Abstract
Objective
: Vitamin D receptor (VDR) mediates vitamin D activity. We examined whether VDR expression in excised melanoma tissues is associated with VDR gene (VDR) polymorphisms.
Methods
: We evaluated VDR protein expression (by monoclonal antibody immunostaining), melanoma characteristics, and carriage of VDR-FokI-rs2228570 (C>T),VDR-BsmI-rs1544410 (G>A),VDR-ApaI-rs7975232 (T>G), andVDR-TaqI-rs731236 (T>C) polymorphisms (by restriction fragment length polymorphism). Absence or presence of restriction site was denoted by a capital or lower letter, respectively: " F” and " f” for FokI, " B” and " b” for BsmI, " A” and " a” for ApaI, and " T” and " t” for TaqI endonuclease. Seventy-four Italian cutaneous primary melanomas (52.1±12.7 years old) were studied; 51.4% were stage I, 21.6% stage II, 13.5% stage III, and 13.5% stage IV melanomas. VDR expression was categorized as follows: 100% positivevs. <100%; over the median 20% (high VDR expression) vs. ≤20% (low VDR expression); absence vs. presence of VDR-expressing cells.
Results
: Stage I melanomas, Breslow thickness of <1.00 mm, level II Clark invasion, Aa heterozygous genotype, and AaTT combined genotype were more frequent in melanomas with high vs. low VDR expression. Combined genotypes BbAA, bbAa, AATt, BbAATt, and bbAaTT were more frequent in 100% vs. <100% VDR-expressing cells. Combined genotype AATT was more frequent in melanomas lacking VDR expression (odds ratio=14.5; P=0.025). VDR expression was not associated with metastasis, ulceration, mitosis >1, regression, tumor-infiltrating lymphocytes, tumoral infiltration of vascular tissues, additional skin and non-skin cancers, and melanoma familiarity.
Conclusions
: We highlighted that VDR polymorphisms can affect VDR expression in excised melanoma cells. Low VDR expression in AATT carriers is a new finding that merits further study. VDR expression possibly poses implications for vitamin D supplementation against melanoma. VDR expression and VDR genotype may become precise medicinal tools for melanoma in the future.
Keywords: Vitamin D receptor, VDR protein expression, VDR polymorphism, cutaneous melanoma, metastatic melanoma, skin cancer, predictive biomarkers, FokI polymorphism
Introduction
Cutaneous melanoma incidence continually increases in developed countries, particularly in fair-skinned individuals1-4. Recent data5 indicated that prevalence of melanoma in Northern Italy is two fold higher than in Southern Italy and with Central Italy showing prevalence at intermediate level. High incidence rates were particularly registered in the Northeast Friuli-Venezia Giulia region (19.6/100,000/year in men; 16.4/100,000/year in women)6, implying that geographically detailed studies should be performed regarding melanoma risk factors7.
Melanoma is the leading cause of mortality among skin cancer patients with low survival rates8. Although new therapeutic treatments are available9, early detection and surgery remain the main treatment options. Therefore, every new discovery on melanoma biological pathways presents opportunity in improving management and treatment options. Considerable preclinical and epidemiologic data suggest that vitamin D may play an important role in cancer pathogenesis and progression10. Numerous preclinical studies specifically indicated that exposure of cancer cells to high concentrations of vitamin D metabolites halts progression through the cell cycle, induces apoptosis, and slows down or stops tumor growth11. Vitamin D also enhances antitumor activity of some cytotoxic anticancer agents in in vivo preclinical models12. Anti-proliferative effects of vitamin D for cancer prevention and treatment were explored in several malignancies, including skin, breast, prostate, colorectal, and other cancers8,13-18. Numerous epidemiological studies supported the hypothesis that individuals with lower serum vitamin D levels feature higher exposure risk to different cancers12. Current literature suggests the chemopreventive role of vitamin D by acting against initiation and progression of tumorigenesis8,12-18. Despite the stronger consensus on protective role of vitamin D against cancer, particularly by reduction of mortality rate, its therapeutic function for cancer patients remains debatable10,19. Remarkably, a recent meta-analysis showed that vitamin D supplementation minimally affects total cancer incidence, even when total cancer mortality is significantly reduced20.
Vitamin D receptor (VDR) is a nuclear transcription factor belonging to the nuclear receptor superfamily that binds 1α, 25-dihydroxyvitamin D (calcitriol) with high affinity and specificity19. Upon binding to the active form of vitamin D, VDR translocates from cytoplasm into the nucleus and binds to vitamin D responsive elements (VDREs), thus up- or down-regulating hundreds of genes directly controlled by vitamin D19,21,22. Increasing evidence showed pleiotropic hormonal effects of vitamin D on calcium and skeletal metabolism, immunological responses, detoxification, oxidative stress, cancer-related metabolic pathways, proliferation, and cell differentiation18,19,21.
VDR is abundantly expressed in the skin19,21. Some VDR expression was also reported to occur in cultured melanoma cells11,13,14,23. Intermittent sun exposure or ultraviolet (UV) radiation and sunburns are known environmental risk factors for melanoma24,25. However, chronic and continuous UV radiation exposure activates vitamin D biosynthesis, which in turn can develop a protective action against tumoral proliferation8,12,16,25. Four recently discovered mechanisms may underlie actions of VDR as a tumor suppressor in the skin10.
Human VDR gene is located on chromosome 12q12-q14 and comprises 11 exons and 11 introns26. Genetic variants of VDR may modulate its actions, with FokI, BsmI, ApaI, and TaqI being the most studied single-nucleotide polymorphisms (SNPs)26-28. VDR-FokI polymorphism is a functional SNP that extends lengths of the receptor protein from 424 to 427 amino acid residues. BsmI, ApaI, and TaqI polymorphisms are located in the 3′ terminal region of the VDR gene and do not affect protein sequence of the VDR receptor. FokI polymorphism is reported not to be in linkage disequilibrium with the other three polymorphisms. Instead, BsmI, ApaI, and TaqI polymorphisms are reported to be in linkage disequilibrium to a variable extent; thus, combined genotypes including two or three of these polymorphisms were investigated in literature26,28,29. Some evidence suggested that genotypes FF, BB, tt, and the combined genotype BBAAtt may be associated with increased expression of VDR, which in turn regulates actions of vitamin D26-28. Roles of VDR polymorphisms in melanoma were evaluated in some recent studies and meta-analyses30-37. However, associations of VDR polymorphisms with skin cancer risk remain insufficiently characterized30,31,36.
At present, no study examined VDR polymorphisms and VDR expression in melanoma cells of excised tissues from patients.
Thus far, only one cohort of 69 Polish melanoma patients was investigated by two studies for VDR expression in tumor tissues38,39. VDR expression progressively decreases from normal skin to melanocytic nevi to melanomas38, suggesting the relationship between VDR expression and melanoma prognosis39. Brożyna and colleagues38 observed reduced expression levels of VDR in skin surrounding nevi and melanomas as opposed to normal skin.
Advances in melanoma treatment can be achieved through developments in understanding of melanoma risk factors, genomics, and molecular pathogenesis31,40.
By immunohistochemical staining of primary cutaneous melanoma tissues, we investigated VDR expression in relation to characteristics, melanoma histological grading, and metastatic stage of patients. We also explored the association of four VDR SNPs- FokI-rs2228570 C>T located in exon 2, BsmI-rs1544410 G>A located in intron 8, ApaI-rs7975232 T>G located in intron 8, and TaqI-rs731236 T>C located in exon 9- with VDR expression levels in cutaneous malignant melanoma tissues.
Patients and methods
Patients
Enrolment and clinical visits of all study participants were performed at the Udine University-Hospital Dermatology Clinic. Diagnostic procedures were conducted according to routine protocols. All participants signed a written informed consent. The Udine Institutional Ethical Committee approved the study protocol in accordance to the Declaration of Helsinki.
Seventy-four (39 males, 35 females, age range: 29-82 years) unrelated patients (hospitalized or outpatients) who consecutively underwent surgical excision of cutaneous melanoma were enrolled based on a retrospective design. Inclusion criteria were as follows: melanoma different from that in in situ only, absence of mucosal melanomas, patient is a resident of Friuli-Venezia Giulia region (Northern Italy), and absence of major chronic diseases, such as autoimmune diseases, and type 1 diabetes.
Assessment of melanoma diagnosis and patient stage classification were performed by clinical/histological findings as described by Balch et al.41. For patients with multiple melanomas, we examined only the first main melanoma according to histological assessment of major primary tumor grading, and primary melanoma characteristics were accounted for study analyses.
Questionnaires were used to collect information from each participant; data obtained included demographic and lifestyle characteristics and medical and family history of melanoma. Body mass index (BMI) was determined by ratio of weight (kg) to squared height (m).
Immunohistochemical staining and evaluation of VDR expression
Slides stained with hematoxylin and eosin were reviewed for each case from formalin-fixed and paraffin-embedded blocks and were selected for VDR immunohistochemical staining. Immunohistochemistry was performed on 5 µm thick paraffin sections as follows: after dewaxing, rehydration and endogenous peroxidase quenching with 3% v/v H 2O 2 in methanol for 15 min, antigen retrieval in 0.01 M citrate buffer at 98°C water bath for 40 min, application and incubation of primary antibody (VDR mouse monoclonal D-6, sc-13133, Santa Cruz Biotechnology, Texas, USA) at 1:200 dilution 1 h at room temperature42,43, incubation with peroxidase-based EnVision+ /Horseradish peroxidase (Dako A/S, Glostrup, Denmark) for 30 min at room temperature, and treatment with diaminobenzidine for 3 min44. The sections were then counterstained with Mayer’s hematoxylin, dehydrated, and mounted. VDR expression was evaluated on tumor cells (nuclear and cytoplasmic staining) of the whole section. Immunolabeled sections were viewed under Nikon Eclipse 80i light microscope at 25× magnification, and a semi-quantitative evaluation was performed to determine VDR expression levels in malignant melanoma cells (nuclear and cytoplasmic staining). Staining of sweat gland cells, which consistently showed strong and diffused positivity throughout all samples, was used as positive reference. The term “emboli” indicates tumoral invasion of vascular cells observed in slide specimens. Melanoma specimens were reviewed by two pathologists involved in the study but were unaware of all other clinical and molecular data during evaluation. Our categorization choices were based on the following considerations: A) when melanomas feature 100% VDR-positive cells, all tumor cells are possibly responsive to vitamin D stimulation; B) when 0% cells are positive for VDR expression, virtually all tumor cells do not respond to vitamin D stimulation; C) aside from extreme conditions, a cut off at median percentage of positive cells can be reasonably used for evaluating the half of samples with highervs. the half of samples with lower VDR expression. Therefore, results were ranked based on percentage of cells positive for VDR expression (irrespective of staining intensity). Cytoplasmic VDR expression ranged from 0% to 100%, with a median value at 20.0%. We categorized variables as follows: 100% cytoplasmic VDR-expression-positive cells versus all remaining melanomas; >20% cytoplasmic VDR-expression-positive cells (high VDR expression) vs. ≤20% positive cells (low VDR expression); and absent (0% positive cells) vs. present (>0% positive cells) VDR expression. Only 11 out of 74 cases (14.9%) showed nuclear VDR-positive immunostaining, and all these cases were categorized into >20% cytoplasmic VDR-expression-positive cells. Given the low number of melanoma specimens showing VDR expression detected in nuclei, this parameter was not further analyzed.
Determination of VDR gene polymorphisms
Determination of SNP VDR-FokI (C>T),VDR-BsmI (G>A),VDR-ApaI (T>G),VDR-TaqI (T>C) was performed as previously described29,45 after extraction of genomic DNA from ethylenediaminetetraacetic-acid -venous blood samples46. Absence or presence of restriction site was denoted by a capital or lower letter, respectively: “F” and “f” for FokI, “B” and “b” for BsmI, “A” and “a” for ApaI, and “T” and “t” for TaqI endonucleases28,29. FokI, BsmI, ApaI and TaqI polymorphisms of VDR were studied using previously tested primers29,45 to amplify appropriate DNA fragments. FokI enzyme (Euroclone, Milano, Italy) digestion of 265 bp amplified DNA was used to determine FokI restriction fragment length polymorphism (RFLP) yielding 196 and 69 bp fragments in the presence of f allele46. To analyze BsmI polymorphism, the resulting amplified 825 bp polymerase chain reaction (PCR) fragment was digested with BsmI restriction enzyme (Euroclone, Milano, Italy), generating 650 and 175 bp fragments in the presence of b allele29. ApaI digestion of the 740 bp amplified DNA was used to determine both ApaI and TaqI RFLP, generating 530 and 210 bp fragments in the presence of a allele. Digestion with TaqI of the 740 bp PCR fragment generated 290, 245, and 205 bp fragments in the presence of t allele and 495 and 245 bp fragments in its absence (T allele) owing to an additional monomorphic TaqI site29. DNA fragments were separated by polyacrylamide gel electrophoresis.
Statistical analysis
Kolmogorov-Smirnov test was used to assess normal data distribution. Percentage of VDR-positive cells by immunohistochemical staining was not normally distributed. Thus, median values and ranges were reported for this variable. Mann-Whitney U test was used to assess differences between groups. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated to determine association of groups with different VDR expression and melanoma characteristics, alleles, genotypes, and combined genotypes. Our study was explorative as no previous investigation determined frequencies of VDR polymorphisms in melanoma patients according to immunohistochemical findings of VDR expression Prior to study enrolment, we evaluated that a number of 70 subjects fitted the 80% power at an alpha level of 0.05 to detect differences between high (above the median) and low VDR expression melanoma groups whether OR value equals 3 or more for a SNP site47. Deviation tests from Hardy-Weinberg equilibrium (HWE) were performed using a separate Chi-square distribution for each SNP29. Linkage disequilibrium (LD) between SNPs was determined as described29. Two-sided significance level was set at 0.05, and P values ≤ 0.10 were considered as a tendency to be significant. Statistical software SPSS (SPSS Inc., Chicago, IL, USA) was used for statistical analyses.
Results
VDR immunohistochemical staining
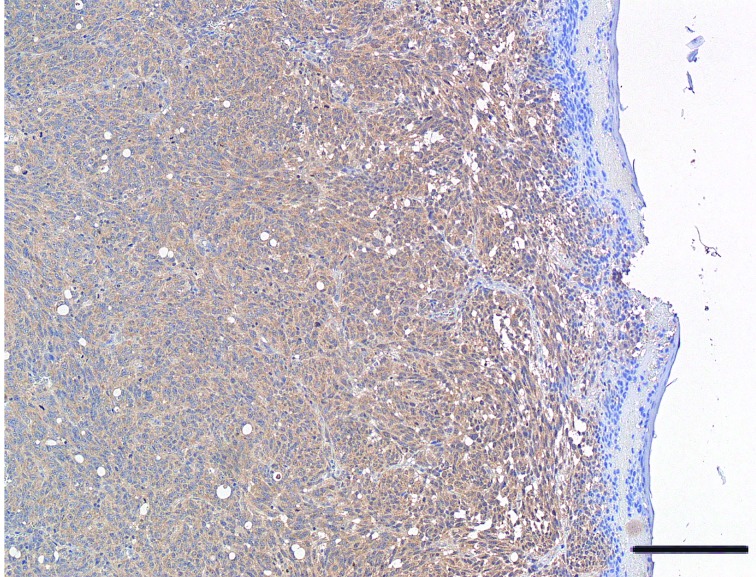
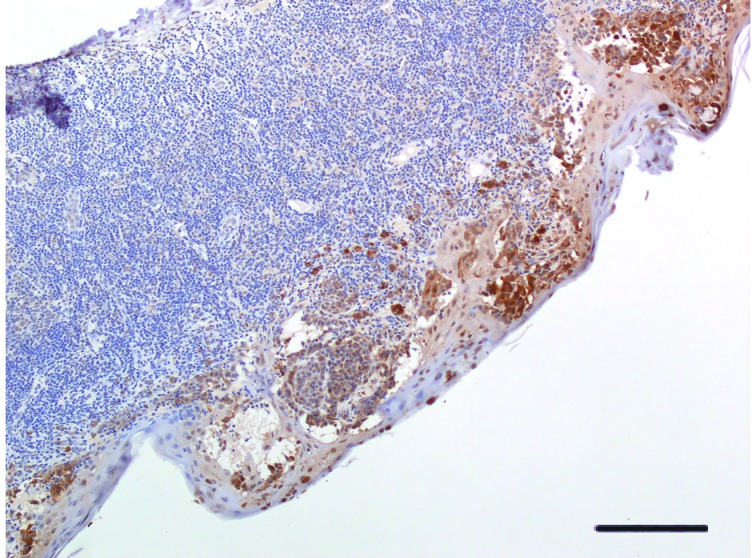
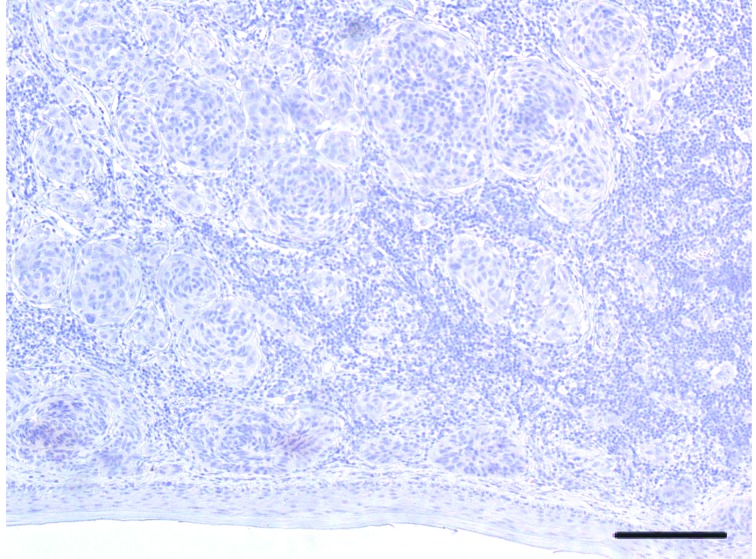
A significant variability in VDR immunohistochemical staining was observed in cytoplasm of melanoma cells, with 16.2% (12/74) of patients tested positive for 100% melanoma cell staining, whereas 20.3% (15/74) were tested negative. Median percentage value of VDR-positive staining reached 20.0%. Figures 1 , 2 , and 3 display representative images of VDR immunohistochemical staining in excised tissues from patients with primary cutaneous melanoma, showing 100% staining, intermediate staining, and no staining, respectively.
1.
Representative image of VDR protein expression in cutaneous melanoma tissue. Melanoma showing diffuse positivity for VDR expression. Eroded epidermis (H&E staining, 25×. Bar indicates 50 μm).
2.
Representative image of VDR protein expression in cutaneous melanoma tissue. Melanoma featuring tumor-infiltrating lymphocytes (TILs). Intraepidermal component of lesion (at the bottom of image) shows strong positivity for VDR expression (H&E staining, 25×. Bar indicates 50 μm).
3.
Representative image of VDR protein expression in cutaneous melanoma tissue. Melanoma featuring TILs. In this case, melanoma cells nesting in the dermis are negative throughout the whole lesion (H&E staining, 25×. Bar indicates 50 μm).
Patients and melanoma characteristics according to VDR expression
All 74 (35 females and 39 males) cutaneous melanoma patients were white residents in Northern Italy. Average age at melanoma diagnosis was 52.1±12.7 years, with 27% (20/74) of patients exhibiting metastatic melanoma (i.e., stages III and IV), and all showed Clark level higher than Clark I invasion.
Table 1 shows the main clinical characteristics of melanoma patients and compares three different binary categories according to percentage of tumor cells positive for VDR staining: a) 100% positivevs. <100% positive, b) over 20% (median value) positive (high VDR expression) vs. ≤20% positive (low VDR expression), and c) absence vs. presence of VDR-positive cells. Complete data on ORs and CIs are reported on Table S1 . Age of melanoma diagnosis, gender, BMI, and smoking did not differ between compared groups. Differences were not significant in comparison of 12 subjects with 100% VDR-positive cells vs. 62 subjects with <100% VDR-positive cells. The following statically significant differences were observed during comparison of 36 melanomas with high VDR expression vs. 38 melanomas with low VDR expression: stage I was more frequent in the former (63.9% vs. 39.5%, OR=2.71, CI=1.06–6.95, P=0.036), whereas stage II was less frequent (11.1% vs. 31.6%, OR=0.27, CI=0.08–0.94, P=0.033); the former presents a lower mean Breslow thickness (1.23±0.88vs. 2.27±1.97 mm,P=0.008) and more frequent Breslow thickness of <1.00 mm (50.0% vs. 26.3%, OR=2.80, CI=1.06–7.41, P=0.036) and less frequent Breslow thickness ≥1.01 mm (50.0% vs. 77.8%, OR=0.36, CI=0.13–0.95, P=0.036). A higher frequency of Clark II invasion (38.9% vs. 15.8%, OR=3.39, CI=1.13–10.2, P=0.025) was observed in high- than low-VDR-expression group. Superficial spreading was present in 61.1% of melanomas with high VDR expression vs. 39.5% of low-VDR-expression melanomas, where this difference did not reach significant P values (P=0.063). In comparing absence vs. presence of VDR-positive immunohistochemical staining of melanoma cells, the only significant finding was a higher frequency of stage II A in the formervs. the latter (26.7% vs. 5.1%, OR=6.79, CI=1.33–34.7, P=0.028) (all sub-stage data are shown in Table S1 ).
1.
Clinical characteristics of 74 consecutively enrolled melanoma patients and comparison between groups of 100% VDR-positive cells (n=12) vs. <100% ( n=62); over the median (>20%) VDR-positive cells (n=36) vs. below or equal the median (≤20%) (n=38); and absence of VDR-positive cell (n=15) vs. remaining cases with detected VDR expression (n=59)
| Characteristics | All melanoma patients (n=74) | 100% VDR- positive cells (n=12) | <100% VDR- positive cells (n=62) | P=100% vs. <100% VDR- positive | >20%a VDR- positive cells (n=36) | ≤20% VDR- positive cells (n=38) | P>20%vs. ≤20% VDR- positive | VDR absence (0% positive) (n=15) | VDR presence (>0% positive) (n=59) | PVDR absence vs. presence |
| Age at melanoma diagnosis, years, mean ± SD | 52.1±12.7 | 49.8±13.0 | 52.7±12.9 | 0.519a | 51.7±13.9 | 52.8±12.0 | 0.770a | 51.5±14.5 | 52.5±12.6 | 0.767a |
| Age <50 years at melanoma diagnosis | 31 (41.9) | 6 (50.0) | 25 (40.3) | 0.534 | 16 (44.4) | 15 (39.5) | 0.665 | 7 (46.7) | 24 (40.7) | 0.675 |
| BMI, kg/m2, mean ± SD | 25.8±4.0 | 25.0±4.6 | 25.9±3.9 | 0.412a | 26.0±4.3 | 25.6±3.8 | 0.804a | 25.7±2.6 | 25.8±4.3 | 0.762a |
| Male | 39 (52.7) | 6 (50.0) | 33 (53.2) | 0.838 | 20 (55.6) | 19 (50.0) | 0.632 | 9 (60.0) | 30 (50.8) | 0.526 |
| Smoker | 11 (14.9) | 2 (16.7) | 9 (14.5) | 1.000 | 4 (11.1) | 7 (18.4) | 0.377 | 2 (13.3) | 9 (15.3) | 1.000 |
| Stage I | 38 (51.4) | 6 (50.0) | 32 (51.6) | 0.919 | 23 (63.9) | 15 (39.5) | 0.036 | 6 (40.0) | 32 (54.2) | 0.325 |
| Stage II | 16 (21.6) | 2 (16.7) | 14 (22.6) | 1.000 | 4 (11.1) | 12 (31.6) | 0.033 | 6 (40.0) | 10 (16.9) | 0.077 |
| Stage III | 10 (13.5) | 3 (25.0) | 7 (11.3) | 0.351 | 6 (16.7) | 4 (10.5) | 0.510 | 1 (6.7) | 9 (15.3) | 0.676 |
| Stage IV | 10 (13.5) | 1 (8.3) | 9 (14.5) | 1.000 | 3 (8.3) | 7 (18.4) | 0.310 | 2 (13.3) | 8 (13.6) | 1.000 |
| Metastatic melanoma (Stage III +IV) | 20 (27.0) | 4 (33.3) | 16 (25.8) | 0.724 | 9 (25.0) | 11 (28.9) | 0.702 | 3 (20.0) | 17 (28.8) | 0.746 |
| Trunk | 47 (63.5) | 5 (41.7) | 42 (67.7) | 0.108 | 24 (66.7) | 23 (60.5) | 0.583 | 7 (46.7) | 40 (67.8) | 0.129 |
| Upper limb | 6 (8.1) | 2 (16.7) | 4 (6.5) | 0.249 | 2 (5.6) | 4 (10.5) | 0.675 | 3 (20.0) | 3 (5.1) | 0.093 |
| >Lower limb | 14 (18.9) | 3 (25.0) | 11 (17.7) | 0.687 | 7 (19.4) | 7 (18.4) | 0.911 | 4 (26.7) | 10 (16.9) | 0.463 |
| Hands/feet | 5 (6.8) | 2 (16.7) | 3 (4.8) | 0.183 | 2 (5.6) | 3 (7.9) | 1.000 | 0 (–) | 5 (8.5) | 0.576 |
| Head/neck | 2 (2.7) | 0 (–) | 2 (3.2) | 1.000 | 1 (2.8) | 1 (2.6) | 1.000 | 1 (6.7) | 1 (1.7) | 0.367 |
| Superficial spreading | 37 (50.0) | 7 (58.3) | 30 (48.4) | 0.528 | 22 (61.1) | 15 (39.5) | 0.063 | 6 (40.0) | 31 (52.5) | 0.386 |
| Nodular | 31 (41.9) | 4 (33.3) | 27 (43.5) | 0.512 | 12 (33.3) | 19 (50.0) | 0.146 | 8 (53.3) | 23 (39.0) | 0.314 |
| Acral lentiginous | 3 (4.1) | 1 (8.3) | 2 (3.2) | 0.417 | 1 (2.8) | 2 (5.3) | 1.000 | 0 (–) | 3 (5.1) | 1.000 |
| Spitzoide | 2 (2.7) | 0 (–) | 2 (3.2) | 1.000 | 1 (2.8) | 1 (2.6) | 1.000 | 1 (6.7) | 1 (1.7) | 0.367 |
| Others | 3 (4.1) | 0 (–) | 3 (4.8) | 1.000 | 0 (–) | 3 (7.9) | 0.240 | 0 (–) | 3 (5.1) | 1.000 |
| Breslow thickness, mm, mean ± SD | 1.77±1.62 | 1.34±0.94 | 1.85±1.71 | 0.416a | 1.23±0.88 | 2.27±1.97 | 0.008a | 2.16±1.88 | 1.66±1.55 | 0.145a |
| Continued | ||||||||||
S1.
Clinical characteristics of 74 consecutively enrolled melanoma patients and comparison between groups of 100% VDR-positive cells (n=12) vs. <100% ( n=62); over the median (>20%) VDR-positive cell (n=36) vs. below or equal the median (≤20%) (n=38); and absence of VDR-positive cell (n=15) vs. remaining cases with detected VDR expression (n=59)
| Characteristics | All melanoma patients (n=74) | 100% VDR-positive cells (n=12) | <100% VDR-positive cells (n=62) | OR (CI)100% vs. < 100% VDR-positive | P=100% vs. < 100% VDR-positive | >20%a VDR-positive cells (n=36) | ≤20% VDR-positive cells (n=38) | OR (CI)>20%vs. ≤ 20% VDR-positive | P>20%vs. ≤ 20% VDR-positive | VDR absence (0% positive)(n=15) | VDR presence (>0% positive)(n=59) | OR (CI)VDR absence vs. presence | PVDR absence vs. presence |
| Age at melanoma diagnosis, years, mean ± SD | 52.1±12.7 | 49.8±13.0 | 52.7±12.9 | – | 0.519a | 51.7±13.9 | 52.8±12.0 | – | 0.770a | 51.5±14.5 | 52.5±12.6 | – | 0.767a |
| Age <50 years at melanoma diagnosis | 31 (41.9) | 6 (50.0) | 25 (40.3) | 1.48 (0.43–5.11) | 0.534 | 16 (44.4) | 15 (39.5) | 1.23 (0.49–3.09) | 0.665 | 7 (46.7) | 24 (40.7) | 1.28 (0.41–3.99) | 0.675 |
| BMI, kg/m2, mean ± SD | 25.8±4.0 | 25.0±4.6 | 25.9±3.9 | – | 0.412a | 26.0±4.3 | 25.6±3.8 | – | 0.804a | 25.7±2.6 | 25.8±4.3 | – | 0.762a |
| Male | 39 (52.7) | 6 (50.0) | 33 (53.2) | 0.88 (0.25–3.03) | 0.838 | 20 (55.6) | 19 (50.0) | 1.25 (0.50–3.12) | 0.632 | 9 (60.0) | 30 (50.8) | 1.45 (0.46–4.59) | 0.526 |
| Smoker | 11 (14.9) | 2 (16.7) | 9 (14.5) | 1.18 (0.22–6.28) | 1.000 | 4 (11.1) | 7 (18.4) | 0.55 (0.15–2.08) | 0.377 | 2 (13.3) | 9 (15.3) | 0.85 (0.16–4.45) | 1.000 |
| Stage I | 38 (51.4) | 6 (50.0) | 32 (51.6) | 0.94 (0.27–3.23) | 0.919 | 23 (63.9) | 15 (39.5) | 2.71 (1.06–6.95) | 0.036 | 6 (40.0) | 32 (54.2) | 0.56 (0.18–1.78) | 0.325 |
| Stage I A | 23 (31.1) | 4 (33.3) | 19 (30.6) | 1.13 (0.30–4.22) | 1.000 | 15 (41.7) | 8 (21.1) | 2.68 (0.96–7.45) | 0.055 | 3 (20.0) | 20 (33.9) | 0.49 (0.12–1.93) | 0.365 |
| Stage I B | 15 (20.3) | 2 (16.7) | 13 (21.0) | 0.75 (0.15–3.87) | 1.000 | 8 (22.2) | 7 (18.4) | 1.26 (0.41–3.94) | 0.684 | 3 (20.0) | 12 (20.3) | 0.98 (0.24–4.03) | 1.000 |
| Stage II | 16 (21.6) | 2 (16.7) | 14 (22.6) | 0.69 (0.13–3.50) | 1.000 | 4 (11.1) | 12 (31.6) | 0.27 (0.08–0.94) | 0.033 | 6 (40.0) | 10 (16.9) | 3.27 (0.95–11.3) | 0.077 |
| Stage II A | 7 (9.5) | 0 (–) | 7 (11.3) | –b | 0.590 | 0 (–) | 7 (18.4) | -b | 0.012 | 4 (26.7) | 3 (5.1) | 6.79 (1.33–34.7) | 0.028 |
| Stage II B | 9 (12.2) | 2 (16.7) | 7 (11.3) | 1.57 (0.28–8.69) | 0.633 | 4 (11.1) | 5 (13.2) | 0.82 (0.20–3.35) | 1.000 | 2 (13.3) | 7 (11.9) | 1.14 (0.21–6.16) | 1.000 |
| Stage II C | 0 (–) | 0 (–) | 0 (–) | –b | –b | 0 (–) | 0 (–) | –b | –b | 0 (–) | 0 (–) | –b | –b |
| Stage III | 10 (13.5) | 3 (25.0) | 7 (11.3) | 2.62 (0.57–12.0) | 0.351 | 6 (16.7) | 4 (10.5) | 1.70 (0.44–6.60) | 0.510 | 1 (6.7) | 9 (15.3) | 0.40 (0.05–3.40) | 0.676 |
| Stage III A | 4 (5.4) | 1 (8.3) | 3 (4.8) | 1.79 (0.17–18.8) | 0.515 | 3 (8.3) | 1 (2.6) | 3.36 (0.33–33.9) | 0.351 | 0 (–) | 4 (6.8) | –b | 0.576 |
| Stage III B | 3 (4.1) | 0 (–) | 3 (4.8) | –b | 1.000 | 0 (–) | 3 (7.9) | –b | 0.240 | 1 (6.7) | 2 (3.4) | 2.04 (0.17–24.1) | 0.499 |
| Stage III C | 3 (4.1) | 2 (16.7) | 1 (1.6) | 12.2 (1.01–147) | 0.067 | 3 (8.3) | 0 (–) | –b | 0.110 | 0 (–) | 3 (5.1) | –b | 1.000 |
| Continued | |||||||||||||
Overall, as shown in Table 1 , none of the tumor markers commonly associated with severe prognosis and metastatic stage were associated with VDR immunohistochemical staining; these markers included ulceration, mitosis >1, absence of tumor-infiltrating lymphocytes (TILs), emboli, and epithelioid variants. The presence of multiple melanomas, additional skin, non-skin cancers, and melanoma familiarity did not correlate with VDR immunohistochemical staining.
By further analysis, median VDR expression did not differ between 20 metastatic melanomas (median: 17.5%, range: 0%–100% VDR-positive cells) and 54 non-metastatic melanomas (median: 25.0%, range: 0%–100% VDR-positive cells), with P=0.796. Significant P value (P=0.095) was not observed in differences in median values of VDR expression of stage I melanomas compared with those of stages II+III+IV. Median values of VDR expression of Clark II (in our cohort, none of melanomas showed Clark I invasion) melanomas were significantly higher compared with Clark III+IV+V levels (median: 70.0%, range: 0%–100% vs. median: 10.0%, range: 0%–100% VDR-positive cells, P=0.019). A significant P value was not observed in differences in median values of VDR expression in superficially spreading melanomas compared with the remaining ones (P=0.075).
Patient and melanoma characteristics according to VDR polymorphisms
Table 2 shows VDR polymorphism genotypes, alleles, and combined genotype frequencies in all 74 patients and in groups of melanomas categorized according to VDR immunohistochemical staining. EachVDR polymorphism of FokI, BsmI, ApaI, and TaqI was in HWE. FokI SNP was not in LD with other SNPs. BsmI was in LD with ApaI and TaqI, and ApaI was in LD with TaqI. Thus, for further analyses, we considered binary and ternary combination of genotypes comprising BsmI, ApaI and TaqI polymorphisms. As observed in other studies26,29, not all theoretically possible binary and ternary combination of genotypes were observed; thus, Table 2 reports only combined genotypes with at least one confirmed finding.
2.
Genotype and allele of ApaI, VDR-polymorphism, and BsmI-ApaI, ApaI-TaqI, BsmI-ApaI-TaqI combined genotypes of 74 melanoma patients and comparisons between groups of 100% VDR-positive cells (n=12) vs. <100% ( n=62); over the median (>20%) VDR-positive cells (n=36) vs. below or equal the median (≤20%) (n=38); and absence of VDR-positive cell (n=15) vs. remaining cases with detected VDR expression (n=59)
| VDR genotype or combined genotype | All melanoma patients (n=74) | 100% VDR-positive cells (n=12) | <100% VDR-positive cells (n=62) | P=100% vs.<100% VDR-positive | >20%a VDR-positive cells (n=36) | ≤20% VDR-positive cells (n=38) | P>20%vs. ≤20% VDR-positive | VDR absence (0% positive)(n=15) | VDR presence (>0% positive)(n=59) | PVDR absence vs. presence |
| a Over the median value of percentage (%) of cells positive for VDR protein. | ||||||||||
| ApaI genotype | ||||||||||
| AA | 30 (40.5) | 8 (66.7) | 22 (35.5) | 0.058 | 13 (36.1) | 17 (44.7) | 0.450 | 8 (53.3) | 22 (37.3) | 0.258 |
| Aa | 36 (48.6) | 4 (33.3) | 32 (51.6) | 0.246 | 22 (61.1) | 14 (36.8) | 0.037 | 5 (33.3) | 31 (52.5) | 0.184 |
| aa | 8 (10.8) | 0 (–) | 8 (12.9) | 0.339 | 1 (2.8) | 7 (18.4) | 0.056 | 2 (13.3) | 6 (10.2) | 0.660 |
| A allele | 96/148 (64.9) | 20/24 (83.3) | 76/124 (61.3) | 0.038 | 48/72 (66.7) | 48/76 (63.2) | 0.655 | 21/30 (70.0) | 75/118 (63.6) | 0.509 |
| a allele | 52/148 (35.1) | 4/24 (16.7) | 48/124 (38.7) | 0.038 | 24/72 (33.3) | 28/76 (36.8) | 0.655 | 9/30 (30.0) | 43/118 (36.4) | 0.509 |
| BsmI-ApaI combined genotype | ||||||||||
| BBAA | 23 (31.1) | 5 (41.7) | 18 (29.0) | 0.498 | 9 (25.0) | 14 (36.8) | 0.271 | 6 (40.0) | 17 (28.8) | 0.533 |
| BbAA | 5 (6.8) | 3 (25.0) | 2 (3.2) | 0.028 | 4 (11.1) | 1 (2.6) | 0.194 | 1 (6.7) | 4 (6.8) | 1.000 |
| BbAa | 31 (41.9) | 1 (8.3) | 30 (48.4) | 0.010 | 18 (50.0) | 13 (34.2) | 0.169 | 5 (33.3) | 26 (44.1) | 0.452 |
| Bbaa | 2 (2.7) | 0 (–) | 2 (3.2) | 1.000 | 0 (–) | 2 (5.3) | 0.494 | 0 (–) | 2 (3.4) | 1.000 |
| bbAA | 2 (2.7) | 0 (–) | 2 (3.2) | 1.000 | 0 (–) | 2 (5.3) | 0.494 | 1 (6.7) | 1 (1.7) | 0.367 |
| bbAa | 5 (6.8) | 3 (25.0) | 2 (3.2) | 0.028 | 4 (11.1) | 1 (2.6) | 0.194 | 0 (–) | 5 (8.5) | 0.576 |
| bbaa | 6 (8.1) | 0 (–) | 6 (9.7) | 0.581 | 1 (2.8) | 5 (13.2) | 0.200 | 2 (13.3) | 4 (6.8) | 0.595 |
| ApaI-TaqI combined genotype | ||||||||||
| AATT | 4 (5.4) | 0 (–) | 4 (6.5) | 1.000 | 0 (–) | 4 (10.5) | 0.115 | 3 (20.0) | 1 (1.7) | 0.025 |
| AATt | 14 (18.9) | 6 (50.0) | 8 (12.9) | 0.008 | 9 (25.0) | 5 (13.2) | 0.194 | 4 (26.7) | 10 (16.9) | 0.463 |
| AAtt | 12 (16.2) | 2 (16.7) | 10 (16.1) | 1.000 | 4 (11.1) | 8 (21.1) | 0.246 | 1 (6.7) | 11 (18.6) | 0.439 |
| AaTT | 12 (16.2) | 3 (25.0) | 9 (14.5) | 0.399 | 9 (25.0) | 3 (7.9) | 0.046 | 1 (6.7) | 11 (18.6) | 0.439 |
| AaTt | 24 (32.4) | 1 (8.3) | 23 (37.1) | 0.089 | 13 (36.1) | 11 (28.9) | 0.511 | 4 (26.7) | 20 (33.9) | 0.761 |
| aaTT | 8 (10.8) | 0 (–) | 8 (12.9) | 0.339 | 1 (2.8) | 7 (18.4) | 0.056 | 2 (13.3) | 6 (10.2) | 0.660 |
| BsmI-ApaI-TaqI combined genotype | ||||||||||
| BBAATT | 1 (1.4) | 0 (–) | 1 (1.6) | 1.000 | 0 (–) | 1 (2.6) | 1.000 | 1 (6.7) | 0 (–) | 0.203 |
| BBAATt | 10 (13.5) | 3 (25.0) | 7 (11.3) | 0.351 | 5 (13.9) | 5 (13.2) | 1.000 | 4 (26.7) | 6 (10.2) | 0.110 |
| BBAAtt | 12 (16.2) | 2 (16.7) | 10 (16.1) | 1.000 | 4 (11.1) | 8 (21.1) | 0.246 | 1 (6.7) | 11 (18.6) | 0.439 |
| BbAATt | 4 (5.4) | 3 (25.0) | 1 (1.6) | 0.012 | 4 (11.1) | 0 (–) | 0.051 | 0 (–) | 4 (6.8) | 0.576 |
| BbAATT | 1 (1.4) | 0 (–) | 1 (1.6) | 1.000 | 0 (–) | 1 (2.6) | 1.000 | 1 (6.7) | 0 (–) | 0.203 |
| BbAaTT | 7 (9.5) | 0 (–) | 7 (11.3) | 0.590 | 5 (13.9) | 2 (5.3) | 0.255 | 1 (6.7) | 6 (10.2) | 1.000 |
| BbAaTt | 24 (32.4) | 1 (8.3) | 23 (37.1) | 0.089 | 13 (36.1) | 11 (28.9) | 0.511 | 4 (26.7) | 20 (33.9) | 0.761 |
| BbaaTT | 2 (2.7) | 0 (–) | 2 (3.2) | 1.000 | 0 (–) | 2 (5.3) | 0.494 | 0 (–) | 2 (3.4) | 1.000 |
| bbAATT | 2 (2.7) | 0 (–) | 2 (3.2) | 1.000 | 0 (–) | 2 (5.3) | 0.494 | 1 (6.7) | 1 (1.7) | 0.367 |
| bbAaTT | 5 (6.8) | 3 (25.0) | 2 (3.2) | 0.028 | 4 (11.1) | 1 (2.6) | 0.194 | 0 (–) | 5 (8.5) | 0.576 |
| bbaaTT | 6 (8.1) | 0 (–) | 6 (9.7) | 0.581 | 1 (2.8) | 5 (13.2) | 0.200 | 2 (13.3) | 4 (6.8) | 0.595 |
Differences between VDR expression groups were not significant for single FokI, BsmI, and TaqI genotypes and alleles, and BsmI-TaqI combined genotypes (data shown in Table S3 ).
S3.
Genotype and allele characteristics of FokI, BsmI, and TaqI VDR-polymorphisms and BsmI-TaqI haplotypes of 74 melanoma patients and comparisons between groups of 100% VDR-positive cells (n=12) vs. <100% ( n=62); over the median (>20%) VDR-positive cells (n=36) versus below or equal the median (≤20%) (n=38); and absence of VDR-positive cell (n=15) versus remaining cases with detected VDR expression (n=59)
| VDR genotype or combined genotype | All melanoma patients (n=74) | 100% VDR-positive cells (n=12) | <100% VDR-positive cells (n=62) | OR (CI) 100% vs. < 100% VDR-positive | P=100% vs. < 100% VDR-positive | >20%a VDR-positive cells (n=36) | ≤20% VDR-positive cells (n=38) | OR (CI)>20%vs. ≤ 20% VDR-positive | P>20%vs. ≤ 20% VDR-positive | VDR absence (0% positive)(n=15) | VDR presence (>0% positive)(n=59) | OR (CI) VDR absence vs. presence | PVDR absence vs. presence |
| FokI genotype | |||||||||||||
| FF | 32 (43.2) | 5 (41.7) | 27 (43.5) | 0.93 (0.26–3.24) | 0.904 | 16 (44.4) | 16 (42.1) | 1.10 (0.44–2.76) | 0.839 | 7 (46.7) | 25 (42.4) | 1.19 (0.38–3.71) | 0.764 |
| Ff | 33 (44.6) | 5 (41.7) | 28 (45.2) | 0.87 (0.25–3.03) | 0.824 | 14 (38.9) | 19 (50.0) | 0.64 (0.25–1.60) | 0.337 | 6 (40.0) | 27 (45.8) | 0.79 (0.25–2.50) | 0.688 |
| ff | 9 (12.2) | 2 (16.7) | 7 (11.3) | 1.57 (0.28–8.69) | 0.633 | 6 (16.7) | 3 (7.9) | 2.33 (0.54–10.1) | 0.302 | 2 (13.3) | 7 (11.9) | 1.14 (0.21–6.16) | 1.000 |
| F allele | 97/148 (65.5) | 15/24 (62.5) | 82/124 (66.1) | 0.85 (0.34–2.11) | 0.732 | 46/72 (63.9) | 51/76 (67.1) | 0.87 (0.44–1.71) | 0.681 | 20/30 (66.7) | 77/118 (65.3) | 1.06 (0.46–2.49) | 0.884 |
| f allele | 51/148 (34.5) | 9/24 (37.5) | 42/124 (33.9) | 1.17 (0.47–2.90 | 0.732 | 26/72 (36.1) | 25/76 (32.9) | 1.15 (0.58–2.27) | 0.681 | 10/30 (33.3) | 41/118 (34.7) | 0.94 (0.40–2.19) | 0.884 |
| BsmI genotype | |||||||||||||
| BB | 23 (31.1) | 5 (41.7) | 18 (29.0) | 1.75 (0.49–6.23) | 0.498 | 9 (25.0) | 14 (36.8) | 0.57 (0.21–1.56) | 0.271 | 6 (40.0) | 17 (28.8) | 1.65 (0.51–5.34) | 0.533 |
| Bb | 38 (51.4) | 4 (33.3) | 34 (54.8) | 0.41 (0.11–1.51) | 0.172 | 22 (61.1) | 16 (42.1) | 2.16 (0.85–5.47) | 0.102 | 6 (40.0) | 32 (54.2) | 0.56 (0.18–1.78) | 0.325 |
| bb | 13 (17.6) | 3 (25.0) | 10 (16.1) | 1.73 (0.40–7.55) | 0.432 | 5 (13.9) | 8 (21.1) | 0.60 (0.18–2.06) | 0.418 | 3 (20.0) | 10 (16.9) | 1.22 (0.29–5.15) | 0.719 |
| Continued | |||||||||||||
As reported in Table 2 (data comprising all ORs and CIs are shown in Table S2 . Table S1 to Table S3 in the supplementary materials, available with the full text of this article at www.cancerbiomed.org), heterozygous Aa genotype was more frequent in melanomas with high than low VDR expression (61.1% vs. 36.8%, OR=2.69, CI=1.05–6.90, P=0.037). A allele was found in 83.3% of 100% VDR-positive melanomas and in 61.3% of those with <100% VDR-positive cells (OR=3.16, CI=1.02–9.80, P=0.038).
S2.
Genotype and allele of ApaI, VDR-polymorphism, and BsmI-ApaI, ApaI-TaqI, BsmI-ApaI-TaqI combined genotypes of 74 melanoma patients and comparisons between groups of 100% VDR-positive cells (n=12) vs. <100% ( n=62); over the median (>20%) VDR-positive cells (n=36) vs. below or equal the median (≤20%) (n=38); and absence of VDR-positive cell (n=15) vs. remaining cases with detected VDR expression (n=59)
| VDR genotype or combined genotype | All melanoma patients (n=74) | 100% VDR-positive cells (n=12) | <100% VDR-positive cells (n=62) | OR (CI) 100% vs. <100% VDR-positive | P=100% vs. <100% VDR-positive | >20%a VDR-positive cells (n=36) | ≤20% VDR-positive cells (n=38) | OR (CI)>20%vs. ≤20% VDR-positive | P>20%vs.≤20% VDR-positive | VDR absence (0% positive)(n=15) | VDR presence (>0% positive)(n=59) | OR (CI) VDR absencevs. presence | PVDR absence vs. presence |
| ApaI genotype | |||||||||||||
| AA | 30 (40.5) | 8 (66.7) | 22 (35.5) | 3.64 (0.98–13.5) | 0.058 | 13 (36.1) | 17 (44.7) | 0.70 (0.27–1.78) | 0.450 | 8 (53.3) | 22 (37.3) | 1.92 (0.61–6.03) | 0.258 |
| Aa | 36 (48.6) | 4 (33.3) | 32 (51.6) | 0.47 (0.13–1.72) | 0.246 | 22 (61.1) | 14 (36.8) | 2.69 (1.05–6.90) | 0.037 | 5 (33.3) | 31 (52.5) | 0.45 (0.14–1.48) | 0.184 |
| aa | 8 (10.8) | 0 (–) | 8 (12.9) | –b | 0.339 | 1 (2.8) | 7 (18.4) | 0.13 (0.01–1.09) | 0.056 | 2 (13.3) | 6 (10.2) | 1.36 (0.24–7.52) | 0.660 |
| A allele | 96/148 (64.9) | 20/24 (83.3) | 76/124 (61.3) | 3.16 (1.02–9.80) | 0.038 | 48/72 (66.7) | 48/76 (63.2) | 1.17 (0.59–2.29) | 0.655 | 21/30 (70.0) | 75/118 (63.6) | 1.34 (0.56–3.18) | 0.509 |
| a allele | 52/148 (35.1) | 4/24 (16.7) | 48/124 (38.7) | 0.32 (0.10–0.98) | 0.038 | 24/72 (33.3) | 28/76 (36.8) | 0.86 (0.44–1.69) | 0.655 | 9/30 (30.0) | 43/118 (36.4) | 0.75 (0.31–1.78) | 0.509 |
| BsmI-ApaI combined genotype | |||||||||||||
| BBAA | 23 (31.1) | 5 (41.7) | 18 (29.0) | 1.75 (0.49–6.23) | 0.498 | 9 (25.0) | 14 (36.8) | 0.57 (0.21–1.56) | 0.271 | 6 (40.0) | 17 (28.8) | 1.65 (0.51–5.34) | 0.533 |
| BbAA | 5 (6.8) | 3 (25.0) | 2 (3.2) | 10.0 (1.46–68.3) | 0.028 | 4 (11.1) | 1 (2.6) | 4.62 (0.49–43.5) | 0.194 | 1 (6.7) | 4 (6.8) | 0.98 (0.10–9.49) | 1.000 |
| BbAa | 31 (41.9) | 1 (8.3) | 30 (48.4) | 0.10 (0.01–0.80) | 0.010 | 18 (50.0) | 13 (34.2) | 1.92 (0.75–4.90) | 0.169 | 5 (33.3) | 26 (44.1) | 0.63 (0.19–2.09) | 0.452 |
| Bbaa | 2 (2.7) | 0 (–) | 2 (3.2) | –b | 1.000 | 0 (–) | 2 (5.3) | –b | 0.494 | 0 (–) | 2 (3.4) | –b | 1.000 |
| bbAA | 2 (2.7) | 0 (–) | 2 (3.2) | –b | 1.000 | 0 (–) | 2 (5.3) | –b | 0.494 | 1 (6.7) | 1 (1.7) | 4.14 (0.24–70.4) | 0.367 |
| bbAa | 5 (6.8) | 3 (25.0) | 2 (3.2) | 10.0 (1.46–68.3) | 0.028 | 4 (11.1) | 1 (2.6) | 4.62 (0.49–43.5) | 0.194 | 0 (–) | 5 (8.5) | –b | 0.576 |
| bbaa | 6 (8.1) | 0 (–) | 6 (9.7) | –b | 0.581 | 1 (2.8) | 5 (13.2) | 0.19 (0.02–1.70) | 0.200 | 2 (13.3) | 4 (6.8) | 2.11 (0.35–12.8) | 0.595 |
| ApaI-TaqI combined genotype | |||||||||||||
| AATT | 4 (5.4) | 0 (–) | 4 (6.5) | –b | 1.000 | 0 (–) | 4 (10.5) | –b | 0.115 | 3 (20.0) | 1 (1.7) | 14.5 (1.39–152) | 0.025 |
| AATt | 14 (18.9) | 6 (50.0) | 8 (12.9) | 6.75 (1.74–26.1) | 0.008 | 9 (25.0) | 5 (13.2) | 2.20 (0.66–7.35) | 0.194 | 4 (26.7) | 10 (16.9) | 1.78 (0.47–6.74) | 0.463 |
| AAtt | 12 (16.2) | 2 (16.7) | 10 (16.1) | 1.04 (0.20–5.48) | 1.000 | 4 (11.1) | 8 (21.1) | 0.47 (0.13–1.72) | 0.246 | 1 (6.7) | 11 (18.6) | 0.31 (0.04–2.63) | 0.439 |
| AaTT | 12 (16.2) | 3 (25.0) | 9 (14.5) | 1.96 (0.44–8.67) | 0.399 | 9 (25.0) | 3 (7.9) | 3.89 (0.96–15.8) | 0.046 | 1 (6.7) | 11 (18.6) | 0.31 (0.04–2.63) | 0.439 |
| AaTt | 24 (32.4) | 1 (8.3) | 23 (37.1) | 0.15 (0.02–1.27) | 0.089^ | 13 (36.1) | 11 (28.9) | 1.39 (0.52–3.68) | 0.511 | 4 (26.7) | 20 (33.9) | 0.71 (0.20–2.51) | 0.761 |
| aaTT | 8 (10.8) | 0 (–) | 8 (12.9) | –b | 0.339 | 1 (2.8) | 7 (18.4) | 0.13 (0.01–1.09) | 0.056 | 2 (13.3) | 6 (10.2) | 1.36 (0.24–7.52) | 0.660 |
| BsmI-ApaI-TaqI combined genotype | |||||||||||||
| BBAATT | 1 (1.4) | 0 (–) | 1 (1.6) | –b | 1.000 | 0 (–) | 1 (2.6) | –b | 1.000 | 1 (6.7) | 0 (–) | –b | 0.203 |
| BBAATt | 10 (13.5) | 3 (25.0) | 7 (11.3) | 2.62 (0.57–12.0) | 0.351 | 5 (13.9) | 5 (13.2) | 1.06 (0.28–4.04) | 1.000 | 4 (26.7) | 6 (10.2) | 3.21 (0.77–13.3) | 0.110 |
| Continued | |||||||||||||
By analyzing combined genotypes ( Table 2 ), six significant differences were observed after comparing 100% vs. <100% VDR-expression-positive groups. Combined genotypes BbAA (OR=10.0, CI=1.46–68.3, P=0.028), bbAa (OR=10.0, CI=1.46–68.3, P=0.028), AATt (OR=6.75, CI=1.74–26.1, P=0.008), BbAATt (OR=20.3, CI=1.90–217, P=0.012), and bbAaTT (OR=10.0, CI=1.46–68.3, P=0.028) were more frequent in the former vs. the latter group, whereas combined genotype BbAa (OR=0.10, CI=0.01–0.80, P=0.010) was less frequent in the former group than the latter.
In comparing the >20% vs. ≤20% VDR-expression-positive groups, combined genotype AaTT (OR=3.89, CI= 0.96–15.8, P=0.046) was more frequent in the former than in the latter group.
In comparing absent- vs. present-VDR-expression groups, the ApaI-TaqI combined genotype AATT (OR=14.5, CI=1.39–152, P=0.025) was more frequent in the melanoma group lacking VDR expression vs. the remaining patients.
Considering continuous median percentage values of VDR-expression-positive cells, with 100% VDR-positive cells, significantly higher VDR expression was observed for bbAa (median: 100.0%, range: 15%–100% vs. median: 20.0%, range: 0%–100%, P=0.029), BbAATt (median: 100.0%, range: 60%–100% vs. median: 20.0%, range: 0%–100%, P=0.011), bbAaTT (median: 100.0%, range: 15%–100% vs. median: 20.0%, range: 0%–100%, P=0.029) combined genotype carriers vs. non-carriers. By contrast, significantly lower VDR expression was noted for carriers of AATT combined genotype (median: 0.0%, range: 0%–10% vs. median; 30.0%, range: 0%–100%, P=0.014) vs. non-carriers.
Discussion
In our study, VDR expression was predominantly assessed in cytoplasm of melanoma cells (79.7%, 59/74 cases), with few tumors (14.9%, 11/74 cases) displaying VDR positivity in the nucleus. Such findings partially contrast those of other studies performed in a Polish cohort of 69 patients with primary cutaneous melanoma (comprising 35 metastatic melanomas and that were classified as follows: 4 Clark I, 6 Clark II, 23 Clark III, 24 Clark IV, and 12 Clark V stage, where 30 were superficial-spreading, 37 were nodular, and two were acral lentiginous melanomas); these previous studies indicated VDR-positive nuclear immunostaining in 84.1% and cytoplasmic immunostaining in 66.7% of patients38,39. The study by Brożyna et al.39, however, showed percentage of melanoma specimens with high nuclear staining at 17.4% (12/69), which is close to the percentage of nuclear staining in our study. Discrepancies between results of Brożyna et al. and our study probably arise from different antibodies employed21, diverse histological characteristics, and/or geographical/genetic backgrounds of melanomas in the respective studies. According to European cancer observatory data 3, the estimated age standardized (European) incidence rate (per 100,000/year) of malignant cutaneous melanoma and mortality are 5.6 and 2.8 in Poland and 13.4 and 2.0 in Italy, respectively.
We further analyzed data regarding cytoplasmic VDR immunohistochemical staining. Once the cytoplasmic VDR binds with 1α, 25-dihydroxyvitamin D ligand, and adequate coreceptor protein, retinoid X receptor, it translocates to the nucleus, and by recruitment of coactivators and corepressors modulates transcription of target genes that encode proteins responsible for final activities induced by vitamin D hormonal signaling18. Consequently, absence or down-regulation of VDR expression may present implications for vitamin D resistance in melanoma tissues38,48,49 and potentially modulates effects of vitamin D supplementation on prevention therapy of melanoma patients49,50.
We did not observe effects of age at melanoma diagnosis and gender on VDR expression in cutaneous melanomas45. Roles of BMI and smoking were focuses of previous melanoma research8,25,51. In the present investigation, we did not observe the effects of BMI and smoking on VDR expression.
Consistent with studies performed by other authors on a Polish cohort38,39, we observed that stage I melanomas were more frequent in tumors with high than with low VDR expression. In our study, stage IIA invasion was particularly more frequent in melanomas lacking VDR expression than in melanomas showing VDR expression. However, we did not observe significant data in relation to metastatic stages III and IV. Causes of these findings will require further enlarged studies.
Overall, metastatic melanomas did not exhibit different VDR expression from non-metastatic melanomas. Such results agreed with observations of some studies13,20,48 but contradicted the findings of other authors38,39. Specific geographical/ethnic backgrounds possibly affected results. Thus, enlarged studies in subjects with different ethnicities should be performed in the future to substantiate this issue.
In our study, localization of melanoma on the body was unrelated with VDR expression. We only observed a tendency (P=0.093) for upper limb melanomas to be more frequent in tumors lacking VDR expression. Thus, despite the expected different exposures to sunlight and environmental factors of different parts of the human body, VDR expression in our study is not associated with body regions in which primary melanomas develop.
Lesion-specific characteristics did not correlate with VDR expression; these characteristics include ulceration, number of mitotic figures, regression, absence of TILs, non-brisk or brisk TILs, tumor emboli, and melanoma subtype (epithelioid and small cell). At variance, in Polish patients studied by Brożyna and colleagues39 cytoplasmic VDR immunostaining was higher in group of brisk TIL-positive vs. that of absent and non-brisk TIL melanomas (P=0.01), and VDR expression was lower in melanomas with ulceration. We noted a tendency for higher frequency of superficial spreading (P=0.063) in melanomas with high than low VDR expression. Similarly, Brożyna and colleagues38 observed higher VDR expression in superficially spreading than nodular melanomas.
Remarkably, VDR expression was related to tumor Breslow thickness and Clark levels in our Italian patients. Melanomas with a thickness below 1.00 mm were more frequently observed in cases with high than low VDR expression, whereas those with thickness of over or equal to 1.01 mm were more frequent in melanomas with low VDR expression. Clark level II (none of the studied melanomas presented a Clark I) was detected more frequently in melanomas with high than low VDR expression (P=0.025). Overall, such findings concur with previous data on Polish patients38,39.
We observed that VDR expression was unrelated with the presence of multiple melanomas, additional non-melanoma skin cancers and non-skin cancers, and melanoma familiarity. To our knowledge, no previous study assessed these issues.
To our knowledge, our study was the first to investigate the relationship between VDR expression of human melanoma cells in excised tissues of patients and VDR polymorphisms. Out of four VDR polymorphisms investigated in our study, individual SNPs of FokI, BsmI, and TaqI did not display any relation with expression of VDR in melanoma. Only the ApaI genotype was correlated to VDR expression in melanoma. The heterozygous genotype Aa was identified in 61.1% of melanomas with high VDR expression vs. 36.8% of melanomas with low VDR expression (OR=2.69, P=0.037). A allele was more frequent in 100% than <100% VDR-positive cells (OR=3.16, P=0.038). To our knowledge, no research studied the role of VDR-ApaI polymorphism in VDR expression in melanoma tissues. VDR-ApaI SNP is located in an intron sequence and thus cannot directly modify the amino acid sequence of VDR protein; however, it participates in VDR RNA processing26. Recent evidence demonstrated that intronic sites of the VDR gene can function as binding sites of transcriptional regulators, such as p5352,53. A meta-analysis study35 indicated that VDR-ApaI polymorphism of the European population features an association with overall skin cancer risk (Aa vs. AA, OR=1.27, CI=1.05–1.53; Aa+aa vs. AA, OR=1.23, CI=1.04–1.47). In a recent Italian study, the Aa heterozygous genotype was associated with increased risk of lumbar pathologies, especially osteochondrosis29.
A number of combined genotypes in our study yielded significant findings according to VDR expression. The AaTT combined genotype was more frequent in melanomas with high than low VDR expression. Combined genotypes BbAA, bbAa, AATt, BbAATt, and bbAaTT were more frequent in 100% VDR-positive cells than <100% VDR-positive cells. The AATT combined genotype was much more frequent in subjects without VDR expression (20%) than in those with VDR expression (1.7%) (OR=14.5, P=0.025). No previous study investigated the relationship of VDR expression with VDR combined genotypes. Lack of VDR expression in excised melanoma tissues has been associated with reduced overall survival of patients38,39. Therefore, melanoma prognosis may be influenced by carrying a VDR combined genotype associated to absent or reduced VDR expression. In our melanoma patients, AATT is a rare combined genotype with a frequency of 5.4%, which is similar to the recent finding in an Italian cohort of 518 non-oncological subjects (6.0%) 29. Further enlarged studies are warranted to assess roles of ApaI, BsmI, and TaqI combined genotypes in VDR expression in melanomas and their prognosis.
Regulation of VDR abundance is an important modulation mechanism of cellular responsiveness to 1α, 25-dihydroxyvitamin D10. Mechanisms underlying regulation of VDR abundance include alterations in transcription rate of VDR gene and/or stability of VDR mRNA and epigenetic changes14,18,54. Interestingly, treatment with calcitriol can enhance VDR mRNA in cultured melanoma cells, showing that increasing vitamin D consumption can induce VDR expression14. On the other hand, enhanced melanogenesis was associated with downregulation of VDR ex-pression38,55,56. Response of melanoma cells to calcitriol corresponds to expression level of VDR mRNA, which in turn may be regulated by VDR miRNAs and by epigenetically modulating drugs50. Remarkably, recent evidence suggests that tumor suppressors, such as p53, are implicated on direct regulation of VDR53. Molecules other than vitamin D, such as curcumin and vitamin E derivatives, were indicated as novel VDR ligands19,57, whereas vitamin A derivatives were suggested as modulators of VDR actions58. Given the wide variety of positive and negative VDR modulators, each individual expression of VDR is highly dynamic in cells, with nuclear translocation of VDR fluctuating upon instant induction18,19,22. We speculate that unresponsiveness of some cutaneous melanomas to anti-proliferative effects of vitamin D possibly resulted from absence or insufficient VDR expression in melanoma and/or melanocytic cells. Further human studies are warranted to assess whether benefits of vitamin D augmentation are modulated by VDR expression in melanoma/melanocytic cells and/or by carriage of a specific VDR genotype and/or combined genotype polymorphisms that affect VDR expression. Such factors can modulate dose requirement of vitamin D treatments59.
Roles of vitamin D in skin cancers still require complete elucidation10,57. All vitamin D actions virtually occur through VDR activation. Recent evidence shows that the effects of 1α, 25-dihydroxyvitamin D and VDR are mediated at least in part by cellular calcium levels; thus, calcium possibly contributes to the suppressive ability of VDR on skin cancer10,57. Deletion of VDR notably results in an increased susceptibility to tumorigenesis and also reduces ability of keratinocytes to clear UVB-induced DNA mutations57. VDR can bind to thousands of VDREs on human genome and up- or downregulate hundreds of genes10,30,57. Based on bioinformatic analysis, almost 15,000 sites in human DNA are bound by VDR, and 16%–21% of these putative binding sites are found at gene promoters10,57,60. Aside from classical VDRE-mediated mechanisms, increasing evidence point to regulatory contribution of several miRNAs14,18,54 and long non-coding RNAs10,57. A recent study on VDR cistrome demonstrated the unexpected complexity of gene regulation, which was examined on a genome-wide scale in target tissues and cells, including a cross-talk between VDR and immune factors60,61. VDR expression is also commonly and significantly down regulated in colon adenocarcinoma. VDR cistrome analyses suggested that reduced VDR expression in colon cancer changes VDR activity by dampening expression of tumor suppressors by either stabilizing or inhibiting down regulation of oncogene expression. In turn, these effects may be associated with severe patient outcomes62. Thus, further research should study complex gene interactions and biological pathways related to vitamin D and melanoma and combine clinical evidence with molecular findings to support further progress57,60,61.
Limitations of our study include the limited number of patients and absence of data on circulating vitamin D levels in patients at the time of melanoma excision. Strengths of this research comprise genetic background restrictions of enrolled patients and determination of demographic, lifestyle, histological, and genetic characteristics.
Given the sample size and multiple comparisons of our study, a validation of independent datasets with larger samples, and multivariable analysis will be necessary to adjust genetic traits for age, sex, tumor location, sun exposure, and smoking. Serum sampling in future studies may also be performed to determine circulating levels of vitamin D at time of melanoma excision.
Conclusions
Current information insufficiently discusses influence of vitamin D oral supplementation to direct VDR modulatory effects in human skin cells, including melanocytes and keratinocytes. Our present findings support the necessity of further studies on this issue by combining clinical and molecular approach.
Our study showed that VDR expression is associated with prognostic parameters of tumor Breslow thickness and Clark level. However, VDR expression was not related to metastatic melanomas. Our immunohistochemical results concur with those of a previous study on VDR expression in colorectal cancer, showing that VDR was not associated with tumor location, stage, and grade42, and with another lung tumor study which demonstrated high variability of VDR expression43. Interestingly, we observed correlation between the Aa genotype and AaTT combined genotype with higher level of VDR expression and between AATT combined genotype and low or absent VDR expression; this new finding will require further validation. Future studies should assess whether VDR expression and VDR combined genotypes affect benefits of vitamin D and can drive an appropriate dose and schedule of calcitriol or other active (low calcemic) vitamin D analogs for melanoma treatment. Future set up of personalized nutrition and behavioral interventions will benefit from molecular studies exploring the connection of biological pathways to bioactive components of food and cancer63. Our study suggests that determination of VDR expression in excised tissues of melanoma and/or determination of VDR genotypes carriage can be used as personalized tool of precision medicine when considering melanoma patients.
Acknowledgments
The authors thank Patrizia Nacci, Martina Linussio, Luca Bazzichetto and Silvia Lolini for their technical assistance.
Conflict of interest statement
No potential conflicts of interest are disclosed.
References
- 1.GLOBOCAN 2012: Estimated cancer incidence, mortality and prevalence worldwide in 2012. World Health Organization. International Agency for Research on Cancer (IARC). Available at: http://globocan.iarc.fr/Default.aspx; accessed March 27, 2017.
- 2.Arnold M, Holterhues C, Hollestein LM, Coebergh JWW, Nijsten T, Pukkala E, et al. Trends in incidence and predictions of cutaneous melanoma across Europe up to 2015. J Eur Acad Dermatol Venereol. 2014;28:1170–8. doi: 10.1111/jdv.12236. [DOI] [PubMed] [Google Scholar]
- 3.EUCAN. Malignant melanoma of skin. Available at: http://eu-cancer.iarc.fr/EUCAN/Cancer.aspx?Cancer=20; accessed March 27, 2017.
- 4.Vos T, Barber RM, Bell B, Bertozzi-Villa A, Biryukov S, Bolliger I, et al. Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2015;386:743–800. doi: 10.1016/S0140-6736(15)60692-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.ITACAN. AIRTUM. The Italian Association of Cancer Registries. I numeri del cancro in Italia - 2016. Available at: htttp://www.registri-tumori.it/itacan; accessed March 27, 2017.
- 6.Registro Tumori del Friuli Venezia Giulia. 2010: www.cro.sanita.fvg.it. accessed March 27, 2017.
- 7.Cecconi L, Busolin A, Barbone F, Serraino D, Chiarugi A, Biggeri A, et al. Spatial analysis of incidence of cutaneous melanoma in the Friuli Venezia Giulia region in the period 1995-2005. Geospat Health. 2016;11:422. doi: 10.4081/gh.2016.422. [DOI] [PubMed] [Google Scholar]
- 8.Newton-Bishop JA, Davies JR, Latheef F, Randerson-Moor J, Chan M, Gascoyne J, et al. 25-Hydroxyvitamin D 2/D 3 levels and factors associated with systemic inflammation and melanoma survival in the Leeds melanoma Cohort . Int J Cancer. 2015;136:2890–9. doi: 10.1002/ijc.29334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pasquali S, Chiarion-Sileni V, Rossi CR, Mocellin S. Immune checkpoint inhibitors and targeted therapies for metastatic melanoma: A network meta-analysis. Cancer Treat Rev. 2017;54:34–42. doi: 10.1016/j.ctrv.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 10.Bikle DD. Vitamin D receptor, a tumor suppressor in skin. Can J Physiol Pharmacol. 2015;93:349–54. doi: 10.1139/cjpp-2014-0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evans SRT, Houghton AM, Schumaker L, Brenner RV, Buras RR, Davoodi F, et al. Vitamin D receptor and growth inhibition by 1,25-dihydroxyvitamin D 3 in human malignant melanoma cell lines . J Surg Res. 1996;61:127–33. doi: 10.1006/jsre.1996.0092. [DOI] [PubMed] [Google Scholar]
- 12.Trump DL, Deeb KK, Johnson CS. Vitamin D: considerations in the continued development as an agent for cancer prevention and therapy. Cancer J. 2010;16:1–9. doi: 10.1097/PPO.0b013e3181c51ee6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Danielsson C, Fehsel K, Polly P, Carlberg C. Differential apoptotic response of human melanoma cells to 1α,25-dihydroxyvitamin D 3 and its analogues . Cell Death Differ. 1998;5:946–52. doi: 10.1038/sj.cdd.4400437. [DOI] [PubMed] [Google Scholar]
- 14.Seifert M, Rech M, Meineke V, Tilgen W, Reichrath J. Differential biological effects of 1,25-dihydroxyvitamin D 3 on melanoma cell lines in vitro . J Steroid Biochem Mol Biol. 2004;89-90:375–9. doi: 10.1016/j.jsbmb.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 15.Feldman D, Krishnan AV, Swami S, Giovannucci E, Feldman BJ. The role of vitamin D in reducing cancer risk and progression. Nat Rev Cancer. 2014;14:342–57. doi: 10.1038/nrc3691. [DOI] [PubMed] [Google Scholar]
- 16.Holick MF. Biological effects of sunlight, ultraviolet radiation, visible light, infrared radiation and vitamin D for health. Anticancer Res. 2016;36:1345–56. [PubMed] [Google Scholar]
- 17.Sehdev A, O'Neil BH. The role of aspirin, vitamin D, exercise, diet, statins, and metformin in the prevention and treatment of colorectal cancer. Curr Treat Options Oncol. 2015;16:43. doi: 10.1007/s11864-015-0359-z. [DOI] [PubMed] [Google Scholar]
- 18.Dou RX, Ng K, Giovannucci EL, Manson JE, Qian ZR, Ogino S. Vitamin D and colorectal cancer: molecular, epidemiological and clinical evidence. Br J Nutr. 2016;115:1643–60. doi: 10.1017/S0007114516000696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haussler MR, Whitfield GK, Kaneko I, Haussler CA, Hsieh D, Hsieh JC, et al. Molecular mechanisms of vitamin D action. Calcif Tissue Int. 2013;92:77–98. doi: 10.1007/s00223-012-9619-0. [DOI] [PubMed] [Google Scholar]
- 20.Keum N, Giovannucci E. Vitamin D supplements and cancer incidence and mortality: a meta-analysis. Br J Cancer. 2014;111:976–80. doi: 10.1038/bjc.2014.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bikle DD. Vitamin D metabolism, mechanism of action, and clinical applications. Chem Biol. 2014;21:319–29. doi: 10.1016/j.chembiol.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee SM, Pike JW. The vitamin D receptor functions as a transcription regulator in the absence of 1,25-dihydroxyvitamin D 3 . J Steroid Biochem Mol Biol. 2016;164:265–70. doi: 10.1016/j.jsbmb.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colston K, Colston MJ, Feldman D. 1,25-dihydroxyvitamin D 3 and malignant melanoma: the presence of receptors and inhibition of cell growth in culture . Endocrinology. 1981;108:1083–6. doi: 10.1210/endo-108-3-1083. [DOI] [PubMed] [Google Scholar]
- 24.Bonin S, Albano A, di Meo N, Gatti A, Stinco G, Zanconati F, et al. Cutaneous melanoma frequencies and seasonal trend in 20 years of observation of a population characterised by excessive sun exposure. Radiol Oncol. 2015;49:379–85. doi: 10.1515/raon-2015-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gandini S, Montella M, Ayala F, Benedetto L, Rossi CR, Vecchiato A, et al. Sun exposure and melanoma prognostic factors. Oncol Lett. 2016;11:2706–14. doi: 10.3892/ol.2016.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uitterlinden AG, Fang Y, Van Meurs JBJ, Pols HAP, Van Leeuwen JPTM. Genetics and biology of vitamin D receptor polymorphisms. Gene. 2004;338:143–56. doi: 10.1016/j.gene.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 27.Selvaraj P, Chandra G, Jawahar MS, Rani MV, Rajeshwari DN, Narayanan PR. Regulatory role of vitamin D receptor gene variants of BsmI, ApaI, TaqI, and FokI polymorphisms on macrophage phagocytosis and lymphoproliferative response to Mycobacterium tuberculosis antigen in pulmonary tuberculosis . J Clin Immunol. 2004;24:523–32. doi: 10.1023/B:JOCI.0000040923.07879.31. [DOI] [PubMed] [Google Scholar]
- 28.Colombini A, Cauci S, Lombardi G, Lanteri P, Croiset S, Brayda-Bruno M, et al. Relationship between vitamin D receptor gene (VDR) polymorphisms, vitamin D status, osteoarthritis and intervertebral disc degeneration . J Steroid Biochem Mol Biol. 2013;138:24–40. doi: 10.1016/j.jsbmb.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 29.Colombini A, Brayda-Bruno M, Lombardi G, Croiset SJ, Ceriani C, Buligan C, et al. BsmI, ApaI and TaqI polymorphisms in the vitamin D receptor gene (VDR) and association with lumbar spine pathologies: An Italian case-control study . PLoS One. 2016;11:e0155004. doi: 10.1371/journal.pone.0155004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gandini S, Gnagnarella P, Serrano D, Pasquali E, Raimondi S. Vitamin D receptor polymorphisms and cancer. Adv Exp Med Biol. 2014;810:69–105. doi: 10.1007/978-1-4939-0437-2_5. [DOI] [PubMed] [Google Scholar]
- 31.Orlow I, Reiner AS, Thomas NE, Roy P, Kanetsky PA, Luo L, et al. Vitamin D receptor polymorphisms and survival in patients with cutaneous melanoma: a population-based study. Carcinogenesis. 2016;37:30–8. doi: 10.1093/carcin/bgv157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santonocito C, Capizzi R, Concolino P, Lavieri MM, Paradisi A, Gentileschi S, et al. Association between cutaneous melanoma, Breslow thickness and vitamin D receptor BsmI polymorphism . Br J Dermatol. 2007;156:277–82. doi: 10.1111/j.1365-2133.2006.07620.x. [DOI] [PubMed] [Google Scholar]
- 33.Randerson-Moor JA, Taylor JC, Elliott F, Chang YM, Beswick S, Kukalizch K, et al. Vitamin D receptor gene polymorphisms, serum 25-hydroxyvitamin D levels, and melanoma: UK case-control comparisons and a meta-analysis of published VDR data. Eur J Cancer. 2009;45:3271–81. doi: 10.1016/j.ejca.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mandelcorn-Monson R, Marrett L, Kricker A, Armstrong BK, Orlow I, Goumas C, et al. Sun exposure, vitamin D receptor polymorphisms FokI and BsmI and risk of multiple primary melanoma . Cancer Epidemiol. 2011;35:e105–10. doi: 10.1016/j.canep.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao XZ, Yang BH, Yu GH, Liu SZ, Yuan ZY. Polymorphisms in the vitamin D receptor (VDR) genes and skin cancer risk in European population: a meta-analysis. Arch Dermatol Res. 2014;306:545–53. doi: 10.1007/s00403-014-1464-8. [DOI] [PubMed] [Google Scholar]
- 36.Lee YH, Gyu Song G. Vitamin D receptor FokI, BsmI, TaqI, ApaI, and EcoRV polymorphisms and susceptibility to melanoma: a meta-analysis . J BUON. 2015;20:235–43. [PubMed] [Google Scholar]
- 37.Hou W, Wan XF, Fan JW. Variants Fok1 and Bsm1 on VDR are associated with the melanoma risk: evidence from the published epidemiological studies. BMC Genet. 2015;16:14. doi: 10.1186/s12863-015-0163-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brożyna AA, Jozwicki W, Janjetovic Z, Slominski AT. Expression of vitamin D receptor decreases during progression of pigmented skin lesions. Hum Pathol. 2011;42:618–31. doi: 10.1016/j.humpath.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brożyna AA, Jóźwicki W, Slominski AT. Decreased VDR expression in cutaneous melanomas as marker of tumor progression: new data and analyses. Anticancer Res. 2014;34:2735–43. [PMC free article] [PubMed] [Google Scholar]
- 40.Erdmann F, Lortet-Tieulent J, Schüz J, Zeeb H, Greinert R, Breitbart EW, et al. International trends in the incidence of malignant melanoma 1953-2008 - are recent generations at higher or lower risk? Int J Cancer. 2013;132:385–400. doi: 10.1002/ijc.27616. [DOI] [PubMed] [Google Scholar]
- 41.Balch CM, Gershenwald JE, Soong SJ, Thompson JF, Atkins MB, Byrd DR, et al. Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol. 2009;27:6199–206. doi: 10.1200/JCO.2009.23.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kure S, Nosho K, Baba Y, Irahara N, Shima K, Ng K, et al. Vitamin D receptor expression is associated with PIK3CA and KRAS mutations in colorectal cancer . Cancer Epidemiol Biomarkers Prev. 2009;18:2765–72. doi: 10.1158/1055-9965.EPI-09-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turner AM, McGowan L, Millen A, Rajesh P, Webster C, Langman G, et al. Circulating DBP level and prognosis in operated lung cancer: an exploration of pathophysiology. Eur Respir J. 2013;41:410–16. doi: 10.1183/09031936.00002912. [DOI] [PubMed] [Google Scholar]
- 44.Di Loreto C, La Marra F, Mazzon G, Belgrano E, Trombetta C, Cauci S. Immunohistochemical evaluation of androgen receptor and nerve structure density in human prepuce from patients with persistent sexual side effects after finasteride use for androgenetic alopecia. PLoS One. 2014;9:e0100237. doi: 10.1371/journal.pone.0100237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Colombini A, Brayda-Bruno M, Ferino L, Lombardi G, Maione V, Banfi G, et al. Gender differences in the VDR-FokI polymorphism and conventional non-genetic risk factors in association with lumbar spine pathologies in an Italian case-control study . Int J Mol Sci. 2015;16:3722–39. doi: 10.3390/ijms16023722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Colombini A, Brayda-Bruno M, Lombardi G, Croiset SJ, Vrech V, Maione V, et al. FokI polymorphism in the vitamin D receptor gene (VDR) and its association with lumbar spine pathologies in the Italian population: a case-control study . PLoS One. 2014;9:e97027. doi: 10.1371/journal.pone.0097027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cauci S, Migliozzi F, Trombetta CS, Venuto I, Saccheri P, Travan L, et al. Low back pain and FokI (rs2228570) polymorphism of vitamin D receptor in athletes. BMC Sports Sci Med Rehabil. 2017;9:4. doi: 10.1186/s13102-017-0069-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reichrath J, Rech M, Moeini M, Meese E, Tilgen W, Seifert M. In vitro comparison of the vitamin D endocrine system in 1,25(OH)2D3-responsive and resistant melanoma cells. Cancer Biol Ther. 2007;6:48–55. doi: 10.4161/cbt.6.1.3493. [DOI] [PubMed] [Google Scholar]
- 49.Pervin S, Hewison M, Braga M, Tran L, Chun R, Karam A, et al. Down-regulation of vitamin D receptor in mammospheres: implications for vitamin D resistance in breast cancer and potential for combination therapy. PLoS One. 2013;8:e53287. doi: 10.1371/journal.pone.0053287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Essa S, Reichrath S, Mahlknecht U, Montenarh M, Vogt T, Reichrath J. Signature of VDR miRNAs and epigenetic modulation of vitamin D signaling in melanoma cell lines. Anticancer Res. 2012;32:383–9. [PubMed] [Google Scholar]
- 51.Jiang AJ, Rambhatla PV, Eide MJ. Socioeconomic and lifestyle factors and melanoma: a systematic review. Br J Dermatol. 2015;172:885–915. doi: 10.1111/bjd.13500. [DOI] [PubMed] [Google Scholar]
- 52.Maruyama R, Aoki F, Toyota M, Sasaki Y, Akashi H, Mita H, et al. Comparative genome analysis identifies the vitamin D receptor gene as a direct target of p53-mediated transcriptional activation. Cancer Res. 2006;66:4574–83. doi: 10.1158/0008-5472.CAN-05-2562. [DOI] [PubMed] [Google Scholar]
- 53.Reichrath J, Reichrath S, Heyne K, Vogt T, Roemer K. Tumor suppression in skin and other tissues via cross-talk between vitamin D- and p53-signaling. Front Physiol. 2014;5:166. doi: 10.3389/fphys.2014.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Essa S, Denzer N, Mahlknecht U, Klein R, Collnot EM, Tilgen W, et al. VDR microRNA expression and epigenetic silencing of vitamin D signaling in melanoma cells. J Steroid Biochem Mol Biol. 2010;121:110–3. doi: 10.1016/j.jsbmb.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 55.Janjetovic Z, Brozyna AA, Tuckey RC, Kim TK, Nguyen MN, Jozwicki W, et al. High basal NF-κB activity in nonpigmented melanoma cells is associated with an enhanced sensitivity to vitamin D 3 derivatives . Br J Cancer. 2011;105:1874–84. doi: 10.1038/bjc.2011.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wasiewicz T, Szyszka P, Cichorek M, Janjetovic Z, Tuckey RC, Slominski AT, et al. Antitumor effects of vitamin D analogs on hamster and mouse melanoma cell lines in relation to melanin pigmentation. Int J Mol Sci. 2015;16:6645–67. doi: 10.3390/ijms16046645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bikle DD, Oda Y, Tu CL, Jiang Y. Novel mechanisms for the vitamin D receptor (VDR) in the skin and in skin cancer. J Steroid Biochem Mol Biol. 2015;148:47–51. doi: 10.1016/j.jsbmb.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Danielsson C, Törmä H, Vahlquist A, Carlberg C. Positive and negative interaction of 1,25-dihydroxyvitamin D3 and the retinoid CD437 in the induction of human melanoma cell apoptosis. Int J Cancer. 1999;81:467–70. doi: 10.1002/(sici)1097-0215(19990505)81:3<467::aid-ijc22>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 59.Bischoff-Ferrari HA, Shao A, Dawson-Hughes B, Hathcock J, Giovannucci E, Willett WC. Benefit-risk assessment of vitamin D supplementation. Osteoporos Int. 2010;21:1121–32. doi: 10.1007/s00198-009-1119-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pike JW, Meyer MB, Lee SM, Onal M, Benkusky NA. The vitamin D receptor: contemporary genomic approaches reveal new basic and translational insights. J Clin Invest. 2017;127:1146–54. doi: 10.1172/JCI88887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Singh PK, van den Berg PR, Long MD, Vreugdenhil A, Grieshober L, Ochs-Balcom HM, et al. Integration of VDR genome wide binding and GWAS genetic variation data reveals co-occurrence of VDR and NF-κB binding that is linked to immune phenotypes. BMC Genomics. 2017;18:132. doi: 10.1186/s12864-017-3481-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Long MD, Campbell MJ. Integrative genomic approaches to dissect clinically-significant relationships between the VDR cistrome and gene expression in primary colon cancer. J Steroid Biochem Mol Biol. 2016 doi: 10.1016/j.jsbmb.2016.12.013. [DOI] [PubMed] [Google Scholar]
- 63.Rescigno T, Micolucci L, Tecce MF, Capasso A. Bioactive nutrients and nutrigenomics in age-related diseases. Molecules. 2017;22:105. doi: 10.3390/molecules22010105. [DOI] [PMC free article] [PubMed] [Google Scholar]