Abstract
The nucleocytoplasmic (N/C) ratio plays a prominent role in the maternal-to-zygotic transition (MZT) in many animals. The effect of the N/C ratio on cell-cycle lengthening and zygotic genome activation (ZGA) has been studied extensively in Drosophila, where haploid embryos experience an additional division prior to completing cellularization and triploid embryos cellularize precociously by one division. In this study, we set out to understand how the obligate difference in ploidy in the haplodiploid wasp, Nasonia, affects the MZT and which aspects of the Drosophila MZT are conserved. While subtle differences in early embryonic development were observed in comparisons among haploid, diploid, and triploid embryos, in all cases embryos cellularize at cell cycle 12. When ZGA was inhibited, both diploid female, and haploid male, embryos went through 12 syncytial divisions and failed to cellularize before dying without further divisions. We also found that key players of the Drosophila MZT are conserved in Nasonia but have novel expression patterns. Our results suggest that zygotically expressed genes have a reduced role in determining the timing of cellularization in Nasonia relative to Drosophila, and that a stronger reliance on a maternal timer is more compatible with species where variations in embryonic ploidy are obligatory.
Keywords: maternal-to-zygotic-transition, ploidy, cellularization, Nasonia, gastrulation
Introduction
The transition from the maternal to zygotic control of development is conserved in all multicellular eukaryotes. In early animal development, maternal mRNAs and proteins are loaded into a developing egg (Matova and Cooley, 2001). These maternally deposited products include basic components for the initial mitoses and patterning events that take place shortly after egg activation (Tadros and Lipshitz, 2009). In many animals, a large fraction of protein-coding maternal RNAs are degraded during different stages of embryogenesis. Some of these maternal products also include repressors of the zygotic genome, which begin to be eliminated preceding zygotic genome activation (ZGA) (Schier, 2007).
A well-known explanation for this phenomenon is the nucleocytoplasmic (N/C) ratio model. In this model, zygotic transcription is repressed by maternally deposited factors, whose presence is diluted by the increase of DNA with each successive nuclear division. (Edgar et al., 1986; Pritchard & Schubiger, 1996; Zamir et al., 1997). This has been observed in vertebrate and invertebrate models, where an increased N/C ratio will result in a premature ZGA (Collart et al. 2013; Edgar et al., 1986; Kane & Kimmel, 1993; Mita & Obata, 1984). Conversely, decreasing the N/C ratio results in a delayed ZGA (Collart et al., 2013; Erickson and Quintero, 2007).
The N/C ratio model has been studied extensively in Drosophila. The maternal products in the Drosophila embryo drive 13 rapid, simultaneous, syncytial nuclear divisions, each comprised of only an M and S phase (Edgar & O’Farrell, 1989; Foe & Alberts, 1983; Rabinowitz, 1941). As these early divisions proceed, the cell cycles gradually lengthen, eventually allowing enough time for more and more zygotic genes to be completely transcribed (Rothe et al., 1992). By the final syncytial division, there is a pause, which coincides with the activation of a large number of zygotic genes including many involved in cellularization and gastrulation of the embryo.
Evidence that timing of these events is related to the N/C ratio comes from Drosophila embryos of different ploidy. Haploid Drosophila mutants undergo an extra simultaneous nuclear division before completing cellularization at cell cycle 15, whereas triploid Drosophila mutants cellularize precociously at cell cycle 13 (Edgar et al., 1986; Erickson & Quintero, 2007; Shermoen et al., 2010). It has recently been shown that it is the amount of transcriptionally engaged DNA, rather than simply the amount of DNA in cell that serves as the numerator in the N/C ratio (Blythe and Wieschaus, 2015).
The N/C ratio also influences the timely expression of cell cycle regulators in Drosophila. In wild-type fly embryos, the destruction of maternally derived protein of the Cdc25 phosphatase homologs String and Twine is necessary for the inhibitory phosphorylation of Cdk1 kinase, which critical for the introduction of a G2 phase at the end of cell cycle 14 (Edgar & O’Farrell, 1990; Farrell & O’Farrell, 2013). String mRNA and protein levels gradually decline during the early syncytial divisions, while Twine protein is rapidly degraded at cell cycle 14 (Edgar & O’Farrell, 1990; Farrell & O’Farrell, 2013) . The degradation of Twine is carried out by tribbles and other zygotically activated genes, whose timely expression is dependent on the N/C ratio (Farrell and O’Farrell, 2013). The N/C ratio also affects the timely expression of fruhstart (frs), which is required for the cell-cycle pause in the last syncytial division(Jörg Groβhans et al., 2003; Lu et al., 2009a). In haploid Drosophila mutants, the decrease in the N/C ratio leads to delayed activation of fruhstart and tribbles by one nuclear cycle, resulting in an additional syncytial division prior to completing cellularization due to the delayed destruction of Twine protein (Farrell & O’Farrell, 2013; Mata et al., 2000).
A second set of zygotic genes in Drosophila are activated irrespective of the N/C ratio (Lu et al., 2009). These genes are thought to be controlled by a maternal timing mechanism, referred to as the ‘maternal clock’, and their expression is dependent upon the destruction of maternal repressors (Howe et al., 1995; Lee et al., 2014). There is evidence that suggests the SAM domain containing RNA binding protein Smaug (Smg) and the transcription factor Zelda (Zld) may act as components of this timer (Tadros et al., 2003). Upon egg activation, Smaug employs multiple mechanisms to destabilize almost two-thirds of the maternal RNAs present in the early embryo, and clearance of these maternal mRNAs is required for zygotic genome activation (Benoit et al., 2009; Semotok et al., 2005; Smibert et al., 1999; Tadros et al., 2007a). Zelda works in concert with Smaug to activate thousands of zygotic genes, many of which are classified as genes that follow the ‘maternal clock’ model. A loss of either Smaug or Zelda sustains repression of the zygotic genome (Liang et al., 2008; Nien et al., 2011; Tadros and Lipshitz, 2009; Tadros et al., 2007a; Benoit et al., 2009).
The relative roles of N/C ratio or maternal timer are not well characterized in insects outside of Drosophila. A particularly interesting group of insects to examine are the Hymentopera, all of which use haplodiploid sex determination. In this mode, unfertilized eggs develop into haploid males and fertilized eggs develop into diploid females (J.A. Lynch, 2015; Verhulst et al., 2010). In this study, we set out to understand how the obligate difference in ploidy affects early development, and which aspects of the Drosophila MZT are conserved in the haplodiploid wasp, Nasonia vitripennis.
We found that Nasonia male and female embryos cellularize at cell cycle 12, irrespective of differing N/C ratios. Artificially increasing ploidy did not affect cell cycle number prior to cellularization. Obstructing zygotic transcription by injecting RNA polymerase II inhibitor, α-amanitin, in unfertilized and fertilized embryos prevented cellularization at cell cycle 12, with no observable changes in preceding events. Many of the key players of the Drosophila MZT are conserved in the wasp but have novel expression patterns that do not differ between male and female embryos.
Overall, our results indicate that the zygotic genome plays a relatively small role in establishing the onset of cellularization in relation to the number of syncytial divisions in Nasonia. This may indicate that a system relying more strongly on a maternal timer is more compatible with embryos where ploidy is obligately variable.
Results and Discussion
We live-imaged and analyzed the development of male and female embryos through gastrulation. Similar to Drosophila, the early cleavage stages are short, but get increasingly long as cellularization approaches (Fig. 1), with gastrulation occurring as the embryo finishes cellularizing. Both haploid and diploid embryos cellularized at cell cycle 12(Fig. 1,Movie S1, S2), confirming the previous proposition (Buchta et al., 2013). While there were subtle differences in cell cycle length, we observed that the completion of the final syncytial division took longer as ploidy increases (Fig. 1).
Fig. 1. Timing of syncytial nuclear divisions.
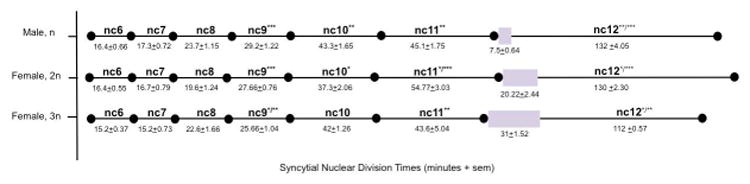
Each row represents the timing between syncytial nuclear division in minutes. Top row, male haploid embryos. Middle row, female diploid embryos. Bottom row, female triploid embryos. (N=9 for n male, N=9 for 2n female, N=5 for 3n female). One asterisk (*) denotes significant difference p<0.05 between haploid comparisons. Two asterisks (**) denotes significant difference p<0.05 between diploid comparisons. Three asterisks (***) denotes significant difference p<0.05 between triploid comparisons. Purple boxes denote wave of nuclear division that was only observable in the last nuclear division preceding cellularization.
We next wondered whether the sex determination pathway could act to “correct” the differences in ploidy between the sexes. We are able to control the phenotypic sex of embryos by knocking down the gene Nv-transformer by pRNAi, (Lynch & Desplan, 2006) which prevents the establishment of a feedback loop necessary to establish femaleness in diploid embryos.. Thus, Nv-transformer knockdowns will produce diploid males from fertilized eggs. (Verhulst et al., 2010). If there were a female-specific factor involved in preventing cellularization of the embryo one cycle earlier than haploid male embryos (as would be expected if the N/C ratio had a role in the wasp), we would expect diploid males to cellularize after cycle 11. We live-imaged diploid, male embryos alongside control, haploid male embryos from Nv-transformer injected virgins. All embryos cellularized at cell cycle 12 (Movie S3). This shows that timing of cellularization in diploids is not dependent on female sex determination.
We also considered that since Hymenoptera are obligate haplodiploids, they may have evolved mechanisms to stabilize the number of cycles so that they are equal between the sexes. We asked whether manipulating the ploidy could overcome any naturally occurring compensatory mechanisms. To create triploid wasps, we allowed a subset of the diploid, male embryos (created as described above) to develop to adults and crossed them to wildtype females. Since there is no meiosis I in male Nasonia, all of the sperm will be diploid, and all fertilized eggs will yield triploid females (Pennypacker, 1958). Embryos from this cross were live-imaged through the onset of gastrulation and triploid females were confirmed by performing Nv-transformer RT-PCR to confirm the sex of each individual embryo. All embryos, including the confirmed triploid females, cellularized at cell cycle 12 (Fig. 1, Movie S4). One notable difference was that the triploid female embryos completed cellularization, 20 minutes earlier compared to wildtype diploid and haploid embryos, on average (Fig. 1).
We did find that one trait that seemed to vary in direct relation to ploidy of the embryo. In Nasonia, the entry into nuclear cycle 12 is observed as an anterior to posterior wave. This wave was 1.5× longer in triploid embryos than in diploids, and 4.1× longer than in haploids. Diploid embryos took approximately 2.6× longer than haploids.
We next asked whether other processes are different between haploid and diploid embryos. It could be that development is buffered enough to initiate cellularization at the same time regardless of ploidy, while earlier processes may be more affected. Nuclear localization of activated RNA polymerase II has been used as a proxy for the onset of zygotic transcription (Nestorov et al., 2013). We examined the distribution of activated RNA polymerase (CTD phospho-serine S5) throughout early embryogenesis in both males and females. While CTD phospho-serine S2 antibodies (ab24758 and ab5095) are more commonly used to assess the onset of zygotic transcription (Chen et al., 2013; Nestorov et al., 2013), we did not see a successful cross-reaction in Nasonia. With a CTD phospho-serine S5 antibody (ab5408), we found nuclear localized active RNA polymerase II is found at all cycles in both sexes (Suppl. Fig. 1a-f;a′-f′). Many embryos in early stages showed nuclear exclusion of the antigen (Suppl. Fig. 1d-d′), but we interpret these as reflecting mitotic state of the nuclei, which is rapidly changing in the earliest stages. The specificity of the antibody used is supported by the conserved reduction in staining in the transcriptionally quiescent pole cells (Suppl. Fig. 1g-g′). The very early onset of zygotic transcription is not unprecedented. Zygotic transcription in insects such as the potato beetle is found as early as the first division (Schenkel and Schnetter, 1979), and in Drosophila, the first zygotic transcripts are detected as early as the second nuclear division.
Given the ambiguity of the result with RNA polymerase II, we considered that histone modifications might better reveal the onset of significant zygotic transcription. We examined the pattern of methylation of lysine 9 on histone H3 (H3K9Me3), which indicates chromatin in a repressed state. This mark was present in all nuclei from the very early cleavage stages, through the blastoderm, and was identical for male and female embryos (Fig. 2a′-c′).
Fig. 2. Patterns of Histone modification in the Nasonia embryo.
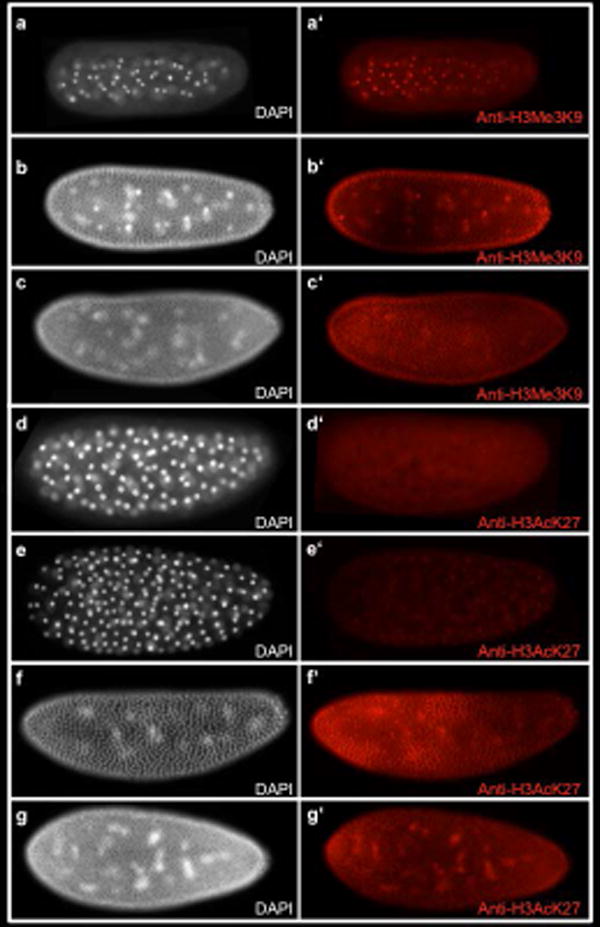
a-g White, DAPI (DNA). Fig a′-c′ red, anti-H3Me3K9 marks repressive chromatin. (a-a′) Embryo in division cycle 5. Anti-H3Me3K9 staining is seen in all nuclei. (b-b′) Embryo in division cycle 9. (c-c′) Embryo in division cycle 12. Fig D-G White, DAPI (DNA) Fig d′-g′ red, anti-H3AcK27 marks active enhancers and promoters. (d-d′) Embryo in division cycle 6 does not show any Anti-H3AcK27 staining. (e-e′) Embryo in division cycle 7 shows nuclear staining of Anti-H3AcK27. (f-f′) Embryo in division cycle 11. (g-g′) Embryo in division cycle 12 shows increased levels of of Anti-H3AcK27 nuclear staining at the anterior and posterior poles of the embryo.
Acetylation of lysine at position 27 of histone H3 (H3AcK27) (Fig. 2d-g;d′-g′), indicates the presence of active promoters and enhancers (Creyghton et al., 2010). Staining of an antibody against this mark was first observed at nuclear cycle 7, when nuclei have arrived at the surface of the embryo (Fig. 2e′). There was no difference between male and female embryos. Thus, we propose that chromatin becomes “open for business” as soon as the syncytial blastoderm forms, and that ploidy has no influence on this event.
We next examined the Nasonia ortholog of two crucial genes that are affected differentially by the N/C ratio in setting the time of cellularization in Drosophila. The paralogous genes string and twine are Drosophila orthologs of the cell cycle regulator Cdc25 that promote mitosis by removing inhibitory phosphates on Cdk1. Only a single ortholog (Nv-string) of these genes exists in Nasonia and most other insects. Nv-string mRNA is provided maternally and is rapidly degraded in the early syncytial cycles (Figure 3a-d;a′-d′). Nv-string mRNA levels begin to rise again at the late blastoderm stage (Figure 3c), and are at very high levels just after the onset of gastrulation (Figure 3d). This pattern is similar to what is seen in Drosophila (Edgar & O’Farrell, 1990; Lehman et al., 1999). Knocking down maternally derived Nv-string with pRNAi did not affect early mitoses, or the timing of cellularization in either males or females (Movie S5). We cannot exclude an incomplete knockdown as the reason for a lack of phenotype at present (Fig. S2).
Fig. 3. Expression of Nv-stg.
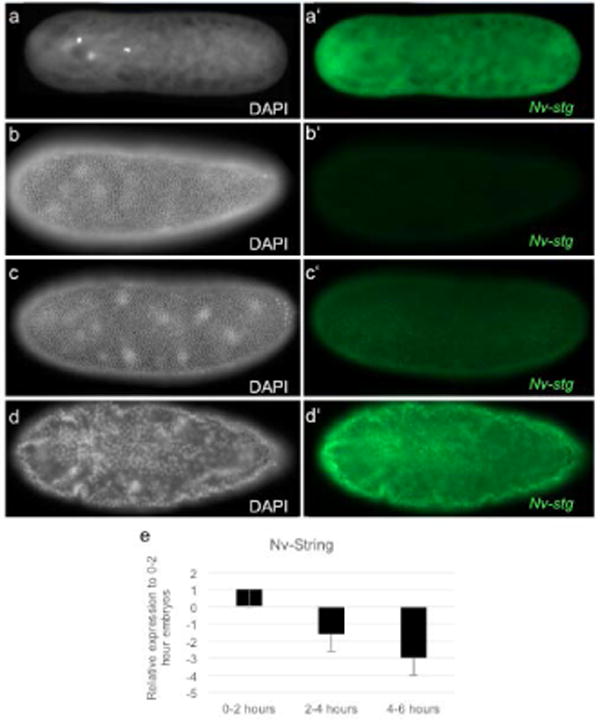
a-d White, DAPI (DNA). a′-d′ green, Nv-stg mRNA. (a-a′) Embryo in division cycle 1. Nv-string mRNA is maternally loaded and is ubiquitous throughout the embryo. (b-b′) Embryo in division cycle 12. Maternal Nv-string mRNA is lost. (c-c′) Embryo in late division cycle 12. Zygotic Nv-string is first seen. (d-d′) Gastrulation. Nv-string mRNA is seen in all nuclei. (e) qRT-PCR of Nv-string normalized to housekeeping gene, Rpo49. Maternal Nv-string levels steadily decrease throughout the syncytial stages. 0-2 hours is representative of pre-blastoderm embryos; 2-4 hours is representative of middle blastoderm embryos; 4-6 hours is representative of late blastoderm and cellularized embryos.
Tribbles is an important regulator of Twine protein, and is an important zygotic component that orchestrates the timing of cell cycle pausing in response to the N/C ratio (Farrell and O’Farrell, 2013; Mata et al., 2000). Nv-tribbles is maternally provided as mRNA, with levels again decreasing in the early cycles, before rising again just before gastrulation (Fig. 4e). The onset of zygotic expression of Nv-tribbles was during cell cycle 12 in both haploid and diploid embryos. The zygotic pattern of Nv-tribbles not completely uniform, but rather is modulated along the AP axis in broad bands (Fig. 4a-a′). Slightly later, we observed strong expression in the presumptive head (Fig. 4b-b′), similar to Nv-tailless (J. A. Lynch et al., 2006). Just before gastrulation, most dorsal and ventral expression disappears, with strong staining remaining in the ventral ectoderm flanking the presumptive mesoderm (Fig. 4c-d, c′-d′). The conspicuous absence of Nv-tribbles in this region is interesting, as Drosophila tribbles is upregulated in the mesoderm, and plays an important role in delaying mitosis in the mesoderm during gastrulation which is crucial for its proper morphogenesis (Groβhans & Wieschaus, 2000). This observation fits an emerging pattern, where several genes required for Drosophila mesoderm morphogenesis are restricted to the ventral neuroectoderm in Nasonia (Pers et al., 2016). Knockdown of Nv-tribbles was only effective against the maternal contributions (Suppl. Fig. 2), while zygotic levels increased to be close to normal (not shown). This partial knockdown did not affect the timing of cellularization in either male or female embryos (Movie S6).
Fig. 4. Expression of Nv-trbl.
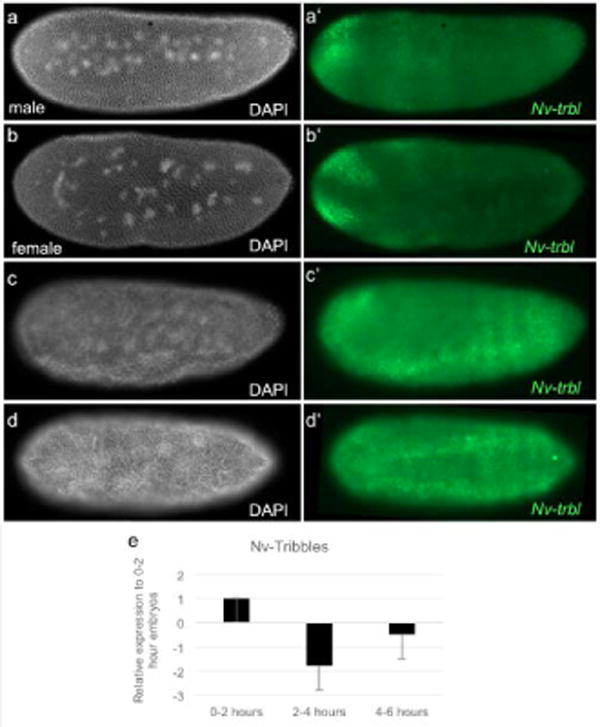
a-d White, DAPI (DNA). Fig a′-d′ green, Nv-trbl mRNA. (a-a′, b-b′) Male and female embryos at division cycle 12. Zygotic Nv-trbl mRNA is expressed at the anterior presumptive head and along broad bands across the anterior-posterior axis. (c-c′) Embryo at the end of division cycle 12, just prior to gastrulation. Zygotic Nv-trbl mRNA is upregulated in the ventral ectoderm. (d-d′) Embryo at the onset of gastrulation. Zygotic Nv-trbl flanks the mesoderm. (e) qRT-PCR of Nv-trbl normalized to housekeeping gene, Rpo49. Maternal Nv-trbl levels decrease throughout the early syncytial stages and zygotic Nv-trbl expression is upregulated in 4-6 hour embryos. 0-2 hours is representative of pre-blastoderm embryos; 2-4 hours is representative of middle blastoderm embryos; 4-6 hours is representative of late blastoderm and cellularized embryos.
fruhstart, another gene whose expression responds to the N/C ratio in Drosophila(Sung et al., 2013) has no clear orthologs outside of the Diptera, and was thus not examined here.
Zelda is a potentially important component of the N/C ratio sensing system, and could also be vital in the N/C independent mechanism. It is a transcription factor that binds to the enhancers of most of the genes expressed during the MZT and is required for their proper expression (Harrison et al., 2011). In addition, some zelda embryos undergo an additional division prior to cellularization (Liang et al., 2008; Sung et al., 2013). There is a maternal mRNA contribution of Nv-zelda, which is degraded in the early cycles (Fig. 5a-a′, 6f). Nv-zelda is strongly upregulated in the syncytial blastoderm stages. It is initially ubiquitous, with a visible upregulation in the middle of the embryo (Fig. 5b-b′). It later shows modulation along the AP axis in stripe-like patterns (Fig. 5c-e,c′-e′), and is also strongly downregulated on the dorsal side where the extra-embryonic membranes will form (Buchta et al., 2013). RNAi against Nv-zelda does not affect the number or timing of nuclear cycles in either haploid or diploid embryos, however, neither cellularization, nor gastrulation occur (Figure 6a-a′, Movie S7), indicating that targets of these processes may be conserved between Nasonia and Drosophila.
Fig. 5. Expression of Nv-zld.
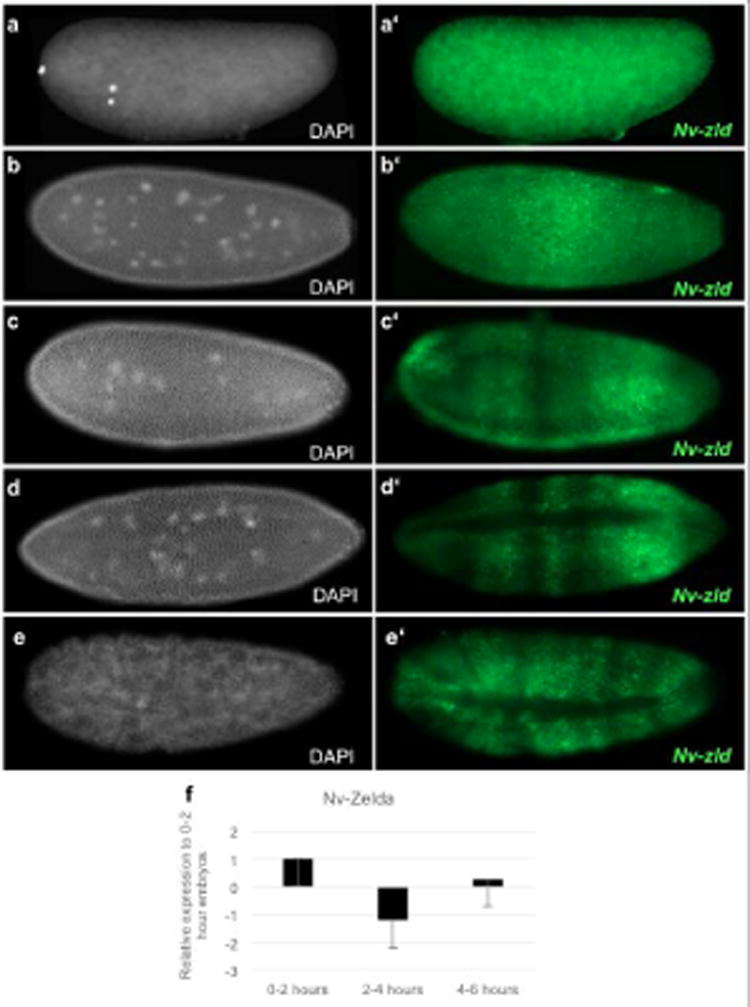
a-d White, DAPI (DNA). a′-d′ green, Nv-zld mRNA. (a-a′) Maternal Nv-zld mRNA is distributed evenly throughout the 2-nuclei staged embryo. (b-b′) At early cell cycle 12, zygotic Nv-zld mRNA is upregulated in the middle of the embryo. (c-c′) During the middle of cell cycle 12, Zygotic Nv-zld mRNA is expressed in stripe-like patterns along the AP axis. (d-d′) At the end of cell cycle 12, Nv-zld mRNA is excluded from the dorsal side of the embryo and is downregulated at the pole of the embryo. (e-e′) At the onset of gastrulation, Nv-zld is excluded from the dorsal side of the embryo and is present in modulating stripes along the anterior-posterior axis. (f) qRT-PCR of Nv-zld normalized to housekeeping gene, Rpo49. Maternal Nv-zld levels decrease throughout the early syncytial stages and zygotic Nv-zld expression is sharply upregulated in 4-6 hour embryos. 0-2 hours is representative of pre-blastoderm embryos; 2-4 hours is representative of middle blastoderm embryos; 4-6 hours is representative of late blastoderm and cellularized embryos.
Fig. 6. Live imaging Nv-zld and Nv-smg pRNAi.
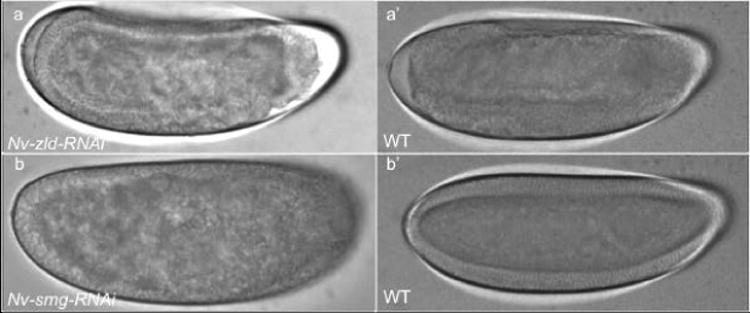
(a-a′) Nv-zld RNAi embryo fails to cellularize and gastrulate compared to WT embryo. (b-b′) Nv-Smg RNAi embryos fail to maintain anchoring of their nuclei to the cortex and cellularize, as compared to WT.
To this point, we could find no evidence that the N/C ratio plays any role in the timing of early embryonic events in Nasonia. To exclude that another, unknown zygotic factor plays a role in controlling the onset of cellularization, we prevented zygotic transcription in collections of unfertilized (100% male) and mostly fertilized (>90% female) embryos by injecting them with the RNA polymerase II inhibitor, α-amanitin. While water-injected embryos of both sexes cellularized after cell cycle 12, all α-amantin –injected embryos injected underwent 12 divisions then failed to cellularize before dying (Figure 7a-b,a′-b′). Since the number of divisions is not affected in either haploid or diploid embryos when zygotic transcription is inhibited, the number of divisions in the Nasonia embryo apparently is not dependent on the zygotic genome.
Fig. 7. Live-imaging of α-amanitin injected embryos.
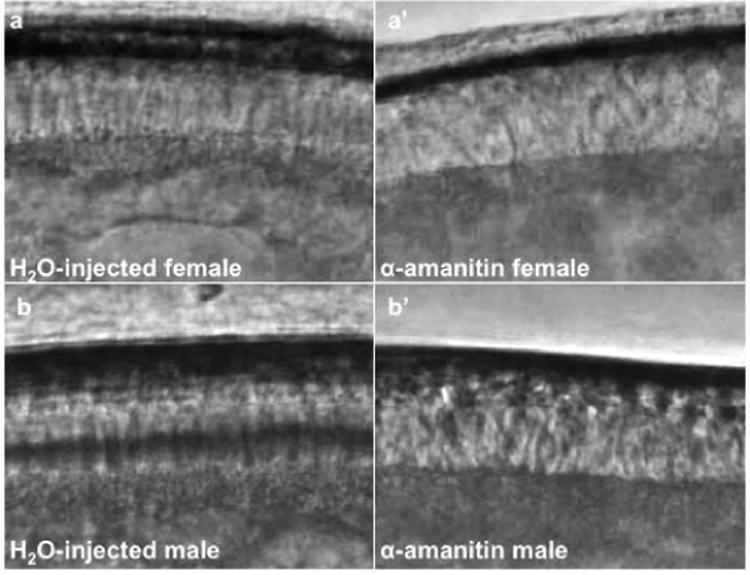
Live-imaging screenshots of alpha amanitin injections in female (a-a′) and male (b-b′) embryos. (a) Water-injected female embryo cellularized at cell cycle 12. (a′) α-amanitin injected female embryo failed to cellularize at cell cycle 12. (b) Water-injected male embryo cellularized at cell cycle 12. (b’) α -amanitin injected male failed to cellularize at cell cycle 12.
A maternally based system has been proposed as an alternative, or complement to the N/C ratio, and the RNA binding protein Smaug has been proposed to be at the center of this system. It is thought to act by destroying maternal mRNAs over the course of early development, which releases repression of the zygotic genome (Lee et al., 2014; Tadros et al., 2007b). This is in parallel to the function of Zelda in preparing the zygotic genes for later expression. As in Drosophila, Nv-smaug is provided at high levels maternally and transcript levels rapidly decrease during the early nuclear divisions(Tadros et al., 2007b)(Fig. 8e). Nv-smaug is almost absent by the blastoderm stage (Fig. 8a-d,a′-d′). Nv-smaug pRNAi leads to major disruption of early embryogenesis, including many embryos where nuclei were not maintained at the cortex, and embryos that do not complete the final two blastoderm divisions (Fig. 6b-b′, Movie S8). This is very similar to the smaug phenotype in Drosophila (Dahanukar et al., 1999), further confirming the conserved role of Smaug in insect embryo early development (Lynch & Desplan, 2010).
Fig. 8. Expression of Nv-smg.
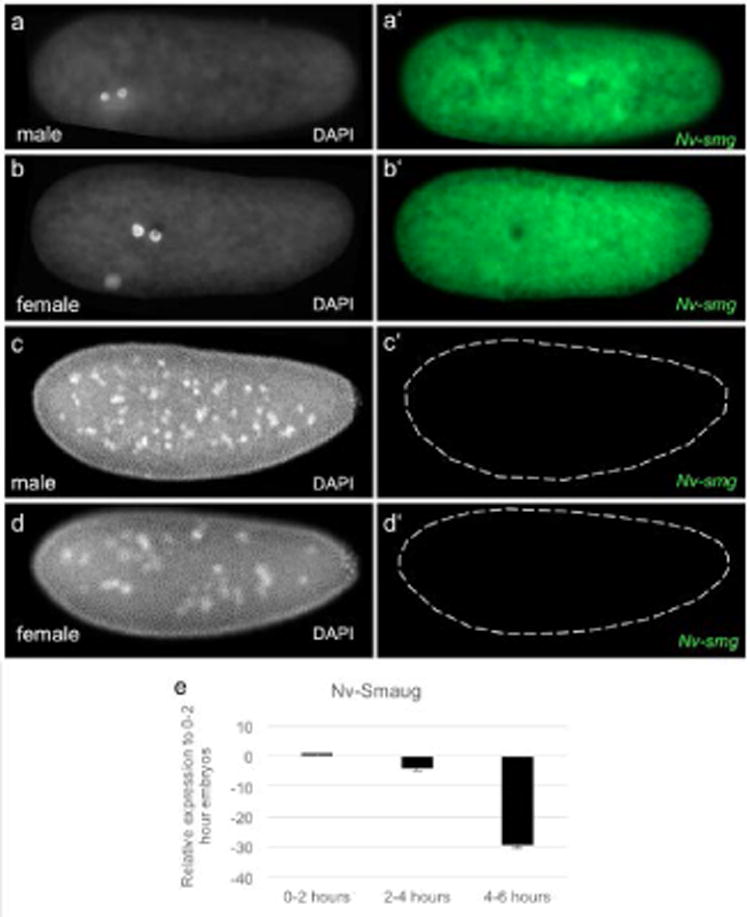
a-d White, DAPI (DNA). Fig a′-d′ green, Nv-smg mRNA. (a-a′, b-b′) Maternal Nv-smg is evenly distributed throughout 2-nuclei male and female embryos. (c-c′, d-d′) Male and female embryos at cell cycle 12, respectively. Dashes outline the embryo in which Nv-smg expression is lost. (e) Maternal Nv-smg decreases sharply throughout the syncytial divisions. 0-2 hours is representative of pre-blastoderm embryos; 2-4 hours is representative of middle blastoderm embryos; 4-6 hours is representative of late blastoderm and cellularized embryos.
Conclusions
Our experiments revealed that the N/C ratio does not affect the number of divisions prior to cellularization and that a maternal program likely controls the number of syncytial divisions in early embryonic Nasonia development. When zygotic transcription in Nasonia is inhibited, the embryo experiences its 12 normal nuclear divisions and fails to cellularize. This is different from Drosophila, where fly embryos will experience an additional rapid division prior to failing cellularization. Additionally, when the ploidy of the Drosophila embryo is artificially manipulated, the number of syncytial divisions prior to cellularization will differ – with triploids cellularizing precociously by one cycle and haploids completing cellularization one cycle later. While triploid Nasonia embryos experience the same number of divisions prior to cellularization and gastrulation, they cellularize and begin to gastrulate after a shorter time spent in cycle 12 than their diploid and haploid counterparts. This may be due to higher levels of zygotic gene expression, that allows a shift in clock-timing (as opposed to relative developmental time).
Our observations have established that the role of the N/C ratio in regulating the major events of early embryogenesis in Nasonia is unclear. What can account for this major difference in Drosophila? There is unlikely to be a single answer for this, given that the major processes, such as cellularization, cell cycle pausing, and ZGA, seem to be affected distinctly by a maternal timer and/or the N/C ratio(Benoit et al., 2009; Jörg et al., 2003; Rose & Wieschaus, 1992). Despite this, our results lead to hypotheses about the nature of the MZT in Nasonia and how this process could change in the course of evolution.
The explanation for why haploid Nasonia embryos do not undergo an additional round of division may be straightforward. In the absence of zygotic transcription, the Drosophila embryo undergoes one extra rapid division before development arrests. This implies that the maternal contribution that drives early divisions is sufficient for up to 15 mitoses, while normally the embryo only undergoes 14, presumably due to the influence of zygotic gene expression(Edgar & Datar, 1996). In contrast, Nasonia embryos lacking zygotic gene expression undergo 12 divisions before arrest, which is also the number of divisions haploids, diploids, and triploids undergo prior to cellularization. This suggests that the Nasonia maternal contribution may not allow additional cycles, regardless of ploidy, while the Drosophila system can drive an extra division if the proper signals from the zygotic genome are not present.
The evolutionary significance of the latent capability to undergo an extra division before cellularization is unclear. It is unlikely to be relevant in natural populations of Drosophila, because haploid embryos do not survive, possibly because of the doubled number of cells interfering with later developmental events. Thus, this excess provision of maternal components appears to be a waste of maternal resources. We propose that the apparent surplus of maternal contribution might be important in driving the exceptionally rapid early divisions in Drosophila, and that the potential for an extra division is an unintended consequence of this adaptation for speed. Since ploidy is normally constant and dependable in Drosophila, the potential extra division made possible by the larger maternal contribution is routinely avoided by the action of the zygotic genome.
On the other hand, every egg produced in Nasonia has the potential to develop into either a haploid male or a diploid female, and therefore, the potential for an extra (and potentially lethal) division in the case of a low N/C ratio must be avoided. Thus, we propose that the restricted maternal contribution in Nasonia should be seen as a trade-off between the speed of early development and the insurance of routine viability of both haploid and diploid embryos. Consistent with this, the cleavage divisions in Nasonia are significantly slower than Drosophila, and it is these early divisions that make up the major difference in embryonic developmental time between the species (Pultz and Leaf, 2003).
The explanation for why triploid Nasonia embryos do not cellularize prematurely is also still elusive. The simplest explanation is that the cell cycle pause and onset of cellularization do not have significant input from the N/C ratio in the wasp. Rather, the exhaustion of the limited maternal contribution provides the “signal” for the cell cycle pause and allows the subsequent activation of zygotic genes required for cellularization and gastrulation. Formally, there is no requirement for an N/C based system if there is a robust maternal timer present. Thus, we propose that the important role for the N/C sensitive system in Drosophila is to counteract the oversupplied maternal system's tendency to drive a deleterious extra cleavage cycle. If this is the case, then the question of how widespread the role of the N/C ratio in regulating early embryonic events arises. It could be that a reliance on the N/C ratio is a feature of organisms with extremely rapid early development driven by strong maternal contribution. This describes the main exemplars of N/C ratio dependence (Drosophila, Xenopus, Danio) quite well, but may not be a feature universal to animals.
Given that there are still major open questions in understanding the N/C based system in Drosophila and that the maternal timer is not well understood (Farrell and O’Farrell, 2014), it will clearly take much further work to understand how the transition from maternal to zygotic control takes place in Nasonia. One approach must include identifying the genes that are up and down regulated as development proceeds in the wasp. Such an experiment was performed in the haplodiploid honeybee Apis (Pires et al. 2016). Their results indicate that much of the early control of the MZT is conserved with flies (except an earlier onset of zygotic transcription), and only seemingly minor differences exist between haploids and diploids (Pires et al., 2016). This only deepens the mystery, and we propose that the ability to easily test gene functions, along with new transcriptomic resources will allow us to begin to unravel the timing of early developmental events in obligate haplo-diploids.
Methods
Embryo collection
All Nasonia vitripennis embryos were collected and fixed as described by Buchta et al., 2013.
Fluorescence In-situ Hybridization and Immunohistochemistry
In-situ hybridization for Nv-smaug, Nv-tribbles, Nv-string, and Nv-zelda were carried out as formerly described in (Lynch et al., 2010, Buchta et al., 2013). In short, we used digoxigenin-labeled probes, which were prepared from PCR amplicons containing T7 RNA polymerase transcriptional start sites. The probes were detected by anti-dig(x02237)POD(Roche, 1:100), and the antibody was detected using Alexa Fluor 488 Tyramide (Invitrogen) as a substrate, which yielded a fluorescent signal.
Immunohistochemistry
Whole embryo immunohistochemistry was performed as described in (Lynch et al., 2010). The antibodies we used in these experiments were: H3me3K9 (abcam ab8898) 1:500, H3AcK27 (ab4729) 1:500, and phosphoS5 RNA polymerase II (4h8, ab5408) 1:500. Secondary antibodies used in this experiment were anti-rabbit(x02237)DyLight 549 (DyLight 1549,1:500).
Parental RNAi
Nasonia vitripennis pupae were injected with 1ug/ul double-stranded RNA (dsRNA) for Nv-smaug, Nv-zelda, Nv-tribbles, Nv-Transformer, and Nv-string as described in (Lynch and Desplan, 2006). RNAi of maternally expressed Nv-smaug and Nv-zelda genes resulted in embryonic lethality. All embryos were live-imaged on an Olympus BX-80 inverted microscope fitted with a Prior controllable stage, under 30× silicone immersion and DIC optics.
Timing Nasonia embryonic divisions
All embryos were live imaged as described above. Each pre-blastoderm embryonic division is observed as a pulse in the embryoplasm. To measure the pre-blastoderm divisions, we measured the minutes in between the apex of one pulse to the apex of the next. During the blastoderm stages, the nuclei are clearly visible at the surface of the embryo. We measured the minutes in between nuclei re-formation and collapse for each blastoderm stage. The divisions and timings were counted before sexing the embryos.
Alpha-amanitin Embryo Injections
We collected 0-1hour Nasonia vitripennis male and female embryos, glued them on a coverslip, and desiccated them for 20 minutes. Embryos injected with 8ug/mL of alpha-amanitin (as described in Edgar et al., 1986) under Halocarbon-200 oil were live-imaged as above.
qRT-PCR
We collected and performed TRIzol RNA extractions on Nasonia embryos at the following times/stages: 0-2hour (pre-blastoderm, nuclear cycles 0-7), 2-4hour (early/middle blastoderm, nuclear cycles 8-10), and 4-6hour (late blastoderm, nuclear cycles 11-12, and through the onset of gastrulation). Using the Protoscript First Strand cDNA synthesis kit (NEB 6300l), we used anchored oligo-d(T) primers to reverse transcribe 40ng of RNA to use in qRT-PCR for the four MZT genes of interest (Nv-smaug, Nv-zelda, Nv-string, and Nv-tribbles) using rpo-49 as a control. These anchored oligo-d(T) primers will always bind to the beginning of the poly-A tail, increasing the likelihood that maternal and zygotic transcripts with varying poly-A tail lengths will be reverse transcribed. The data was analyzed using the delta-delta Ct method (Livak and Schmittgen, 2001). Ct values were normalized to 0-2 hour embryos. Each qRT-PCR was performed on a pool of 30 embryos, and three technical replicates were performed on this pooled cDNA in the same run.
Ploidy manipulation
We used females from a grey eye-color line (oyster) of Nasonia, denoted as NasoniaOy/Oy. These females were injected with dsRNA targeting Nv-transformer and crossed to males with wildtype, brown eye color. Diploid males will express the dominant brown eye-color allele donated from their fathers. The diploid males were then crossed to wildtype females to generate triploid females. We confirmed the sex of each individual embryo that we live-imaged (see Parental RNAi above) by using RT-PCR by looking at the presence of the sex-specific splice isoforms of Nv-transformer.
Primer List:
ZeldaF ggccgcggTAGTAGGCCAGAGCCAAATG
ZeldaR cccggggcACTTTTTCTTGTGCGTCTCG
SmaugF ggccgcggagttcaggctcgatttcttg
SmaugR cccggggcggagacgtagcgttttaagc
TribblesF ggccgcggTTCGGTATCATAGTCGAGCG
TribblesR cccggggcTATCATTTTCGGGGTACGCA
StringF ggccgcggTTCAACGAGCTCATCGACAT
StringR cccggggcGATGAGTCGAGAGAGGGTCT
RT_Smaug_F AGCGCACCCAGAGACAACTT
RT_Smaug_R GCACAGCGACTCCAAACGAG
RT_Zelda F CCCATAATGGCGGGAGTGGT
RT_Zelda_R ACGGCTGATGAGGCATCGAA
RT_String_F AGCCTATCCTCCGGCTACGA
RT_String_R ATGGGCTTCTCCTCGCTGTC
RT_Tribbles_F AGTCATAACCGCGTGAGCCA
RT_Tribbles_R TCTTCTCGTCGCACTCGCAT
RT_Transformer_F GACCAAAAGAGGCACCAAAA
RT_Transformer_R GGCGCTCTTCCACTTCAAT
Supplementary Material
Supplementary Figure 1. Phosphorylated S5 RNA polymerase II is seen throughout all stages in the Nasonia embryo. White, DAPI (DNA); red, anti-4h8 phosphoS5 RNA Polymerase II. (a-a′) Embryo in division cycle 1. (b-b′) Embryo in division cycle 2. (c-c′) Embryo in division cycle 3. (d-d′) Embryo in division cycle 7. RNA polymerase II is excluded from the nuclei (e-e′) Embryo in division cycle 8. (f-f′) Embryo in division cycle 12. (g-g′) Transcriptionally quiescent pole cells have reduced 4h8 staining.
Supplementary Figure 2. qrt-PCR of pRNAi embryos Fold expression change of 0-1 hour Nv-smaug, Nv-string, Nv-tribbles, and Nv-zelda pRNAi embryos. Both wildtype and pRNAi embryos were normalized to a control house-keeping gene, Nv-rp49. pRNAi embryos were compared to wildtype embryos to assess the rate of knockdown.
Acknowledgments
This work was supported under NIH Grant # 1R03HD087476-01, and 1R03HD078578. The authors would like to thank Dr. David O’Brochta and the Insect Genetic Technologies Research Coordination Network for their advice on embryonic injections.
NIH Grant Support: 1R03HD087476-01, 1R03HD078578
References
- Benoit B, He CH, Zhang F, Votruba SM, Tadros W, Westwood JT, Smibert Ca, Lipshitz HD, Theurkauf WE. An essential role for the RNA-binding protein Smaug during the Drosophila maternal-to-zygotic transition. Development. 2009;136:923–932. doi: 10.1242/dev.031815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blythe SA, Wieschaus EF. Zygotic Genome Activation Triggers the DNA Replication Checkpoint at the Midblastula Transition. Cell. 2015:1–13. doi: 10.1016/j.cell.2015.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchta T, Özüak O, Stappert D, Roth S, Lynch Ja. Patterning the dorsal-ventral axis of the wasp Nasonia vitripennis. Dev Biol. 2013;381:189–202. doi: 10.1016/j.ydbio.2013.05.026. [DOI] [PubMed] [Google Scholar]
- Chen K, Johnston J, Shao W, Meier S, Staber C. A global change in RNA polymerase II pausing during the Drosophila midblastula transition. 2013:1–19. doi: 10.7554/eLife.00861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart C, Allen GE, Bradshaw CR, Smith JC, Zegerman P. Titration of four replication factors is essential for the Xenopus laevis midblastula transition. Science. 2013;341:893–6. doi: 10.1126/science.1241530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato Ma, Frampton GM, Sharp Pa, Boyer La, Young Ra, Jaenisch R. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahanukar A, Walker Ja, Wharton RP. Smaug, a novel RNA-binding protein that operates a translational switch in Drosophila. Mol Cell. 1999;4:209–218. doi: 10.1016/s1097-2765(00)80368-8. [DOI] [PubMed] [Google Scholar]
- Edgar Ba, O’Farrell PH. Genetic control of cell division patterns in the Drosophila embryo. Cell. 1989;57:177–187. doi: 10.1016/0092-8674(89)90183-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, Datar SA. Zygotic degradation of two Drosophila ’ s early cell cycle program. 1996:1966–1977. doi: 10.1101/gad.10.15.1966. [DOI] [PubMed] [Google Scholar]
- Edgar BA, Kiehle CP, Schubiger G. Cell cycle control by the nucleo-cytoplasmic ratio in early Drosophila development. Cell. 1986;44:365–372. doi: 10.1016/0092-8674(86)90771-3. [DOI] [PubMed] [Google Scholar]
- Edgar BA, O’Farrell PH. The three postblastoderm cell cycles of Drosophila embryogenesis are regulated in G2 by string. Cell. 1990;62:469–480. doi: 10.1016/0092-8674(90)90012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson JW, Quintero JJ. Indirect effects of ploidy suggest X chromosome dose, not the X:A ratio, signals sex in Drosophila. PLoS Biol. 2007;5:e332. doi: 10.1371/journal.pbio.0050332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell JA, O’Farrell PH. From Egg to Gastrula: How the Cell Cycle Is Remodeled During the Drosophila Mid-Blastula Transition. Annu Rev Genet. 2014;48:269–294. doi: 10.1146/annurev-genet-111212-133531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell JA, O’Farrell PH. Mechanism and regulation of Cdc25/twine protein destruction in embryonic cell-cycle remodeling. Curr Biol. 2013;23:118–126. doi: 10.1016/j.cub.2012.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe VE, Alberts BM. Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J Cell Sci. 1983;61:31–70. doi: 10.1242/jcs.61.1.31. [DOI] [PubMed] [Google Scholar]
- Harrison MM, Li XY, Kaplan T, Botchan MR, Eisen MB. Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genet. 2011;7:e1002266. doi: 10.1371/journal.pgen.1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe JA, Howell M, Hunt T, Newport JW. Identification of a developmental timer regulating the stability of embryonic cyclin A and a new somatic A-type cyclin at gastrulation. Genes Dev. 1995;9:1164–1176. doi: 10.1101/gad.9.10.1164. [DOI] [PubMed] [Google Scholar]
- Groβhans J, Müller HA, Wieschaus E. Control of Cleavage Cycles in Drosophila Embryos by frühstart. Dev Cell. 2003;5:285–294. doi: 10.1016/s1534-5807(03)00208-9. [DOI] [PubMed] [Google Scholar]
- Kane DA, Kimmel CB. The zebrafish midblastula transition. Development. 1993;119:447–456. doi: 10.1242/dev.119.2.447. [DOI] [PubMed] [Google Scholar]
- Lee MT, Bonneau AR, Giraldez AJ. Zygotic Genome Activation During the Maternal-to-Zygotic Transition. Annu Rev Cell Dev Biol. 2014;30:581–613. doi: 10.1146/annurev-cellbio-100913-013027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman DA, Patterson B, Johnston LA, Balzer T, Britton JS, Saint R, Edgar BA. Cis-regulatory elements of the mitotic regulator, string/Cdc25. Development. 1999;126:1793–1803. doi: 10.1242/dev.126.9.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang HL, Nien CY, Liu HY, Metzstein MM, Kirov N, Rushlow C. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. 2008:400–403. doi: 10.1038/nature07388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu X, Li JM, Elemento O, Tavazoie S, Wieschaus EF. Coupling of zygotic transcription to mitotic control at the Drosophila mid-blastula transition. Development. 2009a;136:2101–10. doi: 10.1242/dev.034421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Li JM, Elemento O, Tavazoie S, Wieschaus EF. Coupling of zygotic transcription to mitotic control at the Drosophila mid-blastula transition. Development. 2009b;136:2101–2110. doi: 10.1242/dev.034421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JA. The Expanding Genetic Toolbox of the Wasp Nasonia vitripennis and Its Relatives. Genetics. 2015;199:897–904. doi: 10.1534/genetics.112.147512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JA, Desplan C. A method for parental RNA interference in the wasp Nasonia vitripennis. Nat Protoc. 2006;1:486–494. doi: 10.1038/nprot.2006.70. [DOI] [PubMed] [Google Scholar]
- Lynch JA, Desplan C. Novel modes of localization and function of nanos in the wasp Nasonia. Development. 2010;137:3813–21. doi: 10.1242/dev.054213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JA, Olesnicky EC, Desplan C. Regulation and function of tailless in the long germ wasp Nasonia vitripennis. Dev Genes Evol. 2006;216:493–498. doi: 10.1007/s00427-006-0076-5. [DOI] [PubMed] [Google Scholar]
- Mata J, Curado S, Ephrussi a, Rørth P. Tribbles coordinates mitosis and morphogenesis in Drosophila by regulating string/CDC25 proteolysis. Cell. 2000;101:511–522. doi: 10.1016/s0092-8674(00)80861-2. [DOI] [PubMed] [Google Scholar]
- Matova N, Cooley L. Comparative aspects of animal oogenesis. Dev Biol. 2001;231:291–320. doi: 10.1006/dbio.2000.0120. [DOI] [PubMed] [Google Scholar]
- Mita I, Obata C. Timing of early morphogenetic events in tetraploid starfish embryos. J Exp Zool. 1984;229:215–222. [Google Scholar]
- Nestorov P, Battke F, Levesque MP, Gerberding M. The Maternal Transcriptome of the Crustacean Parhyale hawaiensis Is Inherited Asymmetrically to Invariant Cell Lineages of the Ectoderm and Mesoderm. 2013;8 doi: 10.1371/journal.pone.0056049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nien CY, Liang HL, Butcher S, Sun Y, Fu S, Gocha T, Kirov N, Manak JR, Rushlow C. Temporal coordination of gene networks by Zelda in the early Drosophila embryo. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennypacker MI. The chromosomes of the parasitic wasp, Mormoniella vitripennis. I. In spermatogenesis of haploid and diploid males. Arch Biol (Liege) 1958;69:483–95. [PubMed] [Google Scholar]
- Pers D, Buchta T, Özüak O, Wolff S, Pietsch JM, Memon MB, Roth S, Lynch JA. Global analysis of dorsoventral patterning in the wasp Nasonia reveals extensive incorporation of novelty in a regulatory network. BMC Biol. 2016;14:63. doi: 10.1186/s12915-016-0285-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires CV, De Paula Freitas FC, Cristino AS, Dearden PK, Simões ZLP. Transcriptome analysis of honeybee (Apis Mellifera) haploid and diploid embryos reveals early zygotic transcription during cleavage. PLoS One. 2016;11 doi: 10.1371/journal.pone.0146447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard DK, Schubiger G. Activation of transcription in Drosophila embryos is a gradual process mediated by the nucleocytoplasmic ratio. Genes Dev. 1996;10:1131–1142. doi: 10.1101/gad.10.9.1131. [DOI] [PubMed] [Google Scholar]
- Pultz MA, Leaf DS. The Jewel Wasp Nasonia : Querying the Genome With Haplo-Diploid Genetics. 2003;191:185–191. doi: 10.1002/gene.10189. [DOI] [PubMed] [Google Scholar]
- Rabinowitz M. Studies on the cytology and early embryology of the egg of Drosophila melanogaster. J Morphol. 1941;69 [Google Scholar]
- Groβhans J, Wieschaus EF. A Genetic Link between Morphogenesis and Cell Division during Formation of the Ventral Furrow in Drosophila. Cell. 2000;101:523–531. doi: 10.1016/s0092-8674(00)80862-4. [DOI] [PubMed] [Google Scholar]
- Rose LS, Wieschaus E. The Drosophila cellularization gene nullo produces a blastoderm-specific transcript whose levels respond to the nucleocytoplasmic ratio. Genes Dev. 1992;6:1255–1268. doi: 10.1101/gad.6.7.1255. [DOI] [PubMed] [Google Scholar]
- Rothe M, Pehl M, Taubert H, Jäckle H. Loss of gene function through rapid mitotic cycles in the Drosophila embryo. Nature. 1992;359:156–159. doi: 10.1038/359156a0. [DOI] [PubMed] [Google Scholar]
- Schenkel H, Schnetter W. Transcription During Early Embryogenesis of Leptinotarsa (Coleoptera) 1979;188:179–188. doi: 10.1007/BF00848588. [DOI] [PubMed] [Google Scholar]
- Schier AF. T 406 20. Science (80-) 2007;316:406–407. doi: 10.1126/science.1140693. [DOI] [PubMed] [Google Scholar]
- Semotok JL, Cooperstock RL, Pinder BD, Vari HK, Lipshitz HD, Smibert CA. Smaug recruits the CCR4/POP2/NOT deadenylase complex to trigger maternal transcript localization in the early Drosophila embryo. Curr Biol. 2005;15:284–294. doi: 10.1016/j.cub.2005.01.048. [DOI] [PubMed] [Google Scholar]
- Shermoen AW, McCleland ML, O’Farrell PH. Developmental control of late replication and s phase length. Curr Biol. 2010;20:2067–2077. doi: 10.1016/j.cub.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert Ca, Lie YS, Shillinglaw W, Henzel WJ, Macdonald PM. Smaug, a novel and conserved protein, contributes to repression of nanos mRNA translation in vitro. RNA. 1999;5:1535–1547. doi: 10.1017/s1355838299991392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H, Spangenberg S, Vogt N, Groβhans J. Number of nuclear divisions in the Drosophila blastoderm controlled by onset of zygotic transcription. Curr Biol. 2013;23:133–8. doi: 10.1016/j.cub.2012.12.013. [DOI] [PubMed] [Google Scholar]
- Tadros W, Goldman AL, Babak T, Menzies F, Vardy L, Orr-Weaver T, Hughes TR, Westwood JT, Smibert Ca, Lipshitz HD. SMAUG Is a Major Regulator of Maternal mRNA Destabilization in Drosophila and Its Translation Is Activated by the PAN GU Kinase. Dev Cell. 2007a;12:143–155. doi: 10.1016/j.devcel.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Tadros W, Goldman AL, Babak T, Menzies F, Vardy L, Orr-Weaver T, Hughes TR, Westwood JT, Smibert CA, Lipshitz HD. SMAUG Is a Major Regulator of Maternal mRNA Destabilization in Drosophila and Its Translation Is Activated by the PAN GU Kinase. Dev Cell. 2007b;12:143–155. doi: 10.1016/j.devcel.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Tadros W, Houston SA, Bashirullah A, Cooperstock RL, Semotok JL, Reed BH, Lipshitz HD. Regulation of Maternal Transcript Destabilization During Egg Activation in Drosophila. 2003;1001:989–1001. doi: 10.1093/genetics/164.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development. 2009;136:3033–3042. doi: 10.1242/dev.033183. [DOI] [PubMed] [Google Scholar]
- Verhulst EC, Beukeboom LW, van de Zande L. Maternal control of haplodiploid sex determination in the wasp Nasonia. Science. 2010;328:620–3. doi: 10.1126/science.1185805. [DOI] [PubMed] [Google Scholar]
- Zamir E, Kam Z, Yarden A. Transcription-dependent induction of G1 phase during the zebra fish midblastula transition. Mol Cell Biol. 1997;17:529–536. doi: 10.1128/mcb.17.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Phosphorylated S5 RNA polymerase II is seen throughout all stages in the Nasonia embryo. White, DAPI (DNA); red, anti-4h8 phosphoS5 RNA Polymerase II. (a-a′) Embryo in division cycle 1. (b-b′) Embryo in division cycle 2. (c-c′) Embryo in division cycle 3. (d-d′) Embryo in division cycle 7. RNA polymerase II is excluded from the nuclei (e-e′) Embryo in division cycle 8. (f-f′) Embryo in division cycle 12. (g-g′) Transcriptionally quiescent pole cells have reduced 4h8 staining.
Supplementary Figure 2. qrt-PCR of pRNAi embryos Fold expression change of 0-1 hour Nv-smaug, Nv-string, Nv-tribbles, and Nv-zelda pRNAi embryos. Both wildtype and pRNAi embryos were normalized to a control house-keeping gene, Nv-rp49. pRNAi embryos were compared to wildtype embryos to assess the rate of knockdown.


