Abstract
We isolated a mutant, named coleoptile phototropism1 (cpt1), from γ-ray–mutagenized japonica-type rice (Oryza sativa). This mutant showed no coleoptile phototropism and severely reduced root phototropism after continuous stimulation. A map-based cloning strategy and transgenic complementation test were applied to demonstrate that a NPH3-like gene deleted in the mutant corresponds to CPT1. Phylogenetic analysis of putative CPT1 homologs of rice and related proteins indicated that CPT1 has an orthologous relationship with Arabidopsis thaliana NPH3. These results, along with those for Arabidopsis, demonstrate that NPH3/CPT1 is a key signal transduction component of higher plant phototropism. In an extended study with the cpt1 mutant, it was found that phototropic differential growth is accompanied by a CPT1-independent inhibition of net growth. Kinetic investigation further indicated that a small phototropism occurs in cpt1 coleoptiles. This response, induced only transiently, was thought to be caused by the CPT1-independent growth inhibition. The 3H-indole-3-acetic acid applied to the coleoptile tip was asymmetrically distributed between the two sides of phototropically responding coleoptiles. However, no asymmetry was induced in cpt1 coleoptiles, indicating that lateral translocation of auxin occurs downstream of CPT1. It is concluded that the CPT1-dependent major phototropism of coleoptiles is achieved by lateral auxin translocation and subsequent growth redistribution.
INTRODUCTION
Phototropism is observed ubiquitously in higher plants and is thought to bear significant adaptive values (Iino, 2001). Our knowledge of the mechanism of phototropism has been substantially advanced in the last decade with the use of the dicotyledonous model plant Arabidopsis thaliana. Mutants of Arabidopsis impaired in hypocotyl phototropism were isolated (Khurana and Poff, 1989; Liscum and Briggs, 1996), and molecular genetic analysis of these mutants has lead to the discovery of novel proteins that participate in the early process of phototropism. Most importantly, phototropin 1 (phot1) was uncovered as the major photoreceptor for phototropism (Huala et al., 1997) and phototropin 2 (phot2) as an additional photoreceptor that functions at high fluence rates (Sakai et al., 2001) (for the receptor nomenclature, see Briggs et al., 2001). The protein NPH3 was identified as a signaling component that is essential for hypocotyl phototropism and probably functions by directly interacting with phot1 (Motchoulski and Liscum, 1999). Also identified as a signaling component was RPT2, a homolog of NPH3 (Sakai et al., 2000). It is another important outcome of mutant analysis that phot1, NPH3, and RPT2 also participate in the negative phototropism of primary roots (Liscum and Briggs, 1995, 1996; Sakai et al., 2000).
Before the above achievements with Arabidopsis were made, coleoptiles of Gramineae cereals, such as oats (Avena sativa) and maize (Zea mays), had been the major materials of phototropism research. The results indicated that the phototropism of coleoptiles is multiphasic in its dependence on light fluences and involves spatially and temporally separable response components (Iino, 2001). Detailed molecular genetic analysis of such complexities, which probably bear biological significance, is yet to be made. The role played by auxin in phototropism has also been studied extensively with Gramineae coleoptiles. The major focus of these studies has been the hypothesis known as the Cholodny-Went theory of tropisms. This hypothesis, which states that tropisms are mediated by asymmetrical distribution of auxin generated by its lateral translocation, was supported for phototropism by early bioassay measurement of auxin (Iino, 2001). However, later physicochemical measurement of the native auxin indole-3-acetic acid (IAA), performed mainly by the group of Hasegawa (Togo and Hasegawa, 1991, and references cited therein), showed no such asymmetry in all the materials investigated, including oat and maize coleoptiles. The only exception was the report of Iino (1991), who found an asymmetry in maize coleoptiles. On the other hand, more recent studies with Arabidopsis have provided genetic evidence that auxin is involved in the process of phototropism (Harper et al., 2000; Blakeslee et al., 2004; Tatematsu et al., 2004). For resolving the signaling pathway for phototropism, it is required that the Cholodny-Went hypothesis is firmly established.
Nearly full genome sequences of rice (Oryza sativa) are now available for basic research, and rice is being established as a monocotyledonous model plant. The coleoptile of rice appeared to be less phototropic as compared with those of oats and maize, but it could be resolved that the fluence-response characteristics of rice are similar to those of oats and maize (Neumann and Iino, 1997). Molecular genetic studies of phototropism with rice will provide results that, along with those from Arabidopsis, contribute to our overall understanding of higher-plant phototropism. Furthermore, it is anticipated that such studies allow clarification of the complex and controversial aspects outlined above.
Undoubtedly, mutants of rice altered in coleoptile phototropism will play essential roles in molecular genetic investigation of phototropism. We isolated some phototropism-specific mutants (Iino and Neumann, 2000; Biswas and Iino, 2002) and investigated one of them, named coleoptile phototropism1 (cpt1), in detail. Here, we report on the molecular cloning and characterization of CPT1 as well as on the physiological aspects of coleoptile phototropism investigated with the cpt1 mutant.
RESULTS
Isolation and Characterization of the Rice cpt1 Mutant
The cpt1 mutant was isolated from γ-ray–mutagenized M2 caryopses of cv Nihonmasari as a mutant showing no clear coleoptile phototropism under continuous irradiation with unilateral white light (2 to 5 μmol m−2 s−1) and overhead red light (∼3 μmol m−2 s−1) (see Methods). Investigation with the selfed next generation indicated that the negative phototropism of primary roots, observed in the wild type, is also impaired in the mutant. Seedlings of the F2 population obtained by crossing the cpt1 mutant with the wild type showed segregation of the cpt1 and wild-type phenotypes at a ratio close to 1:3 (62 out of 228 seedlings showed a clear nonphototropic phenotype). This result indicated that the cpt1 phenotype is caused by a recessive mutation at a single locus. The mutant was repeatedly backcrossed to the wild type before used in the experiments.
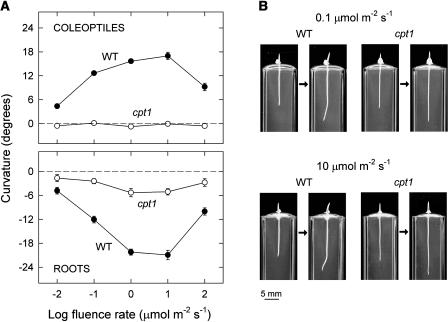
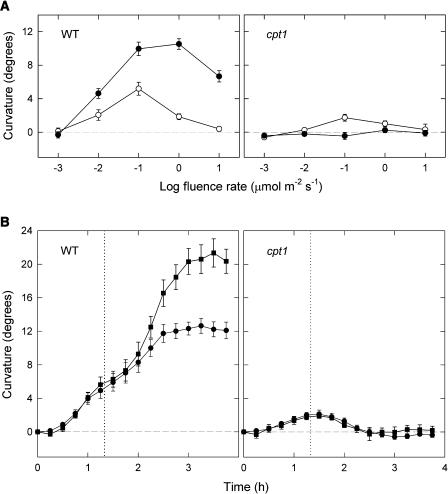
Figure 1A shows a comparison of phototropic fluence rate-response curves between cpt1 and wild-type seedlings. The data were obtained by stimulating seedlings with unilateral blue light for 6 h and measuring curvatures of coleoptiles and primary roots at the end of stimulation. In wild-type seedlings, the fluence rate-response curve for either organ was bell shaped, and the curve for roots was nearly a mirror image of that for coleoptiles. The cpt1 coleoptiles showed no detectable curvature at all the fluence rates examined. However, cpt1 roots were found to show some small curvature. We also confirmed that cpt1 coleoptiles do not show any detectable phototropism even under submerged condition, which is known to enhance the phototropic responsiveness of coleoptiles (Neumann and Iino, 1997). For example, the wild-type coleoptiles submerged 6 h before the onset of blue light showed a mean curvature of 23.6° (se = 1.5°, n = 14) after 6-h blue light stimulation at 1 μmol m−2 s−1, whereas the cpt1 coleoptiles treated in the same way showed a mean curvature of −1.6° (se = 0.5°, n = 14). Together, these results indicated that the cpt1 mutation results in almost a total loss of coleoptile phototropism and in a substantially reduced phototropism of roots.
Figure 1.
Impaired Phototropisms of cpt1 Coleoptiles and Roots.
(A) Phototropic fluence rate-response curves. Seedlings of the cpt1 mutant (open circles) and the wild type Nihonmasari (closed circles), grown for 3 d under red light (2.5 to 3 μmol m−2 s−1), were stimulated continuously with unilateral blue light at the indicated fluence rates. The length of coleoptiles was in the range between 3.5 and 4.5 mm at the onset of blue light. Phototropic curvatures of coleoptiles (top panel) and primary roots (bottom panel) were determined at 6 h of stimulation. The means obtained from 30 to 50 seedlings are shown. The vertical bar indicates ±se.
(B) Photographs illustrating impaired phototropisms of cpt1 coleoptiles and roots. Wild-type and cpt1 seedlings were stimulated with unilateral blue light at a fluence rate of 0.1 μmol m−2 s−1 (top panels) or 10 μmol m−2 s−1 (bottom panels). Each pair of photographs was taken just before and after 6-h blue light stimulation. Other conditions were as described for (A).
Figure 1B shows photographs of wild-type and cpt1 seedlings stimulated with 0.1 and 10 μmol m−2 s−1 blue light. Each pair of photographs was obtained at the onset and the end of 6-h stimulation. These photographs illustrate that cpt1 seedlings were vertically straight, whereas wild-type seedlings showed clear phototropic curvatures of coleoptiles and roots. In wild-type seedlings, the coleoptile showed curvature along its length. On the other hand, the root showed a sharp curvature around the region where the tip was located at the onset of stimulation. The small phototropic curvature of cpt1 roots could not be clearly distinguished in photographs of individual seedlings. As noted in some of the photographs, the root showed a random curvature in its tip (in the apical 1.5- to 2.0-mm zone) that represents circumnutation. The data in Figure 1A were obtained for the steady angle maintained above this zone.
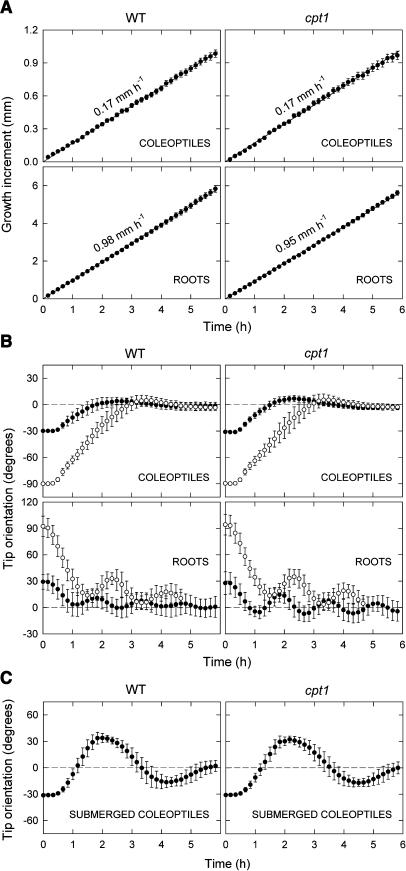
To investigate whether the effect of cpt1 mutation is specific to phototropism, we first compared the growth behaviors of cpt1 and wild-type seedlings. In our daily observations of seedlings, no apparent difference in growth could be noted between the wild type and mutant. To demonstrate this more clearly, the elongation growth of coleoptiles and roots was measured in the linear elongation phase. As shown in Figure 2A, the two genotypes showed no significant difference in the growth rate of either organ. We also analyzed circumnutation observed in the root tip. Wild-type roots circumnutated with a mean period of ∼160 min and a mean amplitude of ∼16°. Mutant roots showed circumnutation with similar period and amplitude.
Figure 2.
Growth and Gravitropism of cpt1 Seedlings.
(A) Growth of coleoptiles and primary roots. Seedlings of the wild type and cpt1 mutant, raised under red light (2.5 μmol m−2 s−1), were monitored for elongation growth of coleoptiles and primary roots. The growth increment (ordinate) indicates the difference in length from the length at time 0. The means ± se from 15 seedlings are shown. Growth rates estimated from the linear regression lines are indicated. The mean initial lengths in millimeters were 3.2 (wild-type coleoptiles), 3.4 (cpt1 coleoptiles), 15.8 (wild-type roots), and 16.3 (cpt1 roots).
(B) Gravitropism of coleoptiles and primary roots. Seedlings of the wild type and the cpt1 mutant, prepared as described for Figure 1A, were displaced by 30° (closed circles) or 90° (open circles) and were monitored for the orientation of the coleoptile tip (top panels) and the root tip (bottom panels). The angle of orientation is 0° when it is parallel to the plumb line. In the case of coleoptiles, the orientation to the side of displacement is indicated by negative angles, and the upward gravitropic curvature of coleoptiles is represented by a movement of the tip from a negative angle to 0°. In the case of roots, the orientation to the side of displacement is indicated by positive angles, and the downward gravitropic curvature of roots is represented by a movement of the tip from a positive angle to 0°. The means obtained from 30 (closed circles) or 20 (open circles) seedlings are shown. The vertical bar indicates ±sd.
(C) Gravitropism of submerged coleoptiles. Seedlings were submerged in water 6 h before they were displaced by 30°. The length of coleoptiles was ∼3 mm at the time of submergence. The means obtained from 12 seedlings are shown. Other details were as described for (B).
A further comparison was made for gravitropism. Figure 2B shows time courses of gravitropic curvatures induced in coleoptiles and roots of the seedlings prepared as for phototropism experiments. Gravitropism was induced by displacing the seedlings by 30° or 90°. The curvature was measured at the tip (not in the subapical zone) because it was initiated in the circumnutating tip zone. This resulted in a greater variation of the measured angles at the onset of stimulation. Wild-type and cpt1 seedlings showed nearly identical gravitropism in both organs. Gravitropically responding wild-type roots oscillated before reaching the vertical position. This oscillation also occurred similarly in cpt1 roots. Gravitropic response of coleoptiles to 30° displacement was further compared between the two genotypes under a submerged condition (Figure 2C). As reported in Iino et al. (1996), wild-type coleoptiles showed a gravitropic curvature to the extent that the tip substantially overshoots the vertical position. Mutant coleoptiles showed a very similar time course. These results indicated that cpt1 mutation does not affect gravitropism.
Under greenhouse and growth cabinet conditions, cpt1 plants grew without any apparent phenotype or abnormality throughout the vegetative and reproductive growth stages. This observation and the results described above demonstrated that the effect of cpt1 mutation is fairly specific for phototropism.
Map-Based Cloning and Analysis of the CPT1 Gene
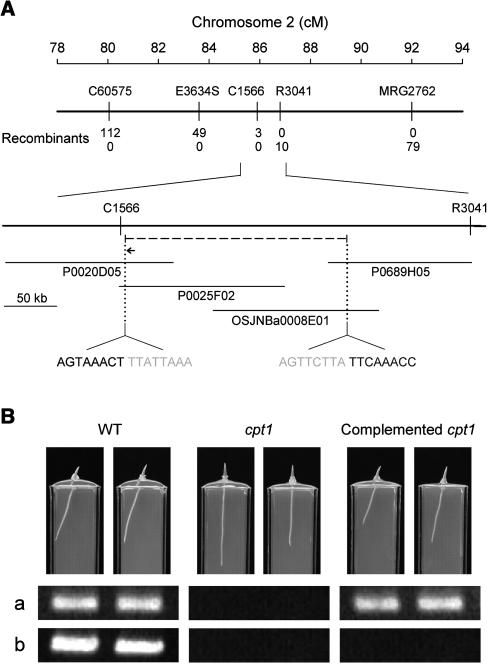
We used a map-based cloning strategy to identify the CPT1 gene. A mapping population of F2 caryopses (∼5000) was generated by crossing the cpt1 mutant to cv Kasalath. Approximately 1400 individuals showing the cpt1 phenotype, presumed to be homozygous for the cpt1 mutant allele, were selected from the F2 population, and genomic DNA samples obtained from these individuals were used for analysis. The first step of mapping was conducted using DNA samples from 150 individuals and restriction fragment length polymorphism (RFLP) markers obtained from the Rice Genome Research Program (RGP) (Harushima et al., 1998). By this analysis, the CPT1 locus was mapped between the markers C449 and R1826 in chromosome 2. The analysis was extended by including the remaining F2 individuals and using additional RFLP markers, cleaved amplified polymorphic sequence (CAPS) markers, and sequence tagged site (STS) markers. Finally, the CPT1 locus was mapped between the RFLP markers C1566 and R3041 with three and 10 recombinants, respectively (Figure 3A).
Figure 3.
Identification of the CPT1 Gene.
(A) Map-based cloning. The CPT1 locus was mapped to chromosome 2. C60575 and MRG2762 are STS markers. E3634S, C1566, and R3041 are RFLP markers. Numbers of recombinants between cpt1 and each marker are shown. P0020D05, P0025F02, and P0689H05 are PAC clones, and OSJNBa0008E01 is a BAC clone. The cpt1 mutant was found to have a deletion of ∼209 kb (dashed line) between C1566 and R3041 markers that enclosed the CPT1 locus. Sequences around the two edges of the deletion are indicated (gray letters represent deletion). Within the deletion, a NPH3-like gene (indicated by a horizontal arrow; 4.5 kb) was predicted. cM, centimorgan.
(B) Transgenic complementation of the cpt1 mutant. A genomic clone (8.2 kb) containing the NPH3-like gene was introduced into the cpt1 mutant by Agrobacterium-mediated transformation. The photographs show 3-d-old seedlings of the wild type, cpt1 mutant, and complemented cpt1 mutant (T3 generation) grown under irradiation with unilateral white light (3 μmol m−2 s−1; from the right side of the photographs) and overhead red light (3 μmol m−2 s−1). The two complemented cpt1 seedlings originated from independent transformants. Bottom panels show ethidium bromide–stained PCR products obtained for the photographed plants. (a) A 428-bp fragment of the NPH3-like gene was amplified with primers specific to this gene (5′-TTGCAGTGCATAGCCAGTAC-3′ and 5′-TTTCCACGTACTTCTCGTCC-3′). (b) A 502-bp genomic fragment located in the deleted region and at a close distance from the NPH3-like gene was amplified (primers 5′-CGTTGTACGACTGGATGGAC-3′ and 5′-CACTCAACCTTCCGCTTCTC-3′).
To construct a physical map that covers the CPT1 locus, the P1-derived artificial chromosome (PAC) genomic library of cv Nipponbare developed at RGP (Baba et al., 2000) was screened with the marker sequences. The PAC clones P0020D05 and P0689H05 were selected as the end clones containing C1566 and R3041, respectively. Because these clones did not overlap each other, we further screened the PAC and BAC libraries with end sequences of these clones and found another two clones, P0025F02 and OSJNBa0008E01, which contained the end sequences of P0020D05 and P0689H05, respectively. With these clones, the physical map between C1566 and R3041 could be completed (Figure 3A). To delimit the CPT1 locus further, we sequenced the clone P0025F02 (155.5 kb) by means of a shotgun strategy (Yano et al., 2000) and developed 16 CAPS markers from C1566 to the other end at ∼10-kb intervals. Unexpectedly, PCR failed to show amplification of cpt1 genomic DNA segments for all markers but C1566. This result indicated that the most part of P0025F02 is absent from the cpt1 mutant.
We used the inverse PCR method to identify the region deleted in the cpt1 mutant (see Supplemental Figure 1 online). At first, genomic DNA gel blot hybridization was conducted to find an appropriate restriction enzyme. Genomic DNAs from the wild type and cpt1 mutant were digested by different restriction enzymes, and the blot was hybridized with the C1566 probe. Digestion with BamHI gave a hybridized band of 16.5 kb for the wild type, in agreement with the wild-type sequence encompassing the probe sequence, but a hybridization band of ∼5 kb for the cpt1 mutant. The result suggested that the 5-kb BamHI fragment of the cpt1 mutant contained the rejoined point after an interstitial deletion. Based on this result, the genomic DNAs of the cpt1 mutant were digested with BamHI, and the fragments were, after self-ligation, subjected to inverse PCR. The sequence of the amplified fragment (∼2 kb) was compared with those of the three clones (P0689H05, P0025F02, and OSJNBa0008E01) and the deletion region of 208,845 bp was determined (Figure 3A).
Thirty-two putative genes were predicted in the deleted region with the Genscan program. A BLAST search of DNA databases indicated, however, that one of the putative genes (Figure 3A) has a high similarity to the Arabidopsis NPH3 gene, which is essential for the phototropism of Arabidopsis hypocotyls (see Introduction). This putative NPH3-like gene was next subjected to transgenic complementation analysis.
One of the subclones generated by RGP for shotgun sequencing of P0025F02 contained the whole sequence of the NPH3-like gene (58D12A1, 8.2 kb). This genomic fragment was introduced into the cpt1 mutant by Agrobacterium tumefaciens–mediated transformation. The comparison with the cDNA sequence described below indicated that the genomic fragment had a 1522-bp extension upstream of the start codon, which is probably long enough to cover the promoter region, and a 2204-bp extension downstream of the stop codon. No separate gene was predicted in the extensions. We obtained 28 T1 transformants containing the hygromycin resistance and target genes. These transformants set seeds normally by self-pollination. Seedlings with normal phototropisms of coleoptiles and roots segregated in the T2 generation. The recovery of phototropisms as well as the presence of the introduced gene could be confirmed in T3 plants (Figure 3B). A genomic fragment located in the deletion could be amplified in the wild type, but not in cpt1 and complemented cpt1 plants, demonstrating that the CPT1 gene was in fact introduced into the cpt1 mutant. These results lead to the conclusion that the CPT1 gene corresponds to the NPH3-like gene investigated.
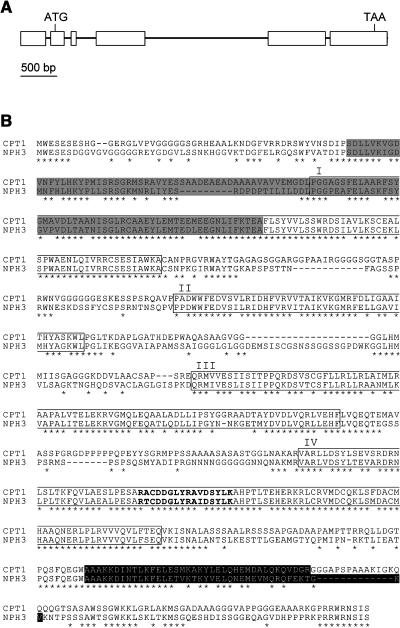
In spite of repeated attempts, we failed to isolate the full-length CPT1 cDNA from cDNA libraries. Therefore, the cDNA clone was obtained by fusing an EST clone, found to correspond to the 5′ half of CPT1 cDNA, and a clone of the 3′ half obtained by performing 3′ rapid amplification of cDNA ends (3′ RACE) (see Methods). Based on the cDNA sequence data, the structure of the genomic CPT1 gene was determined (Figure 4A). The CPT1 gene contained six exons. The deduced amino acid sequence indicated that the CPT1 protein is composed of 762 amino acid residues with a molecular mass of 80.6 kD (Figure 4B).
Figure 4.
Structure of CPT1 and Amino Acid Sequence of CPT1.
(A) Structure of the genomic CPT1 gene. Exons (boxes) and introns (lines) were determined by a comparison of the genomic and cDNA sequences. The positions of the start and stop codons are indicated.
(B) The deduced amino acid sequence of CPT1, shown in comparison with the sequence of NPH3. Asterisks denote residues identical in the two sequences. Gray and black shadings indicate BTB/POZ and coiled-coil domains, respectively. The four regions, I to IV, conserved in the NPH3 family of Arabidopsis and the corresponding regions of CPT1 are boxed. The Tyr phosphorylation site conserved in the NPH3 family of Arabidopsis and the corresponding site of CPT1 are indicated in bold letters.
The amino acid sequences of CPT1 and NPH3 showed 58% identity. As shown for NPH3, coiled-coil and BTB/POZ domains were predicted (Figure 4B), although the prediction significance for the latter domain was low. Furthermore, the four regions of sequence conservation (regions I to IV) shown for the NPH3 family of Arabidopsis (Motchoulski and Liscum, 1999; http://www.biosci.missouri.edu/liscum/research_page/LiscumLab_ResearchPage.html) could be identified in CPT1 (Figure 4B). The identities of regions I to IV between NPH3 and CPT1 were 88.7, 82.6, 73.3, and 89.9%, respectively. The Tyr phosphorylation site conserved in region IV (Motchoulski and Liscum, 1999) could also be identified in CPT1 (Figure 4B). The GC content of the CPT1 cDNA (71.5%) was much higher than that of NPH3 (45.6%). This property explains why it was difficult to isolate the full-length CPT1 cDNA from cDNA libraries.
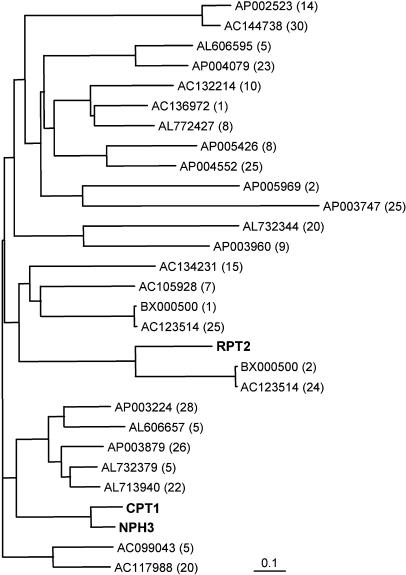
We searched for CPT1 homologs in the japonica rice genome database and obtained predicted amino acid sequences of the homologs identified by this screen. Figure 5 shows the phylogenetic tree of CPT1 and the predicted homologs. Arabidopsis NPH3 and RPT2 are included in the tree. The analysis indicated that rice contains at least 26 homologs of CPT1 and that CPT1 is orthologous to NPH3. It was also predicted that rice has two CPT1 homologs that are orthologous to RPT2.
Figure 5.
Phylogenetic Tree of Rice CPT1 Homologs and the Two Related Arabidopsis Homologs NPH3 and RPT2.
The tree was obtained by the neighbor-joining method using 1000 bootstrap replicates. Homologs of CPT1 were identified in the japonica rice genomic database. Each predicted homolog is indicated by the accession number of a BAC or PAC clone and the number of the predicted gene in the clone (given in parentheses). The accession numbers of CPT1, NPH3, and RPT2 are AB186127, AF180390, and AF181683, respectively. The scale represents 0.1 substitutions per site.
Investigation of Coleoptile Phototropism with the Aid of the cpt1 Mutant
The cpt1 mutant was next used to investigate some physiological aspects of coleoptile phototropism. For this analysis, we adopted blue light conditions that induce time-dependent phototropism (Iino, 1990). Continuous stimulation as practiced to obtain the data shown in Figure 1A is an extreme condition for this type of phototropism.
Figure 6A shows the fluence rate-response curves obtained by stimulating wild-type and cpt1 coleoptiles for 80 min. The phototropic curvature was determined immediately after 80-min simulation and again after another 80-min incubation. Wild-type coleoptiles showed a bell-shaped response curve at either measurement time but with a clear difference. The response at 80 min occurred in a narrower fluence-rate range and peaked at a lower fluence rate as compared with the response at 160 min. The cpt1 coleoptiles showed no detectable response at 160 min (Figure 6A), in agreement with the result obtained with continuous irradiation (Figure 1A). However, a small response was detected at 80 min (Figure 6A). This response peaked at 0.1 μmol m−2 s−1, in agreement with the wild-type response at 80 min.
Figure 6.
Kinetic Investigation of Phototropic Curvature in Wild-Type and cpt1 Coleoptiles.
(A) Phototropic fluence rate-response curves. Wild-type and cpt1 seedlings, prepared as described for Figure 1A, were stimulated with unilateral blue light for 80 min at indicated fluence rates. The curvature was determined immediately (open circles) and 80 min (closed circles) after the end of blue light irradiation.
(B) Time courses of phototropism. Wild-type and cpt1 seedlings were stimulated with unilateral blue light for 80 min (until the time indicated by the vertical dotted line; circles) or continuously (squares) at 0.3 μmol m−2 s−1. The curvature of coleoptiles was monitored after the onset of blue light. The means ± se from 10 (circles) or four (squares) seedlings are shown.
The time course of phototropic response to 80-min stimulation was investigated with wild-type and cpt1 coleoptiles (Figure 6B, circles). The fluence rate of blue light used was 0.3 μmol m−2 s−1. As shown in Figure 6B (circles), wild-type coleoptiles showed a biphasic time course with a shoulder at ∼1.5 h. On the other hand, cpt1 coleoptiles showed a transient curvature response. The peak of the response occurred at ∼1.5 h. This time-course result for cpt1 coleoptiles agrees with the fluence rate-response data described above. The peak of the response in cpt1 coleoptiles and the shoulder of the response in wild-type coleoptiles occurred at comparable times. This agreement suggested that the response found in the former coleoptiles was also induced in the latter coleoptiles, resulting in the observed shoulder.
In addition to measuring the time course of curvature development during and after 80-min starvation, curvature was also measured during 4 h of continuous stimulation (Figure 6B, squares). The time course for cpt1 coleoptiles was essentially identical to that obtained by stimulating for 80 min. In wild-type coleoptiles, continuous stimulation resulted in a greater curvature response as compared with the response to 80-min stimulation. However, the first half of the time course was similar with the latter response and the shoulder could also be resolved. These results indicated that the curvature response identified in cpt1 coleoptiles occurs only transiently after the onset of continuous stimulation.
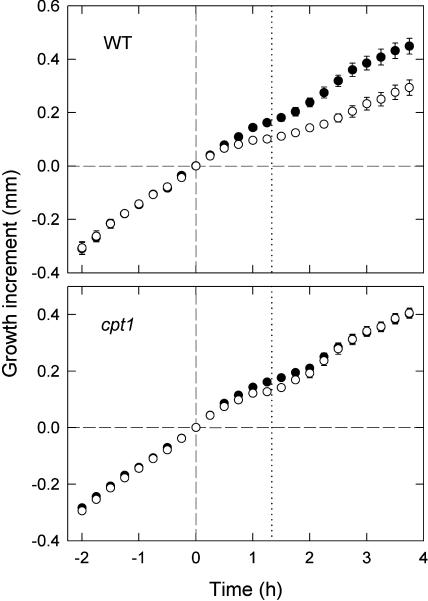
The growth on the irradiated and shaded sides of phototropically stimulated coleoptiles was measured from the photographic images used to obtain the data in Figure 6B (circles). In these experiments, photographs were also obtained for the 2-h period before the onset of stimulation and those were used to measure the growth before stimulation (which served as a control). In wild-type coleoptiles, a clear difference in growth between the two sides, corresponding to the curvature response, could be found (Figure 7). In cpt1 coleoptiles, no major growth difference occurred as anticipated; however, a small growth difference that corresponds to the small transient curvature could be resolved. The growth on the two sides of wild-type coleoptiles appeared to follow complex patterns. It was noted, however, that the net growth (i.e., the averaged growth between the two sides) of wild-type coleoptiles was inhibited by blue light stimulation in the first 1.5-h period, followed by a recovery from this inhibition. This pattern of growth changes was also found in cpt1 coleoptiles. These results suggested that the phototropic differential growth that depends on CPT1 was accompanied by a blue light–induced inhibition of net growth that does not depend on CPT1, resulting in the apparently complex patterns of growth changes observed on the two sides of wild-type coleoptiles. Another interesting result is that the small and transient curvature induced in cpt1 coleoptiles, which is also found as a small growth difference between the two sides, showed an approximate temporal agreement with the blue light–induced growth inhibition. This agreement suggested that the CPT1-independent curvature response and the CPT1-independent growth inhibition are related.
Figure 7.
Phototropic Differential Growth in Wild-Type and cpt1 Coleoptiles.
Coleoptiles were stimulated with unilateral blue light for 80 min (from time 0 to the time indicated by the vertical dotted line) at 0.3 μmol m−2 s−1. The lengths on irradiated (open circles) and shaded (closed circles) sides of the coleoptile were monitored before and after the onset of blue light. The growth increment (ordinate) indicates the difference in coleoptile length from the length at time 0. The means ± se from 10 seedlings are shown.
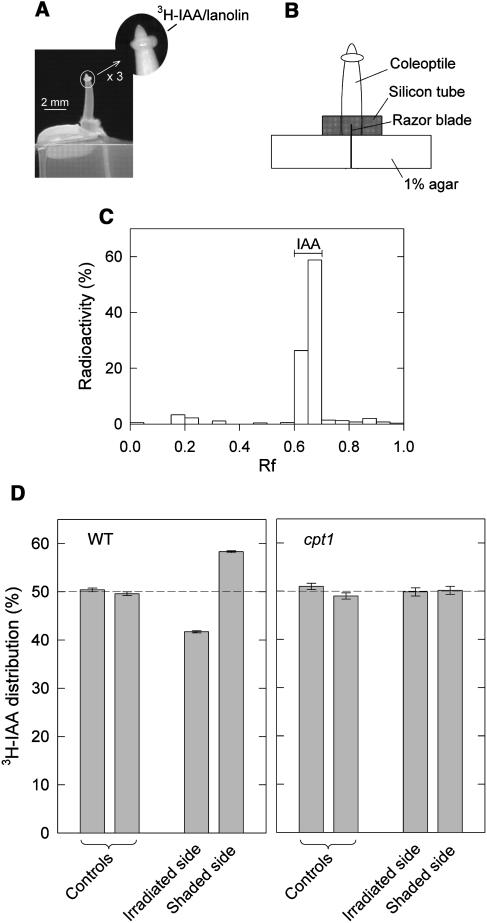
Tracer experiments with 3H-IAA were conducted to investigate the possible contribution of auxin to coleoptile phototropism (Figures 8A and 8B). In brief, a ring of 3H-IAA–containing lanolin was applied to the tip of an intact coleoptile, and the coleoptile was stimulated, 2 h after 3H-IAA application, with blue light for 80 min (the condition used to obtain the above growth data). Right after this stimulation, the coleoptile was excised and placed on a pair of agar blocks for 40 min to collect basipetally diffusing 3H-labeled substances from the irradiated and shaded halves. The control coleoptile was treated similarly but without blue light stimulation. First of all, separation of the collected diffusible substances by thin layer chromatography indicated that most of the recovered radioactivity corresponded to 3H-IAA (Figure 8C). In wild-type coleoptiles, a clear difference in 3H-IAA could be detected between the irradiated and shaded halves (Figure 8D). 3H-IAA was distributed more to the shaded side at a ratio of ∼2:3. By sharp contrast, no significant difference was found in cpt1 coleoptiles.
Figure 8.
Phototropic Lateral Translocation of IAA in Wild-Type and cpt1 Coleoptiles.
The experiment was conducted using wild-type and cpt1 seedlings prepared as described for Figure 1A.
(A) Photographs illustrating the method of 3H-IAA application. Lanolin containing 3H-IAA was applied to the tip region in the form of a ring (∼0.3 mm wide, located ∼0.3 mm below the tip). Two hours after 3H-IAA application, the seedling was irradiated with unilateral blue light as described for Figure 6B. The photograph indicates the side view of a coleoptile to which 3H-IAA was applied. Unilateral blue light was given in the direction perpendicular to this view.
(B) A diagram illustrating the method of recovering diffusible substances from the basal cut surface of 3H-IAA–treated coleoptiles. Immediately after 80-min blue light irradiation, the coleoptile was excised and placed on a pair of agar blocks. A razor blade that separated the two agar blocks also divided the base (∼0.5 mm) of the excised coleoptile into irradiated and shaded halves. A ring of silicon tube served as a guide for placing the coleoptile to the right position. After 40-min incubation, the coleoptile was removed from the assembly, and the radioactivity recovered in the agar blocks was determined.
(C) Identification of the recovered 3H-IAA. The diffusible substances collected as described above (but without blue light irradiation) were separated by thin layer chromatography. The radioactivity in each zone is shown as a percent of the total radioactivity recovered. The position of authentic IAA is shown. Rf, rate of flow.
(D) Distribution of the recovered radioactivity in irradiated and shaded halves of wild-type and cpt1 coleoptiles. The amounts of radioactivity obtained from irradiated and shaded halves were expressed as percentages of the sum from the two halves. Controls were obtained similarly from two halves without blue light irradiation. The means ± se from three (wild type) or four (cpt1) independent experiments (a set of samples per experiment, 12 coleoptiles per sample) are shown.
DISCUSSION
After finding that the cpt1 mutant is impaired specifically in phototropisms of coleoptiles and roots (Figures 1 and 2), we adopted a map-based cloning strategy to identify the CPT1 gene. The CPT1 locus was mapped to a region of chromosome 2 that is largely deleted in the cpt1 mutant (Figure 3A). One of the putative genes deleted in the mutant showed a high similarity to Arabidopsis NPH3. A complementation test has demonstrated that this NPH3-like gene in fact corresponds to the CPT1 gene (Figure 3B).
Arabidopsis has many NPH3 homologs (Motchoulski and Liscum, 1999; Sakai et al., 2000); likewise, rice has many CPT1 homologs (Figure 5). The phylogeny indicates that CPT1 is the rice ortholog of NPH3 (Figure 5). The pioneering work with Arabidopsis (Motchoulski and Liscum, 1999) and this work with rice together provide a generalized view that NPH3/CPT1 is a fundamental component of the phototropic signal transduction in seed plants. The primary structure of CPT1 closely matches that of NPH3 (Figure 4). It is anticipated that CPT1 functions by physically interacting with the photoreceptor phototropin as demonstrated for NPH3 (Motchoulski and Liscum, 1999).
Our detailed physiological analysis has indicated that phototropism of rice seedlings involves, in addition to the major CPT1-dependent response, CPT1-independent responses. This issue and the relationships of phototropism with growth and auxin are discussed below.
CPT1-Dependent and -Independent Phototropisms
In red light–grown rice coleoptiles, a steady phototropic curvature is established by 6 h of continuous stimulation with unilateral blue light (Neumann and Iino, 1997). The fluence rate-response curve obtained under this steady state condition indicated that cpt1 coleoptiles do not show any detectable curvature (Figure 1). This result is in accord with the conclusion that no phototropic curvature occurs in continuously stimulated hypocotyls of Arabidopsis nph3 mutants (Liscum and Briggs, 1996; Sakai et al., 2000).
Unexpectedly, however, cpt1 coleoptiles showed a transient and small phototropic curvature within the first 2.5 h of continuous stimulation (Figure 6B). This response component could be identified as a shoulder in the time course for wild-type coleoptiles. The fluence rate-response curve for the transient response was bell shaped as was that for the steady state response described above. However, the transient response occurred in a narrower range of fluence rates and had its peak at lower fluence rates (Figure 6A; compare the open circles for cpt1 with the closed circles for the wild type). These differences suggest that the CPT1-independent response is fundamentally different from the CPT1-dependent one.
The transient curvature response showed an approximate temporal coincidence with the transient inhibition of net growth (Figure 7; see below). This agreement suggests that the curvature response is mechanistically related to the growth inhibition. A probable explanation is that unilateral blue light induces greater inhibition of growth on the irradiated side than on the shaded side, thereby causing the observed curvature toward the light source. Because no time course has been reported for the phototropism of Arabidopsis nph3 mutants, it is not clear if Arabidopsis hypocotyls also show a similar NPH3-independent phototropism. The transient nature of the CPT1-independent phototropism implies, together with its small response size, that this phototropism bears no major ecological significance.
Although cpt1 coleoptiles showed no phototropic curvature when grown under continuous phototropic stimulation, cpt1 roots showed some curvature that amounted to approximately one-quarter of the curvature found in wild-type roots (Figure 1A). This result contrasts with those reported for Arabidopsis nph3 mutants, which showed no root phototropism (Liscum and Briggs, 1996; Sakai et al., 2000). The phototropic fluence rate-response curve obtained for cpt1 roots resembled that for wild-type roots, the only apparent difference being the amplitude (Figure 1A). This similarity suggests that CPT1-dependent and -independent phototropisms of roots are based on similar mechanisms. A possible explanation is that one or more of CPT1 homologs of rice can substitute the function of CPT1 for root phototropism (but not for coleoptile phototropism). The CPT1-independent response found in coleoptiles and that found in roots differ in their kinetics and fluence-rate dependence; therefore, they are probably mediated by fundamentally different mechanisms. Because the CPT1-independent response of roots is sustained during continuous stimulation, it probably bears ecological significance.
The Relationship between Phototropism and Growth
The net growth of cpt1 coleoptiles was inhibited by unilateral blue light (Figure 7). The major inhibition occurred transiently in the time period 0.5 to 2 h after the onset of irradiation. As described in Results, the apparently complex growth pattern observed on either side of phototropically stimulated wild-type coleoptiles is likely to be caused because the CPT1-independent inhibition of net growth accompanied the CPT1-dependent differential growth. This conclusion is in agreement with the idea that the phototropic differential growth results in principle from a redistribution of growth (Iino and Briggs, 1984; Iino, 2001). On the other hand, as discussed above, it is possible that the transient and small curvature response, isolated in the cpt1 mutant, is caused by the CPT1-indpendent growth inhibition.
The photoreceptor for the CPT1-independent inhibition of growth can be either cryptochrome or phototropin. The growth of red light–grown maize coleoptiles was inhibited transiently after the onset of continuous blue light (Wang and Iino, 1997). This response was thought to be mediated by cryptochrome 1 because it showed some similarities with the protoplast shrinking response mediated by cryptochrome 1 (Wang and Iino, 1998). However, the transient growth inhibition in maize coleoptiles occurred within 10 min of blue light irradiation, and the lag time was shorter than 1 min. Therefore, the response is too rapid to be compared with the CPT1-independent growth inhibition in rice coleoptiles. The rapid and transient growth inhibition in maize, which also requires high fluence rates (Wang and Iino, 1997), is probably distinct from the CPT1-independent growth inhibition identified in this study.
Recent results reported by Folta and Spalding (2001) raise the possibility that phototropin may be the photoreceptor for the CPT1-independent growth inhibition. These authors have demonstrated that the phot1-mediated growth inhibition in Arabidopsis hypocotyls occurs rapidly and transiently after the onset of continuous blue light, whereas the inhibition mediated by cryptochromes 1 and 2 progresses more slowly and is long sustained during irradiation. Furthermore, the same authors found that the phot1-mediated growth inhibition does not involve NPH3. The transient nature of the phot1-mediated growth inhibition and the independence of this response from NPH3 agree with the possibility that rice phototropin plays a photoreceptor role in the transient growth inhibition observed in cpt1 coleoptiles.
The transient phot1-mediated growth inhibition occurred in ∼1 h of blue light irradiation and with a lag time shorter than 5 min (Folta and Spalding, 2001). It appears that the phot1-mediated growth inhibition in Arabidopsis is induced somewhat faster than the CPT1-independet growth inhibition in rice. This might represent the difference in materials. It is also possible that the difference in the blue light fluence rates used is responsible for the difference in response kinetics (80 μmol m−2 s−1 for Arabidopsis and 0.3 μmol m−2 s−1 for rice). Further investigation of the fluence rate dependence of these responses would provide useful information.
The Relationship between Phototropism and Auxin
Although the process of phototropism has long been explained on the basis of the Cholodny-Went hypothesis, this explanation has received repeated criticisms (Iino, 2001; see Introduction). Here, we have demonstrated that the 3H-IAA applied to the tip of wild-type coleoptiles is asymmetrically distributed in the coleoptiles in response to phototropic stimulation and, more importantly, that this response does not occur in cpt1 coleoptiles (Figure 8). It is concluded that lateral translocation of auxin occurs downstream of CPT1, contributing most probably to phototropic signal transduction.
The gradient in 3H-IAA detected in wild-type coleoptiles was 1:1.4 (irradiated:shaded half) (Figure 8D). The gradient in growth rate at the corresponding time (80 to 120 min) was 1:1.9 (irradiated:shaded side) (Figure 7). It is expected that the gradient in 3H-IAA between the two sides is greater than that measured between the two halves. Taking this into consideration and also assuming a linear relationship between auxin concentrations and growth rates (Haga and Iino, 1998), we conclude that the above two ratios are in a reasonable agreement to describe phototropic differential growth in terms of asymmetrical distribution of auxin.
The work on Arabidopsis has provided genetic evidence that phot1, NPH3, and RPT2 participate also in root phototropism (Liscum and Briggs, 1995, 1996; Sakai et al., 2000). We now show that CPT1 also participate in root phototropism (Figure 1A). Furthermore, there is a possibility that a putative auxin transporter is involved in the process of root phototropism (Friml et al., 2002). Together, these lines of evidence suggest that shoot organs and roots share similar mechanisms of phototropism, in which lateral translocation of auxin plays a role.
Further Implications of the Results
This study provided some interesting physiological results that are not directly related to the purposes of the study. Two of them will be considered here. One is the difference in phototropic fluence rate-response curves shown in Figures 1A and 6A. The fluence rate-response curve peaked at 10 μmol m−2 s−1 when measured after 6 h of continuous stimulation (Figure 1). On the other hand, the fluence rate-response curve obtained by stimulating for 80 min and measuring curvature 160 min after the onset of stimulation peaked at 1 μmol m−2 s−1 (Figure 6A). Because the CPT1-independent curvature disappears by the time of curvature measurement (Figure 6B), it is unlikely that this curvature response contributed to the occurrence of the peak at the lower fluence rate. The fluence rate-response curve obtained by stimulating for a shorter period (40 min) showed its peak at a still lower fluence rate (0.1 μmol m−2 s−1), and the threshold fluence rate for 40-min stimulation was similarly lower than that for 80-min stimulation (K.K. Biswas, X. Feng, and M. Iino, unpublished data). The stimulation time-dependent differences in blue light sensitivity probably indicate a participation of sensory adaptation (Iino, 1987).
The other interesting result is the oscillatory movement observed in gravitropically responding roots (Figure 2B). The oscillation occurred before the root tip reached the vertical position. The movement cannot simply be explained in terms of gravitropism. A possibility is that circumnutation was synchronized after gravitropic stimulation as reported for sunflower (Helianthus annuus) hypocotyls (Johnsson and Israelsson, 1969) and dark-grown rice coleoptiles (Yoshihara and Iino, 2005). Another possible explanation is that gravitropic curvature is counteracted by autotropic straightening, and this counteraction results in oscillatory movement as noted in wheat (Triticum aestivum) coleoptiles (Tarui and Iino, 1997). The period of the observed oscillation was significantly shorter than that of circumnutation (for the latter period, see Results). The relatively large standard deviation, which initially represents the random orientation caused by circumnutation, was not reduced during the phase of synchronized oscillation (Figure 2B). These results rather support the explanation based on autotropic straightening.
Concluding Remarks
Molecular genetic study of phototropism has been limited to the model plant Arabidopsis. This work demonstrates the usefulness of rice, another distinct model plant, in supplementing and extending the results from Arabidopsis. Our conclusion that CPT1, the rice ortholog of NPH3, plays a critical role in coleoptile phototropism strongly supports the view that phototropin participates as the photoreceptor for this phototropism. This view, however, needs to be experimentally substantiated and, in particular, rice mutants impaired in phototropin should be made available. Rice homologs of PHOT1 and PHOT2 were cloned by Kanegae et al. (2000). Our search of the japonica rice database indicates that rice has two copies of PHOT1, differing by only four nucleic acids, and one copy of PHOT2 (Iino and Haga, 2005). Therefore, mutants of these three loci are required before we can fully investigate the physiological functions of phototropin in rice as well as the relationship between phototropin and CPT1.
METHODS
Isolation of the cpt1 Mutant and Plant Materials Used in This Study
Mutants of phototropism were screened from the M2 generation of γ-ray–mutagenized japonica-type rice (Oryza sativa cv Nihonmasari). Growth conditions for mutant screening were as described by Biswas et al. (2003). In brief, groups of M2 caryopses, each obtained from a single M1 plant, were surface sterilized and sown on 0.7% agar in acrylic boxes. The sown caryopses were incubated at 25 ± 1°C under continuous irradiation with unilateral white light (2 to 5 μmol m−2 s−1) and overhead red light (2.5 to 3.5 μmol m−2 s−1). Seedlings that appeared to show no or reduced phototropism were selected as candidate mutants and grown in our greenhouse to harvest selfed M3 caryopses for further analysis. The cpt1 mutant was isolated in this screen.
The cpt1 mutant was repeatedly backcrossed to the wild type, and for the experiments described in this report, caryopses of the F2 or F3 generation obtained after three to six times of backcrossing were used. The wild-type caryopses used in each experiment for comparison were those harvested on the same occasion. The indica cultivar Kasalath was used for map-based cloning. The seedlings for physiological studies were grown as described below. The plants for molecular genetic studies were grown in growth cabinets (Biotron LPH-200-RDS; Nippon Medical and Chemical Instruments, Osaka, Japan) or the greenhouse.
Preparation of Seedlings for Physiological Analysis
Rice seedlings were prepared essentially as described in Iino et al. (1996). In brief, surface-sterilized caryopses were sown on 0.7% agar in 4.5-mL acrylic cuvettes (Elkay; Kendall Healthcare, Mansfield, MA). The cuvettes contained agar to the top. Only when submerged seedlings were used, the cuvettes contained agar to 1 cm from the base, and the remaining space was filled with deionized water at indicated times. The sown caryopses were incubated at 25 ± 0.5°C under red light (2.5 to 3.0 μmol m−2 s−1; for the light source, see Liu and Iino, 1996). During incubation, the cuvettes were left on the horizontal surface of an aluminum plate, being covered with a bottom-open acrylic box. Wet paper towels were placed inside the box to maintain a high humidity. During the course of the experiment, seedlings were maintained inside an acrylic box, except the time of handling.
Analyses of Phototropism, Gravitropism, and Growth
Rice seedlings were stimulated for phototropism with unilateral white light or blue light. The source of white light was a white fluorescent tube (FL20S N-EDL; Mitsubishi Electric Osram, Tokyo, Japan). Blue light was obtained by passing light from a slide projector (Kodak Ektagraphic III, EXR 300-W lamp; Tokyo, Japan) or a light source unit (MHF-D100LR; Moritex, Tokyo, Japan) through a Corning blue glass filter (no. 5-50). The former light source was used to obtain the data shown in Figures 1 and 6A, and the latter light source was used in the other cases. The two light sources provided essentially identical photon fluence rate spectra. Seedlings were stimulated for gravitropism by displacing them by 30° or 90° from the vertical (Iino et al., 1996). The direction of either phototropic or gravitropic stimulus was parallel to the plane passing through the two vascular bundles of the coleoptile.
Photographic images, recorded on Kodak technical pan film and expanded with a slide projector, were used to analyze phototropism, gravitropism, and growth of seedlings. Phototropic and gravitropic curvatures of coleoptiles were measured as the changes in tip orientation, which were determined as the angle of the coleoptile tip made with the plumb line. The curvatures of primary roots were measured similarly, but it had to be taken into consideration that the root tip circumnutates. The steady state phototropic curvature of roots established after continuous stimulation (Figure 1) was determined from the angle of the noncircumnutating subapical zone (2 to 4 mm from the tip). Because the gravitropic curvature was initiated in the circumnutating apical zone, the time course of root gravitropism was obtained by measuring the angle of the root tip (Figure 2). The elongation growth of coleoptiles and primary roots was measured from the side view of seedlings (Figure 2A), and the elongation growth on the two opposite sides of phototropically stimulated coleoptiles was measured from the front view of seedlings (Figure 7). The angle and length were determined with a computer-interfaced digitizer.
Analysis of Phototropic IAA Translocation
Tracer experiments with 3H-labeled IAA (3-[5(n)-3H]IAA, 962 GBq mmol−1; Amersham Pharmacia Biotech, Buckinghamshire, UK) were performed to investigate lateral translocation of IAA during phototropism. 3H-IAA was mixed with lanolin as described by Haga and Iino (1998) to a concentration of 3.26 MBq (∼0.6 μg) g−1. The 3H-IAA/lanolin mixture was applied to the coleoptile as shown in Figure 8A. The radioactive substances were recovered in agar blocks from the irradiated and shaded halves of excised coleoptiles (Figure 8B). Four coleoptiles were placed on a pair of agar blocks (each block, 2 × 3.5 × 15 mm3). Agar blocks obtained for either side from three repeats were added to 10 mL of scintillation cocktail (Clearsol I; Nacalai Tesque, Kyoto, Japan) and measured for radioactivity with a liquid-scintillation counter (LS6200; Beckman Coulter, Fullerton, CA).
For identification of 3H-IAA (Figure 8C), the radioactive substances recovered in six agar blocks (eight coleoptiles per block) were extracted with 2 mL of methanol containing cold IAA (100 μg), and the extract was separated by thin layer chromatography using a polyamide plate (0.1 mm, Polyamid-DC 6 UV254; Macherey-Nagel, Doren, Germany) and a solvent system of n-butanol:acetic acid:water (80:5:15, v/v/v). The methanol and solvent system additionally contained butylated hydroxytoluence, an antioxidant, at a concentrations of 0.1 mg mL−1 (Iino et al., 1980). Each zone of the plate was scraped off and added to methanol (0.5 mL) in a scintillation vial. The mixture was left overnight at 4°C and subjected to scintillation counting in 10 mL of the scintillation cocktail.
Map-Based Cloning of CPT1
F2 seedlings obtained by crossing the cpt1 mutant to Kasalath were assayed for phototropism under the mutant screening conditions described above, and those that did not show coleoptile phototropism were selected. The selected seedlings were grown further, and the genomic DNA of each plant (1 to 100 μg) was extracted from leaves as described by Haga and Iino (2004). The RFLP, CAPS, and STS markers used to analyze the polymorphisms between Nipponbare and Kasalath were obtained from the DNA bank at the National Institute of Agrobiological Sciences (http://www.dna.affrc.go.jp/misc/bank.index.html).
The DNA gel blot hybridization for identification of the region deleted in the cpt1 mutant was performed as follows. Genomic DNAs isolated from wild-type and cpt1 leaves were digested with BamHI, EcoRV, HindIII, SacI, SalI, SmaI, SpeI, SphI, or XbaI (Toyobo, Osaka, Japan). The products were electrophoresed on a 0.7% agarose gel, transferred to a nylon membrane, and hybridized with the C1566 probe. The inverse PCR used to identify the exact deletion region was performed as follows. The genomic DNAs isolated from cpt1 leaves were digested with BamHI and circularized with T4 DNA ligase (Toyobo). The circularized DNA fragments were subjected to inverse PCR with the nested primers (first pair, 5′-CCATCCATTCCTCTCTTCACCACCT-3′ and 5′-GCCAGCAACAGGAACTACAGGAGGA-3′; second pair, 5′-CGCATCTCCGTCACCATCCATT-3′ and 5′-GCCTGATGCTACTGCCTCTCATTC-3′) designed from the end sequences of the BamHI fragment in the outward direction. The amplified fragment was subcloned into pGEM-T Easy (Promega, Madison, WI). Rice BLAST (http://riceblast.dna.affrc.go.jp/) was used to determine the deleted region. GENSCAN (http://genes.mit.edu/GENSCAN.html) was used to analyze the putative genes involved in the deletion.
Construction of CPT1 cDNA
The EST clone C1167 (1.7 kb, DNA data bank of Japan accession number D15735), found to correspond to the 5′ half of CPT1 cDNA, was provided by the DNA bank (Yamamoto and Sasaki, 1997). The 3′ half of CPT1 cDNA (1.3 kb) was obtained by 3′ RACE and cloned into Escherichia coli (strain TOP10) with a GeneRacer kit (the kit with AMV RT, TOPO TA cloning; Invitrogen, Carlsbad, CA) according to the manufacture's instructions. The total RNA (5 μg) used for the 3′ RACE was extracted from dark-grown seedling shoots as described by Haga and Iino (2004); the CPT1-specific primer used was 5′-CGCTGGTCACCGAGCTCGAGAA-3′. The full-length cDNA (2.9 kb) was obtained by fusing the above two clones at the BstEII sites present in the overlapping region.
Transgenic Complementation of the cpt1 Mutant
The genomic clone 52D12A1 that contained CPT1 was provided by RGP. The insert DNA was isolated from the clone after digestion with BamHI and KpnI and subcloned into the binary vector pPZP2H-lac (Fuse et al., 2001). The CPT1-containing vector was identified by PCR with CPT1-specific primers (5′-TTGCAGTGCATAGCCAGTAC-3′ and 5′-TTTCCACGTACTTCTCGTCC-3′) and introduced into Agrobacterium tumefaciens strain EHA101. Agrobacterium-mediated transformation of the cpt1 mutant with this vector was performed as described by Hiei et al. (1994). The transformants were grown to yield T2 caryopses. T2 seedlings were assayed for coleoptile phototropism under the mutant screening conditions. Several T2 seedlings that showed clear phototropism were selected for each transformant and grown to yield T3 caryopses.
Sequence Analysis
Rice BLAST (see above) was used to search for homologs of the CPT1 gene. Coding regions of the putative genes were analyzed with Rice GAAS (http://ricegaas.dna.affrc.go.jp/). The predicted protein sequences were aligned with ClustalW (http://www.ddbj.nig.ac.jp/E-mail/homology-j.html). The same program was used to analyze protein phylogeny by the neighbor-joining method. The phylogenetic tree was displayed with TreeView (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html). SMART (http://coot.embl-heidelberg.de/SMART) and COILS (http://www.ch.embnet.org/software/COILS_form.html) were used to analyze BTB/POZ and coiled-coil domains, respectively.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AB186127.
Supplementary Material
Acknowledgments
We thank Osamu Yatoh for the gift of γ-ray–mutagenized rice caryopses, T. Itoh (Research Institute for Sustainable Humanosphere, Kyoto University, Japan) for allowing us to use the Institute's radioisotope research facilities, and RGP for the gift of the shotgun clone 52D12A1. This work was supported by Grant-in-Aid for Scientific Research (10440242) and Special Research (13139204) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan to M.I. and by the Rice Genomic Project (MP1120) to M.T.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Moritoshi Iino (iino@sci.osaka-cu.ac.jp).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.028357.
References
- Baba, T., et al. (2000). Construction and characterization of rice genomic libraries, PAC library of japonica variety Nipponbare, and BAC library of indica variety Kasalath. Bull. Natl. Inst. Agrobiol. Resour. 14, 41–49. [Google Scholar]
- Biswas, K.K., and Iino, M. (2002). A mutant of rice impaired specifically in the phototropism of coleoptiles. J. Plant Res. 115 (suppl.), 71.12884129 [Google Scholar]
- Biswas, K.K., Neumann, R., Haga, K., Yatoh, O., and Iino, M. (2003). Photomorphogenesis of rice seedlings: A mutant impaired in phytochrome-mediated inhibition of coleoptile growth. Plant Cell Physiol. 44, 242–254. [DOI] [PubMed] [Google Scholar]
- Blakeslee, J.J., Bandyopadhyay, A., Peer, W.A., Makam, S.N., and Murphy, A.S. (2004). Relocalization of the PIN1 auxin efflux facilitator plays a role in phototropic responses. Plant Physiol. 134, 28–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs, W.R., et al. (2001). The phototropin family of photoreceptors. Plant Cell 13, 993–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folta, K.M., and Spalding, E.P. (2001). Unexpected roles for cryptochrome 2 and phototropin revealed by high-resolution analysis of blue light-mediated hypocotyl growth inhibition. Plant J. 26, 471–478. [DOI] [PubMed] [Google Scholar]
- Friml, J., Wiśsniewska, J., Benková, E., Mendgen, K., and Palme, K. (2002). Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415, 806–809. [DOI] [PubMed] [Google Scholar]
- Fuse, T., Sasaki, T., and Yano, M. (2001). Ti-plasmid vectors useful for functional analysis of rice genes. Plant Biotechnol. 18, 219–222. [Google Scholar]
- Haga, K., and Iino, M. (1998). Auxin-growth relationships in maize coleoptiles and pea internodes and control by auxin of the tissue sensitivity to auxin. Plant Physiol. 117, 1473–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga, K., and Iino, M. (2004). Phytochrome-mediated transcriptional up-regulation of ALLENE OXIDE SYNTHASE in rice seedlings. Plant Cell Physiol. 45, 119–128. [DOI] [PubMed] [Google Scholar]
- Harper, R.M., Stowe-Evans, E.L., Luesse, D.R., Muto, H., Tatematsu, K., Watahiki, M.K., Yamamoto, K., and Liscum, E. (2000). The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell 12, 757–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harushima, Y., et al. (1998). A high-density rice genetic linkage map with 2275 markers using a single F2 population. Genetics 148, 479–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282. [DOI] [PubMed] [Google Scholar]
- Huala, E., Oeller, P.W., Liscum, E., Han, I.-S., Larsen, E., and Briggs, W.R. (1997). Arabidopsis NPH1: A protein kinase with putative redox sensing domain. Science 278, 2120–2123. [DOI] [PubMed] [Google Scholar]
- Iino, M. (1987). Kinetic modelling of phototropism in maize coleoptiles. Planta 171, 110–126. [DOI] [PubMed] [Google Scholar]
- Iino, M. (1990). Phototropism: Mechanisms and ecological implications. Plant Cell Environ. 13, 633–650. [Google Scholar]
- Iino, M. (1991). Mediation of tropisms by lateral translocation of endogenous indole-3-acetic acid in maize coleoptiles. Plant Cell Environ. 14, 279–286. [Google Scholar]
- Iino, M. (2001). Phototropism in higher plants. In Photomovement: ESP Comprehensive Series in Photosciences, Vol. 1, D. Häder and M. Lebert, eds (Amsterdam: Elsevier), pp. 659–811.
- Iino, M., and Briggs, W.R. (1984). Growth distribution during first positive phototropic curvature of maize coleoptiles. Plant Cell Environ. 7, 97–104. [Google Scholar]
- Iino, M., and Haga, K. (2005). Role played by auxin in phototropism and photomorphogenesis. In Light Sensing in Plants, M. Wada, K. Shimazaki, and M. Iino, eds (Berlin: Springer-Verlag), in press.
- Iino, M., and Neumann, R. (2000). Phototropism of rice seedlings: Characterization and mutant isolation. Plant Cell Physiol. 41 (suppl.), S56. [Google Scholar]
- Iino, M., Tarui, Y., and Uematsu, C. (1996). Gravitropism of maize and rice coleoptiles: Dependence on the stimulation angle. Plant Cell Environ. 19, 1160–1168. [DOI] [PubMed] [Google Scholar]
- Iino, M., Yu, S.-T., and Carr, D.J. (1980). Improved procedure for the estimation of nanogram quantities of indole-3-acetic acid in plant extracts using the indolo-α-pyrone fluorescence method. Plant Physiol. 66, 1099–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson, A., and Israelsson, D. (1969). Phase-shift in geotropical oscillations—A theoretical and experimental study. Physiol. Plant 22, 1226–1237. [DOI] [PubMed] [Google Scholar]
- Kanegae, H., Tahir, M., Savazzini, F., Yamamoto, K., Yano, M., Sasaki, T., Kanegae, T., Wada, M., and Takano, M. (2000). Rice NPH1 homologues, OsNPH1a and OsNPH1b, are differently photoregulated. Plant Cell Physiol. 41, 415–423. [DOI] [PubMed] [Google Scholar]
- Khurana, J.P., and Poff, K.L. (1989). Mutants of Arabidopsis thaliana with altered phototropism. Planta 178, 400–406. [PubMed] [Google Scholar]
- Liscum, E., and Briggs, W.R. (1995). Mutations in the NPH1 locus of Arabidopsis disrupt the perception of phototropic stimuli. Plant Cell 7, 473–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum, E., and Briggs, W.R. (1996). Mutants of Arabidopsis in potential transduction and response components of the phototropic signaling pathway. Plant Physiol. 112, 291–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.J., and Iino, M. (1996). Phytochrome is required for the occurrence of time-dependent phototropism in maize coleoptiles. Plant Cell Environ. 19, 1379–1388. [DOI] [PubMed] [Google Scholar]
- Motchoulski, A., and Liscum, E. (1999). Arabidopsis NPH3: A NPH1 photoreceptor-interacting protein essential for phototropism. Science 286, 961–964. [DOI] [PubMed] [Google Scholar]
- Neumann, R., and Iino, M. (1997). Phototropism of rice (Oryza sativa L.) coleoptiles: Fluence-response relationships, kinetics and photogravitropic equilibrium. Planta 201, 288–292. [DOI] [PubMed] [Google Scholar]
- Sakai, T., Kagawa, T., Kasahara, M., Swartz, T.E., Christie, J.M., Briggs, W.R., Wada, M., and Okada, K. (2001). Arabidopsis nph1 and npl1: Blue light receptors that mediate both phototropism and chloroplast relocation. Proc. Natl. Acad. Sci. USA 98, 6969–6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai, T., Wada, T., Ishiguro, S., and Okada, K. (2000). RPT2: A signal transducer of the phototropic response in Arabidopsis. Plant Cell 12, 225–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarui, Y., and Iino, M. (1997). Gravitropism of oat and wheat coleoptiles: Dependence on the stimulation angle and involvement of autotropic straightening. Plant Cell Physiol. 38, 1346–1353. [DOI] [PubMed] [Google Scholar]
- Tatematsu, K., Kumagai, S., Muto, H., Sato, A., Watahiki, M.K., Harper, R.M., Liscum, E., and Yamamoto, K.T. (2004). MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 16, 379–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togo, S., and Hasegawa, K. (1991). Phototropic stimulation does not induce unequal distribution of indole-3-acetic acid in maize coleoptiles. Physiol. Plant 81, 555–557. [Google Scholar]
- Wang, X., and Iino, M. (1997). Blue-light-induced shrinking of protoplasts from maize coleoptiles and its relationship to coleoptile growth. Plant Physiol. 114, 1009–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., and Iino, M. (1998). Interaction of cryptochrome 1, phytochrome, and ion fluxes in blue-light-induced shrinking of Arabidopsis hypocotyls protoplasts. Plant Physiol. 117, 1265–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, K., and Sasaki, T. (1997). Large-scale EST sequencing in rice. Plant Mol. Biol. 35, 135–144. [PubMed] [Google Scholar]
- Yano, M., Katayose, Y., Ashikari, M., Yamanouchi, U., Monna, L., Fuse, T., Baba, T., Yamamoto, K., Umehara, Y., Nagamura, Y., and Sasaki, T. (2000). Hd1, a major photoperiodic sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12, 2473–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara, T., and Iino, M. (2005). Circumnutation of rice coleoptiles: Its occurrence, regulation by phytochrome, and relationship with gravitropism. Plant Cell Environ., in press. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.