Using Multidimensional Protein Identification Technology (MuDPIT) CSH protocols. 2006;2006 doi: 10.1101/pdb.prot4555. [DOI] [PubMed] [Google Scholar]
- 35.Li XJ, Zhang H, Ranish JA, Aebersold R. Automated statistical analysis of protein abundance ratios from data generated by stable-isotope dilution and tandem mass spectrometry. Anal Chem. 2003;75:6648–6657. doi: 10.1021/ac034633i. [DOI] [PubMed] [Google Scholar]
- 36.Ranish JA, et al. Identification of TFB5, a new component of general transcription and DNA repair factor IIH. Nat Genet. 2004;36:707–713. doi: 10.1038/ng1385. [DOI] [PubMed] [Google Scholar]
- 37.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Montojo J, et al. GeneMANIA Cytoscape plugin: fast gene function predictions on the desktop. Bioinformatics. 2010;26:2927–2928. doi: 10.1093/bioinformatics/btq562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morris JH, et al. Affinity purification-mass spectrometry and network analysis to understand protein-protein interactions. Nat Protoc. 2014;9:2539–2554. doi: 10.1038/nprot.2014.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pavlopoulos GA, Hooper SD, Sifrim A, Schneider R, Aerts J. Medusa: A tool for exploring and clustering biological networks. BMC research notes. 2011;4:384. doi: 10.1186/1756-0500-4-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X, et al. Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell. 2006;127:803–815. doi: 10.1016/j.cell.2006.09.043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. A. Sequence coverage of SMG1, GC and Tet2 indicating identified peptides (green). B. Sequence coverage of Calnexin indicating identified peptides (green), transmembrane domain (orange) and cleaved signal peptide.
Figure S2. High salt concentrations are necessary for elution of hydrophobic CFTR peptides from SCX. The graph shows the distribution of Kyte-Dolittle (KD) values indicating hydrophobicity of identified ΔF508 CFTR peptides over the different MudPIT steps and corresponding salt concentrations. Error bars indicate mean with s.e.m.