Abstract
SPINDLY (SPY) is a negative regulator of gibberellin (GA) responses; however, spy mutants exhibit various phenotypic alterations not found in GA-treated plants. Assaying for additional roles for SPY revealed that spy mutants are resistant to exogenously applied cytokinin. GA also repressed the effects of cytokinin, suggesting that there is cross talk between the two hormone-response pathways, which may involve SPY function. Two spy alleles showing severe (spy-4) and mild (spy-3) GA-associated phenotypes exhibited similar resistance to cytokinin, suggesting that SPY enhances cytokinin responses and inhibits GA signaling through distinct mechanisms. GA and spy repressed numerous cytokinin responses, from seedling development to senescence, indicating that cross talk occurs early in the cytokinin-signaling pathway. Because GA3 and spy-4 inhibited induction of the cytokinin primary-response gene, type-A Arabidopsis response regulator 5, SPY may interact with and modify elements from the phosphorelay cascade of the cytokinin signal transduction pathway. Cytokinin, on the other hand, had no effect on GA biosynthesis or responses. Our results demonstrate that SPY acts as both a repressor of GA responses and a positive regulator of cytokinin signaling. Hence, SPY may play a central role in the regulation of GA/cytokinin cross talk during plant development.
INTRODUCTION
In the last two decades, information has begun to accumulate on the molecular events involved in conveying the gibberellin (GA) signal from an as yet unidentified receptor, through the cytoplasm to the nucleus (Sun, 2000; Olszewski et al., 2002; Sun and Gubler, 2004). Studies of the GA-signaling pathway in various plants, including Arabidopsis thaliana, led to the identification of several positively and negatively acting components (Olszewski et al., 2002; Sun and Gubler, 2004). Mutations at the Arabidopsis SPINDLY (SPY) locus result in phenotypes resembling that of wild-type plants treated with exogenous GA. The spy mutant also suppresses phenotypes associated with the GA-deficient mutant ga1, including inhibition of seed germination, reduced stem elongation, delayed flowering, and male sterility (Wilson and Somerville, 1995; Filardo and Swain, 2003). Overexpression of SPY in Arabidopsis (Swain et al., 2001) and petunia (Petunia hybrida) (Izhaki et al., 2001) produced phenotypes consistent with reduced GA action. This suggests that SPY functions as a negative regulator of GA-signal transduction.
The SPY protein exhibits significant similarity to animal tetratricopeptide repeat (TPR)–containing Ser and Thr O-linked N-acetylglucosamine (O-GlcNAc) transferase (OGT). OGT transfers a single GlcNAc from UDP-GlcNAc to specific Ser/Thr residues via an O-linkage (Wells et al., 2001). O-GlcNAc modifications of animal cytosolic and nuclear proteins affect their nuclear localization, phosphorylation, interaction with other proteins, and/or stability (Wells et al., 2001). Deletion of the mouse OGT gene results in embryo lethality (Shafi et al., 2000), indicating that O-GlcNAcylation of proteins is essential in animals. A body of evidence, including the fact that many of the animal proteins that are modified by OGT have regulatory functions, suggests that this modification plays a role in numerous signaling pathways.
Much less is known about plant OGTs. Recently, a second OGT gene, SECRET AGENT (SEC), was characterized in Arabidopsis (Hartweck et al., 2002) and found to have high similarity to SPY and to animal OGTs. Both SPY and SEC proteins exhibited OGT activity in an in vitro assay, and both can modify themselves (Thornton et al., 1999; Hartweck et al., 2002); however, their targets in planta are still unknown. All OGTs, including SPY, have TPR motifs at their N-terminal end of the protein. These motifs are known to participate in protein–protein interactions (Das et al., 1998). Thus, it was speculated that SPY's TPRs are involved in substrate recognition and/or in the generation of active complexes (Izhaki et al., 2001; Swain et al., 2001; Filardo and Swain, 2003).
Whereas the involvement of SPY in GA-related processes is indisputable, it appears that it is also involved in other cellular processes. spy mutants exhibit short hypocotyls, smaller leaves, and deviant phylotaxy (Swain et al., 2001), all of which are absent in plants treated with GA. Moreover, although the mutation in SEC does not exhibit any obvious phenotypic alteration, the sec spy double mutant is lethal (Hartweck et al., 2002). Because GA has not been reported to cause lethality, the double mutant phenotype further supports the hypothesis that SPY has an unidentified function(s) in processes unrelated to GA signaling. SPY may regulate various signaling pathways via interaction with different proteins through its TPR domains. Using a yeast two-hybrid screen with the barley (Hordeum vulgare) HvSPY as bait, two transcriptional regulators were identified: MYB and NAC-like proteins (Robertson, 2004). These proteins interact with the TPR domain of HvSPY and inhibit GA responses in barley aleurone cells. A yeast two-hybrid screen with the Arabidopsis SPY's TPR as bait identified GIGANTEA (GI). Analysis of spy and gi spy mutant phenotypes implied that both GI and SPY play roles in red-light inhibition of hypocotyl elongation, circadian cotyledon movements, and flowering in response to long days (Tseng et al., 2004).
The regulation of growth and development by GA is affected by other phytohormones and environmental signals. A negative interaction between abscisic acid and GA activity in the regulation of seed germination and gene expression is well established (Sun and Gubler, 2004). Abscisic acid seems to act downstream of the GA-signaling repressors, the GAI/RGA DELLA proteins (Gomez-Cadenas et al., 2001). More recently, a promotive effect of auxin and a repressive effect of ethylene on GA regulation of root elongation have been demonstrated (Achard et al., 2003; Fu and Harberd, 2003). Both ethylene and auxin modulate the rate of DELLA protein degradation by GA; auxin decreases and ethylene increases the protein's stability. The interaction between GA and cytokinin is less clear: GA and cytokinin both promote male development in Arabidopsis and tobacco (Nicotiana tabacum) (Huang et al., 2003). On the other hand, GA inhibits cytokinin-induced cell differentiation in culture (Flick et al., 1983). Furthermore, Arabidopsis mutants with reduced GA levels or a block in GA signaling show an increased ability to regenerate shoot meristems from leaves in culture (Ezura and Harberd, 1995).
In this study, we investigated the role of SPY in various signaling pathways and found that loss of SPY function causes resistance to cytokinin, suggesting that it positively regulates cytokinin signaling. We also show that GA inhibits cytokinin responses and hypothesize that SPY mediates this interaction.
RESULTS
Reduced Cytokinin Responses in spy Mutants
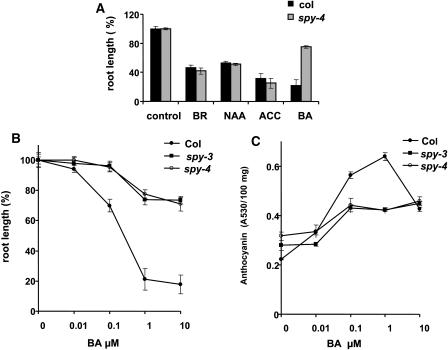
SPY has been shown to act as a negative regulator of GA-signal transduction and to be involved in the transduction of light signal. However, spy mutants exhibit additional phenotypes, not related to these signals, indicating a possible role for SPY in responses to other cues. To test this possibility, we exposed wild-type Columbia and spy-4 seedlings to various phytohormones. Seeds were sown in Petri dishes on MS medium with or without the addition of auxin (α-naphthalene-acetic acid [NAA]), cytokinin (6-benzylamino purine [BA]), an ethylene precursor (1-aminocyclopropane-carboxylic acid [ACC]), or brassinosteroid (24-epibrassinolide [BR]), each at a concentration of 10 μM. Seeds from all treatments geminated at approximately the same time, and after 10 d, root lengths were measured. All tested hormones inhibited root elongation. NAA, ACC, and BR had similar effects on wild-type and spy-4 seedlings. However, the inhibition of wild-type root elongation by BA was greatly suppressed in spy-4 (Figure 1A). Similar results were obtained when zeatin was used instead of BA (data not shown).
Figure 1.
Spy Seedlings Are Resistant to Cytokinin.
(A) Wild-type (Col) and spy-4 seeds were germinated in vertical Petri dishes on MS media with or without brassinosteroid (BR), auxin (NAA), ethylene precursor (ACC), or cytokinin (BA). After 10 d, root length was measured.
(B) and (C) Wild-type (Col), spy-3, and spy-4 seeds were germinated in vertical (B) or horizontal (C) Petri dishes on MS media with or without different BA concentrations. After 10 d, root length (B) and anthocyanin content (C) were measured. The results of root length are expressed as a percentage of control (wild type on MS alone). The results are an average (±se) of 60 seedlings grown in three different plates (20 seeds per plate). Average final root lengths of untreated seedlings were as follows: wild type, 28 ± 0.21 mm; spy-3, 27 ± 0.37 mm; and spy-4, 29 ± 0.44 mm. The experiment was repeated three times with similar results.
To further examine the cytokinin response in spy mutants, we tested the effect of different BA concentrations on root elongation in the wild type and two spy alleles, spy-4 and spy-3. Inhibition of wild-type root elongation was observed at 0.1 μM BA; a 10-fold higher concentration was required to inhibit the elongation of spy-4 and spy-3 roots (Figure 1B). Maximum inhibition of both genotypes was observed with 1 μM BA, which reduced wild-type root elongation by >80% but that of spy-4 and spy-3 by only 20%. In addition to its effect on root elongation, cytokinin induces the accumulation of anthocyanin in seedlings. Figure 1C shows that anthocyanin reached its maximum level in wild-type, spy-4, and spy-3 seedlings at 1 μM BA, with the wild type having almost twice as much as spy-4 and spy-3. Taken together, these results indicate that spy mutants are partially resistant to cytokinin.
GA and spy Inhibit Various Cytokinin Responses
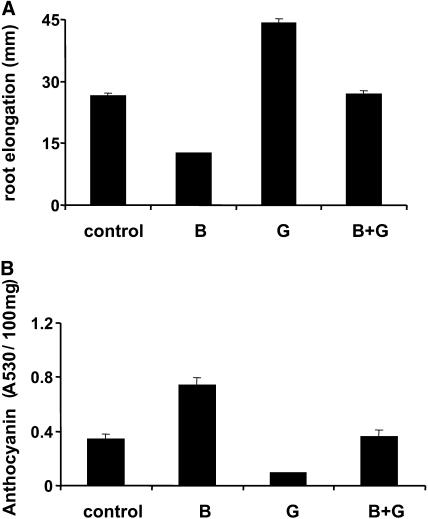
The above results revealed that spy mutants are impaired in their cytokinin responses. Because spy-4 mutants exhibit increased GA responses (Jacobsen and Olszewski, 1993), we tested whether GA also inhibits cytokinin responses. The effect of 5 μM BA, 5 μM GA3, or both on root elongation and anthocyanin accumulation in wild-type Columbia seedlings was examined. Whereas BA reduced root elongation (Figure 2A), GA3 promoted it. Adding GA3 to BA-containing medium inhibited the effect of cytokinin. Similar results were found for anthocyanin content: BA promoted and GA3 inhibited anthocyanin accumulation, and the level of the pigment in the combined treatment was similar to that found in the untreated control (Figure 2B). Analysis of different GA3 concentrations revealed maximum inhibition of cytokinin responses at concentrations between 1 and 10 μM (data not shown).
Figure 2.
GA Represses Cytokinin Responses.
Wild-type (Columbia) seeds were germinated on vertical (A) or horizontal (B) Petri dishes on MS media with or without BA (B), GA3 (G), or both (B + G). After 10 d, root length (A) and anthocyanin content (B) were measured. The results represent an average (±se) of 60 seedlings grown on three different plates (20 seeds per plate). The experiment was repeated three times with similar results.
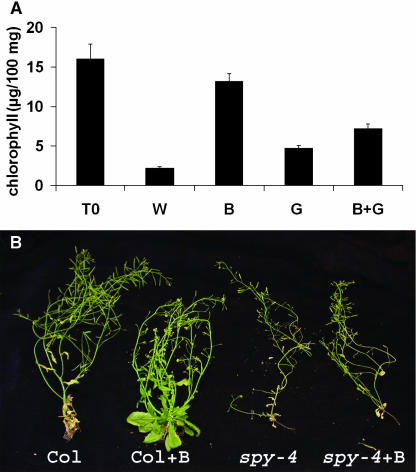
Because GA and BA have opposite effects on root elongation and anthocynin accumulation, the combined treatment may simply be exhibiting the sum of both effects. To examine whether GA inhibits cytokinin signaling or simply affects various cytokinin-regulated processes in an opposite manner, we tested the effect of these two hormones and their combination on leaf senescence. Both cytokinins (Mok and Mok, 2001) and GA (Jacob-Wilk et al., 1999) delay senescence in numerous plant species. We used a chlorophyll-degradation assay in detached leaves (To et al., 2004) to determine the effect of the two hormones on leaf senescence. Mature but not yet senescing rosette leaves from wild-type plants were detached and incubated in water with or without 10 μM BA, 10 μM GA3, or both. The leaves were kept in the dark, and after 10 d, leaf chlorophyll content was measured. Massive loss of chlorophyll occurred in leaves incubated in water alone (Figure 3A), but the addition of cytokinin almost completely blocked pigment degradation. Application of GA3 also reduced chlorophyll loss, but when GA3 was added to the BA-containing solution, it partially inhibited the effect of cytokinin, and the level of chlorophyll found in the combined treatment was approximately half that found with cytokinin alone. These results show that although GA and cytokinin both delayed leaf senescence, GA inhibited the effect of cytokinin on this process. We also tested the effect of spy on leaf senescence, and similar results were obtained: spy-4 inhibited the effect of cytokinin on leaf senescence (Figure 3B) and chlorophyll degradation (data not shown).
Figure 3.
GA3 and spy Suppress Cytokinin's Effect on Leaf Senescence.
(A) Mature wild-type leaves were detached and incubated in water (W) with or without BA (B), GA3 (G), or both (B + G) in the dark. At the beginning of the experiment (T0) and after 10 d in the dark, chlorophyll was extracted and measured. The results are the average (±se) of 30 leaves incubated in three different plates (10 leaves per plate). The experiment was repeated three times with similar results.
(B) Wild-type (Col) and spy-4 plants were treated repeatedly with 10 μM BA (B). Representative plants were photographed 45 d after germination.
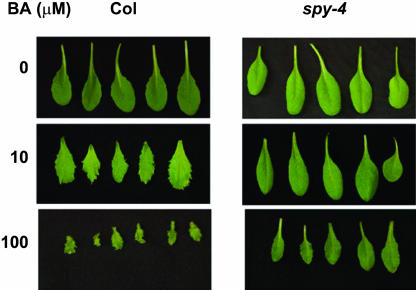
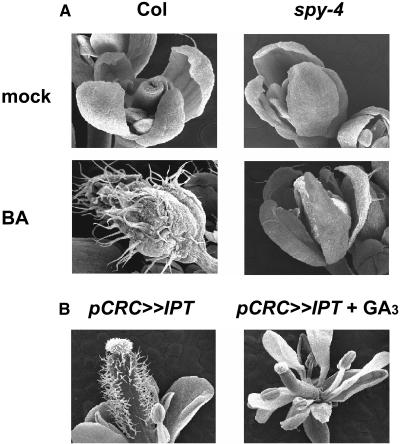
To study the effect of GA and spy on cytokinin responses during Arabidopsis development, we sprayed young wild-type and spy-4 seedlings twice a week with 10 or 100 μM BA. Wild-type plants treated with 10 μM BA exhibited highly serrated rosette and cauline leaves. Leaves of spy-4 plants, on the other hand, were not affected (Figure 4). This treatment also promoted the development of lateral inflorescences in wild-type plants. The number of inflorescences initiated from the rosette leaf axis of BA-treated plants was twice that found in untreated wild-type plants (10.25 ± 1.37 versus 5.4 ± 0.4, respectively). spy-4 plants on the other hand, were hardly affected by the treatment, and the number of lateral inflorescences initiated after BA treatment was only slightly higher than that found in the untreated spy-4 plants (4.1 ± 0.7 versus 3.2 ± 0.25, respectively). BA treatment (10 μM) also inhibited the elongation of inflorescence stems (main and lateral), and again this effect was more pronounced in the wild type than in spy-4 plants (Figure 3B). After treatment with the higher BA concentration (100 μM), wild-type plants produced very small, serrated rosette and cauline leaves (Figure 4); inflorescences failed to elongate, sepals developed a large number of trichomes (Figure 5A), and flower maturation was impaired, with most flowers failing to reach anthesis. In spy-4 plants, the effects of 100 μM BA were suppressed. Leaves were much larger than those of treated wild-type plants and much less serrated (Figure 4). Inflorescence stems were elongated, though they were shorter than untreated spy-4 inflorescences. The number of trichomes developed on spy-4 sepals (Figure 5A) was greatly reduced compared with the wild type. These results further demonstrate spy's partial resistance to cytokinin.
Figure 4.
Suppression of Cytokinin Effects on Leaf Form by spy.
Rosette leaves of wild-type and spy-4 plants treated repeatedly with different concentrations of BA (10 or 100 μM).
Figure 5.
GA and Mutation in SPY Suppress Cytokinin Effects on Flower Morphology.
(A) Young wild-type (Col) and spy-4 plants were treated repeatedly with water (mock) or 100 μM BA, and then inflorescences were detached and analyzed by scanning electron microscopy.
(B) Flowers of a transgenic plant (transactivation line) expressing IPT under the regulation of the carpel-specific CRC promoter (pCRC≫IPT) were treated repeatedly with water or 100 μM GA3. Inflorescences were detached and analyzed by scanning electron microscopy.
GA Can Suppress Trichome Development Induced by Endogenous Cytokinin
To examine the effects of GA on endogenously produced cytokinin, we used transgenic plants expressing the cytokinin biosynthetic gene isopentenyl transferase (IPT) from Agrobacterium tumefaciens under the regulation of the carpel-specific CRABS CLAW (CRC) promoter (Baum et al., 2001). Arabidopsis carpels are hairless (Figure 5A), unlike many other Brassicaceae. However, IPT expression induced numerous ectopic trichomes (Figure 5B), an attribute that has not previously been associated with cytokinin overproduction. Trichome formation was specific to the carpels, as none were observed on the nectaries where the CRC promoter is highly active (Baum et al., 2001). Repeated treatments with 100 μM GA3 completely suppressed the ectopic trichome phenotype (Figure 5B).
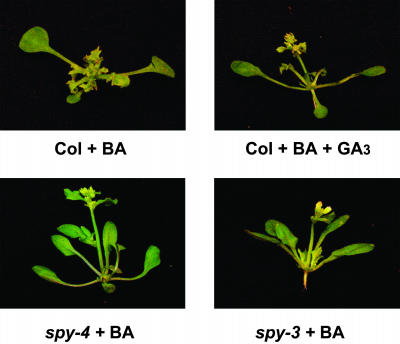
SPY Regulates Cytokinin Responses and GA Signaling through Different Mechanisms
The effect of GA on cytokinin responses in seedlings was similar to that caused by the spy-4 allele. It is therefore possible that elements in GA signal transduction downstream of SPY affect cytokinin responses. Alternatively, SPY itself may act as a positive regulator of cytokinin signal transduction. It is also possible that spy suppresses cytokinin action by both of these mechanisms. To distinguish between these possibilities, we analyzed two different spy alleles showing severe and weak GA-associated phenotypic alterations (Jacobsen et al., 1996). The strong allele, spy-4, is caused by a T-DNA insertion upstream of the first exon that greatly reduces SPY expression. This mutant germinates on the GA biosynthesis inhibitor paclobutrazol and exhibits early flowering, male sterility, and slender inflorescence stems. The weak allele, spy-3, is caused by a single amino acid substitution at the C terminus of the protein. Although spy-3 seeds are able to germinate on paclobutrazol, the plants exhibit very mild GA-related phenotypic alterations: time to flowering is only slightly shorter than in the wild type (Jacobsen and Olszewski, 1993), plants are fertile, internode length is almost normal (Filardo and Swain, 2003), and the inflorescence stem girth is similar to that of the wild type. Despite these differences in GA-associated phenotypes, both alleles inhibited BA-induced anthocyanin accumulation and BA-repressed root elongation equally (Figures 1B and 1C). To examine the effects of the different alleles on cytokinin responses during later stages of plant development, wild-type, spy-4, and spy-3 seedlings were sprayed twice a week with a high BA concentration (100 μM), with or without 100 μM GA3. BA treatment of the wild type caused severe phenotypic changes (Figure 6), as already described. When wild-type plants were treated with BA and GA, the effect of BA was only slightly suppressed. Although the inflorescences of plants treated with GA3 and BA were elongated, these plants still exhibited extremely serrated leaves and their flowers did not reach anthesis. spy-4 and spy-3 showed similar resistance to cytokinin, and both exhibited much higher resistance to BA than that found in the GA3-treated plants. Leaves of spy-3 and spy-4 were much less serrated (Figure 6), and in most cases, flowers developed normally. These results suggest that SPY, and not SPY-regulated elements in the GA signaling pathway, regulates cytokinin responses.
Figure 6.
spy Affects Cytokinin Responses Independently of Its Effect on GA Signal.
Wild-type (Col) seedlings were treated repeatedly until flowering with BA without or with GA3. spy-4 and spy-3 were treated repeatedly until flowering with BA.
GA may affect cytokinin responses through SPY or via SPY-independent pathway. We used spy-3 to distinguish between these possibilities. spy-3 exhibited a similar cytokinin resistance as the null spy-4 allele but retains sensitivity to GA with respect to GA responses. Therefore, if GA inhibits cytokinin responses independently of SPY, we expect that GA treatment of spy-3 will increase the resistance to cytokinin. We thus examined the effect of BA, with or without the addition of GA3, on leaf serration in young spy-3 seedlings. In this experiment, we used 500 μM BA because leaf phenotypes are more pronounced at this concentration. Figure 7 shows that GA did not enhance cytokinin resistance of spy-3. This suggests that GA acts through SPY to suppress cytokinin responses.
Figure 7.
GA Does Not Enhance Cytokinin Resistance of spy-3.
spy-3 seedlings with two true leaves were treated repeatedly (three times) with 500 μM BA with or without 100 μM GA3. For comparison, wild-type seedlings were treated with BA. Representative young leaves were photographed after 10 d.
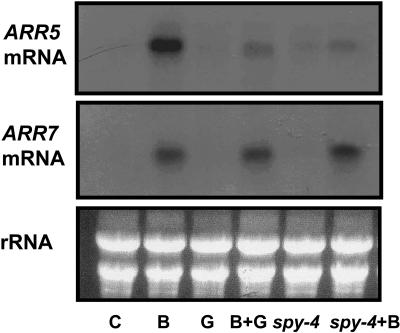
GA and spy Inhibit the Induction of Cytokinin Primary-Response Genes
The repression of cytokinin responses by GA suggests an interaction between GA and cytokinin-signaling pathways. Because GA3 and SPY affect various cytokinin responses, they probably act on a main branch of the cytokinin pathway, common to most cytokinin responses. In-depth studies of the cytokinin signal transduction pathway revealed several positive and negative transduction components. Type-A Arabidopsis response regulators (ARR) are rapidly activated at the transcriptional level by cytokinin (Hutchison and Kieber, 2002) and inhibit cytokinin responses (To et al., 2004). To test whether GA interacts with the cytokinin-signaling pathway upstream of type-A ARRs, we submerged 10-d-old wild-type seedlings in water, 10 μM BA, 10 μM GA3, or both and spy-4 plants in water or 10 μM BA. After 50 min, RNA was extracted, and the abundance of ARR7 and ARR5 transcripts was determined. Figure 8 shows that GA3 treatment and spy-4 suppressed BA's induction of ARR5 but not ARR7 transcript accumulation.
Figure 8.
GA3 and spy-4 Suppress Cytokinin Induction of Type-A ARR Gene Expression.
RNA was extracted from wild-type plants submerged in water (C), BA (B), GA3 (G), or BA and GA3 (B + G) and from spy-4 plants submerged in water or in BA. ARR5 and ARR7 expression was analyzed by RNA gel blots. Ethidium bromide staining of rRNA is presented to show equal loading of RNA. The experiment was repeated three times with similar results.
GA Biosynthesis and Responses Are Not Affected by Cytokinin
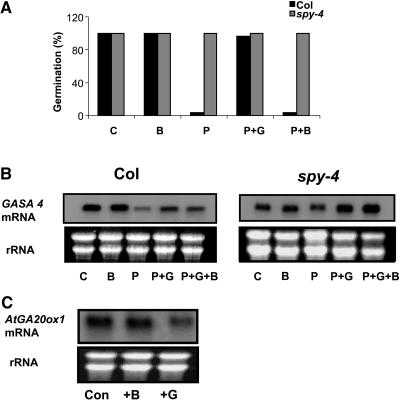
We also tested whether cytokinin affects GA responses, such as germination and flowering. Wild-type and spy-4 seeds were sown in Petri dishes on MS medium with or without 5 μM BA or 10 mg/L paclobutrazol with or without 5 μM BA, 5 μM GA3, or both. Germination rate after 8 d was examined. Paclobutrazol was used in this experiment to study the possible effect of BA on GA signaling. Figure 9A shows that BA did not inhibit wild-type seed germination and had no effect on the germination of spy-4 seeds in the presence of paclobutrazol. This indicates that cytokinin does not affect GA biosynthesis or signaling sufficiently to cause changes in germination under these conditions. We also examined the effect of cytokinin on flowering time. Wild-type and spy-4 seedlings were grown under long-day conditions and sprayed twice a week with 100 μM BA. The number of rosette leaves was counted at bolting. BA treatment did not affect the transition to flowering in wild-type plants (9.8 ± 0.3 versus 10.2 ± 0.3) and had no effect on the early flowering of spy-4 (6.7 ± 0.2 versus 7 ± 0.3).
Figure 9.
Cytokinin Does Not Affect GA Responses or Biosynthesis.
(A) Wild-type (Col) and spy-4 seeds were sown in Petri dishes on MS media with or without BA (B), paclobutrazol (P), paclobutrazol and GA3 (P + G), or paclobutrazol and BA (P + B). After 10 d, germinated seeds were counted. The results are the average percentage of germination of 60 seeds sown on three different plates (20 seeds per plate). The experiment was repeated three times with similar results.
(B) Wild-type and spy-4 plants were grown under long days. When inflorescences started elongating, some of the plants were sprayed with paclobutrazol (P). When first flowers reached anthesis, plants were sprayed with BA (B), GA3 (G), or BA and GA3 (B + G). Six hours after treatment, flowers were detached and RNA was extracted for GASA4 expression analyses. The experiment was repeated twice with similar results.
(C) Wild-type plants were treated with GA3 or BA, and after 8 h, RNA was extracted for AtGA20ox1 expression analyses. The experiment was repeated twice with similar results.
Ethidium bromide staining of rRNA is presented in (B) and (C) to show equal loading of RNA.
We next tested the effect of BA treatment on the expression of the GA-regulated gene GASA4 (Herzog et al., 1995). Wild-type and spy-4 plants were grown under long days and upon bolting, sprayed with 10 mg/L paclobutrazol or water every 4 d. At anthesis, plants were sprayed with 100 μM BA, 100 μM GA3, BA and GA3, or water. Flowers were detached 6 h after treatment, and RNA was extracted for GASA4 expression analyses. Figure 9B shows that paclobutrazol suppressed GASA4 expression in wild-type but not spy-4 flowers. GA3 application to the paclobutrazol-treated wild-type plants partially restored the expression of the gene. BA treatments had no effect on GASA4 expression in wild-type or spy-4 plants, regardless of GA3 treatment.
To determine whether cytokinin affects GA biosynthesis, we examined its effect on the expression of the GA-biosynthetic gene GA 20-oxidase (AtGA20ox1). Plants were grown to flowering and then sprayed with 100 μM BA or 100 μM GA3. After 8 h, RNA levels in inflorescences were determined by RNA gel blot analysis (Figure 9C). As expected from the feedback regulation of GA biosynthetic genes by bioactive GAs (Olszewski et al., 2002), GA3 inhibited the expression of AtGA20ox1. Cytokinin treatment had no effect on AtGA20ox1 expression, further suggesting that the hormone has no effect on GA biosynthesis.
DISCUSSION
SPY is a negative regulator of GA-signal transduction (Filardo and Swain, 2003), but mutations in spy exhibit additional, GA-unrelated, phenotypic alterations. It was therefore suggested that the protein is involved in other signaling pathways (Izhaki et al., 2001; Swain et al., 2001). Recently, Tseng et al. (2004) showed that SPY interacts with the nuclear protein GI and is involved in light-signal transduction controlling flowering, circadian cotyledon movements, and hypocotyl elongation. Here, we present evidence suggesting a positive role for SPY in the transduction of the cytokinin signal.
SPY and Cross Talk between GA- and Cytokinin-Signaling Pathways
Cytokinins and GAs play central roles in the regulation of plant development. Cytokinins act early during shoot initiation to control meristem activity (Schmulling, 2002), and GAs act at later stages, regulating cell division and expansion to control shoot elongation (Richards et al., 2001). Our results suggest cross talk between the two hormones, with GA inhibiting various cytokinin responses at different stages of plant development. Because the GA constitutive signaling mutant spy and GA had similar inhibitory effects, SPY itself or a component downstream of SPY in the GA-signaling pathway may directly control cytokinin signaling. Several pieces of evidence support a direct role for SPY in this interaction. The strong spy-4 and weak spy-3 alleles (with respect to GA signaling; Filardo and Swain, 2003) showed similar resistance to exogenous cytokinin and had similar round, nonserrated leaves, a phenotype associated with the inhibition of cytokinin responses (Figure 4). Furthermore, spy mutants exhibit deviant phylotaxy (Swain et al., 2001), which is also associated with altered cytokinin responses (Giulini et al., 2004) but is not found in GA-treated plants. Finally, for some responses, spy mutants exhibited higher resistance to cytokinin than GA-treated plants, even when GA was applied at high concentrations (Figure 6). All of these observations suggest that SPY, and not SPY-regulated elements in the GA signaling pathway, affects cytokinin responses. They also propose that SPY promotes cytokinin signaling through a distinct mechanism than that involved in the suppression of GA responses.
Because GA and spy displayed similar inhibitory effects on cytokinin responses, GA may suppress cytokinin signaling via inhibition of SPY, independently of SPY, or both. We found that whereas spy-3 retains its sensitivity to GA with respect to GA-associated responses, GA had no significant effect on the resistance to BA conferred by spy-3, with respect to root elongation and anthocyanin accumulation in seedlings (data not shown) and leaf serration (Figure 7). These findings suggest that GA suppresses cytokinin responses at least partially via SPY and is in agreement with the model proposed by Sun and Gubler (2004) that GA inhibit SPY. It was shown previously that GA does not affect SPY mRNA level (Izhaki et al., 2001). In addition, treatments with paclobutrazol or GA3 had no detectable effect on the abundance of a SPY-GFP fusion protein (T.-S. Tseng and N. Olszewski, unpublished data). However, GA may repress SPY activity. A GA effect on SPY function is supported by the finding that application of exogenous GA suppresses the inhibition of GA responses caused by ectopic expression of SPY in transgenic petunia plants (Izhaki et al., 2001).
Although mature spy mutants exhibited greater resistance to exogenously applied cytokinin than GA-treated plants, in seedlings, the effect of GA on cytokinin responses (root elongation and anthocyanin accumulation) was similar to that of spy. Other components of the GA response pathway, in addition to SPY, may interact with the cytokinin-signaling pathway, with their contribution changing in different tissue types and at different developmental stages. It is also possible that the different effects of GA relative to spy result from differential GA sensitivity. Furthermore, because the Arabidopsis genome contains an additional OGT gene, SEC, redundancy may exist. The level/activity of SEC may vary with developmental stage/cell type; therefore, the relative effect of spy versus GA on cytokinin responses may change.
SPY's Role in Cytokinin-Signal Transduction
Whether SPY plays a pivotal role as an activator of cytokinin signal is not yet clear. The fact that the RNA null mutant spy-4 is only partially resistant to cytokinin suggests that SPY does not play a central role in the transduction pathway. Alternatively, the partial effect may result from functional redundancy with SEC. Mutations in SEC do not exhibit any phenotypic alteration; however, the sec spy double mutant is lethal (Hartweck et al., 2002). Because high GA levels or signals do not cause lethality, SEC may also promote cytokinin signaling, and the lack of active SPY and SEC may strongly repress this signal transduction, causing lethality.
Different domains of the SPY protein may be involved in the regulation of cytokinin and GA signals. The spy-3 allele is caused by a substitution of a conserved Gly to Ser at the C terminus of the protein (Jacobsen et al., 1996). Although this substitution has only a slight effect on GA-signal transduction, it affects cytokinin responses similar to the spy-4 null allele. The importance of this specific amino acid to cytokinin signaling is not yet clear. It has been suggested that SPY acts in different signaling pathways through interactions with alternative proteins (Swain et al., 2001). However, because this amino acid is located in the OGT region and not in the TPR domain, its substitution is less likely to affect interactions with other proteins (Tseng et al., 2004), although this possibility cannot be excluded.
Because GA and spy affect cytokinin responses even when the cytokinin is applied exogenously, they most likely regulate cytokinin signaling. We showed that mutations in SPY affect numerous cytokinin responses throughout the life cycle of the plant. These findings are consistent with SPY affecting early steps of the cytokinin-signaling pathway. Cytokinin binds to the CRE1 receptor and induces its autophosphorylation. The phosphate group is transferred through a phosphorelay cascade to the nucleus, where it activates type-B ARRs. Activated type-B ARRs induce the transcription of type-A ARRs (Hutchison and Kieber, 2002). Our results show that GA and spy inhibit the induction of ARR5 (type-A ARR) by cytokinin, suggesting that they affect the phosphorelay cascade. Interestingly, GA3 and spy-4 did not inhibit the induction of another early-response type-A ARR gene, ARR7. This may indicate that SPY affects a subset of type-B ARRs, thus differentiating between different branches of the cytokinin response. SPY may modify specific type-B ARR proteins (O-GlcNac modification). This modification may be required, in addition to phosphorylation, for these proteins' activation and the induction of downstream genes, including specific type-A ARRs. The nuclear colocalization of SPY (Swain et al., 2002) and type-B ARRs is in line with this hypothesis. It should be noted, however, that we do not provide evidence for the mechanism through which GA and SPY regulate ARR5 expression or for where they act on the cytokinin-signaling pathway.
Cytokinin, KNOX Proteins, and GA Biosynthesis
Several previous studies provided clues for cross talk between GA and cytokinin. The meristematic homeodomain KNOX proteins, SHOOT MERISTEMLESS and BP, which play a major role in the regulation of meristem development, were suggested to regulate cytokinin biosynthesis (Ori et al., 1999). Overexpression of KNOX genes in Arabidopsis resulted in the development of serrated and lobed leaves (Chuck et al., 1996), and GA application or a mutation in SPY suppressed this phenotype (Hay et al., 2002). Because cytokinin treatments caused similar phenotypic changes (Figure 4) and mutations in SPY inhibited them, the leaf phenotypes associated with KNOX ectopic expression may result from an increase in cytokinin level, and their suppression by GA, from decreased cytokinin signal because of the suppressed SPY activity.
Because GA suppresses meristematic activities (Hay et al., 2002), factors controlling meristem development, including cytokinin, are expected to downregulate GA level or signal. However, our results clearly show that cytokinin has no major effect on GA biosynthesis or signaling. On the other hand, other factors controlling meristem activity, such as KNOX proteins, suppress GA content (Sakamoto et al., 2001). This effect is probably independent of cytokinin because direct repression of the GA biosynthetic gene AtGA20ox1 by KNOX has been demonstrated (Sakamoto et al., 2001; Chen et al., 2004).
Conclusion
We suggest that SPY acts as both a repressor of GA responses and a positive regulator of cytokinin signaling. Plant development under a changing environment requires a balanced but dynamic ratio between the levels of different growth factors. The two phytohormones, GA and cytokinin, have opposite effects on numerous developmental processes; therefore, a coordination between the two is essential. We hypothesize that SPY acts as a regulator of GA/cytokinin homeostasis and propose (Figure 10) that when GA levels are low, SPY represses typical GA responses and acts as a positive element in the transduction of cytokinin signal. When GA levels increase, SPY activity may be suppressed, GA responses are promoted, and cytokinin signal is inhibited. It is possible that GA suppresses cytokinin response also through SPY-independent pathways. How SPY distinguishes between the two signaling pathways is not yet clear, but interactions with different proteins to create complexes affecting different pathways are possible.
Figure 10.
Hypothetical Model for the Role of SPY in GA- and Cytokinin-Signal Transduction.
At low GA levels, SPY and the DELLA proteins GAI/RGA/RGL repress typical GA responses. At the same time, SPY acts as a positive element in the transduction of cytokinin signal, affecting elements located at the phosphorelay cascade upstream to type-A ARR. When GA level increases, SPY activity and GAI/RGA/RGL levels are suppressed, GA responses are promoted, and cytokinin signal is inhibited. Dashed lines indicate hypothetical interactions suggested by our study.
METHODS
Plant Materials
Arabidopsis thaliana plants, both wild type and mutants, used in this study (except for the transgenic line, see below) were of the Columbia (Col-0) ecotype. Wild-type, spy-3, and spy-4 mutant seeds were sterilized, cold-treated, and germinated on sterile MS media or in pots. Plants were grown in a growth room under controlled temperatures (22°C) and long (16 h light) days. For seed production of spy-4, the plants were grown at 20°C under short days (8 h light) for 30 d and then transferred to long days.
Construction of Transgenic Lines
Transcriptional fusion of the CRC promoter (3.8 kb) in front of the chimeric LhG4 (Moore et al., 1998) in BJ36 was subsequently cloned into the binary vector pMLBART. The IPT cDNA (Gan and Amasino, 1995) was subcloned behind an operator array in BJ36 and subsequently cloned into the binary vector pMLBART. Both constructs were introduced into Agrobacterium tumefaciens ASE and then into plants (Arabidopsis, Landsberg erecta) by floral dip. Selected BAR+ lines were used for the generation of F1 plants where pCRC≫IPT was transactivated.
Hormone-Response Assays in Seedlings
Arabidopsis seeds were grown on vertical plates containing MS medium (Duchefa Biochemie, Haarlem, The Netherlands) with 0.8% (w/v) agar, 3% (w/v) sucrose, and the indicated hormone at the specified concentration. NAA, BA, and ACC were purchased from Sigma-Aldrich (St. Louis, MO), brassinosteroid (BR) from CITECH Research (Plymouth Meeting, PA), and zeatin from Duchefa Biochemie. Plates were kept in a growth room under long-day conditions at 22°C. Root length was marked at days 4 and 9, and on day 10 root growth between days 4 and 9 was measured. For the anthocyanin assay, seeds were grown on horizontal plates containing MS medium with 0.8% agar, 3% sucrose, and the indicated cytokinin and/or GA3 concentration. After 10 d, seedlings were weighed and anthocyanin was extracted and measured spectrophotometrically (Weiss and Halevy, 1989), and the results were normalized to fresh weight.
RNA Extraction and RNA Blot Analyses
Total RNA was extracted using a TRI REAGENT kit (Molecular Reseach Center, Cincinnati, OH). Subsequently, 10 μg of total RNA were fractionated in a 1% (w/v) agarose gel containing formaldehyde and blotted onto Hybond N+ membranes (Amersham-Pharmacia Biotech, Buckinghamshire, UK). The blots were hybridized in 0.263 M Na2HPO4, 7% (w/v) SDS, 1 mM EDTA, and 1% (w/v) BSA at 60°C with 32P-labeled cDNA probes (Rediprime; Amersham-Pharmacia Biotech) for ARR5 and ARR7 (D'Agostino et al., 2000), AtGA20ox1 (Phillips et al., 1995), and GASA4 (Herzog et al., 1995) genes. The membranes were washed twice in 0.1× SSC and 0.1% SDS at 60°C for 20 min each and exposed to x-ray film (Fuji, Tokyo, Japan) with two intensifying screens at −70°C. After autoradiography, filters were washed in boiled 0.1% SDS to remove radioactivity before rehybridization.
Scanning Electron Microscopy
Samples for scanning electron microscopy were fixed in 2.5% (w/v) gluteraldehyde in 0.1 M phosphate buffer, pH 7.2, transferred to ethanol (25% up to 100%), critical-point dried with liquid carbon dioxide in a CPD 750 (Bio-Rad, Hemel Hempstead, UK), sputter-coated with gold, and photographed with a Jeol scanning electron microscope (JSM-5410 LV; Tokyo, Japan).
Senescence Assay
Seedlings were grown in a growth room for 25 d, and then fully expanded leaves (leaf number 7) were detached. To induce senescence, leaves were floated on water in Petri dishes supplemented with 10 μM BA or 10 μM GA at 22°C in the dark for 10 d. Chlorophyll was extracted and measured spectrophotometrically from fresh and senesced leaves and normalized to fresh weight (Arnon, 1949).
Acknowledgments
This research was supported by Research Grant 615/02-1 from the Israel Science Foundation to D.W. and MCB-0112826 from the National Science Foundation to N.O. This work was also supported by the Pearlstein Fund for research in floriculture at the Hebrew University, and we thank the donors for their help. We thank John Bowman (University of California, Davis, CA) for transgenic lines generated in his lab.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: David Weiss (weiss@agri.huji.ac.il).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.028472.
References
- Achard, P., Vriezen, W.H., Van Der Straeten, D., and Harberd, N.P. (2003). Ethylene regulates Arabidopsis development via the modulation of DELLA protein growth repressor function. Plant Cell 15, 2816–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon, D.I. (1949). Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 24, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum, S.F., Eshed, Y., and Bowman, J.L. (2001). The Arabidopsis nectary is an ABC-independent floral structure. Development 128, 4657–4667. [DOI] [PubMed] [Google Scholar]
- Chen, H., Banejrjee, A.K., and Hannapel, D.J. (2004). The tandem complex of BEL and KNOX partners is required for transcriptional repression of ga20ox1. Plant J. 38, 276–284. [DOI] [PubMed] [Google Scholar]
- Chuck, G., Lincoln, C., and Hake, S. (1996). Knat1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 8, 1277–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Agostino, I.B., Deruere, J., and Kieber, J.J. (2000). Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol. 124, 1706–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, A.K., Chohen, P.T., and Barford, D. (1998). The structure of the tetratricopeptide repeats of protein phosphatase 5: Implication for TPR-mediated protein-protein interactions. EMBO J. 17, 1192–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezura, H., and Harberd, N.P. (1995). Endogenous gibberellin levels influence in-vitro shoot regeneration in Arabidopsis thaliana (L.) Heynh. Planta 197, 301–305. [DOI] [PubMed] [Google Scholar]
- Filardo, F.F., and Swain, S.M. (2003). SPYing on GA signaling and plant development. J. Plant Growth Regul. 22, 163–175. [Google Scholar]
- Flick, C.E., Evans, D.E., and Sharp, W.R. (1983). Organogenesis. In Handbook of Plant Cell Culture, Vol 1. D.A. Evans, W.R. Sharp, P.V. Ammirato, and Y. Yamada, eds (New York: Macmillan), pp. 13–81.
- Fu, X.D., and Harberd, N.P. (2003). Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 421, 740–743. [DOI] [PubMed] [Google Scholar]
- Gan, S., and Amasino, R.M. (1995). Inhibition of leaf senescence by autoregulated production of cytokinin. Science 270, 1986–1988. [DOI] [PubMed] [Google Scholar]
- Giulini, A., Wang, J., and Jackson, D. (2004). Control of phyllotaxy by the cytokinin-inducible response regulator homologue ABPHYL1. Nature 430, 1031–1034. [DOI] [PubMed] [Google Scholar]
- Gomez-Cadenas, A., Zentalla, R., Walker-Simmons, M., and Ho, T.-H.D. (2001). Gibberellin/abscisic acid antagonism in barley aleurone cells: Site of action of the protein kinase PKABA1 in relation to gibberellin signaling molecules. Plant Cell 13, 667–679. [PMC free article] [PubMed] [Google Scholar]
- Hartweck, L.M., Scott, C.L., and Olszewski, N.E. (2002). Two O-linked N-acetylglucosamine transferase genes of Arabidopsis thaliana L. Heynh. have overlapping functions necessary for gamete and seed development. Genetics 161, 1279–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay, A., Kaur, H., Phillips, A.S., Hedden, P., Hake, S., and Tsiantis, M. (2002). The gibberellin pathway mediates knotted1-type homeobox function in plants with different body plans. Curr. Biol. 12, 1557–1565. [DOI] [PubMed] [Google Scholar]
- Herzog, M., Dorne, A.M., and Grellet, F. (1995). GASA, a gibberellin-regulated gene family from Arabidopsis thaliana related to the tomato gast1 gene. Plant Mol. Biol. 27, 743–752. [DOI] [PubMed] [Google Scholar]
- Huang, S., Cerny, R.E., Qi, Y.L., Bhat, D., Aydt, C.M., Hanson, D.D., Malloy, K.P., and Ness, L.A. (2003). Transgenic studies on the involvement of cytokinin and gibberellin in male development. Plant Physiol. 131, 1270–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison, C.E., and Kieber, J.J. (2002). Cytokinin signaling in Arabidopsis. Plant Cell 14 (suppl.), S47–S59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izhaki, A., Swain, S.M., Tseng, T., Borochov, A., Olszewski, N.E., and Weiss, D. (2001). The role of SPY and SPY's TPR domains in the regulation of gibberellin action throughout the life cycle of Petunia hybrida plants. Plant J. 28, 181–190. [DOI] [PubMed] [Google Scholar]
- Jacobsen, S.E., Binkowski, K.A., and Olszewski, N.E. (1996). SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc. Natl. Acad. Sci. USA 93, 9292–9296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen, S.E., and Olszewski, N.E. (1993). Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell 5, 887–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob-Wilk, D., Holland, D., Goldschmidt, E.E., Riov, J., and Eyal, Y. (1999). Chlorophyll breakdown by chlorophyllase: Isolation and functional expression of the Chlase1 gene from ethylene-treated Citrus fruit and its regulation during development. Plant J. 20, 653–662. [DOI] [PubMed] [Google Scholar]
- Mok, D.W., and Mok, M.C. (2001). Cytokinins: Chemistry, Activity and Function. (Boca Raton, FL: CRC Press).
- Moore, I., Galweiler, L., Grosskopf, D., Schell, J., and Klaus, P.A. (1998). Transcription activation system for regulated gene expression in transgenic plants. Proc. Natl. Acad. Sci. USA 95, 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski, N., Sun, T.-P., and Gubler, F. (2002). Gibberellin signaling: Biosynthesis, catabolism, and response pathways. Plant Cell 14 (suppl.), S61–S80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori, N., Juarez, M.T., Jackson, D., Yamaguchi, J., Banowetz, G.M., and Hake, S. (1999). Leaf senescence is delayed in tobacco plants expressing the maize homeobox gene knotted1 under the control of a senescence-activated promoter. Plant Cell 11, 1073–1080. [PMC free article] [PubMed] [Google Scholar]
- Phillips, A., Ward, D.A., Uknes, S., Appleford, N.E.J., Lange, T., Huttly, A.K., Caskin, P., Craebe, J.E., and Hedden, P. (1995). lsolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiol. 108, 1049–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, D.E., King, K.E., Ait-ali, T., and Harber, N.P. (2001). How gibberellin regulates plant growth and development: A molecular genetic analysis of gibberellin signaling. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 67–88. [DOI] [PubMed] [Google Scholar]
- Robertson, M. (2004). Two transcription factors are negative regulators of gibberellin responses in the HvSPY-signaling pathway in barley aleurone. Plant Physiol. 135, 2747–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, T., Kamiya, N., Ueguchi-Tanaka, M., Iwahori, S., and Matsuoka, M. (2001). KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Genes Dev. 15, 581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmulling, T. (2002). New insights into the functions of cytokinins in plant development. J. Plant Growth Regul. 21, 40–49. [DOI] [PubMed] [Google Scholar]
- Shafi, R., Iyer, S.P., Ellies, L.G., O'Donnell, N., Marek, K.W., Chui, D., Hart, G.W., and Marth, J.D. (2000). The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc. Natl. Acad. Sci. USA 97, 5735–5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, T.-P. (2000). Gibberellin signal transduction. Curr. Opin. Plant Biol. 3, 418–422. [DOI] [PubMed] [Google Scholar]
- Sun, T.-P., and Gubler, F. (2004). Molecular mechanism of gibberellin signaling in plants. Annu. Rev. Plant Biol. 55, 197–223. [DOI] [PubMed] [Google Scholar]
- Swain, S.M., Tseng, T.-S., and Olszewski, N.E. (2001). Altered expression of SPINDLY affects gibberellin response and plant development. Plant Physiol. 126, 1174–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain, S.M., Tseng, T.S., Thornton, T.M., Gopalraj, M., and Olszewski, N.E. (2002). SPINDLY is a nuclear-localized repressor of gibberellin signal transduction expressed throughout the plant. Plant Physiol. 129, 605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton, T., Kreppel, L., Hart, G., and Olszewski, N. (1999). Genetic and biochemical analysis of Arabidopsis SPY. In Plant Biotechnology and in Vitro Biology in the 21st Century, A. Altman, M. Ziv, and S. Izhar, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 445–448.
- To, J.P.C., Harberer, G., Ferreira, F.J., Deruere, J., Mason, M.G., Schaller, G.E., Alonso, J.M., Ecker, J.R., and Kieber, J.J. (2004). Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16, 658–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng, T.-S., Patrice, A., Salome, P.A., McClung, C.R., and Olszewski, N.E. (2004). SPINDLY and GIGANTEA interact and act in Arabidopsis thaliana pathways involved in light responses, flowering, and rhythms in cotyledon movements. Plant Cell 16, 1550–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, D., and Halevy, A.H. (1989). Stamens and gibberellin in the regulation of corolla pigmentation and growth in Petunia hybrida. Planta 179, 89–96. [DOI] [PubMed] [Google Scholar]
- Wells, L., Vosseller, K., and Hart, G.W. (2001). Glycosylation of nucleocytoplasmic proteins: Signal transduction and O-GlcNac. Science 291, 2376–2378. [DOI] [PubMed] [Google Scholar]
- Wilson, R.N., and Somerville, C.R. (1995). Phenotypic suppression of the gibberellin-insensitive mutant (gai) of Arabidopsis. Plant Physiol. 108, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]