Abstract
Nanomedicines have significant potential for cancer treatment. Although the majority of nanomedicines currently tested in clinical trials utilize simple, biocompatible liposome-based nanocarriers, their widespread use is limited by non-specificity and low target site concentration and thus, do not provide a substantial clinical advantage over conventional, systemic chemotherapy. In the past 20 years, we have identified specific receptors expressed on the surfaces of tumor endothelial and perivascular cells, tumor cells, the extracellular matrix and stromal cells using combinatorial peptide libraries displayed on bacteriophage. These studies corroborate the notion that unique receptor proteins such as IL-11Rα, GRP78, EphA5, among others, are differentially overexpressed in tumors and present opportunities to deliver tumor-specific therapeutic drugs. By using peptides that bind to tumor-specific cell-surface receptors, therapeutic agents such as apoptotic peptides, suicide genes, imaging dyes or chemotherapeutics can be precisely and systemically delivered to reduce tumor growth in vivo, without harming healthy cells. Given the clinical applicability of peptide-based therapeutics, targeted delivery of nanocarriers loaded with therapeutic cargos seems plausible. We propose a modular design of a functionalized protocell in which a tumor-targeting moiety, such as a peptide or recombinant human antibody single chain variable fragment (scFv), is conjugated to a lipid bilayer surrounding a silica-based nanocarrier core containing a protected therapeutic cargo. The functionalized protocell can be tailored to a specific cancer subtype and treatment regimen by exchanging the tumor-targeting moiety and/or therapeutic cargo or used in combination to create unique, theranostic agents. In this review, we summarize the identification of tumor-specific receptors through combinatorial phage display technology and the use of antibody display selection to identify recombinant human scFvs against these tumor-specific receptors. We compare the characteristics of different types of simple and complex nanocarriers, and discuss potential types of therapeutic cargos and conjugation strategies. The modular design of functionalized protocells may improve the efficacy and safety of nanomedicines for future cancer therapy.
Graphical abstract
The modular functionalized protocell is targeted to tumors via peptide ligands or recombinant human single chain variable fragments and contains imaging or therapeutic agents embedded in the mesoporous silica core.
1. Introduction
Limitations of conventional cancer drug efficacy include insolubility, systemic toxicity and drug resistance compounded by debilitating side effects such as nausea, fatigue, neuropathy, and organ failure. An effective solution to circumvent these limitations is to deliver cancer drugs within biocompatible nanocarriers. Simple nanocarriers span diverse materials such as magnetic or colloidal metals, carbon-based structures, silica, liposomes or polymeric formulations. These materials differ in size, shape, loading capacity, payload release, stability, retention and clearance from the body, which impose further restrictions on their efficacy as cancer therapeutics. For example, nanocarrier size is a critical determining parameter since particle sizes <5 nm are cleared in the urine [1] although particles up to 50 nm have been detected as well, and nanoparticles >100 nm are cleared by the mononuclear phagocyte system (MPS), respectively. Ideally, an optimally loaded nanocarrier would be stable in the circulation to protect and deliver its therapeutic cargo to the target site, have good penetrance and retention within the target site so that measured cargo release occurs within a therapeutic window, and ultimately be organically cleared to prevent toxicity from long-term accumulation [2]. By combining features from simple nanocarriers, complex nanocarriers have improved biocharacteristics so that delivery of cancer therapeutics is clinically efficacious.
Although nanocarrier technology has improved, their lack of target specificity limits their widespread use. In solid tumors however, large fenestrations at endothelial cell borders and numerous, loose pericyte attachments are characteristic of rapidly growing tumor blood vessels that allow nanocarriers to passively exit the circulation within tumors and accumulate non-specifically [3–5]. This phenomenon is referred to as the enhanced permeability and retention (EPR) effect [6, 7]. Nevertheless, the EPR effect does not significantly increase payload concentrations at the target site and in fact, increased circulation times dissipate accumulation [8]. So, how could nanocarrier targeting and retention be improved for efficacious tumor treatment?
Since 1996, we and others have used, modified and adapted in vivo and in vitro phage display to identify ligand-receptor or scFv-epitope pairs as a means to specifically deliver a covalently linked apoptotic peptide, chemotherapeutic drug, reporter or suicide gene or imaging agents directly to tumors by intravenous administration [9–30]. Unlike other targeting moieties, peptides identified by in vivo phage display bind only to physiologically accessible receptors and, depending on the selection constraints, can enrich for targeting moieties that are internalized into cells subsequent to ligand binding. Thus, functional selection of targeting peptides embedded within the experimental design circumvents issues such as the EPR effect and non-specific uptake and obviates the need to reassess internalization of tumor-targeted therapeutics during downstream drug development. Additionally, depending on receptor location, i.e., tumor vs. tumor endothelial cells, internalization of nanomedicines will minimize or maximize, respectively, their distribution within the tumor via the bystander effect [31]. Off-target effects are minimized by using targeted liposomes loaded with doxorubicin to treat neuroblastoma [32–35]. Targeting liposomal doxorubicin to cultured human breast cancer or pancreatic adenocarcinoma cells was improved by inserting different targeting peptides purified as fusion proteins of the bacteriophage pVIII major coat proteins [36]. Consequently, one could envision a modular design of a targeted, stable complex nanocarrier consisting of a peptide ligand or monoclonal antibody targeting moiety conjugated to the lipid bilayer coated mesoporous silica nanocarrier, termed a functionalized protocell, which can specifically deliver a protected therapeutic cargo intravenously or locally by peritumor injection or inhalation. The term protocell (also known as a protobiont) is utilized in evolutionary biology to describe a self-organized spherical collection of lipids proposed as a stepping-stone to the creation of life. In the context of nanomedicine (and throughout this review) we use the term protocell to refer to a cell-like nanocarrier composed of a high surface area mesoporous silica nanoparticle core enveloped within a supported lipid bilayer [37–39]. In this construct, the core can be loaded with high concentrations of disparate cargos. The lipid bilayer serves to seal and protect the cargo and provides a biocompatible interface that can be conjugated with polymers to enhance stability and peptides or antibodies to direct specific targeting and intracellular trafficking.
The modular design of functionalized protocells will permit the targeting moiety to be exchanged depending on the tumor of interest. For instance, the targeting moiety can be a peptide or antibody-like moiety such as a single chain variable fragment (scFv) that binds to overexpressed receptor proteins such as interleukin-11 receptor alpha (IL-11Rα) [23, 40–42], or the 78-kDa glucose-regulated protein (GRP78) [43–47] in prostate or breast tumors. EphA5 would be an appropriate surface receptor to target in non-small cell lung tumors due to its high expression [18, 29]. scFvs that exhibit distinct receptor affinities or bind to different epitopes can be used as the binding moiety to elicit a specific therapeutic effect. For example, scFvs can be used to inhibit or modulate receptor function or act synergistically with the delivered therapeutic cargo [48]. Alternatively, binding of the functionalized protocell can elicit receptor internalization for cargo release within the cell. Table 1 lists targeting peptide ligands that have been identified by in vivo and/or in vitro phage display, whereby binding to their target receptor elicits receptor-mediated internalization.
Table 1.
Peptide ligand motifs and corresponding tumor receptors
| Peptide Ligand Motif | Tumor Receptor | In Vitro | In Vivo | Reference |
|---|---|---|---|---|
| RGD-4C, VVISYSMPD | αvβ3 & αvβ5 | Y | Y | [9, 340] |
| TAASGVRSMH, LTLRWVGLMS | NG2 | Y | Y | [341] |
| CTTHWGFTLC | MMP-2, MMP-9 | Y | Y | [342] |
| NGR | APN (CD13) | Y | Y | [74–76, 168, 172] |
| RPL, CTQYAMHLC, CSQYSFNWC, CGFYWLRSC | VEGFR-1, Neuropilin-1 | Y | Y | [78, 80, 88, 343–345] |
| CGRRAGGSC | IL-11Rα | Y | Y | [23, 40–42] |
| CNVSDKSC, WIFPWIQL, WDLAWMFRLPVG, SNTRVAP | GRP78 | Y | Y | [43, 45, 46, 101] |
| CPRECESIC | APA | Y | Y | [84] |
| CKGGRAKDC | Prohibitin (receptor identified in normal adipose tissue) | Y | Y | [19, 346, 347] |
| CVPELGHEC | HSP90 | Y | N | [348] |
| GFE | MDP (metastasis) | Y | Y | [349] |
| CVRAC | EGFR | Y | N | [77] |
| WXDDG | TWEAK | Y | N | [350] |
| CLFMRLAWC, HDERMFLCKS | MUC18 | Y | Y | [351] |
| YRCTLNSPFFWEDMTHECHA | CRKL | Y | Y | [86] |
| CRTIGPSVC | Transferrin | Y | Y | [28] |
| GNFRYLAPP | DNA-PKcs | Y | N | [352] |
| CWKLGGGPC | Human leukocyte proteinase-3 | Y | N | [27] |
| CSGIGSGGC, CRFESSGGC | EphA5 | Y | Y | [18, 29] |
| GLTFKSL | PPP2R1A | Y | Y | [83] |
| CTFAGSSC | Fetulin-A | Y | N | [85] |
| HSYWLRS, YKHSHSYWLRSGGGC | NPTX2/NPTXR | Y | N | [353] |
| YKWYYRGAA-pen | RPL29 | Y | N | [354] |
| CGIYRLRSC | α6 integrin & E-cadherin complex | Y | N | [355] |
Other examples of targeting peptides include tumor-targeting peptides derived from luteinizing hormone/chorionic gonadotropin conjugated to membrane-disrupting lytic peptides to effectively inhibit human breast and prostate xenograft tumor growth and metastases [49–52]. In addition to peptides or antibodies, aptamers, short, single stranded RNA or DNA oligonucleotides have been developed for targeted cancer therapy to treat a variety of tumors in clinical trials by delivering intercalated chemotherapeutics or conjugated directly to nanocarriers containing therapeutic cargos (reviewed in [53, 54]). Combinations of aptamers containing intercalated doxorubicin or a NF-κB decoy were effectively used in vitro to inhibit growth of cultured pancreatic tumor cells by inhibiting nuclear translocation of NF-κB [55].
Similar to chemotherapeutic drugs, targeted therapies are designed to inhibit tumor growth via a dynamic, progressive process. This ensures that toxic cellular byproducts are within physiological limits that can be effectively cleared. Due to the leakiness of tumor blood vessels, there is no doubt that targeted nanocarriers will accumulate in tumors partly due to the EPR effect. Nevertheless, once passive accumulation of targeted nanoparticles occurs, specific binding to tumor-specific receptors, internalization and retention in cells within the tumor microenvironment will ensure effective cargo release and higher, localized therapeutic indices with decreased systemic, collateral damage. Targeted delivery of functionalized protocells may also circumvent problems associated with “binding site inhibition” as this model does not take into account variability in receptor concentrations or turnover at the tumor site [56]. For instance, unless locally administered, intravenous infusion of targeted nanomedicines will be diluted in the circulation so that target site accumulation occurs over time. Unlike passive accumulation, targeted therapies, by definition, can be administered at lower doses due to their increased, effective concentration at the target site as confirmed both experimentally and by modeling and simulation [37, 38, 57]. Furthermore, functionalized protocells have a high cargo loading capacity, so that saturating receptor concentrations are avoided. Finally, the concentration of the targeting moiety can be modulated by varying the composition of functional groups available for conjugation in the protocell lipid bilayer. Given these considerations, selective targeting by functionalized protocells can successfully circumvent binding site inhibition.
Below, we will discuss in detail the advantages of the protocell over other types of nanocarriers. In a similar fashion, a variety of payload cargos or payload combinations will be discussed including non-invasive imaging agents and/or therapeutics, alone or in combination depending on the application. Thus, we envision the modular design of a functionalized protocell may be tailored for a particular tumor type or tumor subtype whose therapeutic payload can be personalized to accommodate a prescribed clinical treatment plan. The objective of this review is to 1) describe how targeting peptides and scFvs are selected using in vivo and in vitro phage or antibody display and examine their clinical utility, 2) compare a variety of simple and complex nanocarriers and types of therapeutic cargoes and 3) review various conjugation strategies to functionalize nanocarriers and optimize therapeutic efficacy. Ultimately, optimization and personalization of targeted nanomedicines developed as cancer therapies will have to be empirically determined.
2. Targeting Strategies
2.1. Peptide phage display
In vitro phage display was originally reported as a novel method to clone genes by using a known antibody to probe phage clones that display peptide epitopes as a fusion protein of the pIII minor coat protein [58]. These studies showed that the correct peptide epitope was enriched by a thousand-fold after a single selection round. Since this initial study, phage display of random peptide libraries displayed on pIII has been used as an unbiased in vivo screening tool [59, 60] to identify numerous ligand-receptor pairs within the physiological context of normal brain, kidney, adipose tissue, lung, skin, pancreas, retina, intestine, uterus, prostate, and adrenal glands [24, 61, 62] and in disease tissues, both in humans and animal models [19, 27, 40, 61, 63–66]. Moreover, in vivo phage display combined with fluorescence laser pressure catapult microdissection revealed endothelial receptors are differentially expressed within specialized sub-cellular regions, such as pancreatic islets, and can be overexpressed in pancreatic islet tumors [67]. These results demonstrate receptors expressed by the vascular endothelia of normal or disease tissues have an inherent and distinctive molecular heterogeneity. This highlights the limitations of other methods to identify clinically relevant cell surface receptors by systematically profiling protein expression, as they do not take anatomical context into account despite the fact that some clinically important endothelial proteins are expressed in restricted locations or become accessible only under specific biologic, physiologically- or pathologically-induced, circumstances.
2.1.1. Selecting peptides by in vivo phage display
In a typical in vivo phage display experiment, a linear or circular peptide library with up to 109 diversity, expressed on the bacteriophage pIII minor coat protein, is injected intravenously so that circulating ligands can preferentially bind to physiologically accessible cell surface receptors (Fig. 1). The organ or tissue of interest is subsequently removed after a period of time and tissue-specific bound phage are recovered by bacterial infection so that the peptide coding sequences can be identified by DNA sequencing [40]. Alternatively, recovered phage from multiple tissues of interest are tagged using PCR-assisted bar-coding followed by high-throughput DNA sequencing [12, 20, 27].
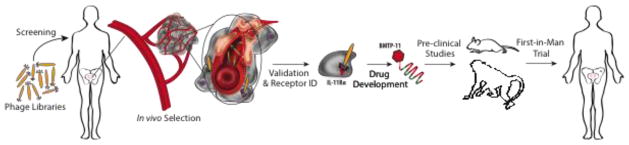
Fig. 1.
Drug development pipeline for BMTP-11. Development of a peptide-based therapeutic, BMTP-11, starting from in vivo phage display using a peptide combinatorial library in a terminal wean patient identified a prostate tumor-specific peptide ligand, which was followed by receptor identification and validation. Drug development of BMTP-11 included toxicological studies in mice and cynomolgus monkeys followed by a first-in-man phase 0 clinical trial, in which BMTP-11 localized and induced apoptosis of tumor cells at a secondary metastatic site [23].
After several iterative rounds of selection, peptide ligands are enriched in the tissue of interest, since each successive round selects for phage recovered from internalized, bound receptors. Bioinformatic analyses of recovered peptide sequences in the forward and reverse directions [68, 69] reveals enriched tripeptide motifs, which define specific protein-protein interactions [70, 71]. Enriched peptide sequences or consensus sequences are cross-referenced against the NIH National Center for Biotechnology Information (NCBI) protein database using the Basic Local Alignment Search Tool (BLAST) to search for putative protein “ligand” identities which in turn may identify their corresponding receptor. Candidate ligand:receptor pairs are verified using in vitro phage binding assays and by ELISA if the putative receptor and appropriate antibodies are commercially available. Binding of individual phage displaying a single peptide ligand is validated in vivo and novel receptor proteins are subsequently isolated by affinity chromatography using purified, synthetic peptide ligands [72].
Thus far, we showed that screening libraries of phage-displayed peptides by intravenous injection into mice, rats, swine, non-human primates or brain-dead patients selects for peptides that bind specifically to normal or diseased organs/injured tissues. Importantly, and unlike untargeted nanocarriers whose fate is affected by serum proteins [8], synthetic peptides bind to the same receptors in these tissues as peptides displayed on phage particles. Moreover, peptides displayed on phage are capable of binding to post-translationally modified receptors that are expressed on cell surfaces as molecular signatures intrinsic to the microenvironment [73–76]. Not only do peptide-displaying phage bind to physiologically accessible receptors, successive rounds of selection enriches for peptides that bind to cell-surface receptors and are internalized. Enrichment of phage in the target tissue increases from 3–35 fold compared to untargeted, control phage [60]. Since 1998, considerable progress has been made in the delivery of targeted peptidomimetic drugs or imaging agents by our group and others as well as the isolation of novel peptide ligands by improvements of in vivo phage display technology and bioinformatic analyses [9, 12, 17, 18, 20, 27–29, 43, 63, 77–82]. Indeed, tissue-specific and angiogenesis-related vascular ligand-receptor pairs have been identified and exploited for targeted delivery of cytotoxic drugs, proapoptotic peptides, fluorophores, or cytokines to tumors, which generally improves selectivity and/or therapeutic windows in preclinical animal models.
2.1.2. Applications of peptide targeting in cancer
The application of in vivo phage display technology is particularly suitable for identifying and exploiting unique vascular receptors in human diseases such as cancer, where tumor cell growth and proliferation are highly dependent upon robust tumor blood vessel growth despite their abnormal molecular signatures and structural morphologies. Due to the leakiness of tumor blood vessels, in vivo phage display has identified unique receptors expressed on tumor endothelial cells as well as on receptors expressed on stromal cells, the extracellular matrix, pericytes, lymphatic endothelial cells and tumor cells [29, 41, 42, 44, 45, 59, 76, 79, 83–85]. Moreover, angiogenic blood vessels acquire unique molecular signatures that can be exploited for specific, targeted delivery of therapeutic agents [9, 19, 22, 23, 28, 42, 43, 45, 86, 87]. Receptors on cultured tumor cells have also been identified using a modified in vitro phage display technique called BRASIL in which phage bound to receptors expressed on the surfaces of cultured cells are separated from unbound phage by centrifugation from a miscible organic phase into an aqueous phase [18, 88]. Analyses of peptide ligands recovered by the BRASIL method identified EphA5 as a putative receptor expressed on the surface of cultured human non-small lung tumor cells [18, 89]. EphA5 expression was subsequently verified as a physiologically accessible, overexpressed receptor in human lung cancer, and its expression correlates with radioinsensitivity. Treatment of lung cancer cells or human lung xenografts with an EphA5 monoclonal antibody (mAb) improved tumor sensitivity to irradiation and prolonged survival in tumor-bearing mice [29].
Phage display in a brain-dead human cancer patient revealed that peptide motifs localize non-randomly to different organs [40, 66]. One selected peptide motif, GRRAGGS, was identified from a prostate biopsy sample that exhibited sequence homology to interleukin 11 (IL-11). Our group as well as others confirmed IL-11 binds to its cognate receptor, IL-11Rα [41, 90, 91], which is overexpressed during tumor progression and metastases in a large cohort of prostate cancer patients [42]. Similar to prostate cancer, expression of IL-11 and IL-11Rα are significantly higher in breast cancer samples compared to healthy mammary tissue [92]. Moreover, IL-11 and IL-11Rα transcript levels are approximately 3-fold greater in node-positive tumor samples compared to node-negative tumor samples [92], indicating that their expression directly correlates with the clinical and pathologic progression of breast cancer. Taken together, the vascular accessibility of overexpressed IL-11Rα and its role in human prostate and breast cancer make it a clinically relevant therapeutic target.
Our studies with IL-11Ra expression in prostate cancer led to the design of a ligand-directed agent, Bone Metastasis Targeting Petidomimetic-11 (BMTP-11), which consists of the IL-11Rα binding peptide motif, CGRRAGGSC (Table 1), conjugated to the apoptotic peptide, D(KLAKLAK)2. D(KLAKLAK)2 is non-immunogenic and nontoxic outside cells but disrupts mitochondrial membranes when internalized [93, 94]. We validated the efficacy of BMTP-11 in pre-clinical models of prostate cancer, and in murine and human osteosarcomas [95]. Mice bearing DU-145, LNCaP prostate tumors or implanted with a patient-derived MDA-PCa-118b tumor, an osteoblastic, androgen receptor-independent prostate tumor [96], and treated with BMTP-11 had significantly smaller tumors compared to tumor-bearing control mice treated with untargeted D(KLAKLAK)2 [23]. Toxicology studies of BMTP-11 in cynomolgus monkeys showed good stability, linear accumulation over time and predictable metabolism [23]. A phase zero clinical trial testing BMTP-11 as an investigational new drug in castrate-resistant prostate cancer patients indicated that BMTP-11-induced apoptosis of secondary bone metastasis. These results illustrate how the IL-11Rα targeting peptide discovered by in vivo phage display in a human subject was translated into a tumor-specific, clinically relevant drug [23] (Fig. 1).
Unlike IL-11Rα expression in tumors, the identification and validation of GRP78 (reviewed in [44, 97]), in breast and prostate tumors was more circuitous due to its association with the unfolded protein response [98–100]. We and other groups showed that WDLAWMFRLPVG, WIFPWIQL and SNTRVAP-displaying phage (Table 1) bind specifically to GRP78 [43, 45, 101]. Expression of GRP78 in the endoplasmic reticulum (ER) or cell surface is induced by acidosis, hypoxia and imbalanced glucose metabolism; its expression serves as a sentinel of ER-related stress in various pathological conditions, including cancer [102, 103]. Anti-GRP78 antibodies were identified in serum from prostate cancer patients by in vitro phage display [46], and retrospective immunohistochemistry (IHC) studies showed that GRP78 expression predicts recurrence in prostate cancer patients [104] and poor survival in advanced breast cancer [102, 105]. Silencing GRP78 expression restored cancer cell sensitivity to chemotherapeutic regimens and established a functional role for GRP78 in cancer cell survival [102, 105]. GRP78 expression correlates with metastatic disease in inflammatory breast cancer, revealing a potential therapeutic target for a disease that currently lacks an effective treatment. mAbs against GRP78 show promise in pre-clinical studies and in early stage clinical trials, thus substantiating the development of anti-GRP78 based therapies [44, 106]. Similar to IL-11Rα, cell surface overexpression and the role of GRP78 in human breast and prostate cancer make it an ideal therapeutic target [45, 47].
2.2. Antibody display
In 1986, the FDA and EMA approved the first therapeutic antibody, the CD3 OKT3 mAb, to prevent organ rejection in kidney transplants. Since then, the clinical use of antibody products has grown steadily, with 38 antibody-based biotherapeutics as of May 2015 and more expected to be approved by the end of 2015 [107]. Antibody-mediated therapeutic interventions have been successful because of their high specificity and because they share the same structural features and catabolic pathways of endogenous circulating antibodies, thereby mitigating potential safety issues in drug development.
2.2.1. Current applications of tumor-targeting antibodies
Antibody based therapies exert anti-tumor effects through a wide variety of mechanisms. Antibody binding to a cell surface receptor is sufficient to trigger antibody-dependent cell-mediated cytotoxicity [108], the effective mechanism of action for rituximab, the anti-CD20 mAb used to treat non-Hodgkin’s lymphoma [109]. Bevacizumab, an anti-VEGF mAb used to treat breast, metastatic colon and rectal carcinoma, arrests tumor angiogenesis by sequestering soluble VEGF and inhibiting its binding to VEGFR-2 [110]. Antibodies that target different epitopes of the same molecule show a potentiated therapeutic effect. For example, binding of transtuzumab and pertuzumab to different domains of the receptor tyrosine kinase HER2 prevents its dimerization and results in enhanced antitumor activity [111, 112]. The development of HER2 antibody-drug conjugates (ADC, reviewed in [113]) represents a step toward personalized medicine. HER2 breast cancer patients who eventually develop resistance to trastuzumab or pertuzumab, possibly due to activation of compensatory mitogenic signaling pathways, can now be treated with the ADC, trastuzumab-DM1, that exploits the tumor-targeting capability of the HER2 mAb to deliver a microtubule-depolymerizing agent (DM1) with improved efficacy, pharmacokinetics and reduced toxicity [114].
More recently, antibodies that engage and activate the host immune system against melanoma cells have pioneered clinical immunotherapy treatment. Antibodies activate the immune system by targeting the T-cell surface receptors CTLA4 (ipilimumab) or PD-1 (nivolumab) and blocking negative regulators of T-cell activation. Combining these separate immunotherapies in a phase 1 trial of advanced stage disease resulted in tumor regression in some, but not all, patients [115]. The efficacy of antibody-based immune checkpoint therapy has been proven in a variety of cancers [116]. Other antibody-based therapies involve conjugating cytokines [117] or bacterial toxins [118] to an antibody component have shown varying degrees of efficacy in clinical trials.
In the following sections, we describe how antibodies that bind to cancer-specific proteins are selected by screening naïve antibody libraries. The design of new cancer therapeutics that utilize targeted antibodies, validated in vivo, to deliver therapeutic cargos in nanocarriers will be discussed, as well as exploiting synergistic combinations of antibodies with their delivered cargos as possible strategies for clinical applications.
2.2.2. Selecting targeting antibodies
In principle, antibodies can be produced from cultured cells [119], and can be engineered [120] or selected [121, 122] against any target protein to regulate its downstream effect. Nevertheless, for decades the hybridoma technique developed by Köhler and Milstein was the only reliable method to produce mAbs from splenocytes isolated from mice immunized with a target antigen. The murine origin of these mAbs however, elicited an immunogenic response in humans and made them unsuitable for therapeutic use. Recombinant antibody technology initially enabled progressive reduction of immunogenicity by producing human/mouse chimeric antibodies by engrafting the mouse-specific complementarity determining regions (CDR) onto a human antibody backbone (see [123] for a comprehensive review). Finally, generation of the genetically engineered Xenomouse [124] allowed producing target-specific human antibodies following antigen immunization.
Parallel paths to generate human antibodies were established through a combination of recombinant antibody and in vitro display technologies to select target-specific human recombinant mAbs (rhAbs) using a high-throughput approach. The concept of rhAb display selection is based on the exploration of large antibody-like diversity spaces (libraries) to obtain target-specific binders. To achieve this, the complexity of antibodies - a molecular complex of 4 polypeptide chains - is reduced to its essential target-binding regions such as the scFv [125], Fab [126] or nanobodies (camelid single variable domains) [127]. These variations of full-length rhAbs are fully capable of binding to target antigens, and are therefore called “antibody-like binders”. All types of antibody-like binders have been explored for antibody display and selection, and can eventually be engineered into full-length immunoglobulin (Ig)-like molecules (reviewed in [128]).
One of the most challenging aspects in antibody display technology is the creation of large, diverse antibody-like libraries so that ideally, virtually any antibody-like binder against any given target molecule can be found. As opposed to peptide libraries, where short random sequences can be generated using degenerate oligonucleotides, antibody libraries suffer from several structural constraints (chain complexity, intra-/inter-chain disulfide bond formation, proper folding of domains and hydrophobic surface interactions) [129], which makes the creation of large, functional libraries a daunting task. Nonetheless, several successful diverse antibody libraries have been produced from naïve human repertoires [25, 130, 131], restricted human antibody scaffolds with natural diversities [126, 132] or by designing synthetic diversities [133]. In general, antibody libraries are created by cloning however; approaches using site-specific recombination [134] facilitate the creation of larger libraries with less effort. Although phage display [135] and its variations (reviewed in [10]) have been the most popular display platform for recombinant antibody libraries to date, ribosome [136] or yeast display [137–139] have also been successfully used to select high-affinity antibody binders against desired target proteins.
By combining in vitro antibody phage display and antibody yeast display [13, 14], a pool of hundreds to thousands of specific antibody binders from a large naïve human library can be fine-tuned to select against a protein of interest. In these studies, pre-selection of a naïve antibody-like library on the desired protein using antibody phage display is followed by antibody yeast display in which limiting amounts of the protein in the 2nd selection phase enriches for high affinity antibody-like binders. We commonly have used in vitro antibody phage and yeast display in successive selection rounds to identify antibody binders to a tumor-specific receptor.
Once the pool of antibody-like binders is selected and enriched from a library, choosing an antibody for tumor targeting presents yet another challenge. Specificity is clearly the driving aspect, but factors such as high tumor interstitial pressure, which influence the distribution of the targeting antibody within the tumor need to be considered as well. For instance, suboptimal concentrations of the targeting antibody to some tumor regions may lead to ineffective treatment and instigate a potential source of resistant, mutant tumor cell populations. We have found that next generation sequencing can identify the best antibodies in a selected pool, providing the selection step is appropriately designed [140].
Affinity and size play another important role in antibody-based targeting strategies. Although tumor uptake of large molecules (full-length antibodies, nanoparticles) is mostly influenced by the EPR effect [7, 141], retention of smaller molecules (peptides, scFvs, Fabs, alternative binding scaffolds) at the tumor site is highly dependent on their binding kinetics [142–146]. Tumor targeting can be achieved either by using small antibody-like binders with very high affinities (pM to low nM) or large antibodies with relatively lower affinities (high nM range) and longer half-lives in the bloodstream. Multivalency (more than one antigen binding site per antibody or antibody-like molecule) is another parameter that favors retention of antibody-like binders or antibodies in tumors and increases their functional affinity [147–149].
Antibody penetration refers to the homogeneous distribution of antibodies within the tumor and is another key factor to consider in selecting solid tumor targeting antibodies for clinical application. Factors that retard tumor penetration are high affinity and internalization. Fujimori and colleagues introduced the concept of “binding site barrier” in which high affinity antigen/antibody interactions reduce the amount of free antibodies available to diffuse into the tumor interstitium, thus spatially limiting the therapeutic effect of the targeting antibody [56]. Additionally, Wittrup and colleagues extensively demonstrated that fast internalization rates and catabolism retard antibody penetration [150]. Despite these studies, internalization following receptor binding has been successfully exploited for delivery of ADCs (reviewed in Section 2.2.1). Furthermore, cell-internalizing antibodies exist for a variety of tumors, and are selected to bind to specifically identified antigens or, using an unbiased approach, to unknown cell surface receptors by screening cancer cells with antibody display libraries [151–153]. The evolution of recombinant antibody and display technologies enable the selection of antibody-like binders with desired properties that can overcome the binding kinetics and distribution hurdles described above.
2.2.3. Validation of antibody-like binders in vivo
Unlike in vivo peptide phage display, which has been used to screen diverse peptide libraries in terminal wean patients or animal models [19, 24, 27, 40, 67, 76, 84], in vivo antibody display has proven to be a more challenging task. To date, only Shukla, Krag and coworkers have shown some degree of success by injecting naïve antibody phage libraries in cancer patients [21, 26], although, the selected antibodies tend to be patient specific. Nevertheless, antibody-like binders to receptor targets identified by in vivo peptide phage display can be isolated by screening a naïve human scFv library using the purified receptor protein. Unpublished data from our group indicate that a combination of in vitro and in vivo antibody display yields tumor-specific antibodies when recombinant tumor receptors (identified by in vivo or in vitro peptide phage display) are used as selection targets. In these studies, an in vitro preselected, enriched antibody phage sub-library containing antibody clones that specifically bind to a known, overexpressed cell surface tumor-specific protein was injected into tumor-bearing mice. Several tumor-localizing rhAbs were recovered from the tumor after assessing their biodistribution using next generation sequencing [140, 154] and immunohistological analyses of tumor sections relative to control organs. Specific therapeutic characteristics such as receptor inhibition by direct or allosteric binding or conjugation to imaging dyes or therapeutic drugs can be evaluated in vivo so that tumor-specific receptors can be fully exploited for treatment by functionalized protocells.
3. Nanocarriers
The experimental design by which tumor-specific peptides and recombinant human scFvs are selected within the physiological setting is a significant improvement for targeted drug delivery. By using tumor-targeting peptides, we and others have successfully demonstrated in vivo tumor growth inhibition using tumor-targeting peptides to deliver: D(KLAKLAK)2 [23, 94], doxorubicin to treat tumor-bearing mice [9], TNF-α [11, 15, 16, 22, 30], and reporter or suicide genes [17, 28]. In this section, the advantages and disadvantages of noteworthy simple and complex nanocarriers will be examined including a discussion of different types of therapeutic cargos.
3.1. Simple nanocarriers
A wide variety of nanoparticles have been developed for the delivery of therapeutic cargos including magnetic and metallic nanoparticles, such as iron oxide or gold nanoparticles, carbon based structures, such as graphene sheets and carbon nanotubes, polymer nanoparticles, dendrimers, quantum dots, hydrogel-based delivery systems, liposomes and silica-based nanoparticles (Fig. 2). Each nanocarrier type has strengths and weaknesses, which can be exploited for specific applications. For example metallic nanoparticles act inherently as contrast agents for imaging while other nanocarriers would require the addition of an imaging agent. The advantages and disadvantages of nanocarriers for therapeutic delivery and repetitive dosing are summarized in Table 2.
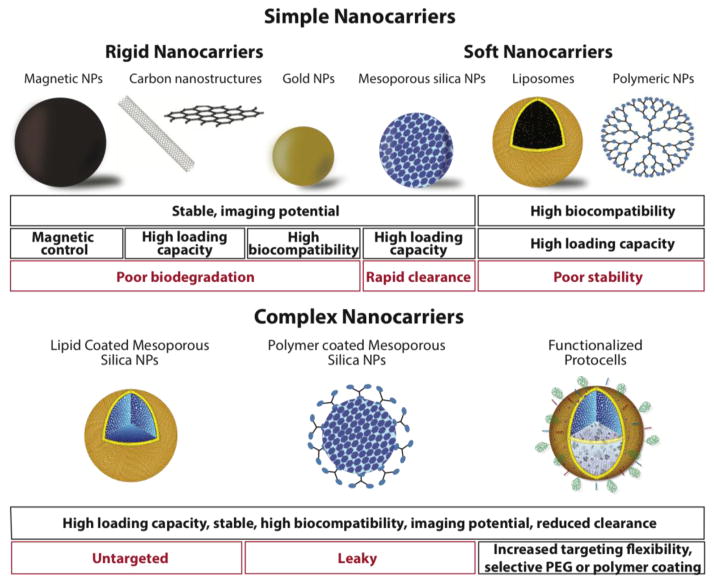
Fig. 2.
Comparison of simple vs. complex nanocarriers. Complex nanocarriers incorporate high loading capacity of a variety of cargos, greater stability and high biocompatibility. The lipid bilayer of the functionalized protocell may contain selective polymers, such as PEG (green) or cholesterol (purple diamonds) to improve membrane fluidity and overall charge. Additional functional moieties for conjugating targeting peptides (red) or fusogenic peptides to promote endosomal escape (blue) may be added. These modifications optimize protocell retention, increase drug concentrations at the tumor site and allow protocells to target different tumors [38].
Table 2.
Advantages and disadvantages of various nanocarrier platforms for therapeutic delivery
| Nanocarrier | Advantages | Disadvantages |
|---|---|---|
| Magnetic nanoparticles | Magnetic targeting [155–157] Imaging potential [158–161] Previous FDA approval [157, 162] Control of size and shape [155] |
No internal loading capacity [155] Requires surface modification to achieve stability [155] Poor biodegradation [156, 157, 163–165] |
| Gold (Au) nanoparticles | Highly biocompatible [157, 163, 166–168] Imaging potential [356] Potential for photothermal therapy [157] |
No internal loading capacity Poor biodegradation [157, 163, 166, 167] |
| Graphene Sheets | Very high loading capacity [159, 171] Stable in suspension [171] |
Dose dependent toxicity in vivo [171] Non-biodegradable [171] |
| Single-walled carbon nanotubes | Very high loading capacity [159] Low synthesis cost [157] |
Potential for pulmonary injury in vivo [157, 172] Immune suppression [172] Accumulation in organs [157, 172, 173] |
| Multi-walled carbon nanotubes | Very high loading capacity [159] Low synthesis cost [157] |
Accumulation in organs [157, 159, 357] Potential for pulmonary damage [157, 175] Immune suppression [157, 174] |
| Mesoporous silica nanoparticles (MSNP) | Biocompatible [189] Biodegradable [178, 188, 190, 191] Very high loading capacity [176, 183, 185, 186] |
Instability in physiological buffers [176] Poor circulation, rapid clearance [178, 192–194] |
| Liposomes | Biocompatible [33, 198–201] Ease of synthesis [33] Flexible formulation [33] FDA approved [195, 197, 198] Potential to add targeting moieties [33, 196, 202, 203] |
Poor universal carrier [204, 205] Invariant size and shape [200, 219, 220] Poorly controllable drug release characteristics [198–201] |
| Polymeric nanoparticles | Biocompatible [157, 159] Easy to manufacture [159] High loading capacity [157] Potential to add targeting moieties [213] |
Limited in vivo stability [159, 218] Poorly controllable drug release characteristics [198–201] Dose dependent toxicity [159, 218] |
Magnetic-based nanoparticles, most commonly iron oxide nanoparticles, have the theoretical advantage of precise therapeutic delivery to the region of interest using a magnet [155–157]. Additionally, metallic nanoparticles have the potential for multimodal, theranostic applications [158–161]. The theranostic potential of magnetic iron oxide particles is supported by FDA approval of a number of iron oxide nanoparticle imaging agents [157, 162]. The non-degradable nature of magnetic and metallic nanoparticles however limits repeated applications for therapeutic efficacy due to their accumulation [156, 163]. For example, iron based nanoparticles degrade slowly in biological systems such that even a single dose of iron oxide nanoparticles shows significant accumulation in the liver, spleen and lungs 90 days post-injection [164], and elimination of accumulated iron through the urine and feces occurs slowly [157, 165]. Finally, the solid nature of the magnetic nanoparticle limits the amount of therapeutic cargo that can be delivered. In addition to iron oxide nanoparticles, the other most commonly proposed metallic therapeutic nanoparticle is gold (Au).
Although Au nanoparticles have the potential to work as imaging agents or in photothermal therapies, are biocompatible, and show no significant toxicity, they accumulate, typically for months post-injection, particularly in the liver and spleen [157, 163, 166–168]. Although the accumulation of Au nanoparticles has not been associated with adverse effects, their lack of biodegradation is of concern for development as therapeutic delivery nanocarriers. Furthermore, the therapeutic loading potential of Au nanoparticles is constrained by their solid structure thereby limiting the therapeutic dose per particle.
To increase therapeutic loading, nanocarriers with very high loading potential such as carbon and silica framework structures including mesoporous silica nanoparticles (MSNP), carbon nanotubes and graphene sheets have been explored. The primary interest in the carbon-based structures is that their extremely large surface area has every atom exposed leading to the possibility of ultra-dense functionalization and therapeutic loading [159]. Despite these advantages, the major disadvantage of carbon-based structures is their limited biodegradability, which causes systemic buildup upon repeated use and potential pulmonary and immune toxicity [159, 169–175]. Similar to carbon-based nanocarriers, MSNPs are characterized by exceptionally high internal surface areas ranging from 500 to over 1200 m2/g due to periodic arrangements of uniformly sized mesopores (ranging in diameter from 2 to >20 nm) embedded within an amorphous silica framework [176]. Methods to synthesize MSNPs allows for a variety of sizes ranging from 25 nm to over 250 nm, and the MSNP shape can vary from prismatic to spherical to torroidal to rod-like [177–182]. Additionally, the MSNP pore diameter can range from 2 nm to over 20 nm and, by using silane coupling chemistry, the pore surface chemistry can be altered to accommodate high concentrations of disparate cargos [183–186]. Another major advantage of MSNPs is that amorphous silica is Generally Recognized As Safe (GRAS) by the FDA and recently, a silica-based nanoparticle was approved for diagnostic applications in a stage I human clinical trial [187]. Although amorphous silica is GRAS, biocompatibility testing of MSNPs has been variable. Occasionally, MSNPs test positive for toxicity, which is most likely due to incomplete removal of residual surfactant used to template the pores [188]. Confirmed removal of the surfactant prior to toxicity testing has shown that very large doses of MSNPs do not adversely affect survival in mice [189]. In addition to its low toxicity, the porous silica framework of MSNPs promotes a high rate of dissolution into soluble, non-toxic silicic acid species that are easily cleared from tested in vivo systems [178, 188, 190, 191]. Nonetheless, disadvantages of MSNPs include instability in physiological buffers and rapid clearance by the mononuclear phagocyte system (MPS) after injection [178, 190, 192–194], which can be mitigated by coating or encapsulation with polymers or lipids vide infra.
To avoid bioaccumulation and uptake by the MPS, highly biocompatible systems such as polymer and lipid based nanostructures have been employed. One of the most successful nanoparticle formulations is a liposomal nanoparticle-based drug delivery of which several FDA approved formulations exist [195–197]. The advantages of liposomes are their high biocompatibility, flexible formulation and easy synthesis [33, 198–201]. Moreover, liposomal formulations can be targeted specifically to tumors by incorporating antibodies such as the GAH, anti-EGFR or anti-HER2 mAbs, small molecules such as folate, transferrin or tumor-targeting peptides such as cyclic RGD [33, 196, 202, 203]. Unfortunately, the success of liposomal formulations varies with the encapsulated drug. Stable lipid formulations of some common chemotherapeutics have been difficult to determine, in particular to limit drug leakage, making liposomes a poor universal carrier [204, 205]. Nevertheless, liposomal encapsulation significantly reduces toxic off-target effects while retaining clinical efficacy for chemotherapeutics such as doxorubicin [206–209], cisplatin [210], camptothecin [211], irinotecan and floxuridine [212]. Polymeric based nanocarriers have also been developed and several novel formulations are currently undergoing clinical trials [196]. In one example, the small cell lung cancer targeting peptide, AHSGMYP, was used to deliver docetaxel loaded into a polylactic acid polymer nanocarrier [213]. Treatment of nude mice bearing small cell lung cancer tumors with AHSGMYP-conjugated docetaxel nanoparticles resulted in higher tumor docetaxel accumulation and survival compared to tumor-bearing mice treated with untargeted docetaxel nanoparticles. Antibodies have also been used to target liposomal drugs [214–216], an approach that requires the addition of lipid tails to the C-terminus of the antibody [215, 217] in order to incorporate into liposomes.
Similar to lipid formulations, many polymer-based nanoparticles are highly biocompatible and easy to produce however, they also suffer from limited stability in in vivo systems and dose-dependent toxicity [159, 218]. Furthermore, both liposomes and polymer-based nanocarriers are subject to invariant size and shape, poorly controllable release profiles and highly interdependent factors whereby altering one parameter, such as size, affects loading efficiency, charge and stability [200, 219, 220].
3.2. Complex nanocarriers
To address the specific limitations described above, complex nanocarriers combine multiple features of simple nanocarriers to exploit their strengths, as well as reduce or eliminate their limitations [37, 38, 194, 221–231] (Fig. 2). For example, both liposomes and polymer-based nanoparticles have good circulation half-lives and biocompatibility but limited stability and drug retention. These limitations can be improved by the inclusion of a stable nanoparticle core within polymeric or liposomal carriers. The combination of polymers or lipids with a stable core can be accomplished with magnetic nanoparticles, Au nanoparticles, carbon based nanocarriers and MSNPs. For example, polygalacturonic acid was used to coat magnetic cobalt spinel ferrite nanoparticles and then conjugated to an EphA2 binding peptide. The resultant complex nanocarrier was used to extract metastatic ovarian cancer cells from the abdominal cavity and circulation [232]. For delivery of therapeutic cargos, the nanoparticle core should have good biocompatibility and biodegradation to allow repeated dosing, a high surface area for high therapeutic loading and a tunable nature to permit loading with a variety of cargos. As described above, MSNPs are biodegradable, biocompatible, stable and porous. Moreover, their facile chemistry allows them to act as a tunable base to load a variety of cargos, as well as a number of covalent and non-covalent coatings. The simple addition of a polymer to the surface of MSNPs, such as PEG, PEG-PEI or NIPAM-co-MAA, greatly increases their circulation time and allows significant accumulation in tumors by utilizing the EPR effect [233, 234] and demonstrates significant delivery of therapeutic drugs in preclinical in vivo models of cancer [233–236]. In addition to polymers, their facile chemistry allows the addition of targeting agents, such as transferrin or folic acid, to their surface [189, 190, 237, 238]. Polymer-coated and -targeted MSNPs particles are currently being used to reassess chemotherapeutics such as selenocysteine, whose clinical efficacy was previously hindered by low selectivity, solubility and stability [189].
Increased flexibility and versatility were achieved by combining liposomes and MSNPs to create a “protocell” [37–39, 176, 224, 239]. Protocells are formed by the encapsulation of the MSNP core within a supported lipid bilayer (SLB), followed by the optional conjugation of polymers, such as PEG, and targeting or trafficking ligands to the surface of the SLB [37, 38, 177, 194, 222, 224, 240–245]. Protocells synergistically combine the advantages of liposomes, low inherent toxicity and immunogenicity, and long circulation times, with the advantages of MSNPs, stability and enormous capacity for multiple cargos and disparate cargo combinations [38, 176]. The adhesion energy between the MSNP and the lipid layer suppresses large-scale membrane bilayer fluctuations, resulting in reduced liposome instability and leakage, and the lipid bilayer permits retention of soluble cargos. Since its inception, the facile chemistry of the MSNP and variability of lipid bilayer formulations have led to a wide variety of protocell designs to include: lipid monolayer encapsulated hydrophobic MSNPs to load hydrophobic cargos [194, 240], covalent attachment of lipids to enable triggered cargo release [246], polymer additives to the lipid layer to enhance circulation times and the EPR effect [240, 245], and native cell membrane encapsulated particles to improve biocompatibility [240, 247] including lipid compositions that mimic red blood cells [246].
3.3. Therapeutic payloads
3.3.1. Imaging agents
The highly modifiable nature and large cargo capacity of MSNPs enables the inclusion of an imaging modality with therapeutics. For example, inclusion of a near infrared (NIR) [242] or fluorescent dyes such as fluoroscein isothiocyanate (FITC) [190] in MSNPs permits imaging in vivo for real time biodistribution analyses of novel nanocarriers after intravenous injection. Labeling MSNPs with NIR dyes enabled the analysis of their biodistribution over time, and the evaluation of various surface coatings such as polyethylene glycol (PEG) or PEG-polyethylenimine (PEI) on their biodistribution and clearance in vivo [234]. Inclusion of fluorescent dyes provides the added advantage of visualizing MSNP localization in tissues after animal dissection to confirm MSNP biodistribution studies [190, 248, 249]. Other encapsulated visualization agents include radioactive nuclides for positron emission tomography (PET) [248] or superparamagnetic iron oxide particles as an MRI contrast agent [250–252]. The structure and facile chemistry of the MSNP platform even allows multiple labels to be incorporated into the same particle, thereby enabling simultaneous confirmation of nanoparticle biodistribution in real time [251]. A similar approach [253] has been proposed to identify patients who may benefit from antibody targeted liposomes, whereby tumors are first tested for their ability to internalize fluorescent liposomes displaying specific antibodies. If internalized fluorescence is noted, liposomes bearing cytotoxic payloads will be similarly internalized, and consequently therapeutic.
In addition to their use in research, MSNPs have a potential benefit in the clinical setting as imaging agents. The ability to readily incorporate imaging agents such as NIR dyes or radioligands makes them ideal for image-guided removal of sentinel lymph nodes or small metastatic foci [254]. This approach was demonstrated using the Cornell dot or “C dot”, which was recently used in a melanoma first-in-human trial [187]. The C dot is an ultra small, 6–7 nm, silica nanoparticle containing Cy5 fluorescent molecules in the core particle, coated with PEG and further modified with radioactive iodine for PET and the cRGDY peptide for integrin-mediated targeting [255]. The dual functionality of C dots facilitated whole body PET imaging and fluorescence optical imaging during sentinel lymph node surgery [256]. In a similar fashion, MSNPs can be targeted using tumor-specific peptides or antibody-like binders, can contain imaging agents, radioligands or therapeutic agents, and can be easily coated with polymers or engineered into functionalized protocells to create biocompatible, targeted imaging agents and/or theranostics [37, 38].
3.3.2. Chemotoxins
The most common area of research for therapeutic MSNPs is chemotherapeutic delivery. Although most studies have focused on systemic delivery, an inhalation delivery study reported significant localization of a luteinizing hormone-releasing hormone peptide targeted-MSNP in a model of lung cancer compared to intravenous administration [257]. These studies demonstrated that localized delivery might be advantageous depending on the location of the tumor.
Many current studies involving systemic delivery of therapeutic MSNPs to tumors have focused on taking advantage of the EPR effect. For example, even bare uncoated MSNPs show therapeutic advantages over free drug in tumor xenografts [190]. Surface modification of MSNPs with PEG, or PEG and PEI enhanced EPR-based accumulation in tumors and resulted in increased drug efficacy and reduced toxicity [234, 235, 258]. In a similar fashion, modifications of protocell constructs have also been utilized to enhance drug delivery via the EPR effect [242, 244, 245]. The addition of the lipid bilayer allows delivery of a hydrophilic drug within the MSNP core and a hydrophobic drug within the lipid bilayer [242]. Polymers associated with the protocell can also exert a therapeutic effect. For instance, the inclusion of Pluronic 123 blocks the action of the breast cancer resistance protein pump and increases the efficacy of the chemotherapeutic cargo in a xenograft breast cancer model [245].
Although the EPR effect can be used by modified nanocarriers to improve drug delivery, recent studies indicate a trend towards adding tumor-targeting moieties to MSNP surfaces [190, 237, 238, 244, 257]. Tumor targeting ensured delivery for tumor types and in patients for which the EPR effect is insufficient for treatment [259, 260]. When directly compared to non-targeted MSNPs, targeted MSNPs showed enhanced therapeutic efficacy and decreased toxicity over non-targeted MSNPs [189, 190, 237, 238]. For example, the addition of the targeting moiety hyaluronan to protocells enhanced their delivery of docetaxel to a xenograft breast cancer model [244].
The use of polymer coated MSNPs has even been explored to treat non-cancer cells to improve vascular access of drugs in difficult cancer types such as pancreatic ductal adenocarcinoma (PDAC). PDAC elicits a dense stromal response that limits vascular access due to pericyte coverage of vascular fenestrations and is a contributing factor to chemotherapy resistance. Tumor-bearing mice treated with an initial delivery of MSNPs containing the TGF-β inhibitor, LY364947, to decrease vascular pericyte coverage, followed by treatment with liposomes containing gemcitabine showed reduced tumor burden compared to treatment with free drug or gemcitabine-loaded liposomes only [236].
Thus, MSNPs show promise for delivering a wide variety of chemotherapy agents with decreased toxicity [233, 235, 238, 244, 245], and may resurrect shelved drugs such as selenocystine [189], whose clinical use has been hindered by low stability or solubility. MSNPs also provide the capability for combinations of therapy agents to be delivered either individually [244] or within a single nanocarrier [242, 258]. New functionalities, such as pH-responsive nanovalves on multifunctional transferrin-modified MSNPs loaded with fluorescent molecules show effective cargo release in vitro and in vivo [237]. These technological advances show the versatility and tunablity of MSNPs in biological systems that capitalize their high loading capacities to ensure targeted, high chemotoxin therapeutic indices at the tumor site.
3.3.3. Reporter and/or suicide genes
Gene delivery to cells in biological systems has been explored using both viral and non-viral vectors. Despite its potential benefit, gene therapy is limited since modified adenoviral vectors may elicit an immune response and cell transduction may be inefficient. To construct a targeted adenoviral vector, we introduced an adeno-associated virus (AAV) bacteriophage chimeric vector termed AAVP [17]. We used the RGD-4C peptide (Table 1) to target AAVP containing the Herpes simplex virus thymidine kinase gene (HSVtk) to human DU145 prostate tumors in tumor-bearing mice. The RGD-4C AAVP-HSVtk vector successfully transduced tumor cells since expressed thymidine kinase enabled tumor imaging by positron emission tomography in the presence of the substrate [18F]-FEAU. Moreover, tumor growth was inhibited in the presence of the thymidine kinase substrate, ganciclovir, compared to tumor-bearing mice treated with untargeted AAVP-HSVtk. By utilizing a MSNP carrier, plasmid DNA is protected from enzymatic degradation in the biological environment [261, 262], facilitating entry of plasmid DNA into cells. In vitro delivery of green fluorescent protein (gfp) reporter plasmids and therapeutic plasmids by polymer-coated MSNPs has been reported [263–268]. Modifying nanocarriers containing genetic material with tumor-targeted peptides or scFvs that are internalized upon binding improves specificity and safety by ensuring only targeted cells will be transduced.
3.3.4. siRNA
Small interfering RNAs (siRNAs) can be targeted to any number of currently undruggable genes and, for instance, to amplified genes in cancer [159, 269]. Given the potential for siRNA to arrest growth of a variety of tumors [270, 271], a number of clinical trials are currently underway which utilize siRNA technology [159, 272]. Unprotected, naked siRNAs are subject to rapid degradation, on the order of 5 minutes, in the extracellular environment and can also lead to systemic inflammation, making the use of a carrier vehicle essential for effective siRNA delivery [159, 269, 272].
A variety of nanocarriers including MSNPs have been utilized to deliver siRNA in vitro and in vivo [159, 218, 273, 274]. The earliest studies utilized the MSNP surface and a protective polymer coating to encapsulate and protect siRNA [268, 275–277]. Surface association limited the amount of siRNA that could be delivered to a level similar to other solid nanocarriers, and the porous MSNP structure could be filled with other therapeutics for dual delivery. Later studies focused on loading siRNA into the MSNP pore structure to facilitate greater loading [278]. Therapeutic delivery of siRNA utilizing MSNPs with polymer coatings has been demonstrated in a variety of in vivo cancer models [250, 258, 279–283]. Co-delivery of therapeutic drugs with siRNA within a single MSNP has been shown both in vitro [277] and in vivo [258]. While the majority of the studies to date have utilized the EPR effect to deliver siRNA to the tumors, targeted delivery has also been demonstrated in vitro [37, 277] and in vivo [282]. We recently reported PCA3, a prostate cancer biomarker, is an antisense intronic long noncoding RNA that controls PRUNE2 levels via a unique regulatory mechanism by forming a PRUNE2/PCA3 double-stranded RNA that undergoes RNA editing [271]. These results established PCA3 as a dominant-negative oncogene and PRUNE2 as an unrecognized tumor suppressor in human prostate cancer. LNCaP prostate tumor-bearing mice treated with a stabilized anti-sense PCA3 siRNA resulted in significant tumor growth inhibition and concomitant decreased serum PSA concentrations compared to tumor-bearing mice injected with a scrambled siRNA control. These studies show that siRNA against PCA3 represents a promising effective nanocarrier cargo to inhibit PCA3 activity and treat prostate tumors.
Nanocarrier cargos can become entrapped in the endosome, and endosomal escape is particularly important for nucleic acid delivery. Endosomal escape can be achieved by adding fusogenic lipids, endosomal escape peptides, membrane disruptive polymers or lysosomotropic agents to the surface of the nanocarrier or with the cargo [284, 285]. For example, efficient delivery of siRNA or DNA during cell transfection is attained by incorporating fusogenic lipids, commonly cationic lipids, into liposomes. Other helper lipids, such as dioleoylphosphatidylethanolamine, are often added to promote fusion of liposomes to endosomal membranes and enhance nucleic acid release into the cytosol [284]. Liposomes containing cationic lipids have successfully delivered siRNA [286–289] and plasmid DNA [290–292] both in vitro and in vivo. In addition to their use in liposomes, fusogenic lipids can also be included into the lipid bilayer of protocells [39, 222, 241].
3.3.5 Aptamers
Although the majority of the aptamer research has focused on extracellular targets, the interactions of aptamers with intracellular proteins may prevent binding of a secondary molecule or alter enzymatic activity or gene expression and therefore, has potential for therapeutic applications. The use of targeted, internalized nanocarriers can transport aptamers into cells and permit clinical development of intracellular aptamers. Additionally, loading aptamers within nanocarriers would alleviate two other challenges, namely nuclease-mediated degradation and rapid renal clearance [293, 294]. Due to their small size, aptamers are rapidly degraded with half-lives as fast as 10–15 minutes [293]. To reduce renal clearance rates, aptamers are currently conjugated to high molecular weight molecules like PEG or cholesterol which increases their half-lives to as long as 12–24 hours [293]. Nevertheless, their protection within a nanocarrier would avoid conjugation to a secondary moiety, which may potentially alter target binding.
RNA based aptamers have been developed that can bind to T-cell factor 1, WT1 and β-catenin and alter their transcriptional activity in colon cancer and Wilms’ tumors [295–297]. Binding of an RNA aptamer to β-catenin inhibits the β-catenin dependent transcription of cyclin D1 and c-myc in colon cancer cells that results in cell cycle arrest and reduced tumor forming potential in colony formation assays [296]. DNA based aptamers have also been designed to interact with transcription factors, such as Amplified in Breast Cancer 1 which is a transcriptional activator and oncogene that is over expressed in a number of human cancers [298]. In addition to transcription factors, DNA based aptamers have been designed to functionally bind to the eIF4e eukaryotic translation initiation factor. Transfection of HeLa and HEK293 cells with eIF4e aptamers reduced cell proliferation that was concentration-dependent [299]. Non-faithful recombination by BCR-ABL1-mediated tyrosine phosphorylation of RAD51 at residue 315 (pY315) may play an important role in the accumulation of chromosomal aberrations and lead to chronic myelogenous leukemia relapse and progression. Peptide aptamers were developed to act as a decoy for RAD51(pY315), and treatment of BCR-ABL1–32Dcl3 cells inhibited non-faithful homologous recombination by approximately 2-fold [300]. Peptide aptamers have also been designed to modify the activity of heat shock proteins 27 (HSP27) and 70 (HSP70) [301, 302]. Over-expression of HSP27 and HSP70 has been associated with chemotherapy insensitivity and decreased tumor cell apoptosis. Treatment of mice bearing B16F10 subcutaneous melanoma with peptide aptamers increased chemosensitivity and reduced tumor burden in vivo [301].
4. Conjugation strategies to functionalize nanocarriers
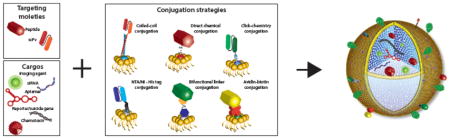
As discussed above, the efficacy of innovative nanocarriers to treat tumors can be significantly improved by targeting. Conjugation strategies that covalently or non-covalently link targeting moieties such as peptides, antibody scFvs or fluorescent molecules to nanocarrier surfaces are detailed in Fig. 3. The selection of an appropriate conjugation strategy is not trivial since the function of the targeting moiety has to be preserved and may be sensitive to alterations in secondary structure integrity during the conjugation process. Other considerations include proper orientation, and density per nanoparticle. As detailed below, direct conjugation strategies utilize existing surface functional groups and a single step process, whereas multistep conjugation strategies employ the addition of a new chemical entity to attach targeting moieties to functional groups on nanocarriers [303, 304].
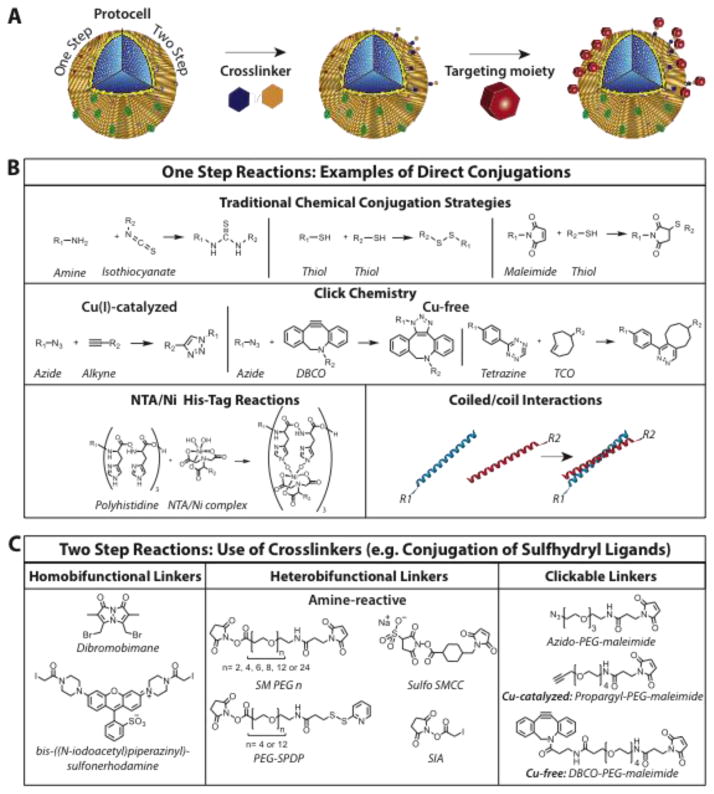
Fig. 3.
Covalent and non-covalent conjugation strategies for nanocarriers. (A) Schematic representation of conjugating targeting moieties to resident functional groups (red or grey spheres) on the phosopholipid head groups of the protocell outer lipid leaflet using a one- or two-step process. (B) Common single-step conjugation strategies including covalent traditional conjugation strategies, click chemistry, NTA/Ni2+-His6 or hydrophobic coiled/coil interactions. (C) Linking targeting moieties that contain sulhydryl groups utilize two-step reactions that require homo- or heterobifunctional crosslinkers or clickable linkers.
4.1. Direct chemical conjugation strategies
4.1.1. Basic conjugation chemistries
Direct conjugation of a targeting moiety or an imaging agent to a nanocarrier may require the addition of functional groups to the surface of the nanocarrier. Surface functional groups on MSNPs can be added by co-condensation during nanoparticle preparation or by post-modification of surface silanols after nanoparticle preparation. This same strategy can be employed for conjugation of targeting moieties to complex nanocarriers such as protocells. Specific lipid compositions can be selected or synthesized to allow direct conjugation of targeting moieties onto liposome or protocell surfaces [305]. For example, amine groups present on the MNSP surface, added either during synthesis or as a post-modification, have high reactivity with isothiocyanates and are used to attach fluorescent probes, such as FITC or rhodamine isothiocyanate (RITC), [306]. Adding thiol functional groups, can also be used to conjugate targeting moieties. A thiol group on the nanocarrier surface can be conjugated to a second thiol group present in the targeting moiety to form a disulfide bond.
Although this reaction is fast and efficient, the disulfide bond is unstable over time under physiological conditions [307]. Nevertheless, a disulfide bond was used to conjugate anti-My9 mAbs onto stealth liposomes containing the cationic ionophore monesin. These antibody-liposomes conjugates bound to CD33 expressed on human HL-60 promyelocytic leukemia cells and potentiated the in vitro cytotoxicity of the anti-My9 immunotoxin by a factor of 2070 [308]. The reduction of cysteine residues is a common method used to conjugate thiol groups on biomolecules with maleimide functional groups on nanocarrier surfaces. These reactions are selective, produce good yields and are stable in human serum for over a day even in the presence of a reducing agent. This type of conjugation strategy has been intensively explored to link anti-HER2 mAbs to liposomes for breast cancer therapy [309–311].
4.1.2. Click Chemistry
In the last decade, the emergence of click chemistry introduced a new set of reactions to conjugate targeting moieties to nanoparticles (reviewed in [312] for liposome conjugation). These new reactions are particularly popular because they are highly specific, efficient, physiologically stable, generate a single reaction product, produce high yields and can be performed under mild reaction conditions in aqueous solutions. Moreover, unreacted functional groups do not result in non-specific binding compared to the amine or thiol group linkages detailed above. Three major classes of reactions are employed: Copper catalyzed Azide-Alkyne Click Chemistry (CuAAC) which involves the reaction between an azide and an alkyne under Cu(I) [313], Strain-promoted Azide - Alkyne Click Chemistry reaction (SPAAC), commonly called Copper free click chemistry, which involves the same components but without a catalyst [314], and Tetrazine – trans-Cyclooctene (TCO) Ligation [315].
4.1.3. Histidine tag
The nitrilotriacetic acid (NTA)/Ni2+ complex was first used to affinity purify proteins containing a polyhistidine tag (His-Tag) of 6 histidine residues, and can comprise up to 14 residues, at either the N or C terminus with 100 fM affinity. Subsequently, the NTA/Ni2+ complex was used to link His6-Tag-biomolecules to nanocarriers since their dissociation constant is stronger than most antibody interactions [316] without non-specific binding [317]. In 1999, Hainfeld et al. presented one of the first applications of His6-Tag on gold nanoparticles in which the NTA/Ni2+ complex was introduced on the surface of gold nanoparticles prior to reaction with a His6-tagged protein [318].
4.1.4. Coiled/Coil
α-helical coiled-coil interactions are naturally occurring tertiary structures in a wide variety of proteins, whereby oligomerization events are energetically favored and are key to many biological functions [319]. The typical primary structure is based on the (a-b-c-d-e-f-g)n repeated amino acid motif, where positions a and d are typically occupied by hydrophobic residues that mediate coil oligomerization, while positions e and g mediate interhelical electrostatic interactions.
Inspired by nature, a variety of coiled-coil pairs were designed and exploited as biosensors, and as protein expression and purification tags. The E and K heterodimer coil pair [320] and its variants [321] are two parallel coils composed of 5 repeats of 7 amino acids. Their interaction affinity is as low as 60 pM and they have been successfully used in a variety of applications, including flow cytometry-based high-throughput screens [322] to display of GFP and its variants on phage particles [323]. Due to the stability, strength and specificity of the coiled/coil interaction, we envision the use of E/K coils as a straightforward and versatile conjugation strategy to functionalize protocells. In the proposed functionalized protocell, the protocell is functionalized with the K-coil and the targeting moiety is expressed, chemically linked or synthesized as a fusion protein product containing the E-coil.
4.2. Multi-step conjugation strategies
4.2.1. Avidin, NeutrAvidin and Streptavidin
The avidin-biotin complex is one of the oldest crosslinker conjugation techniques [324] and also represents one of the strongest non-covalent bonds, with a Kd ~ 10−15 M. The highly specific interaction between avidin and biotin is utilized to decorate avidin-containing liposomes with biotinylated antibodies. Other models have been developed based on the avidin/biotin complex [325] such as streptavidin and neutrAvidin. The characteristics of these proteins include lower molecular weight and the absence of carbohydrates, which decreases isoelectric points and in turn, non-specific binding. Although the avidin-biotin conjugation techniques are very easy to use and produce strong bonds, they have limited utility in in vivo targeting with nanocarriers due to their potential immunogenicity and this restricts their repeated use [326–328]. Research is currently underway to produce low immunogenicity variations of streptavidin to allow continual use of this strategy in therapeutics [326, 328].
4.2.2. Homobifunctional linkers
In the late seventies, homobifunctional crosslinkers such as glutaraldehyde and dimethyl suberimidate were used for amine-amine crosslinking [329, 330] to attach proteins or mannose ligands onto liposomes [331]. Currently, this type of crosslinking is not widely used due to possible homopolymerization during the reaction, which leads to aggregates [332]. Moreover, since a majority of biological ligands contain numerous amine groups, the use of homobifunctional linkers produces a variety of targeting moiety orientations, which may ultimately interfere with specific targeting [303].
Crosslinkers can be used to stabilize direct disulfide bonds formed between a targeting moiety and a variety of nanocarriers. Conjugation between two thiols can be performed by reagents carrying halogens such as bromobimane or bis-((N-iodoacetyl)piperazinyl)-sulfonerhodamine that undergo nucleophilic substitution with thiols. The use of these crosslinkers allows insertion of a fluorescent probe between two biological components [333, 334] but has not yet been used to label nanoparticles to our knowledge.
4.2.3. Heterobifunctional linkers
The use of heterobifunctional crosslinkers represents the future with regards to linking targeting moieties to nanoparticles. These crosslinkers facilitate conjugation reactions because they are available in a multitude of different functional groups. Additionally, they incorporate PEG chains that augment the solubility of nanocarriers in the physiological milieu, thereby increasing end product stability. Three classes of reactions are commonly used: amine to thiol, carboxylic acid to amine and click chemistry [335].
One of the most common uses of heterocrosslinkers is to link a carboxyl group to an amine. This reaction occurs in two stages, initially to create EDC (1-ethyl-3- (3-dimethylaminopropyl) carbodiimide) on the carboxyl group that is present on the nanoparticle surface to form an intermediate reactive species towards primary amines [336]. Recently, other conjugation methods introduced an N-hydroxysulfosuccinimide (NHS) linker in a second step such as sulfo-NHS to produce a more stable intermediate in order to improve reaction efficiency [337]. This approach was used to attach siRNA onto gold nanoparticles to silence the c-myc protooncogene in vitro and in vivo [338].
As described previously (see Section 4.1.1), cysteine residues at the C- or N-terminus in numerous biomolecules can be utilized as functional moieties to conjugate targeting peptides or antibodies. Since nanoparticles can be easily modified to incorporate amines on their surface, most heterobifunctional crosslinkers contain a NHS function on one end to bind to amines, and on the other end to maleimide to link to sulfhydryl groups. Heterobifunctional crosslinkers are water soluble, easy to use, and the reactions are specific and produce high yields. One example of this is the conjugation of the SP94 peptide to protocells containing a drug cocktail to human hepatoma 3B cells [38]. Importantly, these studies demonstrated specific delivery of SP94-protocells loaded with high concentration drug cocktails, and long-term stability with minimal non-specific binding and low toxicity to normal cells.
Finally, click chemistry can be employed with heterobifunctional crosslinkers as well, using the same reactions detailed earlier (see Section 4.1.2) namely, copper-catalyzed, copper-free, and tetrazine - trans-cyclooctene mediated ligation. Most commercially available crosslinkers include a PEG chain in their backbone and allow the insertion of a new functional group by click chemistry. The newly inserted functional group reacts with biomolecules through maleimide (Methyltetrazine-PEG4-maleimide, TCO-PEG3-maleimide), carboxylic acid (DBCO-PEG4-Amine) or even amine groups (Alkyne-PEG4-NHS Ester, Azido-PEG4-NHS Ester). Among recent examples, liposomes containing DBCO labeled lipids were used to conjugate to tetraacetylated N-azidoacetyl-d-mannosamine (Ac4ManNAz) ligands, which resulted in specific binding to A549 cells in vitro and to tumors in vivo [339].
5. Conclusion
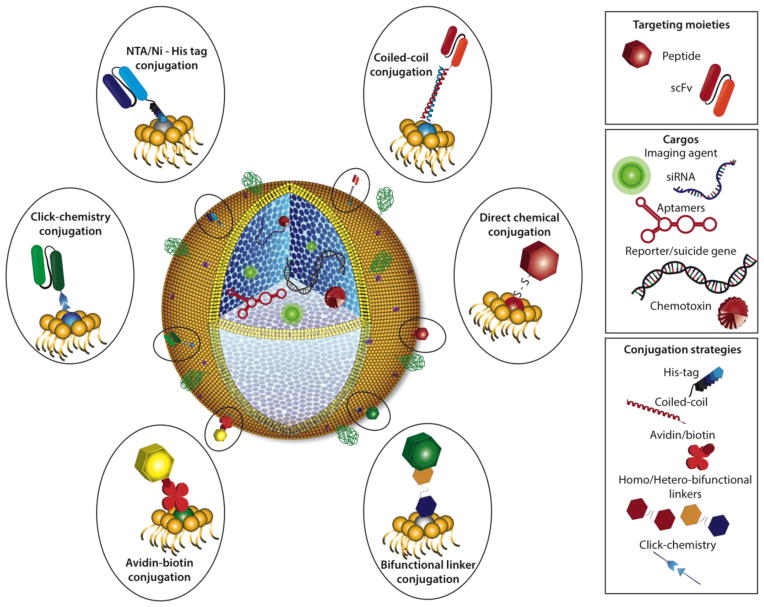
The technological advances in peptide phage display and antibody display, combined with the improved loading and biocompatibility of sophisticated nanocarriers, should facilitate the production of modular, targeted theranostic nanomedicines that specifically treat solid tumors in the near future (Fig. 4). Conjugation of tumor-specific peptide ligands or scFvs to the outer leaflet of the protocell lipid bilayer will depend on the available functional groups and may require the use of homo- or heterobifunctional crosslinkers. The orientation of the targeting moiety can be constrained by adding a His6-Tag, a short α-helical E-coil or a biotin group to the C-terminus for non-covalent association with NTA, a short α-helical K-coil or streptavidin, respectively, present on the surface of the protocell. Additionally, the composition of the protocell lipid bilayer may be adjusted to control the concentration of the targeting moiety, increase its circulation retention time and promote endosomal escape. Each protocell can be loaded with different types of imaging or therapeutic agents depending on the clinical application. Compared to conventional systemic chemotherapy, functionalized protocells present a safe alternative that simultaneously permits real-time, non-invasive imaging to monitor tumor growth inhibition. Taken together, these advantages provide greater clinical flexibility to personalize treatment regimens as dictated by treatment outcomes.
Fig. 4.
Schematic design of a functionalized protocell. Tumor-targeting peptide ligands or recombinant human scFvs can be conjugated directly or indirectly to functional groups on the outer leaflet of the protocell lipid bilayer. Functionalized protocells can be loaded with a wide variety of cargos such as chemotoxins, genes, siRNA, aptamers or imaging agents. The composition of the lipid bilayer can be modified to regulate the concentration of bound peptide ligands or scFvs to minimize binding site inhibition and optimize therapeutic indices, and may also incorporate different polymer coatings (purple dots) to improve circulation retention times.
Acknowledgments
This work was supported by grants from the National Cancer Institute (NCI) grant 2P30CA118100-11, Department of Defense W81XWH-09-1-0224 and the Gillson Longenbaugh Foundation (SD, ASD, WA, RP) and NIH 5U54DK093500–02 (ARMB). CJB and KSB acknowledge support from the NCI Alliance for Nanotechnology in Cancer grant NCI U01CA151792-01, the Leukemia and Lymphoma Society, Oncothyreon Inc, and Sandia National Labortories LDRD. CJB also acknowledges support from the DOE BES Materials Science and Engineering program, who supported fundamental materials science research related to protocells.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Choi HS, Liu W, Misra P, Tanaka E, Zimmer JP, Itty Ipe B, Bawendi MG, Frangioni JV. Renal clearance of quantum dots. Nat Biotechnol. 2007;25:1165–1170. doi: 10.1038/nbt1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nguyen T, Tekrony A, Yaehne K, Cramb DT. Designing a better theranostic nanocarrier for cancer applications. Nanomedicine (Lond) 2014;9:2371–2386. doi: 10.2217/nnm.14.110. [DOI] [PubMed] [Google Scholar]
- 3.Hashizume H, Baluk P, Morikawa S, McLean JW, Thurston G, Roberge S, Jain RK, McDonald DM. Openings between defective endothelial cells explain tumor vessel leakiness. The American journal of pathology. 2000;156:1363–1380. doi: 10.1016/S0002-9440(10)65006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ozawa MG, Yao VJ, Chanthery YH, Troncoso P, Uemura A, Varner AS, Kasman IM, Pasqualini R, Arap W, McDonald DM. Angiogenesis with pericyte abnormalities in a transgenic model of prostate carcinoma. Cancer. 2005;104:2104–2115. doi: 10.1002/cncr.21436. [DOI] [PubMed] [Google Scholar]
- 5.Pasqualini R, Arap W, McDonald DM. Probing the structural and molecular diversity of tumor vasculature. Trends in molecular medicine. 2002;8:563–571. doi: 10.1016/s1471-4914(02)02429-2. [DOI] [PubMed] [Google Scholar]
- 6.Fang J, Nakamura H, Maeda H. The EPR effect: Unique features of tumor blood vessels for drug delivery, factors involved, and limitations and augmentation of the effect. Adv Drug Deliv Rev. 2011;63:136–151. doi: 10.1016/j.addr.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 7.Iyer AK, Khaled G, Fang J, Maeda H. Exploiting the enhanced permeability and retention effect for tumor targeting. Drug Discov Today. 2006;11:812–818. doi: 10.1016/j.drudis.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 8.Mahon E, Salvati A, Baldelli Bombelli F, Lynch I, Dawson KA. Designing the nanoparticle-biomolecule interface for “targeting and therapeutic delivery”. J Control Release. 2012;161:164–174. doi: 10.1016/j.jconrel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 9.Arap W, Pasqualini R, Ruoslahti E. Cancer treatment by targeted drug delivery to tumor vasculature in a mouse model. Science. 1998;279:377–380. doi: 10.1126/science.279.5349.377. [DOI] [PubMed] [Google Scholar]
- 10.Bradbury AR, Marks JD. Antibodies from phage antibody libraries. J Immunol Methods. 2004;290:29–49. doi: 10.1016/j.jim.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 11.Curnis F, Arrigoni G, Sacchi A, Fischetti L, Arap W, Pasqualini R, Corti A. Differential binding of drugs containing the NGR motif to CD13 isoforms in tumor vessels, epithelia, and myeloid cells. Cancer research. 2002;62:867–874. [PubMed] [Google Scholar]
- 12.Dias-Neto E, Nunes DN, Giordano RJ, Sun J, Botz GH, Yang K, Setubal JC, Pasqualini R, Arap W. Next-generation phage display: integrating and comparing available molecular tools to enable cost-effective high-throughput analysis. PloS one. 2009;4:e8338. doi: 10.1371/journal.pone.0008338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferrara F, D’Angelo S, Gaiotto T, Naranjo L, Tian H, Graslund S, Dobrovetsky E, Hraber P, Lund-Johansen F, Saragozza S, Sblattero D, Kiss C, Bradbury AR. Recombinant renewable polyclonal antibodies. mAbs. 2015;7:32–41. doi: 10.4161/19420862.2015.989047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrara F, Naranjo LA, Kumar S, Gaiotto T, Mukundan H, Swanson B, Bradbury AR. Using phage and yeast display to select hundreds of monoclonal antibodies: application to antigen 85, a tuberculosis biomarker. PloS one. 2012;7:e49535. doi: 10.1371/journal.pone.0049535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gregorc V, De Braud FG, De Pas TM, Scalamogna R, Citterio G, Milani A, Boselli S, Catania C, Donadoni G, Rossoni G, Ghio D, Spitaleri G, Ammannati C, Colombi S, Caligaris-Cappio F, Lambiase A, Bordignon C. Phase I study of NGR-hTNF, a selective vascular targeting agent, in combination with cisplatin in refractory solid tumors. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:1964–1972. doi: 10.1158/1078-0432.CCR-10-1376. [DOI] [PubMed] [Google Scholar]
- 16.Gregorc V, Zucali PA, Santoro A, Ceresoli GL, Citterio G, De Pas TM, Zilembo N, De Vincenzo F, Simonelli M, Rossoni G, Spreafico A, Grazia Vigano M, Fontana F, De Braud FG, Bajetta E, Caligaris-Cappio F, Bruzzi P, Lambiase A, Bordignon C. Phase II study of asparagine-glycine-arginine-human tumor necrosis factor alpha, a selective vascular targeting agent, in previously treated patients with malignant pleural mesothelioma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28:2604–2611. doi: 10.1200/JCO.2009.27.3649. [DOI] [PubMed] [Google Scholar]
- 17.Hajitou A, Trepel M, Lilley CE, Soghomonyan S, Alauddin MM, Marini FC, 3rd, Restel BH, Ozawa MG, Moya CA, Rangel R, Sun Y, Zaoui K, Schmidt M, von Kalle C, Weitzman MD, Gelovani JG, Pasqualini R, Arap W. A hybrid vector for ligand-directed tumor targeting and molecular imaging. Cell. 2006;125:385–398. doi: 10.1016/j.cell.2006.02.042. [DOI] [PubMed] [Google Scholar]
- 18.Kolonin MG, Bover L, Sun J, Zurita AJ, Do KA, Lahdenranta J, Cardó-Vila M, Giordano RJ, Jaalouk DE, Ozawa MG, Moya CA, Souza GR, Staquicini FI, Kunyiasu A, Scudiero DA, Holbeck SL, Sausville EA, Arap W, Pasqualini R. Ligand-directed surface profiling of human cancer cells with combinatorial peptide libraries. Cancer research. 2006;66:34–40. doi: 10.1158/0008-5472.CAN-05-2748. [DOI] [PubMed] [Google Scholar]
- 19.Kolonin MG, Saha PK, Chan L, Pasqualini R, Arap W. Reversal of obesity by targeted ablation of adipose tissue. Nature medicine. 2004;10:625–632. doi: 10.1038/nm1048. [DOI] [PubMed] [Google Scholar]
- 20.Kolonin MG, Sun J, Do KA, Vidal CI, Ji Y, Baggerly KA, Pasqualini R, Arap W. Synchronous selection of homing peptides for multiple tissues by in vivo phage display. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2006;20:979–981. doi: 10.1096/fj.05-5186fje. [DOI] [PubMed] [Google Scholar]
- 21.Krag DN, Shukla GS, Shen GP, Pero S, Ashikaga T, Fuller S, Weaver DL, Burdette-Radoux S, Thomas C. Selection of tumor-binding ligands in cancer patients with phage display libraries. Cancer research. 2006;66:7724–7733. doi: 10.1158/0008-5472.CAN-05-4441. [DOI] [PubMed] [Google Scholar]
- 22.Paoloni MC, Tandle A, Mazcko C, Hanna E, Kachala S, Leblanc A, Newman S, Vail D, Henry C, Thamm D, Sorenmo K, Hajitou A, Pasqualini R, Arap W, Khanna C, Libutti SK. Launching a novel preclinical infrastructure: comparative oncology trials consortium directed therapeutic targeting of TNFalpha to cancer vasculature. PloS one. 2009;4:e4972. doi: 10.1371/journal.pone.0004972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pasqualini R, Millikan RE, Christianson DR, Cardó-Vila M, Driessen WH, Giordano RJ, Hajitou A, Hoang AG, Wen S, Barnhart KF, Baze WB, Marcott VD, Hawke DH, Do KA, Navone NM, Efstathiou E, Troncoso P, Lobb RR, Logothetis CJ, Arap W. Targeting the interleukin-11 receptor alpha in metastatic prostate cancer: A first-in-man study. Cancer. 2015 doi: 10.1002/cncr.29344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pasqualini R, Ruoslahti E. Organ targeting in vivo using phage display peptide libraries. Nature. 1996;380:364–366. doi: 10.1038/380364a0. [DOI] [PubMed] [Google Scholar]
- 25.Sblattero D, Bradbury A. Exploiting recombination in single bacteria to make large phage antibody libraries. Nat Biotechnol. 2000;18:75–80. doi: 10.1038/71958. [DOI] [PubMed] [Google Scholar]
- 26.Shukla GS, Krag DN, Peletskaya EN, Pero SC, Sun YJ, Carman CL, McCahill LE, Roland TA. Intravenous infusion of phage-displayed antibody library in human cancer patients: enrichment and cancer-specificity of tumor-homing phage-antibodies. Cancer immunology, immunotherapy : CII. 2013;62:1397–1410. doi: 10.1007/s00262-013-1443-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Staquicini FI, Cardó-Vila M, Kolonin MG, Trepel M, Edwards JK, Nunes DN, Sergeeva A, Efstathiou E, Sun J, Almeida NF, Tu SM, Botz GH, Wallace MJ, O’Connell DJ, Krajewski S, Gershenwald JE, Molldrem JJ, Flamm AL, Koivunen E, Pentz RD, Dias-Neto E, Setubal JC, Cahill DJ, Troncoso P, Do KA, Logothetis CJ, Sidman RL, Pasqualini R, Arap W. Vascular ligand-receptor mapping by direct combinatorial selection in cancer patients. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:18637–18642. doi: 10.1073/pnas.1114503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Staquicini FI, Ozawa MG, Moya CA, Driessen WH, Barbu EM, Nishimori H, Soghomonyan S, Flores LG, 2nd, Liang X, Paolillo V, Alauddin MM, Basilion JP, Furnari FB, Bogler O, Lang FF, Aldape KD, Fuller GN, Hook M, Gelovani JG, Sidman RL, Cavenee WK, Pasqualini R, Arap W. Systemic combinatorial peptide selection yields a non-canonical iron-mimicry mechanism for targeting tumors in a mouse model of human glioblastoma. The Journal of clinical investigation. 2011;121:161–173. doi: 10.1172/JCI44798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Staquicini FI, Qian MD, Salameh A, Dobroff AS, Edwards JK, Cimino DF, Moeller BJ, Kelly P, Nunez MI, Tang X, Liu DD, Lee JJ, Hong WK, Ferrara F, Bradbury AR, Lobb RR, Edelman MJ, Sidman RL, Wistuba, Arap W, Pasqualini R. Receptor tyrosine kinase EphA5 is a functional molecular target in human lung cancer. J Biol Chem. 2015;290:7345–7359. doi: 10.1074/jbc.M114.630525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tandle A, Hanna E, Lorang D, Hajitou A, Moya CA, Pasqualini R, Arap W, Adem A, Starker E, Hewitt S, Libutti SK. Tumor vasculature-targeted delivery of tumor necrosis factor-alpha. Cancer. 2009;115:128–139. doi: 10.1002/cncr.24001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trepel M, Stoneham CA, Eleftherohorinou H, Mazarakis ND, Pasqualini R, Arap W, Hajitou A. A heterotypic bystander effect for tumor cell killing after adeno-associated virus/phage-mediated, vascular-targeted suicide gene transfer. Molecular cancer therapeutics. 2009;8:2383–2391. doi: 10.1158/1535-7163.MCT-09-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cossu I, Bottoni G, Loi M, Emionite L, Bartolini A, Di Paolo D, Brignole C, Piaggio F, Perri P, Sacchi A, Curnis F, Gagliani MC, Bruno S, Marini C, Gori A, Longhi R, Murgia D, Sementa AR, Cilli M, Tacchetti C, Corti A, Sambuceti G, Marchiò S, Ponzoni M, Pastorino F. Neuroblastoma-targeted nanocarriers improve drug delivery and penetration, delay tumor growth and abrogate metastatic diffusion. Biomaterials. 2015;68:89–99. doi: 10.1016/j.biomaterials.2015.07.054. [DOI] [PubMed] [Google Scholar]
- 33.Deshpande PP, Biswas S, Torchilin VP. Current trends in the use of liposomes for tumor targeting. Nanomedicine. 2013;8:1509–1528. doi: 10.2217/nnm.13.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loi M, Di Paolo D, Soster M, Brignole C, Bartolini A, Emionite L, Sun J, Becherini P, Curnis F, Petretto A, Sani M, Gori A, Milanese M, Gambini C, Longhi R, Cilli M, Allen TM, Bussolino F, Arap W, Pasqualini R, Corti A, Ponzoni M, Marchiò S, Pastorino F. Novel phage display-derived neuroblastoma-targeting peptides potentiate the effect of drug nanocarriers in preclinical settings. J Control Release. 2013;170:233–241. doi: 10.1016/j.jconrel.2013.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loi M, Marchiò S, Becherini P, Di Paolo D, Soster M, Curnis F, Brignole C, Pagnan G, Perri P, Caffa I, Longhi R, Nico B, Bussolino F, Gambini C, Ribatti D, Cilli M, Arap W, Pasqualini R, Allen TM, Corti A, Ponzoni M, Pastorino F. Combined targeting of perivascular and endothelial tumor cells enhances anti-tumor efficacy of liposomal chemotherapy in neuroblastoma. J Control Release. 2010;145:66–73. doi: 10.1016/j.jconrel.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 36.Gillespie JW, Gross AL, Puzyrev AT, Bedi D, Petrenko VA. Combinatorial synthesis and screening of cancer cell-specific nanomedicines targeted via phage fusion proteins. Front Microbiol. 2015;6:628. doi: 10.3389/fmicb.2015.00628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ashley CE, Carnes EC, Epler KE, Padilla DP, Phillips GK, Castillo RE, Wilkinson DC, Wilkinson BS, Burgard CA, Kalinich RM, Townson JL, Chackerian B, Willman CL, Peabody DS, Wharton W, Brinker CJ. Delivery of Small Interfering RNA by Peptide-Targeted Mesoporous Silica Nanoparticle-Supported Lipid Bilayers. ACS Nano. 2012;6:2174–2188. doi: 10.1021/nn204102q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ashley CE, Carnes EC, Phillips GK, Padilla D, Durfee PN, Brown PA, Hanna TN, Liu J, Phillips B, Carter MB, Carroll NJ, Jiang X, Dunphy DR, Willman CL, Petsev DN, Evans DG, Parikh AN, Chackerian B, Warton W, Peabody DS, Brinker CJ. The targeted delivery of multicomponent cargos to cancer cells by nanoporous particle-supported lipid bilayers. Nature Materials. 2011;10:389–397. doi: 10.1038/nmat2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J, Stace-Naughton A, Jiang X, Brinker CJ. Porous Nanoparticle Supported Lipid Bilayers (Protocells) as Delivery Vehicles. J Am Chem Soc. 2009;131:1354–1355. doi: 10.1021/ja808018y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arap W, Kolonin MG, Trepel M, Lahdenranta J, Cardó-Vila M, Giordano RJ, Mintz PJ, Ardelt PU, Yao VJ, Vidal CI, Chen L, Flamm A, Valtanen H, Weavind LM, Hicks ME, Pollock RE, Botz GH, Bucana CD, Koivunen E, Cahill D, Troncoso P, Baggerly KA, Pentz RD, Do KA, Logothetis CJ, Pasqualini R. Steps toward mapping the human vasculature by phage display. Nature medicine. 2002;8:121–127. doi: 10.1038/nm0202-121. [DOI] [PubMed] [Google Scholar]
- 41.Cardó-Vila M, Zurita AJ, Giordano RJ, Sun J, Rangel R, Guzman-Rojas L, Anobom CD, Valente AP, Almeida FC, Lahdenranta J, Kolonin MG, Arap W, Pasqualini R. A ligand peptide motif selected from a cancer patient is a receptor-interacting site within human interleukin-11. PloS one. 2008;3:e3452. doi: 10.1371/journal.pone.0003452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zurita AJ, Troncoso P, Cardó-Vila M, Logothetis CJ, Pasqualini R, Arap W. Combinatorial screenings in patients: the interleukin-11 receptor alpha as a candidate target in the progression of human prostate cancer. Cancer research. 2004;64:435–439. doi: 10.1158/0008-5472.can-03-2675. [DOI] [PubMed] [Google Scholar]
- 43.Arap MA, Lahdenranta J, Mintz PJ, Hajitou A, Sarkis AS, Arap W, Pasqualini R. Cell surface expression of the stress response chaperone GRP78 enables tumor targeting by circulating ligands. Cancer cell. 2004;6:275–284. doi: 10.1016/j.ccr.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 44.Lee AS. Glucose-regulated proteins in cancer: molecular mechanisms and therapeutic potential. Nature reviews Cancer. 2014;14:263–276. doi: 10.1038/nrc3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mandelin J, Cardó-Vila M, Driessen WH, Mathew P, Navone NM, Lin SH, Logothetis CJ, Rietz AC, Dobroff AS, Proneth B, Sidman RL, Pasqualini R, Arap W. Selection and identification of ligand peptides targeting a model of castrate-resistant osteogenic prostate cancer and their receptors. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:3776–3781. doi: 10.1073/pnas.1500128112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mintz PJ, Kim J, Do KA, Wang X, Zinner RG, Cristofanilli M, Arap MA, Hong WK, Troncoso P, Logothetis CJ, Pasqualini R, Arap W. Fingerprinting the circulating repertoire of antibodies from cancer patients. Nat Biotechnol. 2003;21:57–63. doi: 10.1038/nbt774. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Y, Tseng CC, Tsai YL, Fu X, Schiff R, Lee AS. Cancer cells resistant to therapy promote cell surface relocalization of GRP78 which complexes with PI3K and enhances PI(3,4,5)P3 production. PloS one. 2013;8:e80071. doi: 10.1371/journal.pone.0080071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bidard FC, Tredan O. Trends in cancer-targeted antibody-drug conjugates. Target Oncol. 2014;9:1–8. doi: 10.1007/s11523-013-0302-9. [DOI] [PubMed] [Google Scholar]
- 49.Morbeck DE, Roche PC, Keutmann HT, McCormick DJ. A receptor binding site identified in the region 81–95 of the beta-subunit of human luteinizing hormone (LH) and chorionic gonadotropin (hCG) Mol Cell Endocrinol. 1993;97:173–181. doi: 10.1016/0303-7207(93)90225-9. [DOI] [PubMed] [Google Scholar]
- 50.Qayum A, Gullick W, Clayton RC, Sikora K, Waxman J. The effects of gonadotrophin releasing hormone analogues in prostate cancer are mediated through specific tumour receptors. Br J Cancer. 1990;62:96–99. doi: 10.1038/bjc.1990.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hansel W, Enright F, Leuschner C. Destruction of breast cancers and their metastases by lytic peptide conjugates in vitro and in vivo. Mol Cell Endocrinol. 2007;260–262:183–189. doi: 10.1016/j.mce.2005.12.056. [DOI] [PubMed] [Google Scholar]
- 52.Hansel W, Leuschner C, Enright F. Conjugates of lytic peptides and LHRH or betaCG target and cause necrosis of prostate cancers and metastases. Mol Cell Endocrinol. 2007;269:26–33. doi: 10.1016/j.mce.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 53.Sun H, Zhu X, Lu PY, Rosato RR, Tan W, Zu Y. Oligonucleotide aptamers: new tools for targeted cancer therapy. Mol Ther Nucleic Acids. 2014;3:e182. doi: 10.1038/mtna.2014.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu J, Huang H, Dong S, Ge L, Zhang Y. Progress in aptamer-mediated drug delivery vehicles for cancer targeting and its implications in addressing chemotherapeutic challenges. Theranostics. 2014;4:931–944. doi: 10.7150/thno.9663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Porciani D, Tedeschi L, Marchetti L, Citti L, Piazza V, Beltram F, Signore G. Aptamer-Mediated Codelivery of Doxorubicin and NF-kappaB Decoy Enhances Chemosensitivity of Pancreatic Tumor Cells. Mol Ther Nucleic Acids. 2015;4:e235. doi: 10.1038/mtna.2015.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fujimori K, Covell DG, Fletcher JE, Weinstein JN. Modeling analysis of the global and microscopic distribution of immunoglobulin G, F(ab’)2, and Fab in tumors. Cancer research. 1989;49:5656–5663. [PubMed] [Google Scholar]
- 57.Pascal J, Ashley CE, Wang Z, Brocato TA, Butner JD, Carnes EC, Koay EJ, Brinker CJ, Cristini V. Mechanistic modeling identifies drug-uptake history as predictor of tumor drug resistance and nano-carrier-mediated response. ACS Nano. 2013;7:11174–11182. doi: 10.1021/nn4048974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 59.Pasqualini R, Koivunen E, Ruoslahti E. Alpha v integrins as receptors for tumor targeting by circulating ligands. Nat Biotechnol. 1997;15:542–546. doi: 10.1038/nbt0697-542. [DOI] [PubMed] [Google Scholar]
- 60.Rajotte D, Arap W, Hagedorn M, Koivunen E, Pasqualini R, Ruoslahti E. Molecular heterogeneity of the vascular endothelium revealed by in vivo phage display. The Journal of clinical investigation. 1998;102:430–437. doi: 10.1172/JCI3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kolonin MG, Pasqualini R, Arap W. Teratogenicity induced by targeting a placental immunoglobulin transporter. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:13055–13060. doi: 10.1073/pnas.162468499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trepel M, Arap W, Pasqualini R. In vivo phage display and vascular heterogeneity: implications for targeted medicine. Current opinion in chemical biology. 2002;6:399–404. doi: 10.1016/s1367-5931(02)00336-8. [DOI] [PubMed] [Google Scholar]
- 63.Barnhart KF, Christianson DR, Hanley PW, Driessen WH, Bernacky BJ, Baze WB, Wen S, Tian M, Ma J, Kolonin MG, Saha PK, Do KA, Hulvat JF, Gelovani JG, Chan L, Arap W, Pasqualini R. A peptidomimetic targeting white fat causes weight loss and improved insulin resistance in obese monkeys. Science translational medicine. 2011;3:108ra112. doi: 10.1126/scitranslmed.3002621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Giordano RJ, Lahdenranta J, Zhen L, Chukwueke U, Petrache I, Langley RR, Fidler IJ, Pasqualini R, Tuder RM, Arap W. Targeted induction of lung endothelial cell apoptosis causes emphysema-like changes in the mouse. J Biol Chem. 2008;283:29447–29460. doi: 10.1074/jbc.M804595200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Koivunen E, Gay DA, Ruoslahti E. Selection of peptides binding to the alpha 5 beta 1 integrin from phage display library. J Biol Chem. 1993;268:20205–20210. [PubMed] [Google Scholar]
- 66.Pentz RD, Flamm AL, Pasqualini R, Logothetis CJ, Arap W. Revisiting ethical guidelines for research with terminal wean and brain-dead participants. The Hastings Center report. 2003;33:20–26. [PubMed] [Google Scholar]
- 67.Yao VJ, Ozawa MG, Trepel M, Arap W, McDonald DM, Pasqualini R. Targeting pancreatic islets with phage display assisted by laser pressure catapult microdissection. The American journal of pathology. 2005;166:625–636. doi: 10.1016/S0002-9440(10)62283-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grant JA, Gallardo MA, Pickup B. A fast method of molecular shape comparison: A simple application of a Gaussian description of molecular shape. J Comput Chem. 1996;17:1653–1666. [Google Scholar]
- 69.Oprea TI, Gottfries J. Chemography: the art of navigating in chemical space. Journal of combinatorial chemistry. 2001;3:157–166. doi: 10.1021/cc0000388. [DOI] [PubMed] [Google Scholar]
- 70.Ung P, Winkler DA. Tripeptide motifs in biology: targets for peptidomimetic design. Journal of medicinal chemistry. 2011;54:1111–1125. doi: 10.1021/jm1012984. [DOI] [PubMed] [Google Scholar]
- 71.Vendruscolo M, Paci E, Dobson CM, Karplus M. Three key residues form a critical contact network in a protein folding transition state. Nature. 2001;409:641–645. doi: 10.1038/35054591. [DOI] [PubMed] [Google Scholar]
- 72.Christianson DR, Ozawa MG, Pasqualini R, Arap W. Techniques to decipher molecular diversity by phage display. Methods in molecular biology. 2007;357:385–406. doi: 10.1385/1-59745-214-9:385. [DOI] [PubMed] [Google Scholar]
- 73.Dondossola E, Corti A, Sidman RL, Arap W, Pasqualini R. Bone marrow-derived CD13 cells sustain tumor progression: A potential non-malignant target for anticancer therapy. Oncoimmunology. 2014;3:e27716. doi: 10.4161/onci.27716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dondossola E, Rangel R, Guzman-Rojas L, Barbu EM, Hosoya H, St John LS, Molldrem JJ, Corti A, Sidman RL, Arap W, Pasqualini R. CD13-positive bone marrow-derived myeloid cells promote angiogenesis, tumor growth, and metastasis. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:20717–20722. doi: 10.1073/pnas.1321139110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guzman-Rojas L, Rangel R, Salameh A, Edwards JK, Dondossola E, Kim YG, Saghatelian A, Giordano RJ, Kolonin MG, Staquicini FI, Koivunen E, Sidman RL, Arap W, Pasqualini R. Cooperative effects of aminopeptidase N (CD13) expressed by nonmalignant and cancer cells within the tumor microenvironment. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:1637–1642. doi: 10.1073/pnas.1120790109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pasqualini R, Koivunen E, Kain R, Lahdenranta J, Sakamoto M, Stryhn A, Ashmun RA, Shapiro LH, Arap W, Ruoslahti E. Aminopeptidase N is a receptor for tumor-homing peptides and a target for inhibiting angiogenesis. Cancer research. 2000;60:722–727. [PMC free article] [PubMed] [Google Scholar]
- 77.Cardó-Vila M, Giordano RJ, Sidman RL, Bronk LF, Fan Z, Mendelsohn J, Arap W, Pasqualini R. From combinatorial peptide selection to drug prototype (II): targeting the epidermal growth factor receptor pathway. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:5118–5123. doi: 10.1073/pnas.0915146107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Giordano RJ, Cardó-Vila M, Salameh A, Anobom CD, Zeitlin BD, Hawke DH, Valente AP, Almeida FC, Nor JE, Sidman RL, Pasqualini R, Arap W. From combinatorial peptide selection to drug prototype (I): targeting the vascular endothelial growth factor receptor pathway. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:5112–5117. doi: 10.1073/pnas.0915141107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pang HB, Braun GB, She ZG, Kotamraju VR, Sugahara KN, Teesalu T, Ruoslahti E. A free cysteine prolongs the half-life of a homing peptide and improves its tumor-penetrating activity. J Control Release. 2014;175:48–53. doi: 10.1016/j.jconrel.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sidman RL, Li J, Lawrence M, Hu W, Musso GF, Giordano RJ, Cardó-Vila M, Pasqualini R, Arap W. The peptidomimetic Vasotide targets two retinal VEGF receptors and reduces pathological angiogenesis in murine and nonhuman primate models of retinal disease. Science translational medicine. 2015;7:309ra165. doi: 10.1126/scitranslmed.aac4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sugahara KN, Teesalu T, Karmali PP, Kotamraju VR, Agemy L, Girard OM, Hanahan D, Mattrey RF, Ruoslahti E. Tissue-penetrating delivery of compounds and nanoparticles into tumors. Cancer cell. 2009;16:510–520. doi: 10.1016/j.ccr.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang S, Placzek WJ, Stebbins JL, Mitra S, Noberini R, Koolpe M, Zhang Z, Dahl R, Pasquale EB, Pellecchia M. Novel targeted system to deliver chemotherapeutic drugs to EphA2-expressing cancer cells. Journal of medicinal chemistry. 2012;55:2427–2436. doi: 10.1021/jm201743s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Christianson DR, Dobroff AS, Proneth B, Zurita AJ, Salameh A, Dondossola E, Makino J, Bologa CG, Smith TL, Yao VJ, Calderone TL, O’Connell DJ, Oprea TI, Kataoka K, Cahill DJ, Gershenwald JE, Sidman RL, Arap W, Pasqualini R. Ligand-directed targeting of lymphatic vessels uncovers mechanistic insights in melanoma metastasis. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:2521–2526. doi: 10.1073/pnas.1424994112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Marchiò S, Lahdenranta J, Schlingemann RO, Valdembri D, Wesseling P, Arap MA, Hajitou A, Ozawa MG, Trepel M, Giordano RJ, Nanus DM, Dijkman HB, Oosterwijk E, Sidman RL, Cooper MD, Bussolino F, Pasqualini R, Arap W. Aminopeptidase A is a functional target in angiogenic blood vessels. Cancer cell. 2004;5:151–162. doi: 10.1016/s1535-6108(04)00025-x. [DOI] [PubMed] [Google Scholar]
- 85.Mintz PJ, Rietz AC, Cardó-Vila M, Ozawa MG, Dondossola E, Do KA, Kim J, Troncoso P, Logothetis CJ, Sidman RL, Pasqualini R, Arap W. Discovery and horizontal follow-up of an autoantibody signature in human prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:2515–2520. doi: 10.1073/pnas.1500097112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mintz PJ, Cardó-Vila M, Ozawa MG, Hajitou A, Rangel R, Guzman-Rojas L, Christianson DR, Arap MA, Giordano RJ, Souza GR, Easley J, Salameh A, Oliviero S, Brentani RR, Koivunen E, Arap W, Pasqualini R. An unrecognized extracellular function for an intracellular adapter protein released from the cytoplasm into the tumor microenvironment. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2182–2187. doi: 10.1073/pnas.0807543105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yao VJ, Ozawa MG, Varner AS, Kasman IM, Chanthery YH, Pasqualini R, Arap W, McDonald DM. Antiangiogenic therapy decreases integrin expression in normalized tumor blood vessels. Cancer research. 2006;66:2639–2649. doi: 10.1158/0008-5472.CAN-05-1824. [DOI] [PubMed] [Google Scholar]
- 88.Giordano RJ, Cardó-Vila M, Lahdenranta J, Pasqualini R, Arap W. Biopanning and rapid analysis of selective interactive ligands. Nature medicine. 2001;7:1249–1253. doi: 10.1038/nm1101-1249. [DOI] [PubMed] [Google Scholar]
- 89.Kuijper S, Turner CJ, Adams RH. Regulation of angiogenesis by Eph-ephrin interactions. Trends Cardiovasc Med. 2007;17:145–151. doi: 10.1016/j.tcm.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 90.Gu C, Liu L, He Y, Jiang J, Yang Z, Wu Q. The binding characteristics of a cyclic nonapeptide, c(CGRRAGGSC), in LNCaP human prostate cancer cells. Oncology letters. 2012;4:443–449. doi: 10.3892/ol.2012.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang W, Ke S, Kwon S, Yallampalli S, Cameron AG, Adams KE, Mawad ME, Sevick-Muraca EM. A new optical and nuclear dual-labeled imaging agent targeting interleukin 11 receptor alpha-chain. Bioconjugate chemistry. 2007;18:397–402. doi: 10.1021/bc0602679. [DOI] [PubMed] [Google Scholar]
- 92.Hanavadi S, Martin TA, Watkins G, Mansel RE, Jiang WG. Expression of interleukin 11 and its receptor and their prognostic value in human breast cancer. Annals of surgical oncology. 2006;13:802–808. doi: 10.1245/ASO.2006.05.028. [DOI] [PubMed] [Google Scholar]
- 93.Javadpour MM, Juban MM, Lo WC, Bishop SM, Alberty JB, Cowell SM, Becker CL, McLaughlin ML. De novo antimicrobial peptides with low mammalian cell toxicity. Journal of medicinal chemistry. 1996;39:3107–3113. doi: 10.1021/jm9509410. [DOI] [PubMed] [Google Scholar]
- 94.Ellerby HM, Arap W, Ellerby LM, Kain R, Andrusiak R, Rio GD, Krajewski S, Lombardo CR, Rao R, Ruoslahti E, Bredesen DE, Pasqualini R. Anti-cancer activity of targeted pro-apoptotic peptides. Nature medicine. 1999;5:1032–1038. doi: 10.1038/12469. [DOI] [PubMed] [Google Scholar]
- 95.Lewis VO, Ozawa MG, Deavers MT, Wang G, Shintani T, Arap W, Pasqualini R. The interleukin-11 receptor alpha as a candidate ligand-directed target in osteosarcoma: consistent data from cell lines, orthotopic models, and human tumor samples. Cancer research. 2009;69:1995–1999. doi: 10.1158/0008-5472.CAN-08-4845. [DOI] [PubMed] [Google Scholar]
- 96.Li ZG, Mathew P, Yang J, Starbuck MW, Zurita AJ, Liu J, Sikes C, Multani AS, Efstathiou E, Lopez A, Wang J, Fanning TV, Prieto VG, Kundra V, Vazquez ES, Troncoso P, Raymond AK, Logothetis CJ, Lin SH, Maity S, Navone NM. Androgen receptor-negative human prostate cancer cells induce osteogenesis in mice through FGF9-mediated mechanisms. The Journal of clinical investigation. 2008;118:2697–2710. doi: 10.1172/JCI33093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sato M, Yao VJ, Arap W, Pasqualini R. GRP78 signaling hub a receptor for targeted tumor therapy. Adv Genet. 2010;69:97–114. doi: 10.1016/S0065-2660(10)69006-2. [DOI] [PubMed] [Google Scholar]
- 98.Lee AS, Bell J, Ting J. Biochemical characterization of the 94- and 78-kilodalton glucose-regulated proteins in hamster fibroblasts. J Biol Chem. 1984;259:4616–4621. [PubMed] [Google Scholar]
- 99.Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- 100.Mori K, Ma W, Gething MJ, Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- 101.Blond-Elguindi S, Cwirla SE, Dower WJ, Lipshutz RJ, Sprang SR, Sambrook JF, Gething MJ. Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell. 1993;75:717–728. doi: 10.1016/0092-8674(93)90492-9. [DOI] [PubMed] [Google Scholar]
- 102.Dong D, Ko B, Baumeister P, Swenson S, Costa F, Markland F, Stiles C, Patterson JB, Bates SE, Lee AS. Vascular targeting and antiangiogenesis agents induce drug resistance effector GRP78 within the tumor microenvironment. Cancer research. 2005;65:5785–5791. doi: 10.1158/0008-5472.CAN-05-0754. [DOI] [PubMed] [Google Scholar]
- 103.Lee AS. The ER chaperone and signaling regulator GRP78/BiP as a monitor of endoplasmic reticulum stress. Methods. 2005;35:373–381. doi: 10.1016/j.ymeth.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 104.Daneshmand S, Quek ML, Lin E, Lee C, Cote RJ, Hawes D, Cai J, Groshen S, Lieskovsky G, Skinner DG, Lee AS, Pinski J. Glucose-regulated protein GRP78 is up-regulated in prostate cancer and correlates with recurrence and survival. Human pathology. 2007;38:1547–1552. doi: 10.1016/j.humpath.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 105.Fu Y, Li J, Lee AS. GRP78/BiP inhibits endoplasmic reticulum BIK and protects human breast cancer cells against estrogen starvation-induced apoptosis. Cancer research. 2007;67:3734–3740. doi: 10.1158/0008-5472.CAN-06-4594. [DOI] [PubMed] [Google Scholar]
- 106.Gonzalez-Gronow M, Cuchacovich M, Llanos C, Urzua C, Gawdi G, Pizzo SV. Prostate cancer cell proliferation in vitro is modulated by antibodies against glucose-regulated protein 78 isolated from patient serum. Cancer research. 2006;66:11424–11431. doi: 10.1158/0008-5472.CAN-06-1721. [DOI] [PubMed] [Google Scholar]
- 107.Reichert JM. Antibodies to watch in 2015. mAbs. 2015;7:1–8. doi: 10.4161/19420862.2015.988944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Clynes RA, Towers TL, Presta LG, Ravetch JV. Inhibitory Fc receptors modulate in vivo cytotoxicity against tumor targets. Nature medicine. 2000;6:443–446. doi: 10.1038/74704. [DOI] [PubMed] [Google Scholar]
- 109.Maloney DG, Grillo-Lopez AJ, White CA, Bodkin D, Schilder RJ, Neidhart JA, Janakiraman N, Foon KA, Liles TM, Dallaire BK, Wey K, Royston I, Davis T, Levy R. IDEC-C2B8 (Rituximab) anti-CD20 monoclonal antibody therapy in patients with relapsed low-grade non-Hodgkin’s lymphoma. Blood. 1997;90:2188–2195. [PubMed] [Google Scholar]
- 110.Ferrara N, Hillan KJ, Gerber HP, Novotny W. Discovery and development of bevacizumab, an anti-VEGF antibody for treating cancer. Nature reviews Drug discovery. 2004;3:391–400. doi: 10.1038/nrd1381. [DOI] [PubMed] [Google Scholar]
- 111.Sliwkowski MX, Mellman I. Antibody therapeutics in cancer. Science. 2013;341:1192–1198. doi: 10.1126/science.1241145. [DOI] [PubMed] [Google Scholar]
- 112.Baselga J, Gelmon KA, Verma S, Wardley A, Conte P, Miles D, Bianchi G, Cortes J, McNally VA, Ross GA, Fumoleau P, Gianni L. Phase II trial of pertuzumab and trastuzumab in patients with human epidermal growth factor receptor 2-positive metastatic breast cancer that progressed during prior trastuzumab therapy. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28:1138–1144. doi: 10.1200/JCO.2009.24.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mack F, Ritchie M, Sapra P. The next generation of antibody drug conjugates. Seminars in oncology. 2014;41:637–652. doi: 10.1053/j.seminoncol.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 114.Lewis Phillips GD, Li G, Dugger DL, Crocker LM, Parsons KL, Mai E, Blattler WA, Lambert JM, Chari RV, Lutz RJ, Wong WL, Jacobson FS, Koeppen H, Schwall RH, Kenkare-Mitra SR, Spencer SD, Sliwkowski MX. Targeting HER2-positive breast cancer with trastuzumab-DM1, an antibody-cytotoxic drug conjugate. Cancer research. 2008;68:9280–9290. doi: 10.1158/0008-5472.CAN-08-1776. [DOI] [PubMed] [Google Scholar]
- 115.Wolchok JD, Kluger H, Callahan MK, Postow MA, Rizvi NA, Lesokhin AM, Segal NH, Ariyan CE, Gordon RA, Reed K, Burke MM, Caldwell A, Kronenberg SA, Agunwamba BU, Zhang X, Lowy I, Inzunza HD, Feely W, Horak CE, Hong Q, Korman AJ, Wigginton JM, Gupta A, Sznol M. Nivolumab plus ipilimumab in advanced melanoma. N Engl J Med. 2013;369:122–133. doi: 10.1056/NEJMoa1302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348:56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 117.List T, Neri D. Immunocytokines: a review of molecules in clinical development for cancer therapy. Clinical pharmacology : advances and applications. 2013;5:29–45. doi: 10.2147/CPAA.S49231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Alewine C, Hassan R, Pastan I. Advances in anticancer immunotoxin therapy. Oncologist. 2015;20:176–185. doi: 10.1634/theoncologist.2014-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kohler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 120.Jones PT, Dear PH, Foote J, Neuberger MS, Winter G. Replacing the complementarity-determining regions in a human antibody with those from a mouse. Nature. 1986;321:522–525. doi: 10.1038/321522a0. [DOI] [PubMed] [Google Scholar]
- 121.Doerner A, Rhiel L, Zielonka S, Kolmar H. Therapeutic antibody engineering by high efficiency cell screening. FEBS letters. 2014;588:278–287. doi: 10.1016/j.febslet.2013.11.025. [DOI] [PubMed] [Google Scholar]
- 122.Nixon AE, Sexton DJ, Ladner RC. Drugs derived from phage display: from candidate identification to clinical practice. mAbs. 2014;6:73–85. doi: 10.4161/mabs.27240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Carter PJ. Potent antibody therapeutics by design. Nature reviews Immunology. 2006;6:343–357. doi: 10.1038/nri1837. [DOI] [PubMed] [Google Scholar]
- 124.Jakobovits A, Amado RG, Yang X, Roskos L, Schwab G. From XenoMouse technology to panitumumab, the first fully human antibody product from transgenic mice. Nat Biotechnol. 2007;25:1134–1143. doi: 10.1038/nbt1337. [DOI] [PubMed] [Google Scholar]
- 125.Huston JS, Levinson D, Mudgett-Hunter M, Tai MS, Novotny J, Margolies MN, Ridge RJ, Bruccoleri RE, Haber E, Crea R, et al. Protein engineering of antibody binding sites: recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:5879–5883. doi: 10.1073/pnas.85.16.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Griffiths AD, Williams SC, Hartley O, Tomlinson IM, Waterhouse P, Crosby WL, Kontermann RE, Jones PT, Low NM, Allison TJ, et al. Isolation of high affinity human antibodies directly from large synthetic repertoires. The EMBO journal. 1994;13:3245–3260. doi: 10.1002/j.1460-2075.1994.tb06626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Decanniere K, Desmyter A, Lauwereys M, Ghahroudi MA, Muyldermans S, Wyns L. A single-domain antibody fragment in complex with RNase A: non-canonical loop structures and nanomolar affinity using two CDR loops. Structure. 1999;7:361–370. doi: 10.1016/s0969-2126(99)80049-5. [DOI] [PubMed] [Google Scholar]
- 128.Frenzel A, Hust M, Schirrmann T. Expression of recombinant antibodies. Frontiers in immunology. 2013;4:217. doi: 10.3389/fimmu.2013.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Worn A, Pluckthun A. Mutual stabilization of VL and VH in single-chain antibody fragments, investigated with mutants engineered for stability. Biochemistry. 1998;37:13120–13127. doi: 10.1021/bi980712q. [DOI] [PubMed] [Google Scholar]
- 130.Glanville J, Zhai W, Berka J, Telman D, Huerta G, Mehta GR, Ni I, Mei L, Sundar PD, Day GM, Cox D, Rajpal A, Pons J. Precise determination of the diversity of a combinatorial antibody library gives insight into the human immunoglobulin repertoire. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:20216–20221. doi: 10.1073/pnas.0909775106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Marks JD, Hoogenboom HR, Bonnert TP, McCafferty J, Griffiths AD, Winter G. Bypassing immunization. Human antibodies from V-gene libraries displayed on phage. Journal of molecular biology. 1991;222:581–597. doi: 10.1016/0022-2836(91)90498-u. [DOI] [PubMed] [Google Scholar]
- 132.Knappik A, Ge L, Honegger A, Pack P, Fischer M, Wellnhofer G, Hoess A, Wolle J, Pluckthun A, Virnekas B. Fully synthetic human combinatorial antibody libraries (HuCAL) based on modular consensus frameworks and CDRs randomized with trinucleotides. Journal of molecular biology. 2000;296:57–86. doi: 10.1006/jmbi.1999.3444. [DOI] [PubMed] [Google Scholar]
- 133.Birtalan S, Zhang Y, Fellouse FA, Shao L, Schaefer G, Sidhu SS. The intrinsic contributions of tyrosine, serine, glycine and arginine to the affinity and specificity of antibodies. Journal of molecular biology. 2008;377:1518–1528. doi: 10.1016/j.jmb.2008.01.093. [DOI] [PubMed] [Google Scholar]
- 134.Sblattero D, Lou J, Marzari R, Bradbury A. In vivo recombination as a tool to generate molecular diversity in phage antibody libraries. J Biotechnol. 2001;74:303–315. doi: 10.1016/s1389-0352(01)00022-8. [DOI] [PubMed] [Google Scholar]
- 135.Clackson T, Hoogenboom HR, Griffiths AD, Winter G. Making antibody fragments using phage display libraries. Nature. 1991;352:624–628. doi: 10.1038/352624a0. [DOI] [PubMed] [Google Scholar]
- 136.Hanes J, Schaffitzel C, Knappik A, Pluckthun A. Picomolar affinity antibodies from a fully synthetic naive library selected and evolved by ribosome display. Nat Biotechnol. 2000;18:1287–1292. doi: 10.1038/82407. [DOI] [PubMed] [Google Scholar]
- 137.Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- 138.Feldhaus MJ, Siegel RW. Yeast display of antibody fragments: a discovery and characterization platform. J Immunol Methods. 2004;290:69–80. doi: 10.1016/j.jim.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 139.Swers JS, Kellogg BA, Wittrup KD. Shuffled antibody libraries created by in vivo homologous recombination and yeast surface display. Nucleic acids research. 2004;32:e36. doi: 10.1093/nar/gnh030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Glanville J, D’Angelo S, Khan TA, Reddy ST, Naranjo L, Ferrara F, Bradbury A. Deep sequencing in library selection projects: what insight does it bring? Curr Opin Struct Biol. 2015;33:146–160. doi: 10.1016/j.sbi.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Matsumura Y, Maeda H. A new concept for macromolecular therapeutics in cancer chemotherapy: mechanism of tumoritropic accumulation of proteins and the antitumor agent smancs. Cancer research. 1986;46:6387–6392. [PubMed] [Google Scholar]
- 142.Adams GP, Schier R, Marshall K, Wolf EJ, McCall AM, Marks JD, Weiner LM. Increased affinity leads to improved selective tumor delivery of single-chain Fv antibodies. Cancer research. 1998;58:485–490. [PubMed] [Google Scholar]
- 143.Adams GP, Schier R, McCall AM, Simmons HH, Horak EM, Alpaugh RK, Marks JD, Weiner LM. High affinity restricts the localization and tumor penetration of single-chain fv antibody molecules. Cancer research. 2001;61:4750–4755. [PubMed] [Google Scholar]
- 144.Rudnick SI, Lou J, Shaller CC, Tang Y, Klein-Szanto AJ, Weiner LM, Marks JD, Adams GP. Influence of affinity and antigen internalization on the uptake and penetration of Anti-HER2 antibodies in solid tumors. Cancer research. 2011;71:2250–2259. doi: 10.1158/0008-5472.CAN-10-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Schmidt MM, Wittrup KD. A modeling analysis of the effects of molecular size and binding affinity on tumor targeting. Molecular cancer therapeutics. 2009;8:2861–2871. doi: 10.1158/1535-7163.MCT-09-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zahnd C, Kawe M, Stumpp MT, de Pasquale C, Tamaskovic R, Nagy-Davidescu G, Dreier B, Schibli R, Binz HK, Waibel R, Pluckthun A. Efficient tumor targeting with high-affinity designed ankyrin repeat proteins: effects of affinity and molecular size. Cancer research. 2010;70:1595–1605. doi: 10.1158/0008-5472.CAN-09-2724. [DOI] [PubMed] [Google Scholar]
- 147.Adams GP, Schier R, McCall AM, Crawford RS, Wolf EJ, Weiner LM, Marks JD. Prolonged in vivo tumour retention of a human diabody targeting the extracellular domain of human HER2/neu. Br J Cancer. 1998;77:1405–1412. doi: 10.1038/bjc.1998.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Cuesta AM, Sainz-Pastor N, Bonet J, Oliva B, Alvarez-Vallina L. Multivalent antibodies: when design surpasses evolution. Trends Biotechnol. 2010;28:355–362. doi: 10.1016/j.tibtech.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 149.McCall AM, Shahied L, Amoroso AR, Horak EM, Simmons HH, Nielson U, Adams GP, Schier R, Marks JD, Weiner LM. Increasing the affinity for tumor antigen enhances bispecific antibody cytotoxicity. J Immunol. 2001;166:6112–6117. doi: 10.4049/jimmunol.166.10.6112. [DOI] [PubMed] [Google Scholar]
- 150.Schmidt MM, Thurber GM, Wittrup KD. Kinetics of anti-carcinoembryonic antigen antibody internalization: effects of affinity, bivalency, and stability. Cancer immunology, immunotherapy : CII. 2008;57:1879–1890. doi: 10.1007/s00262-008-0518-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Liu B, Conrad F, Cooperberg MR, Kirpotin DB, Marks JD. Mapping tumor epitope space by direct selection of single-chain Fv antibody libraries on prostate cancer cells. Cancer research. 2004;64:704–710. doi: 10.1158/0008-5472.can-03-2732. [DOI] [PubMed] [Google Scholar]
- 152.Poul MA, Becerril B, Nielsen UB, Morisson P, Marks JD. Selection of tumor-specific internalizing human antibodies from phage libraries. Journal of molecular biology. 2000;301:1149–1161. doi: 10.1006/jmbi.2000.4026. [DOI] [PubMed] [Google Scholar]
- 153.Zhou Y, Zou H, Zhang S, Marks JD. Internalizing cancer antibodies from phage libraries selected on tumor cells and yeast-displayed tumor antigens. Journal of molecular biology. 2010;404:88–99. doi: 10.1016/j.jmb.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.D’Angelo S, Glanville J, Ferrara F, Naranjo L, Gleasner CD, Shen X, Bradbury AR, Kiss C. The antibody mining toolbox: an open source tool for the rapid analysis of antibody repertoires. mAbs. 2014;6:160–172. doi: 10.4161/mabs.27105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Reddy LH, Arias JL, Nicolas J, Couvreir P. Magnetic Nanoparticles: Design and Characterization, Toxicity and Biocompatibility, Pharmaceutical and Biomedical Applications. Chemical Reviews. 2012;112:5818–5878. doi: 10.1021/cr300068p. [DOI] [PubMed] [Google Scholar]
- 156.Park JW, Bae KH, Kim C, Park TG. Clustered Magnetite Nanocrystals Cross-Linked with PEI for Efficient siRNA Delivery. Biomacromolecules. 2011;12:457–465. doi: 10.1021/bm101244j. [DOI] [PubMed] [Google Scholar]
- 157.Kunzmann A, Andersson B, Thurnherr T, Krug H, Scheynius A, Fadeel B. Toxicology of engineered nanomaterials: Focus on biocompatibility, biodistribution and biodegradation. Biochimica et Biophysica Acta. 2011;1810:361–373. doi: 10.1016/j.bbagen.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 158.Boisselier E, Astruc D. Gold nanoparticles in nanomedicine: preparations, imaging, diagnostics, therapies and toxicity. Chemical Society Reviews. 2009;38:1759–1782. doi: 10.1039/b806051g. [DOI] [PubMed] [Google Scholar]
- 159.Draz MS, Fang BA, Zhang P, Hu Z, Gu S, Weng KC, Gray JW, Chen FF. Nanoparticle-mediated Systemic Delivery of siRNA for Treatment of Cancers and Viral Infections. Theranostics. 2014;4:872–892. doi: 10.7150/thno.9404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Khlebtsov N, Bogatyrev V, Dykman L, Khlebtsov B, Staroverov S, Shirokov A, Matora L, Khanadeev V, Pylaev T, Tsyganova N, Ter-entyuk G. Analytical and Theranostic Applications of Gold Nanoparticles and Multifunctional Nanocomposites. Theranostics. 2013;3:167–180. doi: 10.7150/thno.5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Tong L, Zhao M, Zhu S, Chen J. Synthesis and application of superparamagnetic iron oxide nanoparticles in targeted therapy and imaging of cancer. Frontiers in Medicine. 2011;5:379–387. doi: 10.1007/s11684-011-0162-6. [DOI] [PubMed] [Google Scholar]
- 162.Wang YX. Superparamagnetic iron oxide based MRI contrast agents: Current status of clinical application. Quant Imaging Med Surg. 2011;1:35–40. doi: 10.3978/j.issn.2223-4292.2011.08.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Khlebtsov B, Dykman L. Biodistribution and toxicity of engineered gold nanoparticles: a review of in vitro and in vivo studies. Chemical Society Reviews. 2011;40:1647–1671. doi: 10.1039/c0cs00018c. [DOI] [PubMed] [Google Scholar]
- 164.Mejías R, Gutiérrez L, Salas G, Pérez-Yagüe S, Zotes TM, Lázaro FJ, Morales MP, Barber DF. Long term biotransformation and toxicity of dimercaptosuccinic acid-coated magnetic nanoparticles support their use in biomedical applications. Journal of Controlled Release. 2013;171:225–233. doi: 10.1016/j.jconrel.2013.07.019. [DOI] [PubMed] [Google Scholar]
- 165.Mahmoudi M, Hofmann H, Rothen-Ritishauser B, Petri-Fink A. Assessing the In Vitro and In Vivo Toxicity of Superparamagnetic Iron Oxide Nanoparticles. Chemical Reviews. 2012;112:2323–2338. doi: 10.1021/cr2002596. [DOI] [PubMed] [Google Scholar]
- 166.Wang L, Li Y-F, Zhao L, Liu Y, Meng L, Zhang K, Wu X, Zhang L, Li B, Chen C. Characterization of gold nanorods in vivo by integrated analytical techniques: their uptake, retention, and chemical forms. Analytical and Bioanalytical Chemistry. 2010;396:1105–1114. doi: 10.1007/s00216-009-3302-y. [DOI] [PubMed] [Google Scholar]
- 167.Goel R, Shah N, Visaria R, Paciotti GF, Bischof JC. Biodistribution of TNF-α-coated gold nanoparticles in an in vivo model system. Nanomedicine. 2009;4:401–410. doi: 10.2217/nnm.09.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Jensen SA, Day ES, Ko CH, Hurley LA, Luciano JP, Kouri FM, Merkel TJ, Luthi AJ, Patel PC, Cutler JI, Daniel WL, Scott AW, Rotz MW, Meade TJ, Giljohann DA, Mirkin CA, Stegh AH. Spherical Nucleic Acid Nanoparticle Conjugates as an RNAi-Based Therapy for Glioblastoma. Science translational medicine. 2013;5:209ra152. doi: 10.1126/scitranslmed.3006839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ali-Boucetta H, Kosarelos K. Pharmacology of carbon nanotubes:Toxicokinetics, excretion and tissue accumulation. Advanced Drug Delivery Reviews. 2013;65:2111–2119. doi: 10.1016/j.addr.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 170.Schipper ML, Nakayama-Ratchford N, Davis CR, Kam NWS, Chu P, Liu Z, Sun X, Dai H, Gambhir SS. A pilot toxicology study of single-walled carbon nanotubes in a small sample of mice. Nature Nanotechnology. 2008;3:216–221. doi: 10.1038/nnano.2008.68. [DOI] [PubMed] [Google Scholar]
- 171.Seabra AB, Paula AJ, de Lima R, Alves OL, Dura N. Nanotoxicity of Graphene and Graphene Oxide. Chemical Research in Toxicology. 2014;27:159–168. doi: 10.1021/tx400385x. [DOI] [PubMed] [Google Scholar]
- 172.Chou C-C, Hsiao H-Y, Hong Q-S, Chen C-H, Peng Y-W, Chen H-W, Yang P-C. Single-Walled Carbon Nanotubes Can Induce Pulmonary Injury in Mouse Model. Nano Letters. 2008;8:437–445. doi: 10.1021/nl0723634. [DOI] [PubMed] [Google Scholar]
- 173.Liu J-H, Yang S-T, Wang X, Wang H, Liu Y, Luo PG, Liu Y, Sun Y-P. Carbon Nanoparticles Trapped in VivoSimilar to Carbon Nanotubes in Time-Dependent Biodistribution. ACS Applied Materials & Interfaces. 2014;6:14672–14678. doi: 10.1021/am504022s. [DOI] [PubMed] [Google Scholar]
- 174.Mitchell LA, Lauer FT, Burchiel SW, McDonald JD. Mechanisms for how inhaled multiwalled carbon nanotubes suppress systemic immune function in mice. Nature Nanotechnology. 2009;4:451–456. doi: 10.1038/nnano.2009.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Poland CA, Duffin R, Kinloch I, Maynard A, Wallace WAH, Seaton A, Stone V, Brown S, MacNee W, Donaldson K. Carbon nanotubes introduced into the abdominal cavity ofmice show asbestoslike pathogenicity in a pilot study. Nature Nanotechnology. 2008;3:423–428. doi: 10.1038/nnano.2008.111. [DOI] [PubMed] [Google Scholar]
- 176.Tarn D, Ashley CE, Xue M, Carnes EC, Zink JI, Brinker CJ. Mesoporous Silica Nanoparticle Nanocarriers: Biofunctionality and Biocompatibility. Accounts of Chemical Research. 2013;46:792–801. doi: 10.1021/ar3000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Du L, Liao SJ, Khatib HA, Stoddart JF, Zink JI. Controlled-Access Hollow Mechanized Silica Nanocontainers. J Am Chem Soc. 2009;131:15136–15142. doi: 10.1021/ja904982j. [DOI] [PubMed] [Google Scholar]
- 178.Huang X, Li L, Liu T, Hao N, Liu H, Chen D, Tang F. The Shape Effect of Mesoporous Silica Nanoparticles on Biodistribution, Clearance, and Biocompatibility in Vivo. ACS Nano. 2011;5:5390–5399. doi: 10.1021/nn200365a. [DOI] [PubMed] [Google Scholar]
- 179.Lu Y, Fan H, Stump A, Ward TL, Rieker T, Brinker CJ. Aerosol-assisted self-assembly of mesostructured spherical nanoparticles. Nature. 1999;398:223–226. [Google Scholar]
- 180.Han L, Zhou Y, He T, Song G, Wu F, Jiang F, Hu J. One-pot morphology-controlled synthesis of various shaped mesoporous silica nanoparticles. Journal of Materials Science. 2013;48:5718–5726. [Google Scholar]
- 181.Meng H, Yang S, Li Z, Xia T, Chen J, Ji Z, Zhang H, Wang X, Lin S, Huang C, Zhou H, Zink JI, Nel AE. Aspect Ratio Determines the Quantity of Mesoporous Silica Nanoparticle Uptake by a Small GTPase-Dependent Macropinocytosis Mechanism. ACS Nano. 2011;5:4434–4447. doi: 10.1021/nn103344k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Trewyn BG, Nieweg JA, Zhao Y, Lin VS-Y. Biocompatible mesoporous silica nanoparticles with different morphologies for animal cell membrane penetration. Chemical Engineering Journal. 2008;137:23–29. [Google Scholar]
- 183.Xiong L, Du X, Shi B, Bi J, Kleitz F, Qiao SZ. Tunable stellate mesoporous silica nanoparticles for intracellular drug delivery. Journal of Materials Chemistry B. 2015;3:1712–1721. doi: 10.1039/c4tb01601g. [DOI] [PubMed] [Google Scholar]
- 184.Du X, Qiao SZ. Dendritic Silica Particles with Center-Radial Pore Channels: Promising Platforms for Catalysis and Biomedical Applications. Small. 2015;11:392–413. doi: 10.1002/smll.201401201. [DOI] [PubMed] [Google Scholar]
- 185.Huh S, Wiench JW, Yoo J-C, Pruski M, Lin VS-Y. Organic Functionalization and Morphology Control of Mesoporous Silicas via a Co-Condensation Synthesis Method. Chemistry of Materials. 2003;15:4247–4256. [Google Scholar]
- 186.Nandiyanto ABD, Kim S-G, Iskandar F, Okuyama K. Synthesis of spherical mesoporous silica nanoparticles with nanometer-size controllable pores and outer diameters. Microporous and Mesoporous Materials. 2009;120:447–453. [Google Scholar]
- 187.Phillips E, Penate-Medina O, Zanzonico PB, Carvajal RD, Mohan P, Ye Y, Humm J, Gonen M, Kalaigian H, Schoder H, Strauss HW, Larson SM, Wiesner U, Bradbury MS. Clinical translation of an ultrasmall iorganic optical-PET imaging nanoparticle probe. Science translational medicine. 2014;6:260ra149. doi: 10.1126/scitranslmed.3009524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.He Q, Zhang Z, Gao Y, Shi J, Li Y. Intracellular Localization and Cytotoxicity of Spherical Mesoporous Silica Nano- and Microparticles. Small. 2009;5:2722–2729. doi: 10.1002/smll.200900923. [DOI] [PubMed] [Google Scholar]
- 189.He L, Lai H, Chen T. Dual-function nanosystem for synergetic cancer chemo-/radiotherapy through ROS-mediated signaling pathways. Biomaterials. 2015;51:30–42. doi: 10.1016/j.biomaterials.2015.01.063. [DOI] [PubMed] [Google Scholar]
- 190.Lu J, Liong M, Li Z, Zink JI, Tamanoi F. Biocompatibility, Biodistribution, and Drug-Delivery Efficiency of Mesoporous Silica Nanoparticles for Cancer Therapy in Animals. Small. 2010;6:1794–1805. doi: 10.1002/smll.201000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Souris JS, Lee C-H, Cheng S-H, Chen C-T, Yang C-S, Ho JA, Mou C-Y, Lo L-W. Surface charge-mediate rapid hepatobiliary excretion of mesoporous silica nanoparticles. Biomaterials. 2010;31:5564–5574. doi: 10.1016/j.biomaterials.2010.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Fu C, Liu T, Li L, Liu H, Chen D, Tang F. The absorption, distribution, excretion and toxicity of mesoporous silica nanoparticles in mice following different exposure routes. Biomaterials. 2013;34:2565–2575. doi: 10.1016/j.biomaterials.2012.12.043. [DOI] [PubMed] [Google Scholar]
- 193.He Q, Zhang Z, Gao F, Li Y, Shi J. In vivo Biodistribution and Urinary Excretion of Mesoporous Silica Nanoparticles: Effects of Particle Size and PEGylation. Small. 2011;7:271–280. doi: 10.1002/smll.201001459. [DOI] [PubMed] [Google Scholar]
- 194.Wang L-S, Wu L-C, Lu S-Y, Chang L-L, Teng IT, Yang C-M, Ho J-aA. Biofunctionalized Phospholipid-Capped Mesoporous Silica Nanoshuttles for Targeted Drug Delivery: Improved Water Suspensibility and Decreased Nonspecific Protein Binding. ACS Nano. 2010;4:4371–4379. doi: 10.1021/nn901376h. [DOI] [PubMed] [Google Scholar]
- 195.Allen TM, Cullis PR. Drug Delivery Systems: Entering the Mainstream. Science. 2004;303:1818–1822. doi: 10.1126/science.1095833. [DOI] [PubMed] [Google Scholar]
- 196.Egusquiguirre SP, Igartua M, Hernandez RM, Pedraz JL. Nanoparticle delivery systems for cancer therapy: advances in clinical and preclinical research. Clinical and Translational Oncology. 2012;14:83–93. doi: 10.1007/s12094-012-0766-6. [DOI] [PubMed] [Google Scholar]
- 197.Iwamoto T. Clinical Application of Drug Delivery Systems in Cancer Chemotherapy: Review of the Efficacy and Side Effects of Approved Drugs. Biological and Pharmacuetical Bulletin. 2013;36:715–718. doi: 10.1248/bpb.b12-01102. [DOI] [PubMed] [Google Scholar]
- 198.Davis ME, Chen Z, Shin DM. Nanoparticle therapeutics: an emerging treatment modality for cancer. Nature Reviews Drug Discovery. 2008;7:771–782. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]
- 199.Farokhzad OC, Langer R. Impact of Nanotechnology on Drug Delivery. ACS Nano. 2009;3:16–20. doi: 10.1021/nn900002m. [DOI] [PubMed] [Google Scholar]
- 200.Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R. Nanocarriers as an emerging platform for cancer therapy. Nature Nanotechnology. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 201.Torchilin VP. Recent Advances with Liposomes as Pharmaceutical Carriers. Nature Reviews Drug Discovery. 2005;4:145–160. doi: 10.1038/nrd1632. [DOI] [PubMed] [Google Scholar]
- 202.Reynolds JG, Geretti E, Hendriks BS, Lee H, Leonard SC, Klinz SG, Noble CO, Lücker PB, Zandstra PW, Drummond DC, Olivier KJ, Jr, Nielsen UB, Niyikiza C, Agresta SV, Wickham TJ. HER2-targeted liposomal doxorubicin displays enhanced anti-tumorigenic effects without associated cardiotoxicity. Toxicology and Applied Pharmacology. 2012;262:1–10. doi: 10.1016/j.taap.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 203.Sapra P, Allen TM. Ligand-targeted liposomal anticancer drugs. Prog Lipid Res. 2003;42:439–462. doi: 10.1016/s0163-7827(03)00032-8. [DOI] [PubMed] [Google Scholar]
- 204.Drummond DC, Noble CO, Guo Z, Hayes ME, Connolly-Ingram C, Gabriel BS, Hann B, Liu B, Park JW, Hong K, Benz CC, Marks JD, Kirpotin DB. Development of a highly stable and targetable nanoliposomal formulation of topotecan. Journal of Controlled Release. 2010;141:13–21. doi: 10.1016/j.jconrel.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 205.Noble CO, Guo Z, Hayes ME, Marks JD, Park JW, Benz CC, Kirpotin DB, Drummond DC. Characterization of highly stable liposomal and immunoliposomal formulations of vincristine and vinblastine. Cancer Chemotherapy and Pharmacology. 2009;64:741–751. doi: 10.1007/s00280-008-0923-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Batist G, Ramakrishnan G, Rao CS, Chandrasekharan A, Gutheil J, Guthrie T, Shah P, Khojasteh A, Nair MK, Hoelzer K, Tkaczuk K, Park YC, Lee LW. Reduced cardiotoxicity and preserved antitumor efficacy of liposome-encapsulated doxorubicin and cyclophosphamide compared with conventional doxorubicin and cyclophosphamide in a randomized, multicenter trial of metastatic breast cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2001;19:1444–1454. doi: 10.1200/JCO.2001.19.5.1444. [DOI] [PubMed] [Google Scholar]
- 207.Harris L, Batist G, Belt R, Rovira D, Navari R, Azarnia N, Welles L, Winer E T.D.S. Group. Liposome-encapsulated doxorubicin compared with conventional doxorubicin in a randomized multicenter trial as first-line therapy of metastatic breast carcinoma. Cancer. 2002;94:25–36. doi: 10.1002/cncr.10201. [DOI] [PubMed] [Google Scholar]
- 208.Northfelt DW, Dezube BJ, Thommes JA, Miller BJ, Fischl MA, Friedman-Kien A, Kaplan LD, Du Mond C, Mamelok RD, Henry DH. Pegylated-liposomal doxorubicin versus doxorubicin, bleomycin, and vincristine in the treatment of AIDS-related Kaposi’s sarcoma: results of a randomized phase III clinical trial. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 1998;16:2445–2451. doi: 10.1200/JCO.1998.16.7.2445. [DOI] [PubMed] [Google Scholar]
- 209.O’Brien ME, Wigler N, Inbar M, Rosso R, Grischke E, Santoro A, Catane R, Kieback DG, Tomczak P, Ackland SP, Orlandi F, Mellars L, Alland L, Tendler C C.B.C.S. Group. Reduced cardiotoxicity and comparable efficacy in a phase III trial of pegylated liposomal doxorubicin HCl (CAELYX/Doxil) versus conventional doxorubicin for first-line treatment of metastatic breast cancer. Ann Oncol. 2004;15:440–449. doi: 10.1093/annonc/mdh097. [DOI] [PubMed] [Google Scholar]
- 210.Stathopoulos GP, Boulikas T. Lipoplatin formulation review article. J Drug Deliv. 2012;2012:581363. doi: 10.1155/2012/581363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Zamboni WC, Ramalingam S, Friedland DM, Edwards RP, Stoller RG, Strychor S, Maruca L, Zamboni BA, Belani CP, Ramanathan RK. Phase I and pharmacokinetic study of pegylated liposomal CKD-602 in patients with advanced malignancies. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15:1466–1472. doi: 10.1158/1078-0432.CCR-08-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Batist G, Gelmon KA, Chi KN, Miller WH, Jr, Chia SK, Mayer LD, Swenson CE, Janoff AS, Louie AC. Safety, pharmacokinetics, and efficacy of CPX-1 liposome injection in patients with advanced solid tumors. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15:692–700. doi: 10.1158/1078-0432.CCR-08-0515. [DOI] [PubMed] [Google Scholar]
- 213.Liang Z, Yang N, Jiang Y, Hou C, Zheng J, Shi J, Zhang R, Li D, Liu Y, Zuo P. Targeting docetaxel-PLA nanoparticles simultaneously inhibit tumor growth and liver metastases of small cell lung cancer. Int J Pharm. 2015;494:337–345. doi: 10.1016/j.ijpharm.2015.08.042. [DOI] [PubMed] [Google Scholar]
- 214.Kirpotin DB, Noble CO, Hayes ME, Huang Z, Kornaga T, Zhou Y, Nielsen UB, Marks JD, Drummond DC. Building and characterizing antibody-targeted lipidic nanotherapeutics. Methods Enzymol. 2012;502:139–166. doi: 10.1016/B978-0-12-416039-2.00007-0. [DOI] [PubMed] [Google Scholar]
- 215.Laukkanen ML, Teeri TT, Keinanen K. Lipid-tagged antibodies: bacterial expression and characterization of a lipoprotein-single-chain antibody fusion protein. Protein Eng. 1993;6:449–454. doi: 10.1093/protein/6.4.449. [DOI] [PubMed] [Google Scholar]
- 216.Park JW, Hong K, Kirpotin DB, Meyer O, Papahadjopoulos D, Benz CC. Anti-HER2 immunoliposomes for targeted therapy of human tumors. Cancer Lett. 1997;118:153–160. doi: 10.1016/s0304-3835(97)00326-1. [DOI] [PubMed] [Google Scholar]
- 217.de Kruif J, Storm G, van Bloois L, Logtenberg T. Biosynthetically lipid-modified human scFv fragments from phage display libraries as targeting molecules for immunoliposomes. FEBS letters. 1996;399:232–236. doi: 10.1016/s0014-5793(96)01335-x. [DOI] [PubMed] [Google Scholar]
- 218.Williford J-M, Wu J, Ren Y, Archang MM, Leong KW, Mao H-Q. Recent Advances in Nanoparticle-Mediated siRNA Delivery. Annual Reviews in Biomedical Engineering. 2014;16:347–370. doi: 10.1146/annurev-bioeng-071813-105119. [DOI] [PubMed] [Google Scholar]
- 219.Irvine DJ. One nanoparticle, one kill. Nature Materials. 2011;10:342–343. doi: 10.1038/nmat3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Stark B, Pabst G, Prassl R. Long-term stability of sterically stabilized liposomes by freezing and freeze-drying: Effects of cryoprotectants on structure. European Journal of Pharmaceutical Sciences. 2010;41:546–555. doi: 10.1016/j.ejps.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 221.Amstad E, Kohlbrecher J, Muller E, Schweizer T, Textor M, Reimhult E. Triggered Release from Liposomes through Magnetic Actuation of Iron Oxide Nanoparticle Containing Membranes. Nano Letters. 2011;11:1664–1670. doi: 10.1021/nl2001499. [DOI] [PubMed] [Google Scholar]
- 222.Cauda V, Engelke H, Sauer A, Arcizet D, Bräuchle C, Rädler J, Bein T. Colchicine-Loaded Lipid Bilayer-Coated 50 nm Mesoporous Nanoparticles Efficiently Induce Microtubule Depolymerization upon Cell Uptake. Nano Lett. 2010;10:2484–2492. doi: 10.1021/nl100991w. [DOI] [PubMed] [Google Scholar]
- 223.Chen B, Liu M, Zhang L, Huang J, Yao J, Zhang Z. Polyethylenimine-functionalized graphene oxide as an efficient gene delivery vector. Journal of Materials Chemistry. 2011;21:7736–7741. [Google Scholar]
- 224.Epler K, Padilla D, Phillips G, Crowder P, Castillo R, WIlkinson D, WIlkinson B, Burgard C, Kalinich R, Townson J, Chackerian B, Willman C, Peabody D, Wharton W, Brinker CJ, Ashley C, Carnes E. Delivery of Ricin Toxin A-Chain by Peptide-Targeted Mesoporous Silica Nanoparticle-Supported Lipid Bilayers. Advanced Healthcare Materials. 2012;1:348–353. doi: 10.1002/adhm.201200022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Feng L, Zhang S, Zhuang L. Graphene based gene transfection. Nanoscale. 2011;3:1252–1257. doi: 10.1039/c0nr00680g. [DOI] [PubMed] [Google Scholar]
- 226.Kim H, Namgung R, Singha K, Oh I-K, Kim WJ. Graphene Oxide–Polyethylenimine Nanoconstruct as a Gene Delivery Vector and Bioimaging Tool. Bioconjugate chemistry. 2011;22:2558–2567. doi: 10.1021/bc200397j. [DOI] [PubMed] [Google Scholar]
- 227.Liu L, Wei Y, Zhai S, Chen Q, Xing D. Dihydroartemisinin and transferrin dual-dressed nano-graphene oxide for a pH-triggered chemotherapy. Biomaterials. 2015;62:35–46. doi: 10.1016/j.biomaterials.2015.05.036. [DOI] [PubMed] [Google Scholar]
- 228.Mattingly SJ, O’Toole M, James KT, Clark GJ, Nantz MH. Magnetic Nanoparticle- Supported Lipid Bilayers for Drug Delivery. Langmuir. 2015;31:3326–3332. doi: 10.1021/la504830z. [DOI] [PubMed] [Google Scholar]
- 229.Savarala S, Ahmed S, Ilies MA, Wunder SL. Formation and Colloidal Stability of DMPC Supported Lipid Bilayers on SiO2 Nanobeads. Langmuir. 2010;26:12081–12088. doi: 10.1021/la101304v. [DOI] [PubMed] [Google Scholar]
- 230.Swain AK, Pradhan L, Bahadur D. Polymer Stabilized Fe3O4-Graphene as an Amphiphilic Drug Carrier for Thermo-Chemotherapy of Cancer. Applied Materials & Interfaces. 2015;7:8013–8022. doi: 10.1021/acsami.5b02536. [DOI] [PubMed] [Google Scholar]
- 231.Tam NCM, Scott BMT, Voicu D, Wilson BC, Zheng G. Facile Synthesis of Raman Active Phospholipid Gold Nanoparticles. Bioconjugate chemistry. 2010;21:2178–2182. doi: 10.1021/bc100386a. [DOI] [PubMed] [Google Scholar]
- 232.Scarberry KE, Dickerson EB, McDonald JF, Zhang ZJ. Magnetic nanoparticle-peptide conjugates for in vitro and in vivo targeting and extraction of cancer cells. J Am Chem Soc. 2008;130:10258–10262. doi: 10.1021/ja801969b. [DOI] [PubMed] [Google Scholar]
- 233.Chen Y, Yang W, Chang B, Hua H, Fang X, Sha X. In vivo distribution and antitumor activity of doxorubicin-loaded N-isopropylacrylamide-co-methacrylic acid coated mesoporous silica nanoparticles and safety evaluation. European Journal of Pharmaceutics and Biopharmaceutics. 2013;85:406–412. doi: 10.1016/j.ejpb.2013.06.015. [DOI] [PubMed] [Google Scholar]
- 234.Meng H, Xue M, Xia T, Ji Z, Tarn DY, Zink JI, Nel AE. Use of Size and a Copolymer Design Feature to Improve the Biodistribution and the Enhanced Permeability and Retention Effect of Doxorubicin-Loaded Mesoporous Silica Nanoparticles in a Murine Xenograft Tumor Model. ACS Nano. 2011;5:4131–4144. doi: 10.1021/nn200809t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Li L, Tang F, Liu H, Liu T, Hao N, Chen D, Teng X, He J. In Vivo Delivery of Silica Nanorattle Encapsulated Docetaxel for Liver Cancer Therapy with Low Toxicity and High Efficacy. ACS Nano. 2010;4:6874–6882. doi: 10.1021/nn100918a. [DOI] [PubMed] [Google Scholar]
- 236.Meng H, Zhao Y, Dong J, Xue M, Lin Y-S, Ji Z, Mai WX, Zhang H, Chang CH, Brinker CJ, Zink JI, Nel AE. Two-Wave Nanotherapy To Target the Stroma and Optimize Gemcitabine Delivery To a Human Pancreatic Cancer Model in Mice. ACS Nano. 2013;7:10048–10065. doi: 10.1021/nn404083m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Hwang AA, Lu J, Tamanoi F, Zink JI. Functional Nanovavles on Protein-Coated Nanoparticles for In vitro and In vivo Controlled Drug Delivery. Small. 2015;11:319–328. doi: 10.1002/smll.201400765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Lu J, Li Z, Zink JI, Tamanoi F. In vivo tumor suppression efficacy of mesoporous silica nanoparticles-based drug-delivery system: enhanced efficacy by folate modification. Nanomedicine: NBM. 2012;8:212–220. doi: 10.1016/j.nano.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Lin Y-S, Haynes CL. Impacts of Mesoporous Silica Nanoparticle Size, Pore Ordering, and Pore Integrity on Hemolytic Activity. J Am Chem Soc. 2010;132:4834–4842. doi: 10.1021/ja910846q. [DOI] [PubMed] [Google Scholar]
- 240.Han N, Zhao Q, Wan L, Wang Y, Gao Y, Wang P, Wang Z, Zhang J, Jiang T, Wang S. Hybrid Lipid-Capped Mesoporous SIlica for Stimuli-Resposive Drug Release and Overcoming Multidrug Resistance. Applied Materials & Interfaces. 2015;7:3342–3351. doi: 10.1021/am5082793. [DOI] [PubMed] [Google Scholar]
- 241.Mackowiak SA, Schmidt A, Weiss V, Argyo C, von Schirnding C, Bein T, Bräuchle C. Targeted Drug Delivery in Cancer Cells with Red-Light Photoactivated Mesoporous Silica Nanoparticles. Nano Lett. 2013;13:2576–2583. doi: 10.1021/nl400681f. [DOI] [PubMed] [Google Scholar]
- 242.Meng H, Wang M, Liu H, Liu X, Situ A, Wu B, Ji Z, Chang CH, Nel AE. Use of a lipid-coated mesoporous silica nanoparticle platform for synergistic gemcitabine and Paclitaxel delivery to human pancreatic cancer in mice. ACS Nano. 2015;9:3540–3557. doi: 10.1021/acsnano.5b00510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Porotto M, Yi F, Moscona A, LaVan DA. Synthetic Protocells Interact with Viral Nanomachinery and Inactivate Pathogenic Human Virus. PloS one. 2011;6:e16874. doi: 10.1371/journal.pone.0016874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Wang D, Huang J, Wang X, Yu Y, Zhang H, Chen Y, Liu J, Sun Z, Zou H, Sun D, Zhou G, Zhang G, Lu Y, Zhong Y. The eradication of breast cancer cells and stem cells by 8-hydroxyquinoline-loaded hyaluronan modified mesoporous silica nanoparticle-supported lipid bilayers containing docetaxel. Biomaterials. 2013;34:7662–7673. doi: 10.1016/j.biomaterials.2013.06.042. [DOI] [PubMed] [Google Scholar]
- 245.Zhang X, Li F, Guo S, Chen X, Wang X, Li J, Gan Y. Biofunctionalized polymer-lipid supported mesoporous silica nanoparticles for release of chemotherapeutics in multidrug resistant cancer cells. Biomaterials. 2014;35:3650–3665. doi: 10.1016/j.biomaterials.2014.01.013. [DOI] [PubMed] [Google Scholar]
- 246.Roggers RA, Lin VS-Y, Trewyn BG. Chemically Reducible Lipid Bilayer Coated Mesoporous Silica Nanoparticles Demonstrating Controlled Release and Hela and Normal Mouse Liver Biocompatibility and Cellular Internalization. Molecular Pharmaceutics. 2012;9:2770–2777. doi: 10.1021/mp200613y. [DOI] [PubMed] [Google Scholar]
- 247.Hu C-MJ, Zhang L, Aryal S, Cheung C, Fang RH, Zhang L. Erythrocyte membrane-camouflaged polymeric nanoparticles as a biomimetic delivery platform. Proceedings of the National Academy of Sciences. 2011;108:10980–10985. doi: 10.1073/pnas.1106634108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Kumar R, Roy I, Ohulchanskky TY, Vathy LA, Bergey EJ, Sajjad M. In Vivo Biodistribution and Clearance Studies Using Multimodal Organically Modified Silica Nanoparticles. ACS Nano. 2010;4:699–708. doi: 10.1021/nn901146y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Liu T, Li L, Teng X, Huang X, Liu H, Chen D, Ren J, He J, Tang F. Single and repeated dose toxicity of mesoporous hollow silica nanoparticles in intravenously exposed mice. Biomaterials. 2011;32:1657–1668. doi: 10.1016/j.biomaterials.2010.10.035. [DOI] [PubMed] [Google Scholar]
- 250.Chen Y, Wang X, Liu T, Zhang DS, Wang Y, GH, Di W. Highly effective antiangiogenesis via magnetic mesoporous silica-based siRNA vehicle targeting the VEGF gene for orthotopic ovarian cancer therapy. International Journal of Nanomedicine. 2015;10:2579–2594. doi: 10.2147/IJN.S78774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Kim J, Kim HS, Lee N, Kim T, Kim H, Yu T, Song IC, Moon WK, Hyeon T. Multifunctional Uniform Nanoparticles Composed of a Magnetite Nanocrystal Core and a Mesoporous Silica Shell for Magnetic Resonance and Fluorescence Imaging and for Drug Delivery. Angew Chem Int Ed. 2008;47:8438–8443. doi: 10.1002/anie.200802469. [DOI] [PubMed] [Google Scholar]
- 252.Wu S-H, Lin Y-S, Hung Y, Chou Y-H, Hsu Y-H, Chang C, Mou C-Y. Multifunctional Mesoporous Silica Nanoparticles for Intracellular Labeling and Animal Magnetic Resonance Imaging Studies. ChemBioChem. 2008;9:53–57. doi: 10.1002/cbic.200700509. [DOI] [PubMed] [Google Scholar]
- 253.Nielsen UB, Kirpotin DB, Pickering EM, Drummond DC, Marks JD. A novel assay for monitoring internalization of nanocarrier coupled antibodies. BMC Immunol. 2006;7:24. doi: 10.1186/1471-2172-7-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Mérian J, Gravier J, Navarro F, Texier I. Fluorescent Nanoprobes Dedicated to in Vivo Imaging: From Preclinical Validations to Clinical Translation. Molecules. 2012;17:5564–5591. doi: 10.3390/molecules17055564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Benezra M, Penate-Medina O, Zanzonico PB, Schaer D, Ow H, Burns A, DeStanchina E, Longo V, Herz E, Iyer S, Wolchok J, Larson SM, Wiesner U, Bradbury MS. Multimodal silica nanoparticles are effective cancer-targeted probes in a model of human melanoma. Journal of Clinical Investigation. 2011;121:2768–2780. doi: 10.1172/JCI45600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Bradbury MS, Phillips E, Montero PH, Cheal SM, Stambuk H, Durack JC, Sofocleous CT, Meester RJ, Wiesner U, Patel S. Clinically-translated silica nanoparticles as dual-modality cancer-targeted probes for image-guided surgery and interventions. Integrative Biology. 2013;5:74–86. doi: 10.1039/c2ib20174g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Taratula O, Garbuzenko OB, Chen AM, Minko T. Innovative strategy for treatment of lung cancer: targeted nanotechnology-based inhalation co-delivery of anticancer drugs and siRNA. Journal of Drug Targeting. 2011;19:900–914. doi: 10.3109/1061186X.2011.622404. [DOI] [PubMed] [Google Scholar]
- 258.Meng H, Mai WX, Zhang H, Xue M, Xia T, Lin S, Wang X, Zhao Y, Ji Z, Zink JI, Nel AE. Codelivery of an Optimal Drug/siRNA Combination Using Mesoporous Silica Nanoparticles To Overcome Drug Resistance in Breast Cancer in Vitro and in Vivo. ACS Nano. 2013;7:994–1005. doi: 10.1021/nn3044066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Nehoff H, Parayath NN, Domanovitch L, Taurin S, Greish K. Nanomedicine for drug targeting: strategies beyond the enhanced permeability and retention effect. International Journal of Nanomedicine. 2014;9:2539–2555. doi: 10.2147/IJN.S47129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Prabhakar U, Maeda H, Jain RK, Sevick-Muraca EM, Zamboni W, Farokhzad OC, Barry ST, Gabizon A, Grodzinski P, Blakey DC. Challenges and Key Considerations of the Enhanced Permeability and Retention Effect for Nanomedicine Drug Delivery in Oncology. Cancer research. 2013;73:2412–2417. doi: 10.1158/0008-5472.CAN-12-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Gao F, Botella P, Corma A, Blesa J, Dong L. Monodispersed mesoporous silica nanoparticles with very large pores for enhanced adsorption and release of DNA. J Phys Chem B. 2009;113:1796–1804. doi: 10.1021/jp807956r. [DOI] [PubMed] [Google Scholar]
- 262.Yang H, Zheng K, Zhang Z, Shi W, Jing S, Wanga L, Zheng W, Zhao D, Xu J, Zhang P. Adsorption and protection of plasmid DNA on mesoporous silica nanoparticles modified with various amounts of organosilane. Journal of Colloid and Interface Science. 2012;369:317–322. doi: 10.1016/j.jcis.2011.12.043. [DOI] [PubMed] [Google Scholar]
- 263.Bhattarai SR, Muthuswamy E, Wani A, Brichacek M, Castañeda AL, Brock SL, Oupicky D. Enhanced gene and siRNA delivery by polycation-modified mesoporous silica nanoparticles loaded with chloroquine. Pharmaceutical Research. 2010;27:2556–2568. doi: 10.1007/s11095-010-0245-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Chen W, Tsai P-H, Hung Y, Chiou S-H, Mou C-Y. Nonviral Cell Labeling and Differentiation Agent for Induced Pluripotent Stem Cells Based on Mesoporous Silica Nanoparticles. ACS Nano. 2013;7:8423–8440. doi: 10.1021/nn401418n. [DOI] [PubMed] [Google Scholar]
- 265.Du X, Shi B, Tang Y, Dai S, Qiao SZ. Label-free dendrimer-like silica nanohybrids for traceable and controlled gene delivery. Biomaterials. 2014;35:5580–5590. doi: 10.1016/j.biomaterials.2014.03.051. [DOI] [PubMed] [Google Scholar]
- 266.Kim MH, Na HK, Kim YK, Ryoo SR, Cho HS, Lee KE, Jeon H, Ryoo R, Min DH. Facile synthesis of monodispersed mesoporous silica nanoparticles with ultralarge pores and their application in gene delivery. ACS Nano. 2011;5:3568–3576. doi: 10.1021/nn103130q. [DOI] [PubMed] [Google Scholar]
- 267.Radu DR, Lai CY, Jeftinija K, Rowe EW, Jeftinija S, Lin VS. A Polyamidoamine Dendrimer-Capped Mesoporous Silica Nanosphere-Based Gene Transfection Reagent. Journal of the American Chemical Society. 2004;126:13216–13217. doi: 10.1021/ja046275m. [DOI] [PubMed] [Google Scholar]
- 268.Xia T, Kovochich M, Liong M, Meng H, Kabehie S, George S, Zink JI, Nel AE. Polyethyleneimine coating enhances the cellular uptake of mesoporous silica nanoparticles and allows safe delivery of siRNA and DNA constructs. ACS Nano. 2009;3:3273–3286. doi: 10.1021/nn900918w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Wang Z, Liu G, Zheng H, Chen X. Rigid nanoparticle based delivery of anti-cancer siRNA: challenges and opportunities. Biotechnology advances. 2014;32:831–843. doi: 10.1016/j.biotechadv.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Guo W, Tian H, Dong X, Bai J, Yang X. Knockdown of Gli1 by small-interfering RNA enhances the effects of BCNU on the proliferation and apoptosis of glioma U251 cells. Int J Clin Exp Pathol. 2015;8:7762–7773. [PMC free article] [PubMed] [Google Scholar]
- 271.Salameh A, Lee AK, Cardó-Vila M, Nunes DN, Efstathiou E, Staquicini FI, Dobroff AS, Marchiò S, Navone NM, Hosoya H, Lauer RC, Wen SJ, Salmeron CC, Hoang A, Newsham I, Lima LA, Carraro DM, Oliviero S, Kolonin MG, Sidman RL, Do KA, Troncoso P, Logothetis CJ, Brentani RR, Calin GA, Cavenee WK, Dias-Neto E, Pasqualini R, Arap W. PRUNE2 is a human prostate cancer suppressor regulated by the intronic long noncoding RNA PCA3. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:8403–8408. doi: 10.1073/pnas.1507882112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Miele E, Spinelli GP, Miele E, Di Fabrizio E, Ferretti E, Tomao S, Gulino A. Nanoparticle-based delivery of small interfering RNA: challenges for cancer therapy. International Journal of Nanomedicine. 2012;7:3637–3657. doi: 10.2147/IJN.S23696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Lee SJ, Yook S, Yhee JY, Yoon HY, Kim MG, Ku SH, Kim SH, Park JH, Jeong JH, Kwon IC, Lee S, Lee H, Kim K. Co-delivery of VEGF and Bcl-2 dual-targeted siRNA polymer using a single nanoparticle for synergistic anti-cancer effects in vivo. J Control Release. 2015 doi: 10.1016/j.jconrel.2015.08.032. [DOI] [PubMed] [Google Scholar]
- 274.Ku SH, Kim K, Choi K, Kim SH, Kwon IC. Tumor-Targeting Multifunctional Nanoparticles for siRNA Delivery: Recent Advances in Cancer Therapy. Advanced Healthcare Materials. 2014;3:1182–1193. doi: 10.1002/adhm.201300607. [DOI] [PubMed] [Google Scholar]
- 275.Chen AM, Zhang M, Wei D, Stueber D, Taratula O, Minko T, He H. Co-delivery of doxorubicin and Bcl-2 siRNA by mesoporous silica nanoparticles enhances the efficacy of chemotherapy in multidrug-resistant cancer cells. Small. 2009;5:2673–2677. doi: 10.1002/smll.200900621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Hom C, Lu J, Liong M, Luo H, Li Z, Zink JI, Tamanoi F. Mesoporous Silica Nanoparticles Facilitate Delivery of siRNA to Shutdown Signaling Pathways in Mammalian Cells. Small. 2010;6:1185–1190. doi: 10.1002/smll.200901966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Ma X, Zhao Y, Ng KW, Zhao Y. Integrated Hollow Mesoporous Silica Nanoparticles for Target Drug/siRNA Co-Delivery. Chemistry: A European Journal. 2013;19:15593–15603. doi: 10.1002/chem.201302736. [DOI] [PubMed] [Google Scholar]
- 278.Li X, Xie QR, Zhanga J, Xia W, Gu H. The packaging of siRNA within the mesoporous structure of silica nanoparticles. Biomaterials. 2011;32:9546–9556. doi: 10.1016/j.biomaterials.2011.08.068. [DOI] [PubMed] [Google Scholar]
- 279.Finlay J, Roberts CM, Dong J, Zink JI, Tamanoi F, Glackin CA. Mesoporous silica nanoparticle delivery of chemically modified siRNA against TWIST1 leads to reduced tumor burden. Nanomedicine: Nanotechnology, Biology, and Medicine. 2015;11:1657–1666. doi: 10.1016/j.nano.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Li X, Chen Y, Wang M, Ma Y, Xia W, Gu H. A mesoporous silica nanoparticle e PEI e Fusogenic peptide system for siRNA delivery in cancer therapy. Biomaterials. 2013;34:1391–1401. doi: 10.1016/j.biomaterials.2012.10.072. [DOI] [PubMed] [Google Scholar]
- 281.Morry J, Ngamcherdtrakul W, Gu S, Goodyear SM, Castro DJ, Reda MM, Sangvanich T, Yantasee W. Dermal delivery of HSP47 siRNA with NOX4-modulating mesoporous silica-based nanoparticles for treating fibrosis. Biomaterials. 2015;66:41–52. doi: 10.1016/j.biomaterials.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Ngamcherdtrakul W, Morry J, Gu S, Castro DJ, Goodyear SM, Sangvanich T, Reda MM, Lee R, Mihelic SA, Beckman BL, Hu Z, Gray JW, Yantasee W. Cationic Polymer Modified Mesoporous Silica Nanoparticles for Targeted SiRNA Delivery to HER2+ Breast Cancer. Advanced Functional Materials. 2015;25:2646–2659. doi: 10.1002/adfm.201404629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Shen J, Kim H-C, Su H, Wang F, Wolfram J, Kiriu D, Mai J, Mu C, Ji L-N, Mao Z-W, Shen H. Cyclodextrin and Polyethylenimine Functionalized Mesoporous Silica Nanoparticles for Delivery of siRNA Cancer Therapeutics. Theranostics. 2014;4:487–497. doi: 10.7150/thno.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.El-Sayed A, Futaki S, Harashima H. Delivery of macromolecules using arginine-rich cell-penetrating peptides: ways to overcome endosomal entrapment. AAPS J. 2009;11:13–22. doi: 10.1208/s12248-008-9071-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Hou KK, Pan H, Schlesinger PH, Wickline SA. A role for peptides in overcoming endosomal entrapment in siRNA delivery - A focus on melittin. Biotechnol Adv. 2015;33:931–940. doi: 10.1016/j.biotechadv.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Hattori Y, Nakamura A, Arai S, Kawano K, Maitani Y, Yonemochi E. siRNA delivery to lung-metastasized tumor by systemic injection with cationic liposomes. J Liposome Res. 2015:1–8. doi: 10.3109/08982104.2014.992024. [DOI] [PubMed] [Google Scholar]
- 287.Mudhakir D, Akita H, Tan E, Harashima H. A novel IRQ ligand-modified nano-carrier targeted to a unique pathway of caveolar endocytic pathway. J Control Release. 2008;125:164–173. doi: 10.1016/j.jconrel.2007.10.020. [DOI] [PubMed] [Google Scholar]
- 288.Nakamura Y, Kogure K, Futaki S, Harashima H. Octaarginine-modified multifunctional envelope-type nano device for siRNA. J Control Release. 2007;119:360–367. doi: 10.1016/j.jconrel.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 289.Taetz S, Bochot A, Surace C, Arpicco S, Renoir JM, Schaefer UF, Marsaud V, Kerdine-Roemer S, Lehr CM, Fattal E. Hyaluronic acid-modified DOTAP/DOPE liposomes for the targeted delivery of anti-telomerase siRNA to CD44-expressing lung cancer cells. Oligonucleotides. 2009;19:103–116. doi: 10.1089/oli.2008.0168. [DOI] [PubMed] [Google Scholar]
- 290.Hyndman L, Lemoine JL, Huang L, Porteous DJ, Boyd AC, Nan X. HIV-1 Tat protein transduction domain peptide facilitates gene transfer in combination with cationic liposomes. J Control Release. 2004;99:435–444. doi: 10.1016/j.jconrel.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 291.Kale AA, Torchilin VP. Enhanced transfection of tumor cells in vivo using “Smart” pH-sensitive TAT-modified pegylated liposomes. J Drug Target. 2007;15:538–545. doi: 10.1080/10611860701498203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Yu DD, Wang CT, Shi HS, Li ZY, Pan L, Yuan QZ, Leng F, Wen Y, Chen X, Wei YQ. Enhancement of cisplatin sensitivity in Lewis Lung carcinoma by liposome-mediated delivery of a survivin mutant. J Exp Clin Cancer Res. 2010;29:46. doi: 10.1186/1756-9966-29-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Keefe AD, Pia S, Ellington A. Aptamers as therapeutics. Nature Reviews Drug Discovery. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Sundaram P, Kurniawan H, Byrne M, Wower J. Therapuetic RNA aptamers in clinical trials. European Journal of Pharmaceutical Sciences. 2013;48:259–271. doi: 10.1016/j.ejps.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 295.Lee SK, Park MW, Yang EG, Yu J, Jeong S. An RNA aptamer that binds to the b-catenin interaction domain of TCF-1 protein. Biochemical and Biophysical Research Communications. 2005;327:294–299. doi: 10.1016/j.bbrc.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 296.Lee HK, Choi YS, Park YA, Jeong S. Modulation of Oncogenic Transcription and Alternative Splicing by B-Catenin and an RNA Aptamer in Colon Cancer Cells. Cancer research. 2006;66:10560–10566. doi: 10.1158/0008-5472.CAN-06-2526. [DOI] [PubMed] [Google Scholar]
- 297.Bardeesy N, Pelletier J. Overlapping RNA and DNA binding domains of the wt1 tumor suppressor gene product. Nucleic acids research. 1998;26:1784–1792. doi: 10.1093/nar/26.7.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.An Y, Wu J, Yang B, Zhu Z, Gao M, Yu C, Yang CJ. Selection and Application of DNA Aptamer Against Oncogene Amplified in Breast Cancer 1. Journal of Molecular Evolution. 2015;81:179–185. doi: 10.1007/s00239-015-9703-y. [DOI] [PubMed] [Google Scholar]
- 299.Guo WM, Kong KW, Brown CJ, Quah ST, Yeo HL, Hoon S, Seow Y. Identification and Characterization of an eIF4e DNA Aptamer That Inhibits Proliferation With High Throughput Sequencing Molecular Therapy. Nucleic Acids. 2014;3:e217. doi: 10.1038/mtna.2014.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Slupianek A, Dasgupta Y, Ren S, Gurdek E, Donlin M, Nieborowska-Skorska M, Fleury F, Skorski T. Targeting RAD51 phosphotyrosine-315 to prevent unfaithful recombination repair in BCR-ABL1 leukemia. Blood. 2011;118:1062–1068. doi: 10.1182/blood-2010-09-307256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Rérole A, Gobbo J, De Thonel A, Schmitt E, de Barros JPP, Hammann A, Lanneau D, Fourmaux E, Deminov O, Micheau O, Lagrost L, Colas P, Kroemer G, Garrido C. Peptides and Aptamers Targeting HSP70: A Novel Approach for Anticancer Chemotherapy. Cancer research. 2011;71:484–495. doi: 10.1158/0008-5472.CAN-10-1443. [DOI] [PubMed] [Google Scholar]
- 302.Gibert B, Simon S, Dimitrova V, Diaz-Latoud C, Arrigo A-P. Peptide aptamers: tools to negatively or positively modulate HSPB1(27) function. Philosophical Transactions of the Royal Society B. 2013;368:20120075. doi: 10.1098/rstb.2012.0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Jølck R, Feldborg L, Andersen S, Moghimi SM, Andresen T. Engineering Liposomes and Nanoparticles for Biological Targeting. In: Nyanhongo GS, Steiner W, Gübitz G, editors. Biofunctionalization of Polymers and their Applications. Springer; Berlin Heidelberg: 2011. pp. 251–280. [Google Scholar]
- 304.Nobs L, Buchegger F, Gurny R, Allémann E. Current methods for attaching targeting ligands to liposomes and nanoparticles. Journal of Pharmaceutical Sciences. 2004;93:1980–1992. doi: 10.1002/jps.20098. [DOI] [PubMed] [Google Scholar]
- 305.Sperling RA, Parak WJ. Surface modification, functionalization and bioconjugation of colloidal inorganic nanoparticles. 2010 doi: 10.1098/rsta.2009.0273. [DOI] [PubMed] [Google Scholar]
- 306.Santra S, Yang H, Dutta D, Stanley JT, Holloway PH, Tan W, Moudgil BM, Mericle RA. TAT conjugated, FITC doped silica nanoparticles for bioimaging applications. Chemical Communications. 2004:2810–2811. doi: 10.1039/b411916a. [DOI] [PubMed] [Google Scholar]
- 307.Martin FJ, Hubbell WL, Papahadjopoulos D. Immunospecific targeting of liposomes to cells: a novel and efficient method for covalent attachment of Fab’ fragments via disulfide bonds. Biochemistry. 1981;20:4229–4238. doi: 10.1021/bi00517a043. [DOI] [PubMed] [Google Scholar]
- 308.Shaik MS, Kanikkannan N, Singh M. Conjugation of anti-My9 antibody to stealth monensin liposomes and the effect of conjugated liposomes on the cytotoxicity of immunotoxin. Journal of Controlled Release. 2001;76:285–295. doi: 10.1016/s0168-3659(01)00450-3. [DOI] [PubMed] [Google Scholar]
- 309.Kirpotin D, Park JW, Hong K, Zalipsky S, Li W-L, Carter P, Benz CC, Papahadjopoulos D. Sterically Stabilized Anti-HER2 Immunoliposomes: Design and Targeting to Human Breast Cancer Cells in Vitro. Biochemistry. 1997;36:66–75. doi: 10.1021/bi962148u. [DOI] [PubMed] [Google Scholar]
- 310.Park JW, Hong K, Carter P, Asgari H, Guo LY, Keller GA, Wirth C, Shalaby R, Kotts C, Wood WI. Development of anti-p185HER2 immunoliposomes for cancer therapy. Proceedings of the National Academy of Sciences. 1995;92:1327–1331. doi: 10.1073/pnas.92.5.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Park JW, Hong K, Kirpotin DB, Colbern G, Shalaby R, Baselga J, Shao Y, Nielsen UB, Marks JD, Moore D, Papahadjopoulos D, Benz CC. Anti-HER2 Immunoliposomes: Enhanced Efficacy Attributable to Targeted Delivery. Clinical Cancer Research. 2002;8:1172–1181. [PubMed] [Google Scholar]
- 312.Marqués-Gallego P, de Kroon AIPM. Ligation Strategies for Targeting Liposomal Nanocarriers. BioMed Research International. 2014;2014:12. doi: 10.1155/2014/129458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Kolb HC, Finn MG, Sharpless KB. Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angewandte Chemie International Edition. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 314.Baskin JM, Prescher JA, Laughlin ST, Agard NJ, Chang PV, Miller IA, Lo A, Codelli JA, Bertozzi CR. Copper-free click chemistry for dynamic in vivo imaging. Proceedings of the National Academy of Sciences. 2007;104:16793–16797. doi: 10.1073/pnas.0707090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Blackman ML, Royzen M, Fox JM. Tetrazine Ligation: Fast Bioconjugation Based on Inverse-Electron-Demand Diels–Alder Reactivity. Journal of the American Chemical Society. 2008;130:13518–13519. doi: 10.1021/ja8053805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Schmitt J, Hess H, Stunnenberg H. Affinity purification of histidine-tagged proteins. Mol Biol Rep. 1993;18:223–230. doi: 10.1007/BF01674434. [DOI] [PubMed] [Google Scholar]
- 317.Janknecht R, de Martynoff G, Lou J, Hipskind RA, Nordheim A, Stunnenberg HG. Rapid and efficient purification of native histidine-tagged protein expressed by recombinant vaccinia virus. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:8972–8976. doi: 10.1073/pnas.88.20.8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Hainfeld JF, Liu W, Halsey CMR, Freimuth P, Powell RD. Ni NTA Gold Clusters Target His-Tagged Proteins. Journal of Structural Biology. 1999;127:185–198. doi: 10.1006/jsbi.1999.4149. [DOI] [PubMed] [Google Scholar]
- 319.Parry DA, Fraser RD, Squire JM. Fifty years of coiled-coils and alpha-helical bundles: a close relationship between sequence and structure. J Struct Biol. 2008;163:258–269. doi: 10.1016/j.jsb.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 320.Litowski JR, Hodges RS. Designing heterodimeric two-stranded alpha-helical coiled-coils. Effects of hydrophobicity and alpha-helical propensity on protein folding, stability, and specificity. J Biol Chem. 2002;277:37272–37279. doi: 10.1074/jbc.M204257200. [DOI] [PubMed] [Google Scholar]
- 321.Mortier J, Nyakatura EK, Reimann O, Huhmann S, Daldrop JO, Baldauf C, Wolber G, Miettinen MS, Koksch B. Coiled-coils in phage display screening: insight into exceptional selectivity provided by molecular dynamics. J Chem Inf Model. 2015;55:495–500. doi: 10.1021/ci500689c. [DOI] [PubMed] [Google Scholar]
- 322.Ayriss J, Woods T, Bradbury A, Pavlik P. High-throughput screening of single-chain antibodies using multiplexed flow cytometry. J Proteome Res. 2007;6:1072–1082. doi: 10.1021/pr0604108. [DOI] [PubMed] [Google Scholar]
- 323.Velappan N, Fisher HE, Pesavento E, Chasteen L, D’Angelo S, Kiss C, Longmire M, Pavlik P, Bradbury AR. A comprehensive analysis of filamentous phage display vectors for cytoplasmic proteins: an analysis with different fluorescent proteins. Nucleic acids research. 2010;38:e22. doi: 10.1093/nar/gkp809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Hiller Y, Gershoni JM, Bayer EA, Wilchek M. Biotin binding to avidin. Oligosaccharide side chain not required for ligand association. 1987 doi: 10.1042/bj2480167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Green NM. Avidin and streptavidin. Methods Enzymol. 1990;184:51–67. doi: 10.1016/0076-6879(90)84259-j. [DOI] [PubMed] [Google Scholar]
- 326.Kawato T, Mizohata E, Shimizu Y, Meshizuka T, Yamamoto T, Takasu N, Matsuoka M, Matsumura H, Kodama T, Kanai M, Doi H, Inoue T, Sugiyama A. Structure-based design of a streptavidin mutant specific for an artificial biotin analogue. J Biochem. 2015;157:467–475. doi: 10.1093/jb/mvv004. [DOI] [PubMed] [Google Scholar]
- 327.Weir C, Hudson AL, Moon E, Ross A, Alexander M, Peters L, Langova V, Clarke SJ, Pavlakis N, Davey R, Howell VM. Streptavidin: a novel immunostimulant for the selection and delivery of autologous and syngeneic tumor vaccines. Cancer Immunol Res. 2014;2:469–479. doi: 10.1158/2326-6066.CIR-13-0157. [DOI] [PubMed] [Google Scholar]
- 328.Yumura K, Ui M, Doi H, Hamakubo T, Kodama T, Tsumoto K, Sugiyama A. Mutations for decreasing the immunogenicity and maintaining the function of core streptavidin. Protein Sci. 2013;22:213–221. doi: 10.1002/pro.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Torchilin VP, Goldmacher VS, Smirnov VN. Comparative studies on covalent and noncovalent immobilization of protein molecules on the surface of liposomes. Biochemical and Biophysical Research Communications. 1978;85:983–990. doi: 10.1016/0006-291x(78)90640-x. [DOI] [PubMed] [Google Scholar]
- 330.Torchilin VP, Khaw BA, Smirnov VN, Haber E. Preservation of antimyosin antibody activity after covalent coupling to liposomes. Biochemical and Biophysical Research Communications. 1979;89:1114–1119. doi: 10.1016/0006-291x(79)92123-5. [DOI] [PubMed] [Google Scholar]
- 331.Mitra M, Mandal AK, Chatterjee TK, Das N. Targeting of mannosylated liposome incorporated benzyl derivative of Penicillium nigricans derived compound MT81 to reticuloendothelial systems for the treatment of visceral leishmaniasis. Journal of Drug Targeting. 2005;13:285–293. doi: 10.1080/10611860500233306. [DOI] [PubMed] [Google Scholar]
- 332.Bäumert HG, Fasold H. Methods in Enzymology. Academic Press; 1989. [32] Cross-linking techniques; pp. 584–609. [DOI] [PubMed] [Google Scholar]
- 333.Kim E, Wriggers W, Phillips M, Kokabi K, Rubenstein PA, Reisler E. Cross-linking constraints on F-actin structure1. Journal of molecular biology. 2000;299:421–429. doi: 10.1006/jmbi.2000.3727. [DOI] [PubMed] [Google Scholar]
- 334.Peng H, Chen W, Cheng Y, Hakuna L, Strongin R, Wang B. Thiol Reactive Probes and Chemosensors. Sensors. 2012;12:15907. doi: 10.3390/s121115907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Yu MK, Park J, Jon S. Targeting Strategies for Multifunctional Nanoparticles in Cancer Imaging and Therapy. Theranostics. 2012;2:3–44. doi: 10.7150/thno.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Werengowska-Ciecwierz K, Wisniewski M, Terzyk AP, Furmaniak S. The chemistry of bioconjugation in nanoparticles-based drug delivery system. Advances in Condensed Matter Physics. 2015;2015 [Google Scholar]
- 337.Conde J, Dias JT, Grazú V, Moros M, Baptista PV, De La Fuente JM. Revisiting 30 years of Biofunctionalization and Surface Chemistry of Inorganic Nanoparticles for Nanomedicine. Frontiers in Chemistry. 2014;2 doi: 10.3389/fchem.2014.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Conde J, Ambrosone A, Sanz V, Hernandez Y, Marchesano V, Tian F, Child H, Berry CC, Ibarra MR, Baptista PV, Tortiglione C, de la Fuente JM. Design of Multifunctional Gold Nanoparticles for In Vitro and In Vivo Gene Silencing. ACS Nano. 2012;6:8316–8324. doi: 10.1021/nn3030223. [DOI] [PubMed] [Google Scholar]
- 339.Koo H, Lee S, Na JH, Kim SH, Hahn SK, Choi K, Kwon IC, Jeong SY, Kim K. Bioorthogonal Copper-Free Click Chemistry In Vivo for Tumor-Targeted Delivery of Nanoparticles. Angewandte Chemie International Edition. 2012;51:11836–11840. doi: 10.1002/anie.201206703. [DOI] [PubMed] [Google Scholar]
- 340.Cardó-Vila M, Arap W, Pasqualini R. Alpha v beta 5 integrin-dependent programmed cell death triggered by a peptide mimic of annexin V. Mol Cell. 2003;11:1151–1162. doi: 10.1016/s1097-2765(03)00138-2. [DOI] [PubMed] [Google Scholar]
- 341.Burg MA, Pasqualini R, Arap W, Ruoslahti E, Stallcup WB. NG2 proteoglycan-binding peptides target tumor neovasculature. Cancer research. 1999;59:2869–2874. [PubMed] [Google Scholar]
- 342.Koivunen E, Arap W, Valtanen H, Rainisalo A, Medina OP, Heikkila P, Kantor C, Gahmberg CG, Salo T, Konttinen YT, Sorsa T, Ruoslahti E, Pasqualini R. Tumor targeting with a selective gelatinase inhibitor. Nat Biotechnol. 1999;17:768–774. doi: 10.1038/11703. [DOI] [PubMed] [Google Scholar]
- 343.Giordano RJ, Anobom CD, Cardó-Vila M, Kalil J, Valente AP, Pasqualini R, Almeida FC, Arap W. Structural basis for the interaction of a vascular endothelial growth factor mimic peptide motif and its corresponding receptors. Chem Biol. 2005;12:1075–1083. doi: 10.1016/j.chembiol.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 344.Jaalouk DE, Ozawa MG, Sun J, Lahdenranta J, Schlingemann RO, Pasqualini R, Arap W. The original Pathologische Anatomie Leiden-Endothelium monoclonal antibody recognizes a vascular endothelial growth factor binding site within neuropilin-1. Cancer research. 2007;67:9623–9629. doi: 10.1158/0008-5472.CAN-07-2737. [DOI] [PubMed] [Google Scholar]
- 345.Karjalainen K, Jaalouk DE, Bueso-Ramos CE, Zurita AJ, Kuniyasu A, Eckhardt BL, Marini FC, Lichtiger B, O’Brien S, Kantarjian HM, Cortes JE, Koivunen E, Arap W, Pasqualini R. Targeting neuropilin-1 in human leukemia and lymphoma. Blood. 2011;117:920–927. doi: 10.1182/blood-2010-05-282921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Daquinag AC, Tseng C, Zhang Y, Amaya-Manzanares F, Florez F, Dadbin A, Zhang T, Kolonin MG. Targeted Proapoptotic Peptides Depleting Adipose Stromal Cells Inhibit Tumor Growth. Mol Ther. 2015 doi: 10.1038/mt.2015.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Hossen MN, Kajimoto K, Akita H, Hyodo M, Ishitsuka T, Harashima H. Ligand-based targeted delivery of a peptide modified nanocarrier to endothelial cells in adipose tissue. J Control Release. 2010;147:261–268. doi: 10.1016/j.jconrel.2010.07.100. [DOI] [PubMed] [Google Scholar]
- 348.Vidal CI, Mintz PJ, Lu K, Ellis LM, Manenti L, Giavazzi R, Gershenson DM, Broaddus R, Liu J, Arap W, Pasqualini R. An HSP90-mimic peptide revealed by fingerprinting the pool of antibodies from ovarian cancer patients. Oncogene. 2004;23:8859–8867. doi: 10.1038/sj.onc.1208082. [DOI] [PubMed] [Google Scholar]
- 349.Oh Y, Mohiuddin I, Sun Y, Putnam JB, Jr, Hong WK, Arap W, Pasqualini R. Phenotypic diversity of the lung vasculature in experimental models of metastases. Chest. 2005;128:596S–600S. doi: 10.1378/chest.128.6_suppl.596S. [DOI] [PubMed] [Google Scholar]
- 350.Bover LC, Cardó-Vila M, Kuniyasu A, Sun J, Rangel R, Takeya M, Aggarwal BB, Arap W, Pasqualini R. A previously unrecognized protein-protein interaction between TWEAK and CD163: potential biological implications. J Immunol. 2007;178:8183–8194. doi: 10.4049/jimmunol.178.12.8183. [DOI] [PubMed] [Google Scholar]
- 351.Staquicini FI, Tandle A, Libutti SK, Sun J, Zigler M, Bar-Eli M, Aliperti F, Perez EC, Gershenwald JE, Mariano M, Pasqualini R, Arap W, Lopes JD. A subset of host B lymphocytes controls melanoma metastasis through a melanoma cell adhesion molecule/MUC18-dependent interaction: evidence from mice and humans. Cancer research. 2008;68:8419–8428. doi: 10.1158/0008-5472.CAN-08-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Moeller BJ, Sidman RL, Pasqualini R, Arap W. Discovery of DNA repair inhibitors by combinatorial library profiling. Cancer research. 2011;71:1816–1824. doi: 10.1158/0008-5472.CAN-10-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Bartolini A, Di Paolo D, Noghero A, Murgia D, Sementa AR, Cilli M, Pasqualini R, Arap W, Bussolino F, Ponzoni M, Pastorino F, Marchiò S. The Neuronal Pentraxin-2 Pathway Is an Unrecognized Target in Human Neuroblastoma, Which Also Offers Prognostic Value in Patients. Cancer research. 2015;75:4265–4271. doi: 10.1158/0008-5472.CAN-15-0649. [DOI] [PubMed] [Google Scholar]
- 354.Rangel R, Guzman-Rojas L, le Roux LG, Staquicini FI, Hosoya H, Barbu EM, Ozawa MG, Nie J, KD, Langley RR, Sage EH, Koivunen E, Gelovani JG, Lobb RR, Sidman RL, Pasqualini R, Arap W. Combinatorial targeting and discovery of ligand-receptors in organelles of mammalian cells. Nat Commun. 2012;3:788. doi: 10.1038/ncomms1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Marchiò S, Soster M, Cardaci S, Muratore A, Bartolini A, Barone V, Ribero D, Monti M, Bovino P, Sun J, Giavazzi R, Asioli S, Cassoni P, Capussotti L, Pucci P, Bugatti A, Rusnati M, Pasqualini R, Arap W, Bussolino F. A complex of alpha6 integrin and E-cadherin drives liver metastasis of colorectal cancer cells through hepatic angiopoietin-like 6. EMBO Mol Med. 2012;4:1156–1175. doi: 10.1002/emmm.201101164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Lee J, Chatterjee DK, Lee MH, Krishnana S. Gold nanoparticles in breast cancer treatment: Promise and potential pitfalls. Cancer Letters. 2014;347:46–53. doi: 10.1016/j.canlet.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Li R, Wu R, Zhao L, Qin H, Wu J, Zhang J, Bao R, Zou H. In vivo detection of magnetic labeled oxidized multi-walled carbon nanotubes by magnetic resonance imaging. Nanotechnology. 2014;25:495102. doi: 10.1088/0957-4484/25/49/495102. [DOI] [PubMed] [Google Scholar]